Abstract
Autophagy is associated with multiple biological processes and has protective and defensive functions with respect to immunity, inflammation, and resistance to microbial infection. In this experiment, we wished to investigate whether autophagy is a factor in the midgut cell response of Bombyx mori to infection by the B. mori cytoplasmic polyhedrosis virus (BmCPV). Our results indicated that the expression of three autophagy-related genes (BmAtg8, BmAtg5, and BmAtg7) in the midgut did not change greatly after BmCPV infection in B. mori. Basal ATG8/ATG8PE protein expression was detected in different B. mori tissues by using western blot analysis. Immunohistochemistry showed that the ATG8/ATG8PE proteins were located mainly in the cytoplasm. ATG8/ATG8PE protein levels decreased at 12 and 16 h after BmCPV infection. Our results indicate that autophagy responded slightly to BmCPV infection, but could not prevent the invasion and replication of the virus.
Keywords: autophagy, silkworm (Bombyx mori) midgut, cytoplasmic polyhedrosis virus (CPV), western blotting, immunohistochemistry
Cell autophagy is a process of self-digestion, and it degrades and recycles long-lived proteins and entire organelles to maintain cell homeostasis. These degradative processes are particularly important during development and under certain environmental stress conditions, such as cell death or starvation (Klionsky and Emr 2000). Massive autophagosomes are present when autophagic cell death occurs, and these then fuse with lysosomes to generate auto-lysosomes, whose contents are degraded by a variety of lysosomal hydrolases (He and Klionsky 2009). In addition to the capital homeostatic function, autophagy has an important role in multiple biological processes, including development, differentiation, and tissue remodeling (Levine and Klionsky 2004; Rabinowitz and White 2010).
Autophagy and apoptosis are involved in the cell death of larval organs and tissues during the metamorphic process of insects in the order Lepidoptera (Dai and Gilbert 1997; Uwo et al. 2002; Parthasarathy and Palli 2007; Tettamanti et al. 2007; Sumithra et al. 2010). The silkworm, Bombyx mori, has been used as a model for studying the autophagic process in vivo, that is, degradation of larval tissues (fat body, silk glands, and midgut epithelium) during metamorphosis. Morphological features of autophagy have been observed in different tissues and cells (Dai and Gilbert 1997; Mpakou et al. 2006; Sumithra et al. 2010), and several autophagy-related genes have been analyzed at the transcription level (Zhang et al. 2009; Li et al. 2011; Franzetti et al. 2012). Studies have revealed that autophagy and apoptosis are triggered by ecdysteroids during insect metamorphosis (Dai and Gilbert 1997; Silva-Zacarin et al. 2007). In the fat body of B. mori, 20-hydroxyecdysome upregulates Atg genes to induce autophagy (Tian et al. 2013).
Recently, increasing attention has been paid to the protective and defensive mechanisms of autophagy with respect to immunity, inflammation, and resistance to microbial infection (Deretic 2011; Levine et al. 2011). Diseases and microbial infections could be prevented by activating autophagy (Levine 2005; Lee and Iwasaki 2008). The B. mori cytoplasmic polyhedrosis virus (BmCPV) is a common pathogen that directly affects midgut cells and causes serious losses in sericulture. In this study, we found three autophagy-related genes that showed different expression patterns in the midgut of B. mori during BmCPV infection, and the expression of BmATG8/BmATG8PE proteins, which are located in the cytoplasm, decreased slightly at 12 and 16 h after BmCPV infection. Our study provides information on autophagy functions in B. mori.
Materials and Methods
Animals and Feeding Protocol
B. mori larvae (Jingsong × Haoyue) were provided by the Research Institute of Sericulture and Silk (Shaanxi, China), and they were fed fresh mulberry leaves in the laboratory and maintained at 25 ± 1°C, 60–70% relative humidity, and a photoperiod of 14:10 (L:D) h photoperiod. For reverse transcription-polymerase chain reaction (RT-PCR) and quantitative real-time PCR (qRT–PCR), six samples of the midgut were obtained from the B. mori larvae at 2, 4, 8, 12, 24, 36, 48, 60, 72, 96, 120, and 144 h after BmCPV infection and stored at −80°C until analysis. For western blot (WB) and immunohistochemical analyses, three midgut tissue samples were collected separately every 4 h after BmCPV infection until 24 h.
