Abstract
Since the first report of induced pluripotent stem cells (iPSCs) by Takahashi and Yamanaka, numerous attempts have been made to derive iPSCs from other species via the ectopic expression of defined factors. Sheep iPSCs (siPSCs) have significant potential for biotechnology and agriculture. Although several groups have described siPSCs, the reprogramming efficiency was extremely low. The exogenous transgenes could be not silenced in the iPSCs, which hampered their development and application. Here, we report that p53 knockdown and antisilencing function 1A (ASF1A) overexpression promoted iPSC generation from sheep kidney cells (SKCs). Compared with transduction with eight human defined transcription factors (Oct4, Sox2, Klf4, c-Myc, Nanog, Lin28, hTERT, and SV40LT), the additional introduction of p53 RNA interference (RNAi) and/or ASF1A in the presence of small-molecule compounds [vitamin C (Vc) and valproic acid (VPA)] greatly improved the efficiency of sheep iPSC generation. The siPSCs exhibited morphological features similar to mouse embryonic stem cells (ESCs) and were positive for alkaline phosphatase and, pluripotent marker genes (Oct4, Nanog, Sox2, Rex1, TRA-1-60, TRA-1-81, and E-cadherin). Furthermore, these cells exhibited a normal karyotype of 54 chromosomes and were able to differentiate into all three germ layers both in vitro and in vivo. Moreover, the exogenous genes were silenced in siPSCs when p53 small hairpin RNA (shRNA) and ASF1A were added. Our results may help to reveal the role of p53 and ASF1A in sheep somatic cell reprogramming and provide an efficient approach to reprogramming sheep somatic cells.
Introduction
Since 2006, induced pluripotent stem cells (iPSCs) have opened a new era of reprogramming, providing a powerful tool in regenerative medicine and for the study of the molecular mechanisms of reprogramming (Park et al., 2008; Takahashi and Yamanaka, 2006a; Wernig et al., 2007; Yamanaka, 2012; Yamanaka and Blau, 2010; Yu et al., 2007). Sheep iPSCs (siPSCs) have tremendous promise for both basic biotechnology research and agricultural applications, such as improving the production traits and disease resistance of sheep as well as biopharming (Malaver-Ortega et al., 2012; Plews et al., 2012). At present, reprogramming sheep somatic cells to iPSCs can be achieved by virus-mediated transduction of defined transcription factors.
The first siPSCs were described by two groups in early January of 2011. Bao et al. obtained siPSCs from sheep primary ear fibroblasts using a doxycycline (DOX)-inducible lentivirus expressing the human defined factors Oct4, Sox2, c-Myc, Klf4, Nanog, Lin28, SV40 large T, and human telomerase reverse transcriptase (OSCKNL + T + hTERT). These siPSCs resembled mouse embryonic stem cells (ESCs) in morphology and properties (Bao et al., 2011). However, Li et al. used a cocktail of DOX-controlled tetracycline (TET)-on-inducible lentiviruses containing mouse-derived defined factors [Oct4, Sox2, c-Myc, and Klf4 (OSCK)] to generate siPSCs, and these cells resembled human ESCs (Li et al., 2011). In these two cases, the siPSC lines could not maintain pluripotency without exogenous transgenes, indicating partial reprogramming. The efficiency was very low (∼0.041% by Bao et al.). Two other groups used retroviruses with four human transcription factors to reprogram sheep fibroblasts to pluripotency (Liu et al., 2012b; Sartori et al., 2012). These siPSCs partially contributed to liveborn chimeric lambs (Sartori et al., 2012), but the efficiency remained low (Liu et al., 2012b, Sartori et al., 2012). Therefore, it was necessary to explore an efficient method to improve the generation of siPSCs for development and potential applications.
The tumor suppressor p53 (TP53 in humans and Trp53 in mice), known as the “guardian of the genome,” was recently shown to play vital roles in suppressing pluripotency and cellular dedifferentiation (Zhao and Xu, 2010). Several groups identified the role of the p53 pathway in iPSC formation and found that inhibiting the p53 pathway dramatically increased the efficiency of iPSC generation (Hong et al., 2009; Kawamura et al., 2009; Li et al., 2009; Marión et al., 2009; Menendez et al., 2010; Spike and Wahl, 2011). Furthermore, antisilencing function 1 (ASF1), the most conserved histone 3 and histone 4 chaperone, has been implicated in nucleosome assembly, transcriptional silencing, and the cellular response to DNA damage (Gonzalez-Muñoz et al., 2014; Mousson et al., 2005; Tagami et al., 2004; Tyler et al., 1999). Recent studies have focused on the necessary role of ASF1A, a histone-remodeling chaperone specifically enriched in metaphase II human oocytes, for cellular reprogramming of somatic cells into undifferentiated iPSCs. Overexpression of ASF1A and OCT4 alone in human adult dermal fibroblasts (hADFs) exposed to the oocyte-specific growth factor growth differentiation factor 9 (GDF9) reprogrammed hADFs into pluripotent cells (Gonzalez-Muñoz et al., 2014). However, information on the effect of p53 inhibition and ASF1A on somatic cell reprogramming in sheep is lacking.
In our study, p53 small hairpin RNA (shRNA) and/or ASF1A together with eight human defined transcription factors were introduced into the kidney cells of a Chinese Merino sheep fetus. The efficiency of sheep somatic cell reprogramming to pluripotent cells was significantly improved compared with transduction with eight defined factors alone. Furthermore, these iPSCs are similar to ESCs in terms of marker gene expression, pluripotency, karyotype, epigenetic status, and differentiation into all three germ layers through embryoid bodies (EBs) and teratomas.
