Abstract
Chitin deacetylases (CDAs) convert chitin into chitosan, the N-deacetylated form of chitin, which influences the mechanical and permeability properties of structures such as the cuticle and peritrophic matrices. In this article, a new CDA encoding gene, Hacda2, was cloned by reverse transcription-polymerase chain reaction method in Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae), with an open reading frame of 1,611 bp. The deduced protein composed of 536 amino acid residues with a signal peptide, a chitin-binding domain, a low-density lipoprotein receptor class A domain, and a polysaccharide deacetylase-like catalytic domain. The highest expression level of Hacda2 was detected in fat body among tissues tested in the fifth-instar larvae using real-time quantitative polymerase chain reaction method. Feeding of Bacillus thuringiensis (Bt) (Bacillales: Bacillaceae) diet changed the expression level of Hacda1, Hacda2, Hacda5a, and Hacda5b significantly and differentially in the third-instar larvae. Hacda5a and Hacda5b expression were initially down-regulated and then up-regulated, whereas, the expression level of Hacda1 and Hacda2 was suppressed constantly postfeeding on Bt diet. These results suggested that HaCDAs may be involved in the response of H. armigera larvae to Bt and may be helpful to elucidate the roles of HaCDAs in the action of Bt cry toxin. The potential of HaCDAs to be used as synergists of Bt insecticidal protein needs to be further tested.
Keywords: chitin deacetylase, RT-qPCR, synergist, Bacillus thuringiensis, Helicoverpa armigera
Chitin, the β-1,4-linked homopolymer of N-acetylglucosamine, is an abundant natural biopolymer, such as cellulose. It is one of the main components of insect cuticle and the peritrophic membrane (PM) and plays a critical role in modeling tissues of various shapes, sizes, and mechanical properties (Zhao et al. 2010). Chitin deacetylases (CDAs, EC 3.5.1.41) convert chitin into chitosan, the N-deacetylated form of chitin, and belong to a carbohydrate esterase family 4 of the carbohydrate-active enzymes database (http://www.cazy.org) (Kramer and Muthukrishnan 1997).
To date, cDNAs encoding CDAs have been characterized in at least 12 insect species, including Anopheles gambiae (Dixit et al. 2008), Apis mellifera (Dixit et al. 2008), Drosophila melanogaster (Luschnig et al. 2006, Wang et al. 2006), Tribolium castaneum (Dixit et al. 2008), Mamestra configurata (Toprak et al. 2008), Trichoplusia ni (Guo et al. 2005), Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae) (Campbell et al. 2008, Jakubowska et al. 2010), Agrotis ipsilon Hufngel (FJ899541), Bombyx mori (Zhong et al. 2014), Mamestra brassicae (AEI30868.1), Choristoneura funiferana (Quan et al. 2013), and Nilaparvata lugens (Xi et al. 2014). Insect CDAs can be divided into five major groups, i.e., CDA groups I–V (Dixit et al. 2008). All members of group I CDA contain a chitin-binding domain (ChBD), a low-density lipoprotein receptor class A domain (LDLa), and a polysaccharide deacetylase-like catalytic domain (CDAD). The domain organization mode of group I and group II is similar but with substantially different amino acid sequences. Group III and IV CDAs contain CDAD and ChBD domains, and group V CDAs contain only CDAD domain.
Insect CDAs genes have been found to be expressed in either gut or nongut tissues. In gut tissues, CDAs will not only alter chitin fibril structure but also affect the binding of PM proteins, PM integrity, and porosity (Arakane et al. 2009, Jakubowska et al. 2010). CDAs that are mainly expressed in the integument play critical roles in molting, old cuticle degradation, and new cuticle formation, as revealed by dsRNA-mediated RNAi experiments in the C. fumiferana, N. lugens, and T. castaneum (Dixit et al. 2008, Quan et al. 2013, Xi et al. 2014). Moreover, in the D. melanogaster, two CDA-like proteins, CDA1 and CDA2, have been found to be associated with the tracheal extracellular matrix and limited tube elongation (Luschnig et al. 2006, Wang et al. 2006).
Bacillus thuringiensis (Bt) (Bacillales: Bacillaceae) crystal proteins (Cry toxins), as the potent bioinsecticides, kill the majority of pests, including Lepidoptera, Diptera, and Coleoptera but cause little or no harm to vertebrates and most other organisms (Tabashnik et al. 2008). Although alternative model in action of Bt Cry toxin is available, the pore-forming model that Bt Cry toxins act by forming pores which disrupts midgut epithelial cells inside the larval midgut and causes damage to insects is still widely accepted (Vachon et al. 2012). Cry1Ab intoxication of C. fumiferana and Manduca sexta causes induction of immune, structural, and stress responsive genes and repression of most metabolic genes in larval stage (Van Munster et al. 2007). Similarly, genes related to synthesis of structural proteins were reported to be up-regulated and degradation enzyme (e.g., cellulase) genes to be down-regulated for Diabrotica virgifera fed with the Cry3Bb1 corn diet (Sayed et al. 2010). Thus, it may be natural to suppose that genes that can be up- or down-regulated would be critical in the larval defense machinery after ingestion of Bt toxins. CDAs are one of the key enzymes required for chitin degradation and are responsible for either the homeostasis of the PM structure or the molting (Jakubowska et al. 2010), and then their overall gene expression levels may be changed after ingestion of Bt intoxication. In this study, we cloned a new CDA encoding gene, Hacda2, and examined the expression of four CDA genes, including Hacda1, Hacda2, Hacda5a, and Hacda5b, in H. armigera larvae in response to Bt diet feeding.
