Abstract
We report a novel strategy for regiospecific hetero-assembly of DNA-modified gold nanoparticles (DNA-AuNPs) onto upconversion nanoparticles (UCNPs) into hybrid lab-on-a-particle systems. The DNA-AuNPs have been assembled onto the hexagonal plate-like UCNPs with well-regulated stoichiometry and controlled organization onto the different facets of UCNP, forming various addressable superstructures. The fine-tuning of stoichiometry and organization is realized by biorecognition specificity of DNA toward specific crystal facets of UCNPs. Such a hetero-assembled DNA-AuNP/UCNP system maintains both plasmonic resonance of AuNPs and fluorescent properties of UCNPs, allowing targeted dual-modality imaging of cancer cells using an aptamer.
Nanoparticles (NPs) with excellent controlled size, shape, composition, and geometry-dependent properties have been well developed in the past decade. Discrete assembling these NPs into well-defined superstructure is a major focus of current nanoscale science and engineering, as it holds promises for both fundamental investigation of NP–NP interactions and practical applications in areas such as optoelectronics and medicine.1 However, rationally controlling the self-assembly from the “bottom-up” while maintaining precise control over the stoichiometry and spatial organization of the NPs at nanometer scale represents a major challenge in this area.1 Recently, DNA has attracted great attention as an ideal programmable assembly agent for controllable self-assembly of NPs into well-defined superstructures, because of the high programmability.2 However, the method was mainly applied to assembly of the same type of nanomaterial building blocks such as gold nanoparticles (AuNPs),3 quantum dots,4 and carbon nanotubes.5 Despite much effort to explore hetero-assembly of different types of NPs (e.g., QDs and AuNPs) into controllable hybrid superstructures,6 a significant gap remains in the ability to achieve strict control over the organization of two or more kinds of NPs into spatially addressable configurations, which often requires controlling of the “valency” of the NPs (i.e., the number of DNA molecules on the surface of NPs), and the process is time- and cost-consuming and lacks high yields.4,6
Among the various types of potential NP building blocks, lanthanide-doped upconversion nanoparticles (UCNPs) is becoming an exciting candidate, because of their unique optical properties (emitting tunable shorter-wavelength luminescence under near-infrared excitation) and diverse applications such as medical imaging, therapeutics, and photovoltaics.7 Although some efforts have been made to construct UCNPs-based multifunctional materials through direct attachment of other types of NPs or core–shell fabrication,8 the primary hurdle in assembling UCNPs-based superstructures is to control the composition and binding sites on its surface because they are normally capped with hydrophobic ligands that lacks any functional groups for surface modification. Herein, we report for the first time a new strategy for hetero-assembly of UCNPs and AuNPs into controlled sophisticated superstructures (Scheme 1). Compared with previously reported DNA-mediated NPs assembly based on sequence-specific base pairing, our present strategy relies on the controlled anisotropic surface property of UCNP and its facet-selected binding with DNA molecules. By deliberately controlling the biorecognition specificity of DNA toward specific crystal facets of upconversion nanoplates and the ratios of Au-to-UCNPs, the AuNPs could be selectively assembled onto the different facets of a hexagonal UCNP. Such hybrid nanostructures with both plasmonic and upconversion luminescent properties enable dual-modal cell imaging.
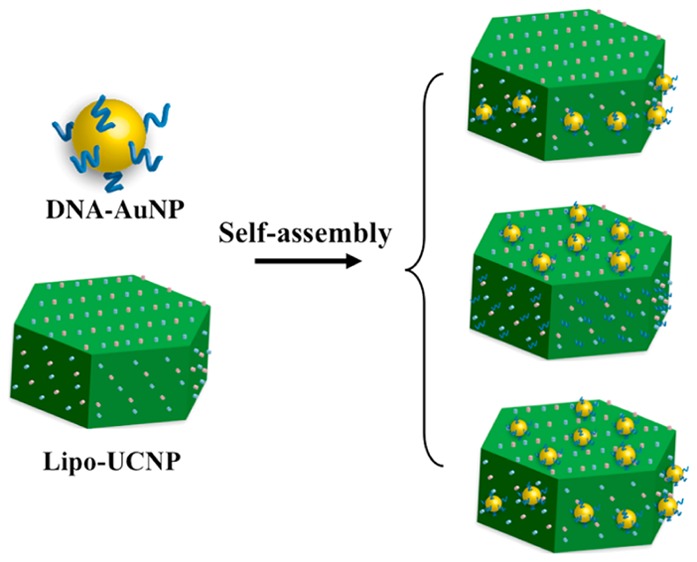
Scheme 1. Schematic Illustrations of the Hetero-Assembly of Lab-on-a-Particle Superstructures.