Materials and Methods
Cell culture
Mouse embryonic fibroblasts (MEFs) were derived from CF-1 stain mice (Slaccas, China) at 12.5 days after fertilization and served as feeder layers. The kidney was removed from a Chinese Merino fetus (∼45 day), and its cortex was divided into approximately 1-mm3 tissue pieces, washed twice with culture medium, and placed into a 60-mm cell culture dish containing a small amount of Dulbecco's Modified Eagle Medium (DMEM) plus 10% fetal bovine serum (FBS; Gibco). Tissue pieces were removed when a significant number of primary sheep kidney cells (SKCs) were apparent. MEFs and SKCs were cultured in DMEM supplemented with 10% FBS and 2 mM nonessential amino acids (NEAA) (Gibco). siPSCs were cultured on X-ray–irradiated CF-1 MEFs in DMEM/F12 medium plus 20% KnockOut Serum Replacement (Gibco), 1 mM GlutaMAX (Gibco), 2 mM NEAA, 0.1 mM β-mercaptoethanol (Sigma), 1 μg/ml DOX (Chemicon), 4 ng/mL basic fibroblast growth factor (bFGF; Peprotech), and the small molecules [50 μg/mL vitamin C (Vc) and 1 mM valproic acid (VPA)].
Plasmids construction
shRNAs targeting the sheep p53 gene (GenBank acc. no. FJ855223) were designed using the clontech RNAi Target Sequence Selector and cloned into the lentivirus pLL3.7 vector. The sheep ASF1A gene (GenBank accession number GQ221052) was amplified from the cDNA of SKCs and cloned into the lentiviral expression vector pLEX-MCS. The lentiviral vectors (pLL3.7 and pLEX-MCS) and the corresponding helper plasmids were kindly provided by Mingjun Liu (Liu et al., 2014). Primers for RNA interference (RNAi) and ASF1A are provided in Table S1 (Supplementary Data are available at www.liebertpub.com/cell/).
Lentivirus generation and transduction
The DOX-inducible lentivirus system containing eight human defined transcription factors (Oct4, Sox2, Klf4, c-Myc, Nanog, Lin28, hTERT, and SV40LT). The helper plasmids (VSVG and Δ8.9) were kindly provided by Prof. Lei Xiao (Bao et al., 2011). The lentiviruses were packaged in 293FT cells and concentrated with an Amicon Ultra-15 centrifugal filter unit (6000 × g, 0.5 h), and titrated by infecting 293FT cells with a series of dilutions. The generation of the p53 RNAi lentivirus and ASF1A lentivirus were performed as previously described (Liu et al., 2012a, 2014). Approximately 5 × 104 SKCs per well were plated into a six-well plate and infected with viral supernatant at 10 multiplicity of infections (MOI) in the presence of Polybrene (8 μg/mL). The medium was replaced by fresh SKC medium at 12 h postinfection.
Generation of iPSCs
At 48 h postinfection, infected SKCs were dissociated with TrypLE (Gibco) and plated on a six-well plate coated with feeders. Culture medium was replaced by ESC medium plus 1 μg/mL DOX. At 11 and 17 days postinfection, these cells were trypsinized and passaged twice. ESC-like colonies were picked and transferred onto a new feeder-coated 96-well plate supplemented with ESC medium and DOX at 23 days postinfection as described previously (Takahashi et al., 2007a). The remaining plates were subjected to alkaline phosphatase (AP) staining, and the number of colonies was counted to calculate the efficiency of colony generation. The efficiency of sheep iPSC generation was calculated using the ratio of AP-positive colonies from 5 × 104 fibroblasts.
AP staining
AP staining was performed using a nitro-blue tetrazolium and 5-bromo-4-chloro-3′-indolyphosphate (NBT/BCIP) AP color development kit (Beyotime, China) according to the manufacturer's instructions.
Western blotting
Western blotting was performed as previously described (Fu et al., 2014) to determine the protein levels of p53, p21, and ASF1A. The primary antibodies used were as follows: Rabbit anti-p53 (1:2000; BS6437, Bioworld), rabbit anti-p21 (1:1500; BS6561, Bioworld), and goat anti-ASF1A (1:2500; ab65942, Abcam).
Real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen) and reverse transcribed to cDNA using a PrimeScript™ First Strand cDNA Synthesis kit (TaKaRa, China). Real-time PCR was performed on a Roche Light-Cycler Roche 480 with the Roche Light-Cycler Roche 480 master kit. The 2−ΔΔCt method (Livak and Schmittgen, 2001) was applied to analyze the transcription levels of genes, which were normalized to the internal control [glyceraldehyde 3-phosphate dehydrogenase (GAPDH)] levels.
Immunocytochemistry
The iPSCs were fixed with 4% paraformaldehyde for 15 min at room temperature and blocked with nonprotein blocking solution plus 0.1% Triton X-100 for 1 h (intracellular proteins) and incubated with primary antibodies at 4°C overnight and secondary antibodies for 1 h. The primary antibodies against Oct4 (1:100; Santa Cruz), Sox2 (1:1000; Millipore), Nanog (1:100; Santa Cruz), SSEA1 (Ascites, 1:400; Developmental Studies Hybridoma Bank), Tra-1-60 (1:100; Millipore), Tra-1-81 (1:100; Millipore), Rex1 (1:200; Santa Cruz), E-Cadherin (1:100; BD), βIII-Tubulin (1:100; Santa Cruz), SMA (1:150; Santa Cruz), and Sox17 (1:100; R&D) were used in immunocytochemistry analysis. The Cy3-labeled donkey anti-mouse immunoglobulin G (IgG), anti-rabbit IgG, anti-mouse IgM, and anti-rat IgM were purchased from Jackson Immuno-Research Laboratories and served as the secondary antibodies.
Bisulfite genomic sequencing
Bisulfite treatment was performed using an EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA, USA) according to the manufacturer's instructions for the bisulfite genomic sequencing of the Nanog promoter regions as previously described (Bao et al., 2011).
Karyotyping
The siPSCs at passage 20 were incubated with 0.3 mg/mL colcemid for 3 h and trypsinized for karyotype analysis as previously described (Li et al., 2011).