Materials and Methods
Experimental Insects and Bacterial Strain
H. armigera larvae were reared on an artificial diet (Li et al. 2012) and maintained at 28 ± 2°C, 60-70% relative humidity, under a photoperiod of 16:8 (L:D) h. Bt strain HW33-1 producing Cry1Ac (LC50, 0.02 μg/ml) (Chakrabarti et al. 1998) was kindly provided by Professor Shuliang Feng, the Plant Protection Institute, Hebei Academy of Agricultural Sciences.
Cloning of a H. armigera CDA cDNA
Total RNA was isolated from the fifth-instar larvae using the TRIzol Reagent (CWbio, Beijing, China). For in vitro reverse transcription, PrimeScript RT reagent Kit with gDNA Eraser (Takara, Dalian, China) was used to prepare cDNA without genomic DNA contamination according to the manufacturer’s instructions. A pair of gene-specific primers (CDA2F and CDA2R) was designed according to conserved regions of insect CDAs (Table 1). The polymerase chain reaction (PCR) products were recovered and cloned into a pUCm-T TA cloning vector (Sangon, Shanghai, China) and transformed into DH5α competent cells, followed by plasmid DNA purification and sequencing (Genewiz, Suzhou, China).
Table 1.
Primers used during this study
| Primer | Sequence (5'–3') | Primer | Sequence (5'–3') |
|---|---|---|---|
| CDA2F | AGCGTCATGGCTCGACACGCG | CDA5aqR | TACTCCAAGCCATATTCCTG |
| CDA2R | TCACTTCTTCACGTTGAAGCCTTC | CDA5bqF | CTTACGATGAACTGAAAGAGGA |
| CDA2qF | GGTCAGGTCTTAAACAAATCAC | CDA5bqR | GTCTGTGAAAGTGTTGGTGG |
| CDA2qR | ATTCCTTCTCGATACAGTCTC | β-ActinF | CTGACCGTATGCAGAAGGAG |
| CDA1qF | ATCCAGTACAAGAAAGCCTC | β-ActinR | CACAAGCGTAATTTGAGCC |
| CDA1qR | CAGTTCTACAACTTCCTCAACC | EF-F | GACAAACGTACCATCGAGAAG |
| CDA5aqF | AGAACTCCTCAAACCTACTG | EF-R | GATACCAGCCTCGAACTCAC |
Sequence Analysis
The amino acid sequence of HaCDA2 was deduced by the DNAMAN 7.0 software (Lynnon, Biosoft, Vaudreuil, Quebec, Canada). Prediction of putative transmembrane helices, the signal peptide, protein domain, and N-glycosylation sites were carried out with the TMHMM (http://www.cbs.dtu.dk/services/TMHMM/), SignalP 4.0 (http://www.cbs.dtu.dk/services/SignalP/), BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and NetOGlyc (http://www.cbs.dtu.dk/services/NetOGlyc/) programs, respectively. Phylogenetic analyses were performed using the neighbor-joining method with 1,000 bootstraps, using the ClustalX 2.0 and MEGA 6.0 programs. Distances were corrected for multiple substitutions according to the method of Kimura (1980).
Tissue Expression Analysis
To obtain various tissues of H. armigera larvae, the fifth-instar larvae were paralyzed and surface sterilized for 1 min in 95% ethanol and then rinsed three times using sterile distilled water prior to dissection. The larvae were pinned on a wax plate and washed with sterile artificial hemolymph solution (Palli and Locke 1987). H. armigera larvae were dissected and various tissues, including foregut, midgut, hindgut, fat body, malpighian tubules, and integument, were collected and stored in liquid nitrogen prior to RNA isolation. Total RNA isolation and the first-strand cDNA synthesis were carried out as previously described.