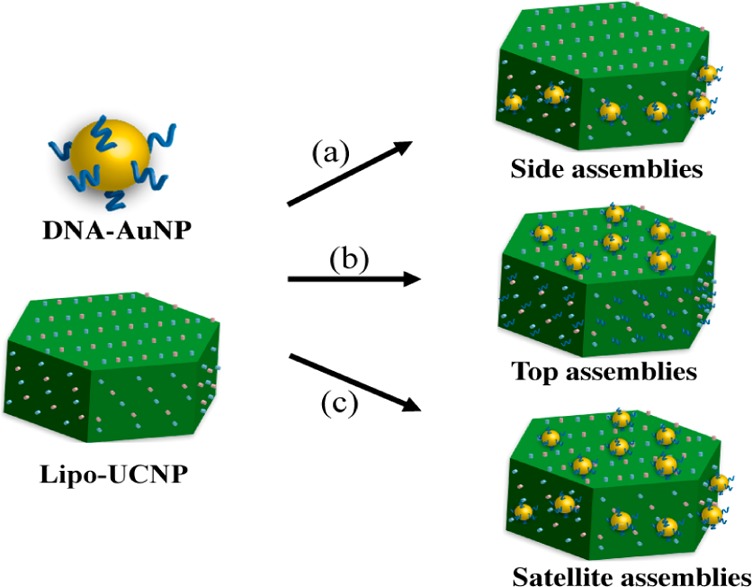
(a) Forming side assemblies with low ratio of DNA-AuNPs to Lipo-UCNP. (b) Forming top assemblies through incubation Lipo-UCNP with DNA molecules first and then assembly with DNA-AuNPs. (c) Forming satellite assemblies with high ratio of DNA-AuNPs to Lipo-UCNP.
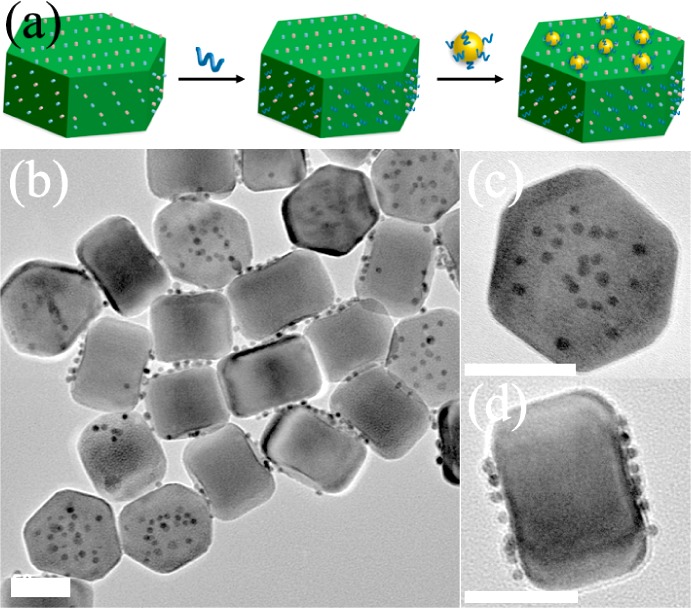
The hetero-assembly involves two essential building blocks: DNA-modified AuNPs and water-soluble UCNPs. DNA-modified 5 nm AuNPs (DNA-AuNPs) were prepared using a well-established method based on the thiol adsorption on gold surface.9 Oleic acid-capped hexagonal-phase NaYF4:18%Yb/2%Er UCNPs were synthesized according to the literature methods,10 which were then transferred into water-soluble ones based on a phospholipids (DSPE-PEG) coating (Lipo-UCNPs), as reported previously.7j As shown in Figures 1a–c and S1, the Lipo-UCNPs used for the assembly display uniform hexagonal plate-like shape with a flat hexagonal top surface (edge length of ∼50 nm) and six rectangular side surfaces (∼50 × 55 nm), standing on the TEM grids on either the bottom face or the side face. In a typical hetero-assembly process, a small volume of concentrated DNA-AuNPs solution was added into the buffer solution of Lipo-UCNPs with a final DNA-AuNPs to Lipo-UCNPs molar ratio of 26:1. This mixture was then kept at room temperature for more than 2 h with shaking. The resulted DNA-AuNPs/UCNP assemblies were collected by centrifugation, washed with water to remove unassembled DNA-AuNPs, and then characterized by transmission electron microscopy (TEM). As shown in Figures 1d–f and S1, the discrete DNA-AuNPs/UCNP assemblies could be directly formed in high yields (92%). Interestingly, the DNA-AuNPs were found to locate exclusively on the six adjacent side faces of the Lipo-UCNP, and few DNA-AuNPs were found on the hexagonal top surfaces of the Lipo-UCNP. These results clearly