BmCPV Infection
BmCPV was maintained at 4°C in the laboratory (Supplementary Fig. S1). Virus polyhedra were counted using a haemocytometer under an optical microscope and diluted to 1 × 107 individuals/ml with saline. BmCPV suspensions (10 μl) were smeared onto 1-cm2 mulberry leaves and provided to each B. mori larva (total, 150) at the fourth instar stage on day 1. The negative control group was provided with mulberry leaves smeared with 10 µl of saline.
To confirm that the B. mori larvae were infected with BmCPV, a pair of primers was designed on the basis of the RDRP gene sequence of the virus (GenBank No.: AY496445)—forward: 5′-CTCAGGGTAAACAAGCAGG-3′ and reverse: 5′-GACTTCCACATTCTTAGCGT-3′—with a length of 215 bp. RT-PCR was performed using a C1000 thermal cycler (Bio-Rad, USA) with a reaction volume of 20 μl, which consisted of 10 μl of 2× Taq PCR master mix (Bioteke, Beijing, China), 1 μl of forward and reverse primers, 200 ng of template cDNA, and 7 μl of ddH2O. The following PCR conditions were used: 95°C for 3 min, followed by 20 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 10 min and 4°C thereafter.
RNA Isolation, cDNA Synthesis, and qRT-PCR
Total RNA was isolated from the midgut tissue of B. mori larvae by using Trizol Reagent (Tiangen, Beijing) and treated with RNase-free DNase. RNA quality and concentration were determined using a spectrophotometer (NanoDrop, USA). Total RNA (500 ng) was used for first-strand cDNA synthesis with a PrimeScript RT Reagent Kit (TaKaRa, Kyoto, Japan).
Primers for qRT-PCR were designed according to the target gene sequences. The primer sequences used in this experiment were as follows: BmAtg8 (FJ_416330) forward: 5′-GAAGAACATTCATTTGAGAAGAG-3′ and reverse: 5′-AATCAGACGGAACTAAATACTTC-3′; BmAtg5 (NM_001142487) forward: 5′-TGGGCTATCAACAGACGT-3′ and reverse: 5′-TTAGGACAGACAAGGCGT-3′; BmAtg7 (XP_004929628) forward: 5′-ATGCGGCGTTAGGTTTCG-3′ and reverse: 5′-ACAGAAATAGCAACCGAGG-3′; BmActin3 (NM_001126254) was used as a housekeeping gene (Zhang et al. 2009; Li et al. 2011): forward: 5′-AGAGGTTCCGTTGTCCCG-3′ and reverse: 5′-GGCGGTGATCTCCTTCTGCA-3′.
qRT-PCR was performed using a 20 -μl reaction volume with an IQ5 Real-time Quantitative PCR system (Bio-Rad) and SYBR-premix Ex Taq II (TaKaRa), according to the manufacturers’ instructions. PCR efficiency was calculated on the basis of the slope of the standard curve and the following equation was used: E% = (10−1/slope − 1) × 100%. Relative gene expression (2−△Ct, △Ct = Cttarget gene − CtActin3) was calculated on the basis of three independent replicates. The Wilcoxon test was used to determine significant differences between the negative control and BmCPV infection.
BmATG8 Antibody Preparation, Total Protein Isolation, and WB Analysis
The pET32a-BmATG8 vector was constructed and transfected into Escherichia coli Rosetta cells. The expressed recombinant protein was purified and used to immunise 2 New Zealand rabbits. Then, the antiserum was collected and purified after three immunisations, and the antibody titre was measured using enzyme-linked immunosorbent assay (ELISA; GenScript, Nanjing, China). Total proteins were extracted from larval midgut by using a tissue or cell total protein extraction kit (Sangon Biotech, Shanghai, China).
Then, 15% sodium dodecyl sulphate-polyacrylamide gel electrophoresis was performed with 10 μg of proteins per lane. The proteins were transferred to polyvinylidene difluoride membranes (GE, Uppsala, Sweden) after electrophoresis. The membranes were blocked with 10% skim milk (diluted with phosphate-buffered saline) and then incubated with anti-BmATG8 antisera (1:2000) at 25°C for 3 h. After washing three times with phosphate buffer saline (PBS), the membranes were incubated with goat anti-rabbit IgG (Beyotime, Shanghai, China) conjugated with horseradish peroxidase at a dilution of 1:2000 for 1 h and washed three times with PBS. Finally, immunoreactivity was detected using a MicroChemi Bio-imaging System (DNR, Israel) with an ECL WB Kit (Biotech, Beijing, China).