Real-time quantitative PCR (RT-qPCR) was used to determine the transcription level Hacda2 gene in various tissues, following the MIQE guidelines (Bustin et al. 2009). RT-qPCR was carried out on a LightCycler 96 (Roche Diagnostics, Mannheim, Germany) machine. PCR reactions were performed in a volume of 20 μl containing 10 μl of GoTaq qPCR Master Mix (Promega, Madison, WI), 1 μl of cDNA templates (corresponding to 50 ng of starting total RNA template) and forward and reverse primers designed using Perlprimer software were added to a final concentration of 0.6 μM (Table 1). PCR conditions were 95°C for 5 min for initial denaturation, followed by 45 cycles of 95°C for 10 s, 55°C for 15 s, and 72°C for 10 s. The melting curves of amplicons were measured by taking continuous fluorescence reading, while increasing temperature from 65 to 97°C. After melting curve analysis, the relative quantities of Hacda2 transcripts were assessed using the 2-△△Ct method (Schmittgen and Livak 2008), with H. armigera β-actin as the reference gene and template-free as a control. All RT-qPCR experiments were repeated three times. Means and standard errors were calculated and analyzed by analysis of variance followed by the Tukey–Kramer test, using SPSS 16.0 for Windows (SPSS, Chicago, IL).
Effects of Toxin on HaCDA Expression
The Cry1Ac-producing Bt strain HW33-1 was cultured in liquid Luria-Bertani medium (tryptone 1%, yeast extract 0.5%, NaCl 1%, pH 7–7.2) for 3 d at 30°C, 220 r/min. The production of parasporal crystals in spores was confirmed by microscopic observation, and the concentration of Bt was quantified using a specific gravity method. Twenty milliliters of the spore-crystal mixture were placed in plastic tubes, which was weighted as P1, and then was centrifuged at 8,000 × g for 20 min at 4°C. The supernatant was decanted, and the pellet was thoroughly dried in tubes under room temperature and weighted as P2. Thus, the concentration of Bt bacteria was calculated by formula, C (g/ml) = (P1 − P2)/20.
The third-instar larvae were starved for 12 h and then were placed individually in glass tubes (1 cm in diameter and 6 cm in height) containing 1 g artificial diet without Bt (200 µl H2O) as a control or Bt (200 µl, 18.6 g/liter) as a treatment. Feeding experiments were conducted at 28 ± 2°C, 60–70% relative humidity, under a photoperiod of 12:12 (L:D) h. Each treatment was repeated three times using 24 larvae for each replicate. For each replicate, three larvae fed on Bt diet or non-Bt diet were randomly collected at 12 h, 36 h, 60 h, and 72 h postfeeding until the larvae either died or pupated. Total RNA was isolated from whole body and cDNA was synthesized. The expression levels of Hacdas were analyzed by RT-qPCR as described previously. Primers used for the RT-qPCR analysis are given in Table 1.