In vitro and in vivo differentiation
In vitro differentiation of siPSCs through EB formation with the pendant drop method was performed as previously described (Li et al., 2011; Takahashi et al., 2007b; Takahashi and Yamanaka, 2006b). PCR analysis of various differentiation markers for the three germ layers in EBs derived from siPSCs compared with the parental SKCs was performed as previously described (Bao et al., 2011). After 8 days of floating culture, EBs were transferred to gelatin-coated plates and cultured in 10% fetal bovine serum (FBS) DMEM medium for an additional 8 days. Immunofluorescence staining for the three germ layers was carried out as described above to detect differentiation markers (βIII-Tubulin, Sox17, and SMA). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI).
To assess further the pluripotency of siPSCs in vivo, each siPSC was individually injected subcutaneously into five nonobese diabetes/severe-combined immunodeficient (NOD/SCID) mice (four injected sites—double hind limbs, dorsum, and neck, approximately 107 cells/per site). Teratomas formed 4 to 6 weeks postinjection, and palpable tumors were subjected to histological examination as previously described (Bao et al., 2011).
Statistical analysis
Statistical analysis was performed using SPSS 17.0 for Windows (SPSS Inc. Chicago, IL, USA). Values of *p < 0.05 and **p < 0.01 were determined using the Student two-tailed t-test for independent samples and considered to be significant and highly significant, respectively.
Results
RNAi targeting p53 downregulates the expression of p53 and p21
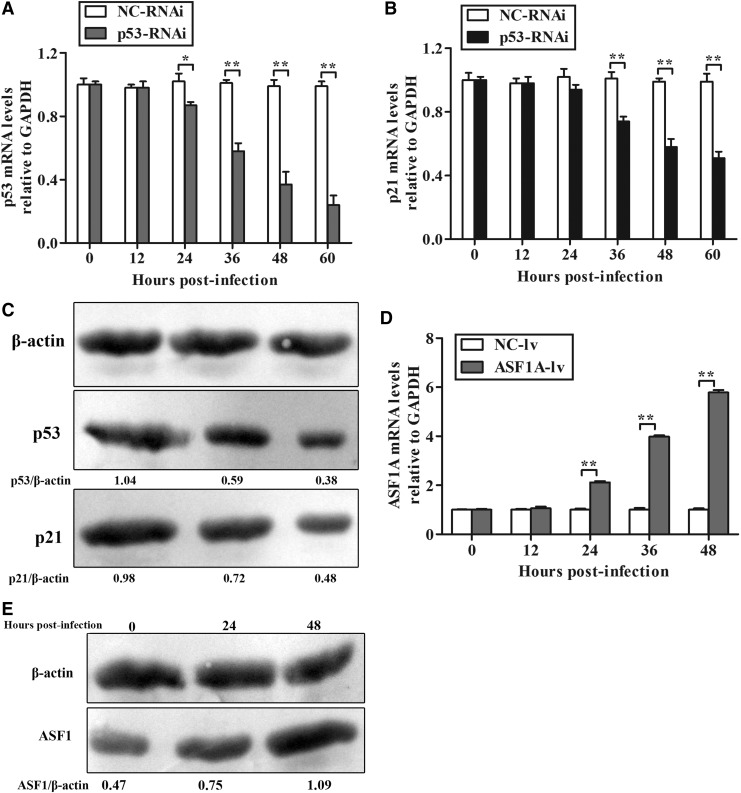
The shRNA was designed and cloned into a lentivirus vector to downregulate p53 expression. As shown in Figure 1A, p53 mRNA levels were decreased significantly in p53 RNAi lentivirus-infected cells at 24 h postinfection compared with the negative control. With increased postinfection intervals, the p53 mRNA levels decreased consistently. Most significantly, the p53 mRNA level in the p53 RNAi lentivirus-infected sample was reduced approximately 0.75 units compared with the negative control sample at 60 h postinfection. Western blotting further demonstrated that p53 RNAi inhibited p53 expression (Fig. 1C). Collectively, these results demonstrate that p53 expression was significantly inhibited by RNAi targeting p53.
FIG. 1.
Detection of p53, p21, and ASF1A expression levels. (A, B, and D) p53, p21 and ASF1A mRNA levels, respectively, relative to GAPDH were detected by quantitative real-time PCR at different time points after lentiviral infection. The results are shown as the mean ± standard deviation (SD) (error bars), n = 3, (*) p < 0.05, (**) p < 0.01. (C and E) Protein levels of p53, p21, and ASF1A were detected by western blotting (n = 3). NC-lv, negative control lentivirus.
Previous studies demonstrated that the p53/p21 pathway serves as a barrier in iPSC generation (Hong et al., 2009). To investigate further the effects of p53 downregulation on the p53/p21 pathway, p21 mRNA and protein levels were determined by real-time PCR and western blot analysis. Over a series of postinfection time intervals, the p21 mRNA and protein levels were analyzed. Compared to the negative control, p21 mRNA levels were significantly decreased in p53 RNAi lentivirus-infected cells within 36 and 60 h postinfection (Fig. 1B). Western blotting was performed to further examine whether the inhibition of p21 expression corresponded to a reduction of p53 expression and p21 protein levels were significantly decreased in p53 RNAi lentivirus-infected cells within 36 and 60 h postinfection (Fig. 1C). Collectively, these results suggested that RNAi targeting p53 inhibited p53 expression and further suppressed p21 expression.