Preparation of Paraffin Sections and Immunohistochemistry
Three replicate midgut tissues were quickly obtained from B. mori larvae (as described earlier) and fixed in neutral formalin (10%, pH 7.4) at 4°C for 24 h. The midgut tissues were dehydrated using an ascending ethanol series, treated with xylene, and then embedded in paraffin. Paraffin sections (4 μm) were obtained using a Leica section machine (RM2235; Leica, Germany).
Tissue antigen retrieval was performed using 0.01 mol/l citric acid-citrate buffer for 3 min in a microwave oven after dewaxing and hydration. The tissues were blocked with goat serum (0.1 mg/ml) for 20 min, and ATG8 antibody was then used to combine endogenous ATG8 at 4°C overnight. The tissues were incubated with 20% egg white to exclude interference by endogenous biotin and labeled using the Ultrasensitive S-P Hypersensitivity Kit (biotinylated goat anti-rabbit antibody and streptavidin-peroxidase) and then stained using the DAB Detection Kit (streptavidin-biotin) for 5–10 s. Next, the midgut tissues were stained with haematoxylin for 30 s and washed with PBS for 5 min. Glass slides were mounted with 60% neutral gum (dissolved in xylene) after dehydration and observed under an optical microscope (XSZ-HS7; Xinqing, Chengdu). The procedure used for immunohistochemistry has been explained in detail in the Supplementary Text.
Results
Confirmation of BmCPV Infection
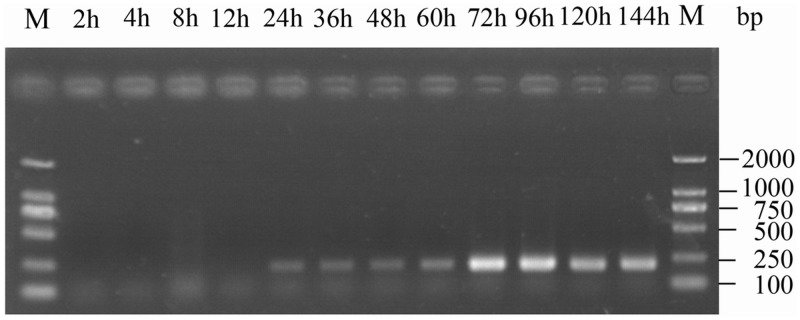
The virions were present both outside and inside the midgut cells at 3 h post-inoculation (Liu et al. 2012), at which time the viral genome may have begun self-replication. In this experiment, the viral RDRP gene could be detected at 24 h after BmCPV infection by using RT-PCR, and a larger amount was observed at 72 h (Fig. 1). Similar results were obtained using six duplicate samples.
Fig. 1.
Proliferation of the viral RDRP gene after BmCPV infection in Bombyx mori. The number of PCR cycles was 20, and polyhedra were detected at 60 h.
Autophagy-Related Gene Expression in B. mori Midgut During BmCPV Infection
To calculate the amplification efficiency, concentration gradient PCR was performed. The correlation coefficients of the standard curves were higher than 0.99, and the PCR efficiency was between 95 and 105%.