Results
Cloning of a H. armigera CDA cDNA
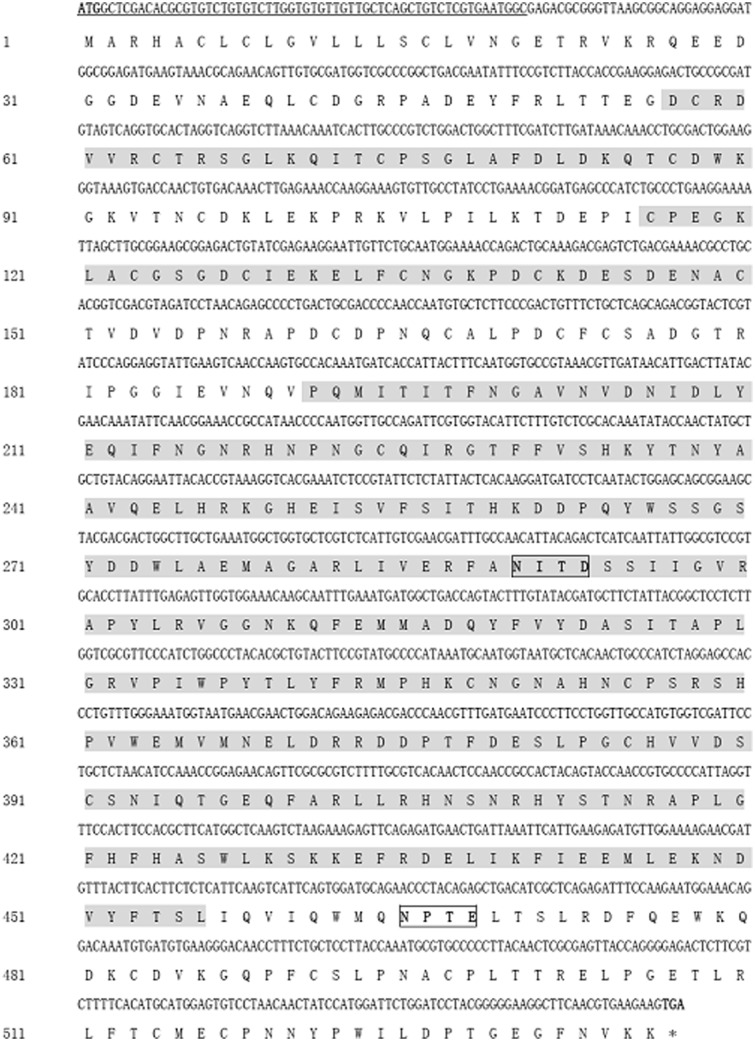
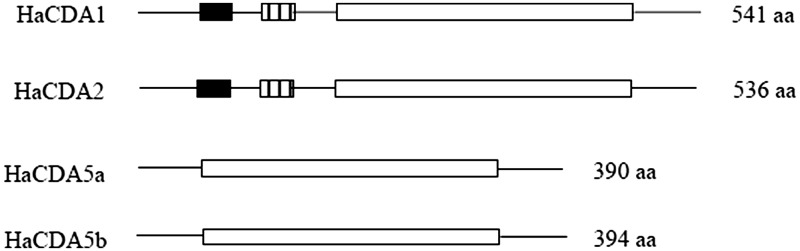
A cDNA fragment of Hacda2 gene was obtained by reverse transcription-PCR (Fig. 1), with an intact open reading frame of 1,611 nucleotides. The encoded protein was 536 amino acids in length with a predicted molecular weight of approximately 60.78 kDa. The predicted isoelectric point (pI) of HaCDA2 was 5.31. Two N-glycosylation sites were found in HaCDA2 protein by NetOGlyc analysis, indicating that the protein was a glycoprotein (Fig. 2). In addition, analysis of deduced amino acid sequence using TMHMM demonstrated that there were no transmembrane helices in the protein, suggesting that the putative protein was not an integral membrane protein. Domain analysis indicated that the HaCDA2 has a putative signal peptide domain, a chitin-binding domain, a low-density LDLa, and a CDAD (Fig. 2). This domain organization mode of HaCDA2 was the same to HaCDA1 but different from HaCDA5a and HaCDA5b (Fig. 3).
Fig. 1.
Electrophoresis of reverse transcription-PCR product of Hacda2. Lane 1, the cDNA fragment of Hacda2 gene. M, DL 2000 marker (Takara, Dalian, China).
Fig. 2.
Nucleotide and deduced amino acid sequences of HaCDA2. Stop codon is asterisked and the signal peptide region is underlined. The three domains, a CDAD (191–456 aa), a chitin-binding domain (ChBD) (57-90 aa), and a low-density LDLa (116–150 aa), as predicted by the NCBI protein BLAST tool, are shaded gray. Potential N-glycosylation sites are boxed.
Fig. 3.
Schematic diagram of domain architectures of four known H. armigera CDA genes. The deduced amino acid sequences were used to predict the domain architectures of four HaCDAs with NCBI protein BLAST tool. The black boxes indicate chitin-binding domains deacetylase catalytic domains, the stripe boxes indicate low-density LDLa, and the white boxes indicate deacetylase catalytic domains.
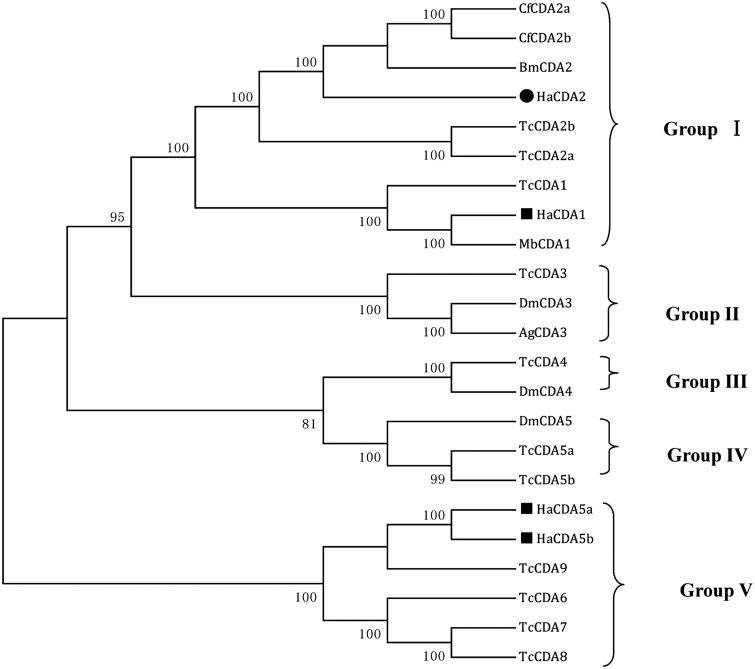
To investigate the evolutionary relationship of HaCDA2, a phylogenetic analysis was performed with the orthologs of the other six insect species, using the ClustalX 2.0 and MEGA 6.0 programs. HaCDA2 was clearly distant from the HaCDA5a and HaCDA5b, together with HaCDA1, falling into CDA group I whose members contain all three domains (CDAD, ChBD, and LDLa) (Fig. 4).
Fig. 4.
