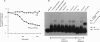Abstract
The AraC protein, which regulates the L-arabinose operons in Escherichia coli, was dissected into two domains that function in chimeric proteins. One provides a dimerization capability and binds the ligand arabinose, and the other provides a site-specific DNA-binding capability and activates transcription. In vivo and in vitro experiments showed that a fusion protein consisting of the N-terminal half of the AraC protein and the DNA-binding domain of the LexA repressor dimerizes, binds well to a LexA operator, and represses expression of a LexA operator-beta-galactosidase fusion gene in an arabinose-responsive manner. In vivo and in vitro experiments also showed that a fusion protein consisting of the C-terminal half of the AraC protein and the leucine zipper dimerization domain from the C/EBP transcriptional activator binds to araI and activates transcription from a PBAD promoter-beta-galactosidase fusion gene. Dimerization was necessary for occupancy and activation of the wild-type AraC binding site.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann E., Ochs B., Abel K. J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988 Sep 30;69(2):301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985 Dec;43(3 Pt 2):729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Brunelle A., Schleif R. Determining residue-base interactions between AraC protein and araI DNA. J Mol Biol. 1989 Oct 20;209(4):607–622. doi: 10.1016/0022-2836(89)90598-6. [DOI] [PubMed] [Google Scholar]
- Carra J. H., Schleif R. F. Variation of half-site organization and DNA looping by AraC protein. EMBO J. 1993 Jan;12(1):35–44. doi: 10.1002/j.1460-2075.1993.tb05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J. Regulation of the regulatory gene for the arabinose pathway, araC. J Mol Biol. 1976 Jul 5;104(3):557–566. doi: 10.1016/0022-2836(76)90120-0. [DOI] [PubMed] [Google Scholar]
- Godowski P. J., Picard D., Yamamoto K. R. Signal transduction and transcriptional regulation by glucocorticoid receptor-LexA fusion proteins. Science. 1988 Aug 12;241(4867):812–816. doi: 10.1126/science.3043662. [DOI] [PubMed] [Google Scholar]
- Granger-Schnarr M., Benusiglio E., Schnarr M., Sassone-Corsi P. Transformation and transactivation suppressor activity of the c-Jun leucine zipper fused to a bacterial repressor. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4236–4239. doi: 10.1073/pnas.89.10.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. C. A structural taxonomy of DNA-binding domains. Nature. 1991 Oct 24;353(6346):715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Hendrickson W., Schleif R. F. Regulation of the Escherichia coli L-arabinose operon studied by gel electrophoresis DNA binding assay. J Mol Biol. 1984 Sep 25;178(3):611–628. doi: 10.1016/0022-2836(84)90241-9. [DOI] [PubMed] [Google Scholar]
- Hu J. C., O'Shea E. K., Kim P. S., Sauer R. T. Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science. 1990 Dec 7;250(4986):1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- Jordan S. R., Pabo C. O. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988 Nov 11;242(4880):893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., Adashi E. Y., Graves B. J., McKnight S. L. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988 Jul;2(7):786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- Lee N., Francklyn C., Hamilton E. P. Arabinose-induced binding of AraC protein to araI2 activates the araBAD operon promoter. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8814–8818. doi: 10.1073/pnas.84.24.8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L. L., Little J. W. Autodigestion and RecA-dependent cleavage of Ind- mutant LexA proteins. J Mol Biol. 1989 Dec 5;210(3):439–452. doi: 10.1016/0022-2836(89)90121-6. [DOI] [PubMed] [Google Scholar]
- Little J. W., Hill S. A. Deletions within a hinge region of a specific DNA-binding protein. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2301–2305. doi: 10.1073/pnas.82.8.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell R. B., Schleif R. F. DNA looping and unlooping by AraC protein. Science. 1990 Oct 26;250(4980):528–532. doi: 10.1126/science.2237403. [DOI] [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- Menon K. P., Lee N. L. Activation of ara operons by a truncated AraC protein does not require inducer. Proc Natl Acad Sci U S A. 1990 May;87(10):3708–3712. doi: 10.1073/pnas.87.10.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón A., Wolberger C., Harrison S. C. Structure of phage 434 Cro protein at 2.35 A resolution. J Mol Biol. 1989 Jan 5;205(1):179–188. doi: 10.1016/0022-2836(89)90374-4. [DOI] [PubMed] [Google Scholar]
- Ogden S., Haggerty D., Stoner C. M., Kolodrubetz D., Schleif R. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R. E. The nucleotide sequence of pACYC177. Nucleic Acids Res. 1988 Jan 11;16(1):356–356. doi: 10.1093/nar/16.1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schevitz R. W., Otwinowski Z., Joachimiak A., Lawson C. L., Sigler P. B. The three-dimensional structure of trp repressor. 1985 Oct 31-Nov 6Nature. 317(6040):782–786. doi: 10.1038/317782a0. [DOI] [PubMed] [Google Scholar]
- Schmidt-Dörr T., Oertel-Buchheit P., Pernelle C., Bracco L., Schnarr M., Granger-Schnarr M. Construction, purification, and characterization of a hybrid protein comprising the DNA binding domain of the LexA repressor and the Jun leucine zipper: a circular dichroism and mutagenesis study. Biochemistry. 1991 Oct 8;30(40):9657–9664. doi: 10.1021/bi00104a013. [DOI] [PubMed] [Google Scholar]
- Somers W. S., Phillips S. E. Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands. Nature. 1992 Oct 1;359(6394):387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- Steffen D., Schleif R. Overproducing araC protein with lambda-arabinose transducing phage. Mol Gen Genet. 1977 Dec 9;157(3):333–339. doi: 10.1007/BF00268671. [DOI] [PubMed] [Google Scholar]
- Wilcox G., Meuris P. Stabilization and size of araC protein. Mol Gen Genet. 1976 Apr 23;145(1):97–100. doi: 10.1007/BF00331563. [DOI] [PubMed] [Google Scholar]
- Wolberger C., Dong Y. C., Ptashne M., Harrison S. C. Structure of a phage 434 Cro/DNA complex. Nature. 1988 Oct 27;335(6193):789–795. doi: 10.1038/335789a0. [DOI] [PubMed] [Google Scholar]