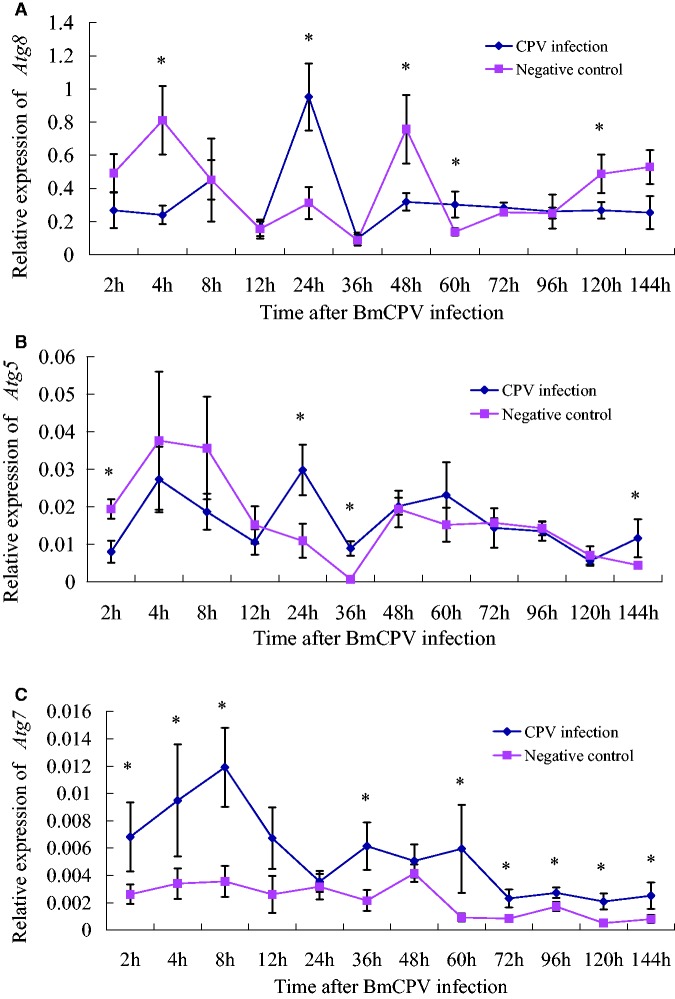
Nonparametric tests showed that, compared with the negative control, relative expressions of BmAtg8 and BmAtg5 were higher or lower at some time points after BmCPV infection (Fig. 2A and B). Compared to the negative control, the BmAtg8 gene showed higher expression at 24 and 60 h after BmCPV infection and lower expression at 4, 48, and 120 h. BmAtg5 expression was higher at 24, 36, and 144 h, yet lower at 2 h. In other words, BmAtg8 and BmAtg5 were both down-regulated at the beginning of BmCPV infection (2–4 h) and showed a high expression at 24 h, when the BmCPV genome started to amplify (Fig. 1). Further, the two genes have similar expression patterns, possibly because Atg8 and Atg5 are both located in ubiquitin-like protein conjugation systems (Zhang et al. 2009).
Fig. 2.
Relative expressions of three autophagy-related genes in the midgut tissues after BmCPV infection. A: BmAtg8, B: BmAtg5, C: BmAtg7.
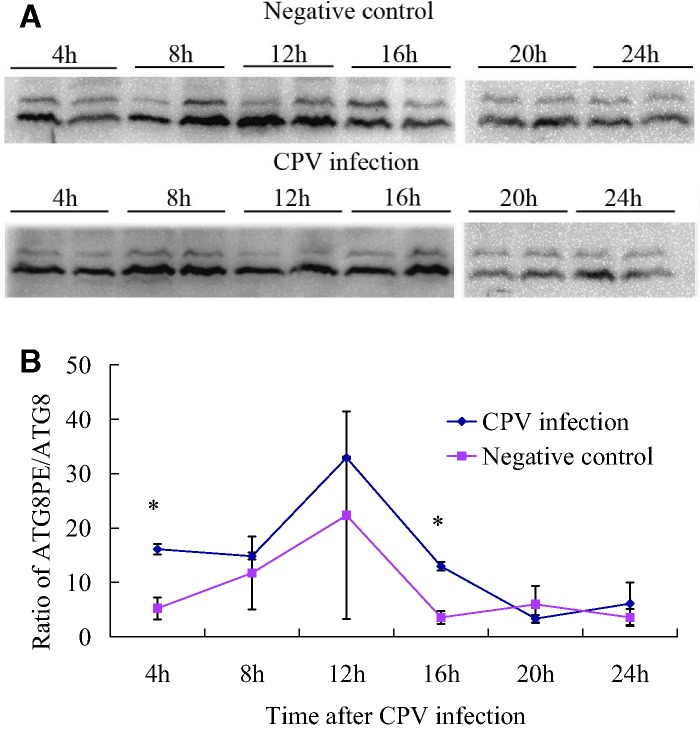
Interestingly, Atg7 gene expression was up-regulated after BmCPV infection, indicating the role of Atg7 in the recognition of BmCPV (Fig. 2C). The main role of the ATG7 protein is to activate ATG8 for the formation of autophagosomes (Mizushima et al. 1998). Therefore, the ATG8PE/ATG8 ratios were higher in the BmCPV infection group (Fig. 5B), but only significant at 4 and 16 h. However, Atg8 expression was not significantly increased after BmCPV infection; thus, the autophagy pathway did not specifically respond to BmCPV infection at a transcriptional level.