ASF1A overexpression after lentivirus transduction
To overexpress ASF1A in sheep somatic cells, the ASF1A gene was cloned into the lentivirus pLEX-MCS vector to generate the ASF1A-lentivirus. Twenty-four h postinfection, ASF1A mRNA levels significantly increased in ASF1A-lentivirus–infected cells compared with the negative control (Fig. 1D). Western blot results further suggested that ASF1A expression was increased significantly (Fig. 1E).
p53 RNAi and ASF1A overexpression improves the efficiency of siPSCs generation
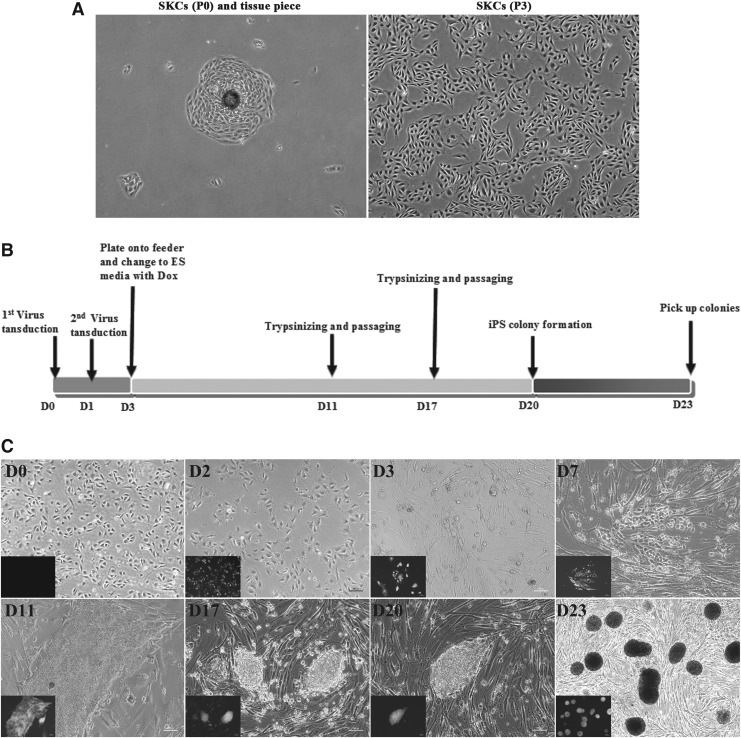
The tabular and slabstone-like kidney cells began to grow from minced fragments in 3–4 days (Fig. 2A). When a sufficient number of cells were noted (80–90%), the cells were passaged with TrypLE and plated into a 100-mm culture dish to expand the culture. After passaging for three to four generations, the cells with the best conditions were used for siPSC induction. The TET-on inducible enhanced green fluorescent protein (eGFP)-expressing lentiviral system carrying Oct4, Sox2, Klf4, c-Myc, Nanog, Lin28, hTERT, and SV40LT, respectively, was introduced to reprogram sheep somatic cells to pluripotent cells.
FIG. 2.
Schematic diagram and representative image of SKC reprogramming. (A) Tabular and slabstone-like kidney cells grew from the minced fragments and were expanded in culture to induce siPSC. (B) The paradigm used for siPSC induction from sheep kidney cells (SKCs). (C) Cell morphology and expression of enhanced green fluorescent protein (eGFP) induced by DOX in the lentiviral vector-based TET-on system at different times after lentivirus transduction.
As shown in Figure 2, B and C, 5 × 104 SKCs cells per well (D0) were plated into a six-well plate and infected with the defined factors lentivirus (MOI = 10) along with p53 RNAi and/or ASF1A lentivirus in the presence of Polybrene. At 48 h postinfection, lentivirus-infected cells (D3) expressing GFP at high levels were trypsinized and plated into a feeder-coated six-well plate. The medium was changed—ESC medium plus DOX, Vc, and VPA. At 3–7 days after lentivirus transduction, infected SKCs (D7) exhibited morphology changes and proliferated rapidly. The infected cells (D11 and D17) were trypsinized with TrypLE and passaged twice. Numerous siPSC colonies were observed at 20 days postinfection. At D23, colonies with clear boundaries were stochastically selected and placed into a 96-well plate to harvest with TrypLE for 5 min. Subsequently the cells were transferred onto a new feeder-coated 96-well plate for further expansion.
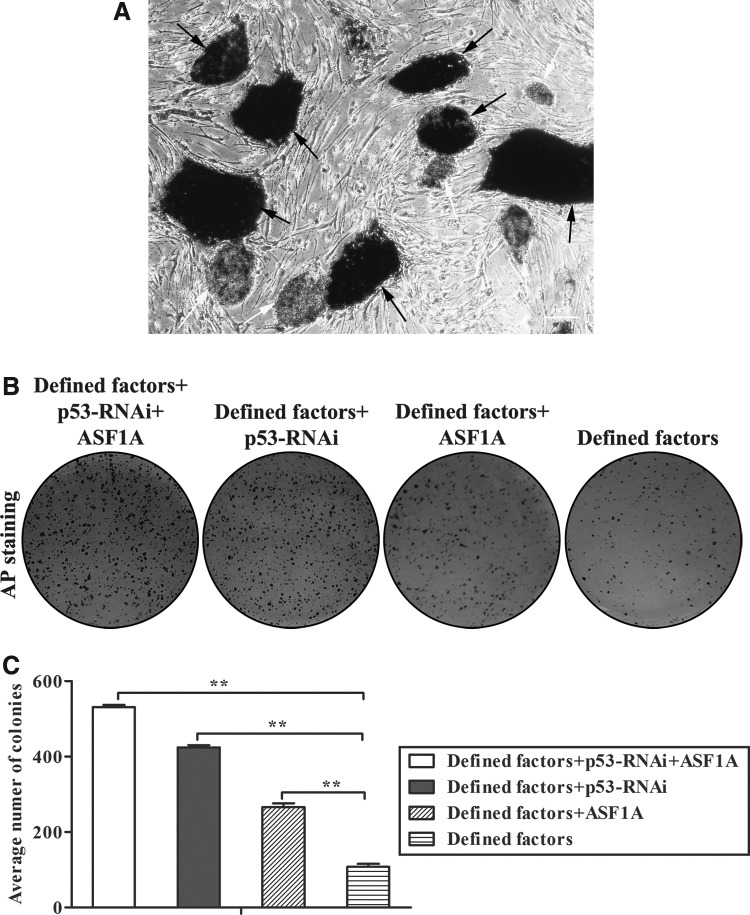
The remaining cells were subjected to AP staining, and the number of colonies was counted to calculate the efficiency of sheep somatic cell reprogramming. As shown in Figure 3A, AP-positive siPSC colonies were stained purple or dark purple. Approximately 531 AP-positive colonies were derived from 5 × 104 sheep somatic cells infected with a cocktail of lentiviruses (defined factors lentivirus, p53-RNAi-lv and ASF1A-lv), which is five-fold more than in the defined factors lentivirus treated group (about 107 AP-positive colonies from 5 × 104 fibroblasts) (Fig. 3B, C). Moreover, the introduction of p53-shRNA or ASF1A with defined factors also greatly improved the efficiency of AP-positive colonies (Fig. 3B, C). Collectively, these results indicated that p53 RNAi and ASF1A overexpression significantly increased the efficiency of siPSC generation.