The phylogenetic analysis of CDAs from H. armigera and other five insect species. CDA group I–V are illustrated. Amino acid sequences used in the alignment were as follows: H. armigera CDAs HaCDA1 (ADB43610.1), HaCDA2 (KM598637), HaCDA5a (ADB43611.1), and HaCDA5b (ADB43612.1), C. funiferan CDAs CfCDA2a (AGT28748.1), CfCDA2b (AGT28749.1), T. castaneum CDAs, TcCDA1 (NP_001095946.1), TcCDA2a (NP_001096047.1), TcCDA2b (NP_001116303.1), TcCDA3 (NP_001104011.1), TcCDA4 (NP_001103903.1), TcCDA5a (NP_001103739.1), TcCDA5b (NP_001107799.1), and TcCDA6-9 (NP_001103905.1, NP_001104012.1, NP_001103906.1, NP_001103904.1), B. mori CDA BmCDA2 (AD024154.1), D. melanogaster CDAs, DmCDA3 (NP_609806.1), DmCDA4 (XP_003703278.1), and DmCDA5 (NP_001245807.1), M. brassicae CDA MbCDA1 (AEI30868.1), and A. gambiae CDA, AgCDA3 (XP_317336.3). Sequences were aligned using ClustalX 2.0. A bootstrap analysis of 1,000 replications was used, and bootstrap values are shown in the cladogram.
Tissue Expression Analysis
In our work, to test the efficacy of reference gene used in RT-qPCR, we compared β-actin (X97614.1) and H. armigera elongation factor 1-alpha (HaEF1α, U20129.1). No significant difference was found between them at least in RT-qPCR analysis in this work (data not shown). Hence, β-actin was used as the reference gene for normalization of Hacda2 transcription level in RT-qPCR analysis. To ensure the specificity of amplification of Hacdas and β-actin, dissociation curves were checked for the presence of a unique peak.
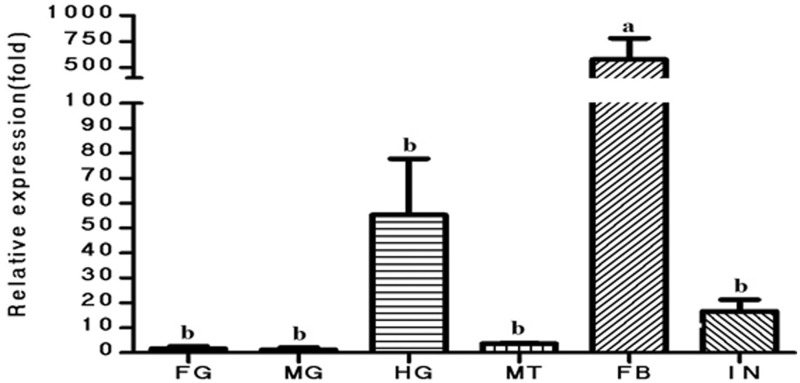
The relative mRNA expression level of Hacda2 in foregut, midgut, hindgut, fat body, malpighian tubules, and integument in the fifth-instar larvae of H. armigera were determined by RT-qPCR. As shown in Fig. 5, the highest expression level of Hacda2 was detected in fat body among tissues tested.
Fig. 5.
The relative mRNA expression level of Hacda2 as determined by RT-qPCR in foregut (FG), midgut (MG), hindgut (HG), fat body (FB), malpighian tubules (MT), and integument (IN) in the fifth-instar larvae of H. armigera. β-actin was used as an internal reference gene. The relative expression (fold) was calculated based on the value of the lowest expression (midgut), which was ascribed an arbitrary value of 1. The data were given as means ± SE and were analyzed by analysis of variance followed by the Tukey–Kramer test, using SPSS 16.0 for Windows (SPSS). Different letters on the columns mean significant difference (P < 0.05).
Effects of Bt Cry Toxin on Hacda Expression
In the Bt diet feeding experiment, reduced food ingestion, decreased the amount of exercise, and blocked growth were observed among the third-instar larvae of H. armigera fed on Bt cry toxin diet.
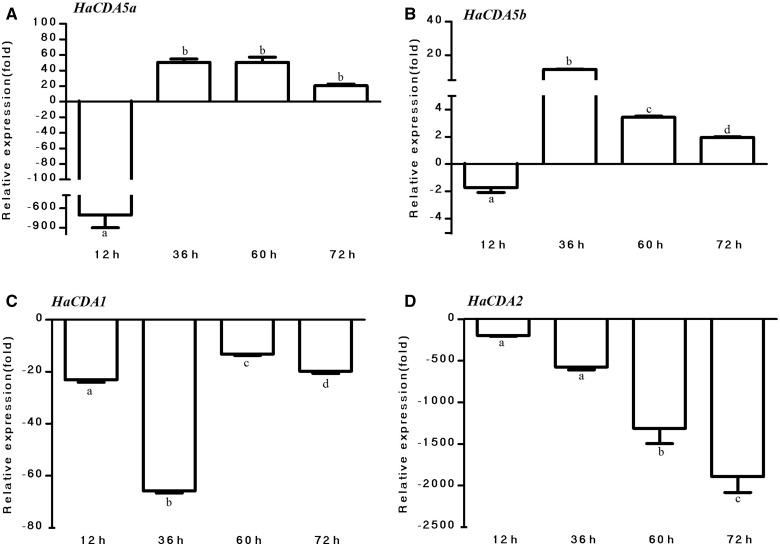
To identify the effects of Bt cry toxin on the expression levels of Hacda2 and other published HaCDA genes, we checked the changes in expression of HaCDA genes in response to Bt cry toxin. The RT-qPCR results showed that the expression of the Hacda1, Hacda2, Hacda5a, and Hacda5b was differentially affected by Bt diet. As shown in Fig. 6A, Hacda5a expression was down-regulated as early as 12 h and then remained up-regulated before dying in Bt diet feeding group. The influence of Bt cry toxin on the Hacda5b expression was similar to Hacda5a shown in Fig. 6B.