Fig. 5.
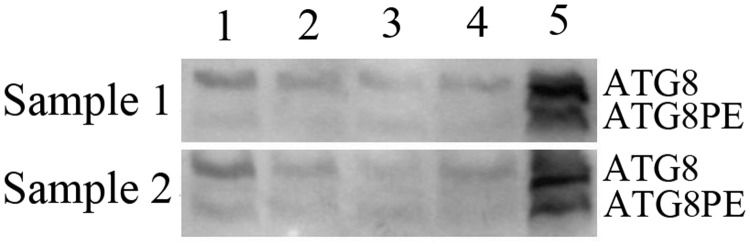
ATG8-PE/ATG8 blots of total midgut proteins at 4–24 h after BmCPV infection. (A) Results of the WB analysis; two parallel samples obtained from separate B. mori larvae at each time point were used for WB analysis; (B) The ATG8-PE/ATG8 ratio based on the grey value of the blots in Fig. 5A.
WB Analysis of the BmATG8 Protein in the Midgut After BmCPV Infection
ATG8PE is a major component of the autophagic membrane, and, therefore, its expression reflects the autophagy level (Kabeya et al. 2000). We prepared polyclonal antibodies for BmATG8, and high antibody titres were detected using ELISA (Yang et al. 2014). The polyclonal antibodies were used to detect the expression of ATG8 and ATG8-PE proteins in B. mori with the WB method and to determine the localization of ATG8 in cells with immunohistochemistry.
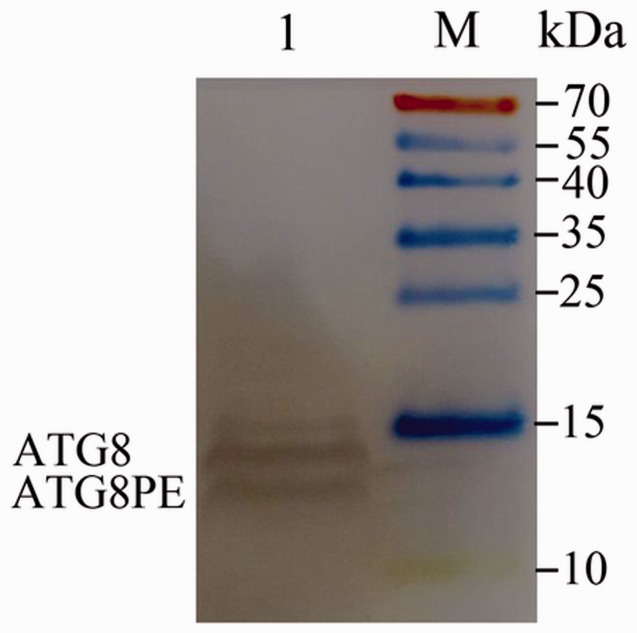
Reference antibodies from three mouse proteins, actin, tubulin, and GAPDH, were used to bind B. mori proteins in this experiment, but no WB band was obtained. Therefore, another method for normalization was used: the concentration of total midgut proteins was determined and diluted to ensure that the same amount was loaded for each sample. Similar to the autophagy protein of LC3 in mammalian cells (Kabeya et al. 2000), the BmATG8 protein (about 14 kDa) was modified and conjugated with phosphatidyl ethanolamine (PE) to form an active BmATG8PE protein. Although the molecular weight of BmATG8PE was higher than that of BmATG8, BmATG8-PE moved faster in polyacrylamide gel (approximately located at the position for 12 kDa) because of the higher negative charge of PE (Franzetti et al. 2012), and both were detected in the total midgut proteins of B. mori by using the WB technique (Fig. 3). The expression of the two forms of proteins was the maximum in the silk glands, less in the fat bodies and midgut, and minimum in the abdominal muscles and skin (Fig. 4).
Fig. 3.
Western blot (WB) of total midgut proteins with BmATG8 antibody (Diaminobenzidine staining). Two specific bands were considered as BmATG8 and BmATG8PE.
Fig. 4.
BmATG8 protein expression in different tissues of B. mori measured using WB analysis at the fifth instar stage on day 5. 1: Fat body; 2: Abdominal muscles; 3: Skin; 4: Midgut; 5: Silk gland.