demonstrated formation of anisotropic plasmonic upconversion superassemblies. Such assembly could be performed in solutions of different pH values or salt concentrations (Figure S2). Furthermore, DNA of other sequences (e.g., T27, C27, and G20) could be used by the same method to obtain superassemblies (Figure S3).
Figure 1.
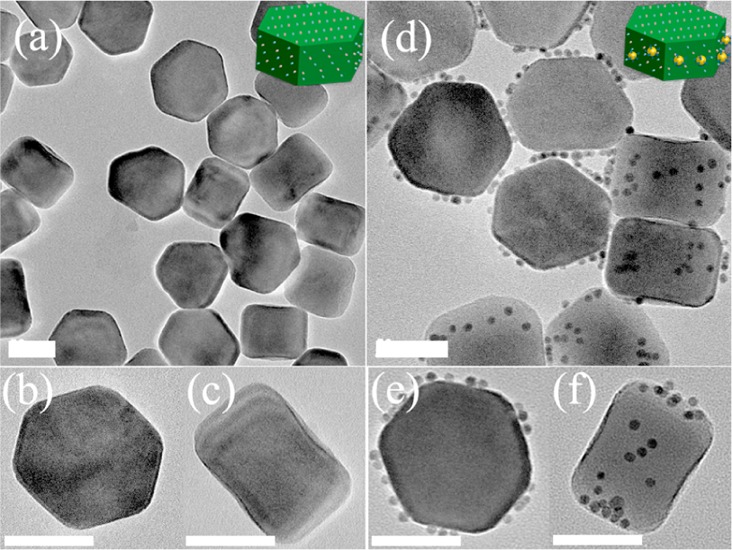
TEM images of the (a–c) as-prepared and (d–f) DNA-AuNPs/UCNP side assemblies, which stand on the TEM grids on either the bottom or side face. Scale bar = 50 nm.
To optimize the conditions under which the anisotropic nanostructures were obtained, we systematically varied the ratio of the two types of NPs. At a molar ratio of DNA-AuNPs to Lipo-UCNP (10:1), the asymmetric assemblies formed with DNA-AuNPs occurred on the six side faces of Lipo-UCNP (Figures 2b–d and S4). When the ratio was increased to 26:1, more DNA-AuNPs were attached on the side faces of Lipo-UCNPs (Figures 1d–f and S1). When the ratio was further increased to 60:1, the side faces of Lipo-UCNP were almost fully covered with DNA-AuNPs, and the DNA-AuNPs were observed starting to assembly on top faces of Lipo-UCNPs (Figures 2e–g and S4). When the ratio further reaches 200:1, the planet-satellite analogues with DNA-AuNPs attached homogeneously on all the eight faces of Lipo-UCNPs can be observed (Figures 2h–j and S4). The formed superstructures were further confirmed by UV–vis absorption. When the ratio was increased, the color of dispersed assemblies changed from colorless to light- and dark-red (Figure 2k), as a result of the characteristic SPR absorption of nanostructured gold along with the formation of discrete assemblies. Indeed, the UV–vis absorption spectra of the resulted assemblies revealed that the SPR band of the superstructures at 520 nm simultaneously increased when the ratio of DNA-AuNPs to Lipo-UCNPs was varied from low to high values (Figure 2l).