Fig. 6.
Expression of different CDAs in H. armigera larvae. Total RNAs were isolated from the larvae at different time post ingestion and then reverse-transcribed, and RT-qPCR was performed using specific primers and normalized relative to the transcription level of reference gene (β-actin). The data were given as means ± SE and were analyzed by analysis of variance followed by the Tukey–Kramer test, using SPSS 16.0 (SPSS) for MS Windows. Different letters on the columns meant significant difference (P < 0.05).
However, the changes in expression of Hacda2 and Hacda1 genes in response to Bt cry toxin were quite different from Hacda5a and Hacda5b. The expression level of Hacda1 and Hacda2, as shown in Fig. 6C and D, was suppressed constantly after ingestion of Bt diet.
Discussion
CDAs are important enzymes that regulate molting, organ formation, and antipathogen immunity during insect development (Luschnig et al. 2006, Wang et al. 2006, Dixit et al. 2008, Quan et al. 2013, Xi et al. 2014). In this article, we cloned a new CDA encoding gene, Hacda2. The putative HaCDA2 protein composed of 536 amino acids, which includes a 20-amino acid residue signal peptide, a chitin-binding domain, a low-density LDLa, and a CDAD (Fig. 2). In comparison with other three published CDA proteins, HaCDA2 showed a high amino acid sequence similarity (59%) to HaCDA1 and a similarity of only 36% to HaCDA5a and HaCDA5b. All the four HaCDAs shared a CDAD domain, while only two of them, HaCDA1 and HaCDA2, contained a ChBD domain and a LDLa domain (Fig. 3). Phylogenetic analysis suggested that HaCDA2 is clearly distant from the HaCDA5a and HaCDA5b and together with HaCDA1, falling into CDA group I (Fig. 4). This group has been further divided into two subgroups (subgroup Ia and Ib) (Xi et al. 2014), with HaCDA1 and HaCDA2 belonging to subgroup Ia and subgroup Ib, respectively. Some studies showed that gene of subgroup Ib usually generates two splicing variants displaying differential expression and function in A. gambiae, Ap. mellifera, D. melanogaster, T. castaneum (Dixit et al. 2008), B. mori (HM450149 and HM450150), and C. fumiferana (Quan et al. 2013). Despite the efforts in cloning the alternative splicing site sequence using reverse transcription-PCR method, no new splicing variant has been detected in our laboratory so far.
According to Jakubowska et al. (2010), Hacda5a was expressed mainly in the gut, with comparatively low expression in the malpighian tubules and fat body. These authors also found that the pathogenicity and lethality were greatly enhanced in baculoviruses expressing HaCDA5a. This may be attributed to the action of HaCDA5a on the PM to change its permeability and thus enable easier entrance of virus into epithelial cells. Hacda5b and Hacda1 were expressed mainly in the fat body and malpighian tubules, and the former also showed limited expression in the midgut. Proteomic analysis indicated that HaCDA1 protein is also present in nondiapausing pupal heads and may be involved in regulating insect molting (Chen et al. 2010). Hacda2 was expressed mainly in the fat body (Fig. 5). These enzymes might be associated with structures of the malpighian tubules, just as two CDA-like proteins, which play important roles in normal tracheal tube morphogenesis in the D. melanogaster (Luschnig et al. 2006, Wang et al. 2006). However, the function of CDAs in the fat body was unknown.
Obvious changes in expression level of HaCDAs genes were detected after larvae were fed on the Bt diet. The expression of all the four HaCDA genes were found to be substantially repressed 12 h post Bt diet ingestion (Fig. 6). Similarly, CDA-like protein was found to be down-regulated due to baculovirus infection in the midguts of H. armigera and Spodoptera exigua larvae (Jakubowska et al. 2010). These authors suggested that the decrease may constitute an early mechanism response to Cry toxins damage, which is probably related to their defense mechanism. After ingested by insects, the Cry protoxin is then cleaved by proteases in the gut to yield an active toxin (Vachon et al. 2012). This process not only produces an activated toxin monomer but also involves the mechanism that may reduce the damage and support a functional gut system. Likewise, Yao et al. (2014) found that the trypsin inhibitor transcript, which may be involved in insect defense against Cry1Ab intoxication by inhibiting proteolytic activation of protoxin to toxin, is up-regulated by 90-fold after ingestion of Bt cry toxin based on the microarray data. Additionally, the down-regulation of the gut chitinase gene and the up-regulation of chitin synthase 2 and Glutamine fructose-6-phosphate amidotransferase gene may act as a defense mechanism to maintain the gut system integrity in response to the Cry1Ab ingestion in the larvae (Yao et al. 2014).