Whether the autophagy pathway can identify the entry of the virus into midgut cells (4 h after infection) and self-replication (within 24 h after infection) would be reflected by the ATG8 protein expression level. Therefore, total midgut proteins from B. mori at 4–24 h after BmCPV infection was used to determine the ATG8 expression with WB analysis. It is worth noting the differences between the control and BmCPV blots at 8–12 h after infection. In the negative controls, the amount of ATG8PE at 12 h was a little higher than that at 8 h, while in the BmCPV group, the amount of ATG8PE was lower at 12 h than at 8 h (Fig. 5). This indicated ATG8PE consumption at 12 h after BmCPV infection and could be regarded as the response of B. mori to BmCPV infection.
Location of BmATG8 Proteins in Midgut Cells
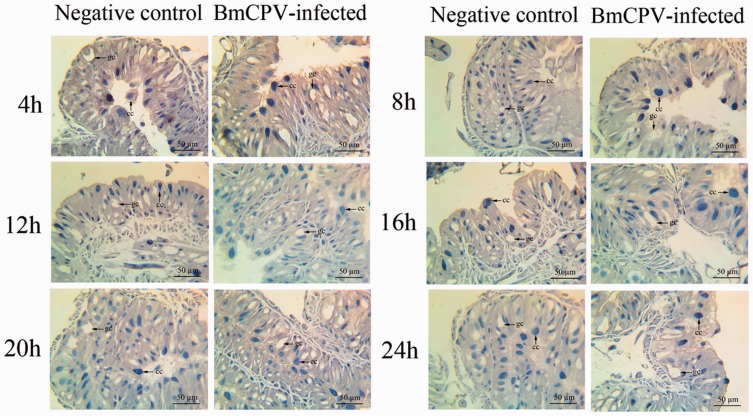
Immunohistochemistry showed the location of endogenous ATG8/ATG8PE in midgut tissues. ATG8/ATG8PE was widely distributed in the cytoplasm outside the chromatin (Fig. 6). In addition, ATG8/ATG8PE protein expression was lower at 12 and 16 h in the BmCPV group than in the control because of BmCPV infection (Fig. 6). This result was similar to that for the WB analysis.
Fig. 6.
Location of endogenous ATG8/ATG8PE in the midgut of B. mori. cc: columnar cell; gc: goblet cell. The brown parts show a higher stained ATG8/ATG8PE distribution. The shallow blue part is unstained. The dark blue parts indicated using arrows are the nuclei of columnar cells. Goblet cells contain secretory vesicles and are displayed as blank. ATG8/ATG8PE proteins are mainly located in the cytoplasm.
Discussion
Autophagy is relatively conservative in eukaryotes, from yeasts to humans, with a cell self-protection function conducive to the stability and development of the body (Klionsky et al. 2007). The ATG8/ATG8PE proteins are structural proteins on autophagic membranes, and therefore, they must have a basal expression level in the cell and increase or decrease according to different stresses. In this experiment, we found that ATG8/ATG8PE had different expression patterns in B. mori tissues, which reflected the cell stability of different tissues. For example, the silk gland grows rapidly in fifth instar larvae and degrades during pupation, and thus, it had higher ATG8/ATG8PE expression. The autophagy pathway can hydrolyse cellular lipids stored as triglycerides in lipid droplets into fatty acids for energy during nutrient deprivation (Singh et al. 2009). Consequently, ATG8/ATG8PE expression was also easily detected in fat bodies. Similarly, the midgut becomes the yellow body during pupation, which still required a degradation mechanism, while abdominal muscles and skin do not.
It is clear that autophagy can not only degrade intracellular pathogens but also has good antiviral immune responses (Yordy and Iwasaki 2011). Many viruses have been shown to affect autophagy pathways. Some viruses suppress this pathway for their survival, but others enhance or exploit it to benefit their replication (Sir and Ou 2010). RT-PCR of the viral RDRP gene confirmed that the B. mori larvae were infected with BmCPV. Inhibiting the expressions of BmAtg8 and BmAtg5 at the beginning of BmCPV infection (2–4 h) showed the function of BmCPV in autophagy prevention. In addition, the high gene expressions of BmAtg8 and BmAtg5 at 24 h indicated the autophagy response to infection. However, fluctuations in the curves for BmAtg8 and BmAtg5 gene expressions and the higher expression of BmAtg7 made it difficult to determine the autophagy function under BmCPV infection, because of the small changes in autophagy-related genes.