Figure 2.
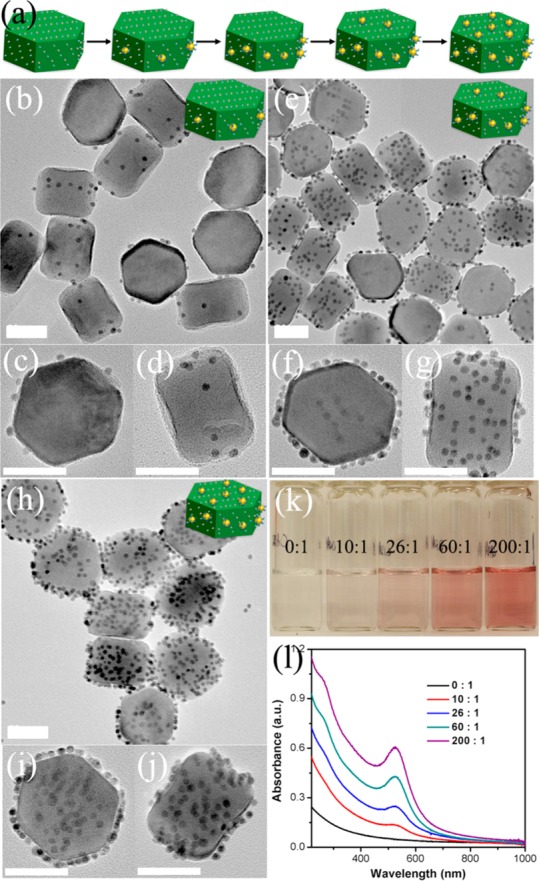
(a) Schematics of the hetero-assembly of DNA-AuNPs/UCNP superstructures with increased NPs ratio of DNA-AuNPs to Lipo-UCNP. TEM images of the DNA-AuNPs/UCNP assemblies produced from the NPs molar ratio of DNA-AuNPs to Lipo-UCNPs of (b–d) 10:1, (e–g) 60:1, and (h–j) 200:1. (k) Photographs and (l) UV–vis absorption spectra of the DNA-AuNPs/UCNP superstructures assembled from different NPs ratio of DNA-AuNPs to Lipo-UCNP. Scale bar = 50 nm.
Organic surfactants that show preferential adsorption to certain crystal facets have been extensively explored to manipulate NP shapes.11 Recently, it is found that some biomolecules show specific recognition toward specific materials and a particular crystal facet.12,13 We explore the self-assembly of DNA-AuNPs on selected faces of a single UCNP based on the recognition of DNA toward specific facets of UCNPs. DNA has shown strong binding affinity for lanthanide ions due to an abundance of negatively charged backbone.14 We have recently demonstrated that DNA molecules show a strong binding on UCNPs surface based on this interfacial interaction.7k For the current system, the binding of DNA with UCNPs was further confirmed by incubation of a FAM dye-labeled DNA with Lipo-UCNPs. After 12 h of incubation, both of the fluorescences from the FAM and UCNPs, respectively, were visualized from the purified nanoparticles (Figure S5). As a control, we also performed assembly using PEG-modified AuNPs to investigate the effect of DNA. Without DNA modification on AuNPs, no attachment of the two kinds of nanoparticles was observed (Figure S6). Another control experiment using DSPE-PEG coated iron oxide NPs indicated a negligible binding with DNA-AuNPs (Figure S7), suggesting that there was little nonspecific binding of DNA-AuNPs to the phospholipids layer. These results confirm that the DNA binding to UCNPs surface is critical in driving the hetero-assembly of DNA-AuNPs and Lipo-UCNPs. Also, it is known that the hexagonal plate-like β-NaYF4 possesses typical crystal planes of top (001) and equivalent six prismatic side planes of (100) families. During the crystal growth, oleic acid (OA) interacted more strongly with (001) facets than (100) facets, resulting in the formation of hexagonal nanoplates with much higher OA ligands coverage on (001) planes.10a Control experiments using OA-free UCNPs resulted in the binding with DNA-AuNPs without facet selectivity (Figure S8). Therefore, the controlled anisotropic surface property of UCNP and its face-selected binding of DNA are key factors in the formation of such superassemblies. At low DNA-AuNPs to Lipo-UCNP ratio, DNAs on the AuNPs prefer to bind to the equivalent six prismatic side facets of UCNPs, because this facet has less OA capping ligands and thus more exposed Ln3+ on these planes. With gradually increasing DNA-AuNPs to Lipo-UCNP ratio, the coverage of DNA-AuNPs on the six prismatic side faces began to saturate, then the DNA-AuNPs began to assemble on the top faces of UCNPs, forming planet-satellite analogues. As a control, incubating DNA-AuNPs with the superstructure (Figure 2e–g) with a DNA-AuNPs to the superstructure ratio of 26:1 resulted in more AuNPs being assembled on top faces of the UCNPs (Figure S9), confirming that high concentration of DNA-AuNPs could lead to more chance for assembling on the top faces.