FIG. 3.
AP staining and reprogramming efficiency calculation. (A) Representative image of AP-positive ESC-like colonies. (B) After selecting ESC-like colonies, the remaining cells treated with different lentiviruses were subjected to AP staining to calculate the efficiency of colony generation. A representative experiment is shown. (C) The number of AP-positive colonies generated from 5 × 104 SKCs was counted to calculate the reprogramming efficiency. The results are shown as the mean ± standard deviation (SD) (error bars), n = 3, (**) p < 0.01.
Sheep iPSCs exhibit typical ESC morphology and marker expression
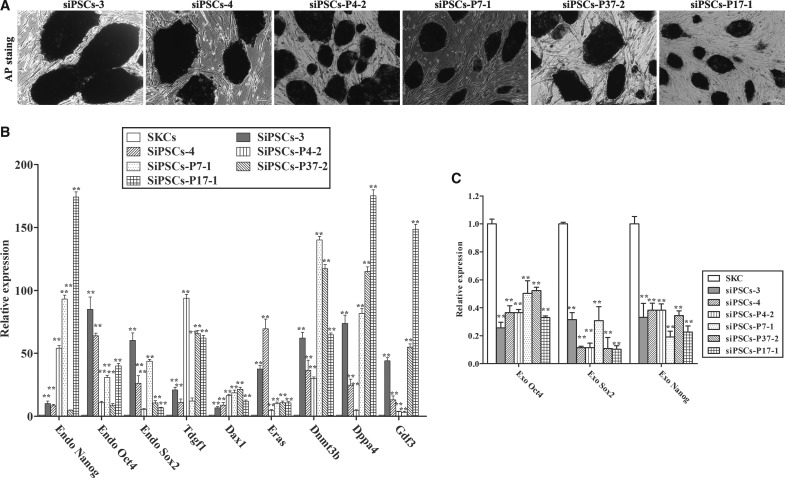
After picking over 200 colonies at 23 days postinfection and further examining whether colonies exhibited typical ESC morphology and markers expression, we successfully established six siPSCs, referred to as siPSC-3, siPSC-4, siPSC-4-2, siPSC-7-1, siPSC-37-2, and siPSC-17-1, which were stably passaged for greater than 30 generations.
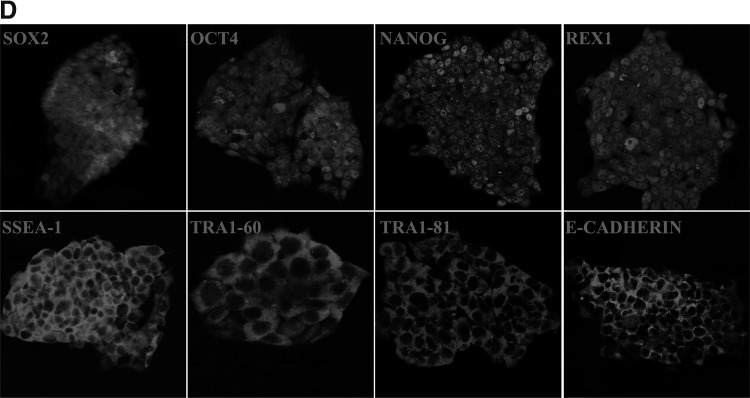
AP staining was performed subsequently to classify siPSC-3, siPSC-4, siPSC-4-2, siPSC-7-1, siPSC-37-2, and siPSC-17-1 as undifferentiated. As shown in Figure 4A, all siPSCs stained positive for AP. Moreover, real-time PCR was performed to evaluate the expression levels of ESC marker genes, including Nanog, Oct4, Sox2, Tdgf1, Dax1, Eras, Dnmt3b, Dppa4, and Gdf3. The results showed that siPSCs at passage 15 expressed these ESC pluripotent marker genes at high levels (Fig. 4B). We analyzed the expression of the three exogenous transgenes Oct4, Sox2, and Nanog in siPSCs at passage 10 by quantitative real-time PCR and found that all three genes were silenced (Fig. 4C). Furthermore, immunofluorescence staining revealed that siPSC colonies at passage 15 expressed ESC pluripotent makers, including SOX2, OCT4, NANOG, REX1, SSEA-1, TRA1-60, TRA1-81, and E-Cadherin (Fig. 4D).
FIG. 4.
Generation of sheep induced pluripotent stem cells (iPSCs) from sheep kidney cells (SKCs) by lentivirus transduction. (A) Representative image of alkaline phosphatase (AP) staining for siPSCs-3, siPSCs-4, siPSCs-P4-2, siPSCs-P7-1, siPSCs-P37-2, and siPSCs-P17-1. (B) Quantitative real-time PCR analyses of different pluripotency markers expressed in sheep iPSCs (siPSCs) relative to parental SKC. The results are shown as the mean ± SD (error bars), n = 3, **p < 0.01. (C) Quantitative real-time PCR analyses of the expression of the Oct4, Sox2, and Nanog exogenous transgenes in siPSCs relative to parental SKCs. The results are shown as the mean ± standard deviation (SD) (error bars), n = 3, **p < 0.01. (D) Immunocytochemistry analyses of pluripotency marker expression in siPSCs.