To the best of our knowledge, there are no preventive measures for BmCPV infection, except for disinfection. Expressions of genes related to valine, isoleucine degradation, retinol metabolism, and vitamin B6 metabolism were down-regulated, and enzyme activities of serine protease, trypsin-like protease, and inhibitor of apoptosis proteins decreased because of BmCPV infection. The expressions of genes involved in ribosome and proteasome pathways were up-regulated (Ping et al. 2009; Wu et al. 2011). In our experiment, the amount of ATG8/ATG8PE decreased at 12 h, which may be caused by low BmAtg8 expression at the beginning of infection (2–4 h) and also the reason for BmAtg8 up-regulation at 24 h (Fig. 2A). This phenomenon showed the homeostatic balance function of autophagy in B. mori midgut cells.
It is noteworthy that differences were detected in the relative abundance of ATG8 and ATG8-PE in this study (higher abundance of ATG8-PE; Fig. 5) and a previous study by Franzetti et al. (2012) (higher abundance of ATG8). A reason for these differences may be that the midgut samples were collected at different instar stages. Low consumption of ATG8-PE by fourth instar larvae resulted in ATG8-PE accumulation, while ATG8-PE was consumed rapidly during pupation.
Autophagy precedes apoptosis in the B. mori midgut during cell remodeling, which shows a regulation function for autophagy in B. mori growth and development. To the best of our knowledge, no studies have described the relationship between autophagy and disease in B. mori. Recognition is the first step in the degradation of the virus. The changes in autophagy-related gene expressions suggested the role of autophagy in BmCPV infection, although the infected B. mori larvae all died at the end of the experiment. However, taking into consideration the immunological analysis of ATG8/ATG8PE, we propose that the autophagy pathway cannot inhabit BmCPV invasion and replication.
Conclusion
Similar to Drosophila, the silkworm (B. mori) of the order Lepidoptera can be used as a model to study autophagy and apoptosis in vivo. The BmCPV-infected B. mori larvae showed fluctuations in expression curves on the basis of the determination of three autophagy-related genes. Both BmATG8 and BmATG8PE proteins were detected in different tissues of B. mori larvae by using WB analysis and were widely distributed in the cytoplasm outside the chromatin. The autophagy pathway in B. mori larvae responded to a limited extent to BmCPV infection. Further studies are required to investigate whether autophagy can prevent infection due to a small number of BmCPV or other pathogenic microbes.
Supplementary Data
Supplementary data are available at Journal of Insect Science online.
Acknowledgments
We are grateful to the modern agricultural technology system (No. CARS-35), public service sectors (agriculture) Special: (201303057), and the Fundamental Research Special of Northwest A & F University (No. ZD2012008, XNY2013-27).
Reference Cited
- Dai J.-D., Gilbert L. I. 1997. Programmed cell death of the prothoracic glands of Manduca sexta during pupal-adult metamorphosis. Insect Biochem. Mol. 27: 69–78. [DOI] [PubMed] [Google Scholar]
- Deretic V. 2011. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol. Rev. 240: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzetti E., Huang Z.-J., Shi Y.-X., Xie K., Deng X.-J., Li J.-P., Li Q.-R., Yang W.-Y., Zeng W.-N., Casartelli M. 2012. Autophagy precedes apoptosis during the remodeling of silkworm larval midgut. Apoptosis, 17: 305–324. [DOI] [PubMed] [Google Scholar]
- He C., Klionsky D. J. 2009. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet., 43: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J., 19: 5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Cuervo A. M., Seglen P. O. 2007. Methods for monitoring autophagy from yeast to human. Autophagy 3: 181–206. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Emr S. D. 2000. Autophagy as a regulated pathway of cellular degradation. Science 290: 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. K., Iwasaki A. 2008. Autophagy and antiviral immunity. Curr. Opin. Immunol. 20: 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. 2005. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 120: 159–162. [DOI] [PubMed] [Google Scholar]