Interestingly, our RT-qPCR results also showed that the expression level of Hacda5a and Hacda5b of H. armigera larvae after being fed on Bt diet for 36 h were up-regulated by 50-fold and 11-fold, respectively (Fig. 6). Since both genes are mainly expressed in gut and are essential for PM integrity, this up-regulation may be associated with the amount of toxin intake. Physiological metabolism of H. armigera may be broken completely as a result of excessive toxins intake, which facilitates the insertion of toxins into the membrane with a concomitant pore formation that result in up-regulated CDAs in the gut (Vachon et al. 2012). However, the expression level of Hacda1 and Hacda2 were not changed after ingestion of toxin by 36 h. The suppression of gene expression may not only be an adaptive strategy in response to Bt ingestion but also a consequence of the metabolic interference of the toxin with the larval growth.
To grow, insects shed their old cuticle and acquire a new, larger one. Chitin is one of the major components of the cuticle. As a result, precise regulation of its synthesis, modification, and degradation is crucial for successful insect development. Insecticides targeting chitin biosynthesis and degradation are safe to humans as the chitin synthesis pathway is absent in vertebrates. Thus, chitin synthesis and degradation have long been considered as promising targets for biorational insecticide development (Cohen 1993, Kramer and Muthukrishnan 1997). In recent years, insecticides, acting on the larval stages of the lepidopteran insects by inhibiting chitin synthesis, have been developed and applied widely, such as diflubenzuron, hexaflumuron, and chlorfluazuron (Li et al. 2013). In addition, chitinase, with the activity of hydrolyzing chitin, has been proved to be a potential synergist of bioinsecticide (Lertcanawanichakul et al. 2004, Thamthiankul et al. 2004). This work suggests that CDAs may be also possible to enhance the insecticidal activity of Bt. However, the feasibility of HaCDAs to be used as synergists of the insecticides needs to be further investigated.
Acknowledgments
This work was supported by Natural Science Foundation of Hebei Province (C2014201056), the Project for Improvement of Integrated Strength of Universities in Central and Western Areas of China (Plant diversity and resources utilization), Cutting-edge and Characteristic Disciplines of Biology (Botany), and the Bioengineering Key Discipline of Hebei Province, China.
References Cited
- Arakane Y., Dixit R., Begum K., Park Y., Specht C. A., Merzendorfer H., Kramer K. J., Muthukrishnan S., Beeman R. W. 2009. Analysis of functions of the chitin deacetylase gene family in Tribolium castaneum. Insect Biochem. Mol. Biol. 39: 355–365. [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M. W., Shipley G. L., et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55: 611–622. [DOI] [PubMed] [Google Scholar]
- Campbell P. M., Cao A. T., Hines E. R., East P. D., Gordon K. H. 2008. Proteomic analysis of the peritrophic matrix from the gut of the caterpillar, Helicoverpa armigera. Insect Biochem. Mol. Biol. 38: 950–958. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S. K., Mandaokar A., Kumar P. A., Sharma R. P. 1998. Efficacy of lepidopteran specific delta-endotoxins of Bacillus thuringiensis against Helicoverpa armigera. J. Invertebr. Pathol. 72: 336–337. [DOI] [PubMed] [Google Scholar]
- Chen L., Ma W., Wang X., Niu C., Lei C. 2010. Analysis of pupal head proteome and its alteration in diapausing pupae of Helicoverpa armigera. J. Insect Physiol. 56: 247–252. [DOI] [PubMed] [Google Scholar]
- Cohen E. 1993. Chitin synthesis and degradation as targets for pesticide action. Arch. Insect Biochem. Physiol. 22: 245–261. [DOI] [PubMed] [Google Scholar]
- Dixit R., Arakane Y., Specht C. A., Richard C., Kramer K. J., Beeman R. W., Muthukrishnan S. 2008. Domain organization and phylogenetic analysis of proteins from the chitin deacetylase gene family of Tribolium castaneum and three other species of insects. Insect Biochem. Mol. Biol. 38: 440–451. [DOI] [PubMed] [Google Scholar]
- Guo W., Li G., Pang Y., Wang P. 2005. A novel chitin-binding protein identified from the peritrophic membrane of the cabbage looper, Trichoplusia ni. Insect Biochem. Mol. Biol. 35: 1224–1234. [DOI] [PubMed] [Google Scholar]