Epigenetic status of siPSCs and karyotyping
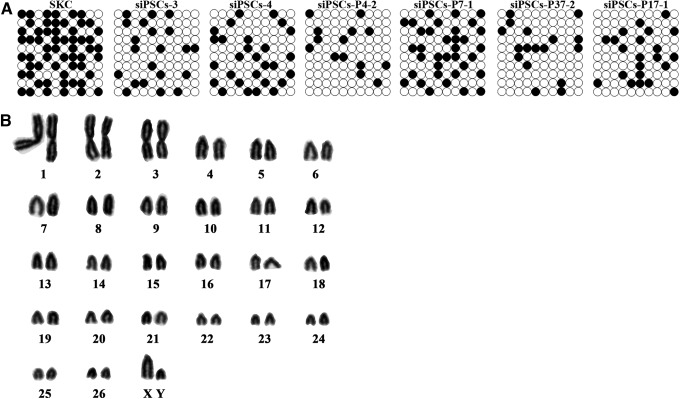
Bisulfite genomic sequencing was performed to evaluate the epigenetic status of siPSCs. The siPSC genomic DNA was extracted, treated with bisulfite, and subsequently subjected to PCR using primers as previously described (Bao et al., 2011). The methylation status of the Nanog promoter in siPSCs at passage 15 was evaluated, and the results indicated that the CpGs were highly unmethylated in siPSCs compared with SKCs (Fig. 5A), suggesting that the Nanog promoter was reactivated during SKC reprogramming. In addition, karyotyping was also conducted on six siPSCs at passage 20, and the siPSCs exhibited a normal chromosomal karyotype. Out of 80 metaphase spreads counted, 68 contained 54 chromosomes (Fig. 5B)
FIG. 5.
The epigenetic status of siPSCs was evaluated using bisulfite genomic sequencing and karyotype analysis. (A) Bisulfite genomic sequencing of the DNA methylation profile of the CpGs in the Nanog promoter region. (Open circles) unmethylated CpGs; (closed circles) methylated CpGs. (B) Karyotype analysis of siPSCs at passage 20. The results indicate that the siPSCs exhibited a normal karyotype of chromosomes. Out of 80 metaphase spreads counted, 68 contained 54 chromosomes.
In vitro and in vivo differentiation of siPSCs
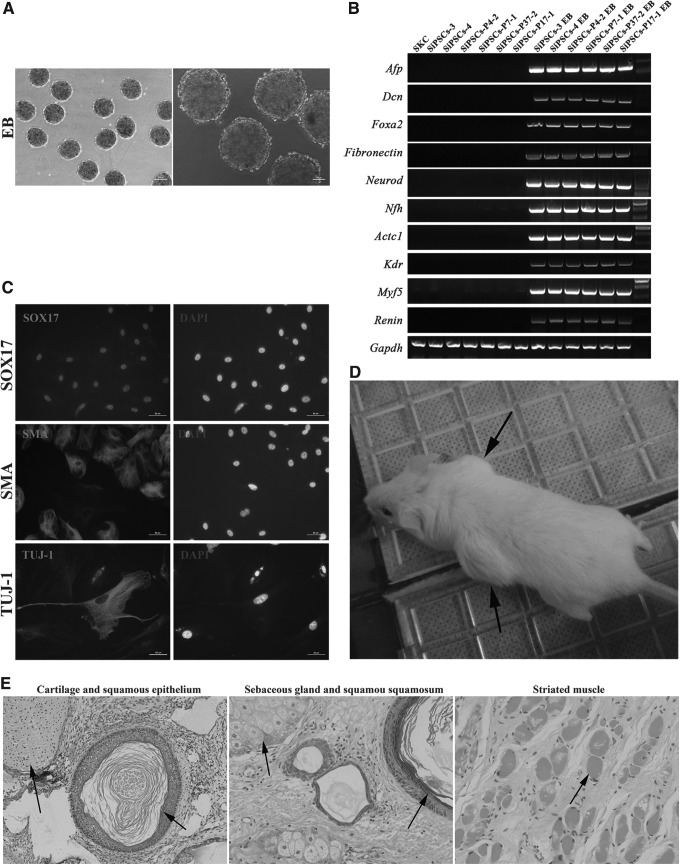
We then addressed whether siPSCs could differentiate into cell types of the three germ layers in vitro through EB-mediated differentiation. All the siPSCs we established effectively formed spherical EBs (Fig. 6A) and differentiated into cells with various morphologies after attachment. Total RNA of the EBs was extracted and reverse-transcribed into cDNA. Then the presence of endoderm (Afp, Dcn, and Foxa2), ectoderm (Fibronectin, Neurod, and Nfh), and mesoderm (Actc1, Kdr, Myf5, and Renin) germ layer markers was assessed. As shown in Figure 6B, the three germ layer markers were expressed at extremely high levels in EBs derived from siPSCs. Furthermore, the in vitro differentiation ability of siPSCs was subsequently analyzed by immunocytochemistry. As shown in Figure 6C, EBs derived from siPSCs were positive for TUJ-1 (βIII-Tubulin, ectoderm), SOX17 (endoderm), and SMA (smooth muscle actin, mesoderm). These results showed that siPSCs could be induced to randomly differentiate cells of the three germ layers.
FIG. 6.
siPSCs can differentiate into three germ layers in vitro and in vivo. (A) siPSCs were differentiated into EBs in vitro. (B) PCR analyses of various differentiation markers for the three germ layers in EBs. (C) Immunocytochemistry for detecting the differentiation markers of the three germ layers. (D) Teratoma formation in NOD/SCID mouse after injection with siPSCs. Black arrows indicate teratomas. (E) Histological sections from teratomas derived from siPSCs. A teratoma is composed of various types of tissues from all three germ layers. (Left) 1, cartilage (mesoderm); 2, squamous epithelium (ectoderm). (Middle) 1, sebaceous gland (endoderm); 2, squamous epithelium (ectoderm). (Right) striated muscle (mesoderm).
To examine further the pluripotency of siPSCs in vivo, NOD/SCID mice (Vital River Laboratories, Beijing, China) were injected subcutaneously with siPSCs. Six weeks later, palpable tumors were observed (Fig. 6D). Histological examination revealed that the representative differentiated derivatives included all three germ layers within the tumors, including squamous epithelium (ectoderm), cartilage (mesoderm), striated muscle (mesoderm), and sebaceous gland (endoderm) (Fig. 6E).