- Levine B., Klionsky D. J. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6: 463. [DOI] [PubMed] [Google Scholar]
- Levine B., Mizushima N., Virgin H. W. 2011. Autophagy in immunity and inflammation. Nature 469: 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Deng X., Huang Z., Zheng S., Tettamanti G., Cao Y., Feng Q. 2011. Expression of autophagy-related genes in the anterior silk gland of the silkworm (Bombyx mori) during metamorphosis. Can. J. Zool. 89: 1019–1026. [Google Scholar]
- Liu Z., Liu S., Cui J., Tan Y., He J., Zhang J. 2012. Transmission electron microscopy studies of cellular responses to entry of virions: one kind of natural nanobiomaterial. Int. J. Cell. Biol. 2012: 596589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M. D., Klionsky D. J., Ohsumi M., Ohsumi Y. 1998. A protein conjugation system essential for autophagy. Nature 395: 395–398. [DOI] [PubMed] [Google Scholar]
- Mpakou V. E., Nezis I. P., Stravopodis D. J., Margaritis L. H., Papassideri I. S. 2006. Programmed cell death of the ovarian nurse cells during oogenesis of the silkmoth Bombyx mori. Dev. Growth Differ. 48: 419–428. [DOI] [PubMed] [Google Scholar]
- Parthasarathy R., Palli S. R. 2007. Developmental and hormonal regulation of midgut remodeling in a lepidopteran insect, Heliothis virescens. Mech. Develop. 124: 23–34. [DOI] [PubMed] [Google Scholar]
- Ping W., Li M.-W., Wang X., Zhao P., Wang X.-Y., Liu T., Qin G.-X. 2009. Differentially expressed genes in the midgut of silkworm infected with cytoplasmic polyhedrosis virus. Afr. J. Biotechnol. 8: 3711–3720. [Google Scholar]
- Rabinowitz J. D., White E. 2010. Autophagy and metabolism. Sci. Signal. 330: 1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Zacarin E., Tomaino G., Brocheto-Braga M., Taboga S., De Moraes R. S. 2007. Programmed cell death in the larval salivary glands of Apis mellifera (Hymenoptera: Apidae). J. Biosci. 32: 309–328. [DOI] [PubMed] [Google Scholar]
- Singh R., Kaushik S., Wang Y., Xiang Y. 2009. Autophagy regulates lipid metabolism. Nature 458: 1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir D., Ou J.-H.J. 2010. Autophagy in viral replication and pathogenesis. Mol. Cells 29: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumithra P., Britto C. P., Krishnan M. 2010. Modes of cell death in the pupal perivisceral fat body tissue of the silkworm Bombyx mori L. Cell Tissue Res. 339: 349–358. [DOI] [PubMed] [Google Scholar]
- Tettamanti G., Grimaldi A., Pennacchio F., de Eguileor M. 2007. Lepidopteran larval midgut during prepupal instar: digestion or self-digestion? Autophagy 3: 630–631. [DOI] [PubMed] [Google Scholar]
- Tian L., Ma L., Guo E., Deng X., Ma S., Xia Q., Cao Y., Li S. 2013. 20-hydroxyecdysone upregulates Atg genes to induce autophagy in the Bombyx fat body. Autophagy 9: 1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwo M. F., Ui-Tei K., Park P., Takeda M. 2002. Replacement of midgut epithelium in the greater wax moth, Galleria mellonela, during larval-pupal moult. Cell Tissue Res. 308: 319–331. [DOI] [PubMed] [Google Scholar]
- Wu P., Wang X., Qin G. X., Liu T., Jiang Y. F., Li M. W., Guo X. J. 2011. Microarray analysis of the gene expression profile in the midgut of silkworm infected with cytoplasmic polyhedrosis virus. Mol. Biol. Rep. 38: 333–341. [DOI] [PubMed] [Google Scholar]
- Yang X. B., Li H. J., Wu Y. P., Liu Q. H., Juan Zhang M., Qian Y. H., Jiao F. 2014. Preparation of polyclonal antibody and tissue expression analysis of BmATG8 in silkworm (Bombyx mori) (in Chinse). Sci. Sericulture 40: 818–823. [Google Scholar]
- Yordy B., Iwasaki A. 2011. Autophagy in the control and pathogenesis of viral infection. Curr. Opin. Virol. 1: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Hu Z.-Y., Li W.-F., Li Q.-R., Deng X.-J., Yang W.-Y., Cao Y., Zhou C.-Z. 2009. Systematic cloning and analysis of autophagy-related genes from the silkworm Bombyx mori. BMC Mol. Biol. 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]