- Jakubowska A. K., Caccia S., Gordon K. H., Ferre J., Herrero S. 2010. Downregulation of a chitin deacetylase-like protein in response to baculovirus infection and its application for improving baculovirus infectivity. J. Virol. 84: 2547–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Kramer K. J., Muthukrishnan S. 1997. Insect chitinases: molecular biology and potential use as biopesticides. Insect Biochem. Mol. Biol. 27: 887–900. [DOI] [PubMed] [Google Scholar]
- Lertcanawanichakul M., Wiwat C., Bhumiratana A., Dean D. H. 2004. Expression of chitinase-encoding genes in Bacillus thuringiensis and toxicity of engineered B. thuringiensis subsp. aizawai toward Lymantria dispar larvae. Curr. Microbiol. 48: 175–181. [DOI] [PubMed] [Google Scholar]
- Li J., Li X., Chen Y., Yang Z., Guo S. 2012. Solexa sequencing based transcriptome analysis of Helicoverpa armigera larvae. Mol. Biol. Rep. 39: 11051–11059. [DOI] [PubMed] [Google Scholar]
- Li Y., Qin Y., Yang N., Sun Y., Yang X., Sun R., Wang Q., Ling Y. 2013. Studies on insecticidal activities and action mechanism of novel benzoylphenylurea candidate NK-17. PLoS One 8: e66251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig S., Batz T., Armbruster K., Krasnow M. A. 2006. Serpentine and vermiform encode matrix proteins with chitin binding and deacetylation domains that limit tracheal tube length in Drosophila. Curr. Biol. 16: 186–194. [DOI] [PubMed] [Google Scholar]
- Palli S. R., Locke M. 1987. The synthesis of hemolymph proteins by the larval midgut of an insect Calpodes ethlius (Lepidoptera:Hesperiidae). Insect Biochem. 17: 561–572. [Google Scholar]
- Quan G., Ladd T., Duan J., Wen F., Doucet D., Cusson M., Krell P. J. 2013. Characterization of a spruce budworm chitin deacetylase gene: stage- and tissue-specific expression, and inhibition using RNA interference. Insect Biochem. Mol. Biol. 43: 683–691. [DOI] [PubMed] [Google Scholar]
- Sayed A., Wiechman B., Struewing I., Smith M., French W., Nielsen C., Bagley M. 2010. Isolation of transcripts from Diabrotica virgifera virgifera LeConte responsive to the Bacillus thuringiensis toxin Cry3Bb1. Insect Mol. Biol. 19: 381–389. [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Tabashnik B. E., Gassmann A. J., Crowder D. W., Carriere Y. 2008. Insect resistance to Bt crops: evidence versus theory. Nat. Biotechnol. 26: 199–202. [DOI] [PubMed] [Google Scholar]
- Thamthiankul S., Moar W. J., Miller M. E., Panbangred W. 2004. Improving the insecticidal activity of Bacillus thuringiensis subsp. aizawai against Spodoptera exigua by chromosomal expression of a chitinase gene. Appl. Microbiol. Biotechnol. 65: 183–192. [DOI] [PubMed] [Google Scholar]
- Toprak U., Baldwin D., Erlandson M., Gillott C., Hou X., Coutu C., Hegedus D. D. 2008. A chitin deacetylase and putative insect intestinal lipases are components of the Mamestra configurata (Lepidoptera: Noctuidae) peritrophic matrix. Insect Mol. Biol. 17: 573–585. [DOI] [PubMed] [Google Scholar]
- Vachon V., Laprade R., Schwartz J. L. 2012. Current models of the mode of action of Bacillus thuringiensis insecticidal crystal proteins: a critical review. J. Invertebr. Pathol. 111: 1–12. [DOI] [PubMed] [Google Scholar]
- Van Munster M., Prefontaine G., Meunier L., Elias M., Mazza A., Brousseau R., Masson L. 2007. Altered gene expression in Choristoneura fumiferana and Manduca sexta in response to sublethal intoxication by Bacillus thuringiensis Cry1Ab toxin. Insect Mol. Biol. 16: 25–35. [DOI] [PubMed] [Google Scholar]
- Wang S., Jayaram S. A., Hemphala J., Senti K. A., Tsarouhas V., Jin H., Samakovlis C. 2006. Septate-junction-dependent luminal deposition of chitin deacetylases restricts tube elongation in the Drosophila trachea. Curr. Biol. 16: 180–185. [DOI] [PubMed] [Google Scholar]
- Xi Y., Pan P. L., Ye Y. X., Yu B., Zhang C. X. 2014. Chitin deacetylase family genes in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Mol. Biol. 23: 695–705. [DOI] [PubMed] [Google Scholar]
- Yao J., Buschman L. L., Lu N., Khajuria C., Zhu K. Y. 2014. Changes in gene expression in the larval gut of Ostrinia nubilalis in Response to Bacillus thuringiensis Cry1Ab protoxin ingestion. Toxins (Basel) 6: 1274–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Park R. D., Muzzarelli R. A. 2010. Chitin deacetylases: properties and applications. Mar. Drugs 8: 24–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X. W., Wang X. H., Tan X., Xia Q. Y., Xiang Z. H., Zhao P. 2014. Identification and molecular characterization of a chitin deacetylase from Bombyx mori peritrophic membrane. Int. J. Mol. Sci. 15: 1946–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]