Discussion
In recent years, iPSCs have been generated with varied efficiency from multiple cell types, including adult or fetal fibroblasts (Mali et al., 2008), adipose tissue–derived cells (Sugii et al., 2010; Sun et al., 2009; Tat et al., 2010), renal epithelial cells (Zhou et al., 2012), keratinocytes (Maherali et al., 2008), bone marrow stromal cells (BMSCs) (Kuroda, 2010), and terminally differentiated B cells (Stadtfeld et al., 2012). In our previous study, we attempted to obtain siPSCs from skin fibroblasts, SKCs, and BMSCs derived from the same fetal Chinese Merino sheep using defined factors plus Vc and/or VPA. SKCs exhibited the potential to be efficiently reprogrammed to pluripotency. From the perspective of reprogramming dynamics, SKCs generated ESC-like colonies at 12 days postinfection, which was earlier than reprogramming with the other two cells types. In terms of the stoichiometry of reprogramming factors during the reprogramming process, increased expression of Oct4 and Klf4 combined with reduced expression of c-Myc and Sox2, the optimum combination to induce reprogramming (Carey et al., 2011). Klf4 is a transcription factor expressed in a variety of tissues, including the kidney epithelium, which may accelerate the conversion of these cells to siPSCs.
Increasing reprogramming efficiency by modulating epigenetic regulators has been highlighted in recent studies. Vc accelerates transcriptome changes during reprogramming and allows the conversion of pre-iPSCs to iPSCs (Esteban et al., 2010). Treatment with the histone deacetylase (HDAC) inhibitor VPA for 1 week improved the percentage of Oct4-GFP–positive cells by greater than 100-fold and 50-fold for three-factor (OSK) and four-factor (OSKM) reprogramming, respectively (Huangfu et al., 2008). Here, the modulatory regulator combination (Vc and VPA) or either agent alone was added to increase the siPSC generation efficiency. Although the growth rate and the amount of colonies improved as a result of Vc (50 μg/mL) addition, increased efficiency in the yield of highly AP-positive colonies was not noted. According to Huangfu et al. (2008), VPA (2 mM) addition improved the reprogramming efficiency of SKCs, but resulted in an increased amount of cell death. Therefore, the VPA concentration was changed to 1 mM and combined with Vc to increase the efficiency of siPSC generation. Interestingly, not only the amount and growth rate of colonies, but also the ratio of AP+ colonies to total colonies increased.
Many research groups have reported that p53 downregulation is closely related to increased reprogramming efficiency. Indeed, among the first genetic events during cell reprogramming is the inactivation of the p53/p21 pathway (Hong et al., 2009; Li et al., 2009). Furthermore, ASF1A, which is highly expressed in oocytes, plays an important role in cell reprogramming and promotes its efficiency. Therefore, we combined these approaches promoting reprogramming and effectively improved the efficiency of siPSC generation. More importantly, exogenous genes could not be silenced in previous studies (Bao et al., 2011; Li et al., 2011). Surprisingly, we found that exogenous transgenes were significantly silenced in this study, which is potentially related to p53 downregulation and/or ASF1A overexpression. Therefore, systematically investigating the molecular mechanisms of ASF1A and p53 in promoting complete reprogramming provides important significance to obtaining siPSCs and sheep chimera. Additionally, Gonzalez-Muñoz have reported that ASF1A and OCT4 overexpression in human adult dermal fibroblasts (hADFs) exposed to the oocyte-specific paracrine growth factor GDF9 can reprogram hADFs into pluripotent cells (Gonzalez-Muñoz et al., 2014). Reducing defined transcription factors and exploring the reprogramming methods with high efficiency, high security, and simplification are urgent problems to be solved.
Supplementary Material
Acknowledgments
We thank Prof. Lei Xiao (College of Animal Science, Zhejiang University, Hangzhou, China) for kindly providing the DOX-inducible lentivirus system. We also thank Lijuan Pang and Weiwei Cao (College of Medicine, Shihezi University, Shihezi, China) for histological examination and technical assistance with confocal microscopy. This work was supported by the Natural Science Foundation of China (grant nos. 31560328, 31260534, and 31201800) and International S&T Cooperation Program of China (grant no. 2013DFR30970).
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Bao L., He L., Chen J., Wu Z., Liao J., Rao L., Ren J., Li H., Zhu H., and Qian L. (2011). Reprogramming of ovine adult fibroblasts to pluripotency via drug-inducible expression of defined factors. Cell Res. 21, 600–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Hanna J.H., Faddah D.A., Buganim Y., Kim J., Ganz K., Steine E.J., Cassady J.P., and Creyghton M.P. (2011). Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell 9, 588–598 [DOI] [PubMed] [Google Scholar]
- Esteban M.A., Wang T., Qin B., Yang J., Qin D., Cai J., Li W., Weng Z., Chen J., and Ni S. (2010). Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 6, 71–79 [DOI] [PubMed] [Google Scholar]
- Fu Q., Shi H., Ren Y., Guo F., Ni W., Qiao J., Wang P., Zhang H., and Chen C. (2014). Bovine viral diarrhea virus infection induces autophagy in MDBK cells. J. Microbiol. 52, 619–625 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Muñoz E., Arboleda-Estudillo Y., Otu H.H., and Cibelli J.B. (2014). Histone chaperone ASF1A is required for maintenance of pluripotency and cellular reprogramming. Science 345, 822–825 [DOI] [PubMed] [Google Scholar]
- Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., and Yamanaka S. (2009). Suppression of induced pluripotent stem cell generation by the p53–p21 pathway. Nature 460, 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A.E., and Melton D.A. (2008). Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 26, 795–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T., Suzuki J., Wang Y.V., Menendez S., Morera L.B., Raya A., Wahl G.M., and Belmonte J.C.I. (2009). Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460, 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda S. (2010). Transplantation of autologous bone marrow stromal cells (BMSC) for CNS disorders—Strategy and tactics for clinical application. J. Stem Cells Regen. Med. 6, 167. [PubMed] [Google Scholar]
- Li H., Collado M., Villasante A., Strati K., Ortega S., Cañamero M., Blasco M.A., and Serrano M. (2009). The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460, 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Cang M., Lee A.S., Zhang K., and Liu D. (2011). Reprogramming of sheep fibroblasts into pluripotency under a drug-inducible expression of mouse-derived defined factors. PLoS One 6, e15947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Li W., Zhang X., Zhang N., He S., Huang J., Ge Y., and Liu M. (2012a). The critical role of myostatin in differentiation of sheep myoblasts. Biochem. Biophys. Res. Commun. 422, 381–386 [DOI] [PubMed] [Google Scholar]
- Liu C., Li W., Zhang X., Zhang N., He S., Huang J., Ge Y., and Liu M. (2014). Knockdown of endogenous myostatin promotes sheep myoblast proliferation. In Vitro Cell Dev. Biol. Anim. 50, 94–102 [DOI] [PubMed] [Google Scholar]
- Liu J., Balehosur D., Murray B., Kelly J.M., Sumer H., and Verma P.J. (2012b). Generation and characterization of reprogrammed sheep induced pluripotent stem cells. Theriogenology 77, 338–346. e331 [DOI] [PubMed] [Google Scholar]
- Livak K.J., and Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Maherali N., Ahfeldt T., Rigamonti A., Utikal J., Cowan C., and Hochedlinger K. (2008). A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell 3, 340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaver-Ortega L.F., Sumer H., Liu J., and Verma P.J. (2012). The state of the art for pluripotent stem cells derivation in domestic ungulates. Theriogenology 78, 1749–1762 [DOI] [PubMed] [Google Scholar]
- Mali P., Ye Z., Hommond H.H., Yu X., Lin J., Chen G., Zou J., and Cheng L. (2008). Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells 26, 1998–2005 [DOI] [PubMed] [Google Scholar]
- Marión R.M., Strati K., Li H., Murga M., Blanco R., Ortega S., Fernandez-Capetillo O., Serrano M., and Blasco M.A. (2009). A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460, 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez S., Camus S., and Belmonte J.C.I. (2010). p53: Guardian of reprogramming. Cell Cycle 9, 3887–3891 [DOI] [PubMed] [Google Scholar]
- Mousson F., Lautrette A., Thuret J.-Y., Agez M., Courbeyrette R., Amigues B., Becker E., Neumann J.-M., Guerois R., and Mann C. (2005). Structural basis for the interaction of Asf1 with histone H3 and its functional implications. Proc. Natl. Acad. Sci. USA 102, 5975–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.-H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., and Daley G.Q. (2008). Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146 [DOI] [PubMed] [Google Scholar]
- Plews J.R., Gu M., Longaker M.T., and Wu J.C. (2012). Large animal induced pluripotent stem cells as pre-clinical models for studying human disease. J. Cell. Mol. Med. 16, 1196–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori C., DiDomenico A.I., Thomson A.J., Milne E., Lillico S.G., Burdon T.G., and Whitelaw C.B.A. (2012). Ovine-induced pluripotent stem cells can contribute to chimeric lambs. Cell. Rep. 14, 8–19 [DOI] [PubMed] [Google Scholar]
- Spike B.T., and Wahl G.M. (2011). P53, stem cells, and reprogramming tumor suppression beyond guarding the genome. Gene. Canc. 2, 404–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Apostolou E., Ferrari F., Choi J., Walsh R.M., Chen T., Ooi S.S., Kim S.Y., Bestor T.H., and Shioda T. (2012). Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat. Genet. 44, 398–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugii S., Kida Y., Kawamura T., Suzuki J., Vassena R., Yin Y.-Q., Lutz M.K., Berggren W.T., Belmonte J.C.I., and Evans R.M. (2010). Human and mouse adipose-derived cells support feeder-independent induction of pluripotent stem cells. Proc. Natl. Acad. Sci. USA 107, 3558–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N., Panetta N.J., Gupta D.M., Wilson K.D., Lee A., Jia F., Hu S., Cherry A.M., Robbins R.C., and Longaker M.T. (2009). Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc. Natl. Acad. Sci. USA 106, 15720–15725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H., Ray-Gallet D., Almouzni G., and Nakatani Y. (2004). Histone H3. 1 and H3. 3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 [DOI] [PubMed] [Google Scholar]
- Takahashi K., and Yamanaka S. (2006a). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- Takahashi K., and Yamanaka S. (2006b). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Okita K., Nakagawa M., and Yamanaka S. (2007a). Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2, 3081–3089 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., and Yamanaka S. (2007b). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- Tat P.A., Sumer H., Jones K.L., Upton K., and Verma P.J. (2010). The efficient generation of induced pluripotent stem (iPS) cells from adult mouse adipose tissue-derived and neural stem cells. Cell Transplant. 19, 525–536 [DOI] [PubMed] [Google Scholar]
- Tyler J.K., Adams C.R., Chen S.-R., Kobayashi R., Kamakaka R.T., and Kadonaga J.T. (1999). The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402, 555–560 [DOI] [PubMed] [Google Scholar]
- Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B.E., and Jaenisch R. (2007). In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448, 318–324 [DOI] [PubMed] [Google Scholar]
- Yamanaka S. (2012). Induced pluripotent stem cells: Past, present, and future. Cell Stem Cell 10, 678–684 [DOI] [PubMed] [Google Scholar]
- Yamanaka S., and Blau H.M. (2010). Nuclear reprogramming to a pluripotent state by three approaches. Nature 465, 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., and Stewart R. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- Zhao T., and Xu Y. (2010). p53 and stem cells: New developments and new concerns. Trends Cell Biol. 20, 170–175 [DOI] [PubMed] [Google Scholar]
- Zhou T., Benda C., Dunzinger S., Huang Y., Ho J.C., Yang J., Wang Y., Zhang Y., Zhuang Q., and Li Y. (2012). Generation of human induced pluripotent stem cells from urine samples. Nat. Protoc. 7, 2080–2089 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.