Considering different geometries for the assembly of AuNPs around a UCNP, there are three basic options: AuNPs can be assembled only to the top (001) facets, the sides (100) facets, and both top and side faces of UCNPs. These three types of superstructures are designated as top, side, and satellite assemblies, respectively. Having demonstrated the controlled preparation of side (Figure 1d–f) and satellite (Figure 2h–j) assemblies, we then explored the controlled assembly of top superstructure based on the DNA mediation. When the UCNPs are incubated with free DNA molecules (A27, 5 μM) first and then with DNA-AuNPs, the side faces of the UCNPs were blocked with DNA molecules, while DNA-AuNPs may only attach on the top faces (Figure 3a). As expected, the top assemblies were produced with high yield based on this design with a DNA-AuNPs to Lipo-UCNPs ratio of 30:1 (Figures 3b–d and S10). The DNA-AuNPs were found to locate on the hexagonal top surfaces of Lip-UCNPs, while there are few DNA-AuNPs observed on the six side faces of Lip-UCNPs. The formation of top assemblies not only further confirmed the DNA-facet directed self-assembly mechanism but also highlight the power of the strategy for controlled tuning the assembly of AuNPs and UCNPs.
Figure 3.
(a) Schematic illustration of hetero-assembly of DNA-AuNPs/UCNP top assemblies. (b–d) TEM images of DNA-AuNPs/UCNP top assemblies. Scale bar = 50 nm.
We then investigated the versatility of our method in assembling of DNA-AuNPs and UCNPs with different particle sizes. The assembly was carried out through a process similar to that used for preparing the sample shown in Figure 1, except that the 13, 20, and 30 nm AuNPs were used. As shown in Figure S11, the hetero-assemblies made of differently sized AuNPs could be easily obtained with high yields (>88%). Furthermore, the UCNPs used can also be independently varied to form the superstructure (Figure S12). Moreover, since DNA is exposed on the AuNPs of the superstructures and can be readily hybridized to complementary DNA (cDNA), the obtained superstructures can further be used to realize more complex designs. As a demonstration of this feature, 20 nm AuNPs modified with cDNA was assembled with satellite assemblies to form ternary NPs superstructure (94% yield) (Figure S13).
Combining plasmonic materials such as gold with fluorescent materials such as quantum dots has been of great interest due to their diverse applications, such as in bioimaging.6,15 We therefore investigate the optical properties of the DNA-AuNPs/UCNP assemblies. Interestingly, as shown in Figure S14, minimal AuNPs-induced luminescence quenching was observed for the superstructures. The results suggest that the nanoassemblies may act as excellent dual-modality imaging probes because of their combined plasmon scattering property of AuNPs and upconversion fluorescent property of UCNPs. Furthermore, the strategy produces a prescribed superstructure with an addressable polyvalent DNA coating, which may display biorecognition ability and hence enable sensitive targeted bioimaging. We first investigated their ability to enter live cells by incubation of satellite assemblies with HeLa cells for 12 h. The resulting HeLa cells exhibited bright NIR-light responsive upconverted fluorescence within the cytoplasm (Figure S15), suggesting that the assemblies are capable of crossing cell membranes without the need of transfection agents. We then explored the use of DNA aptamer functionalized satellite assemblies for targeted dual-modality imaging of cancer cells. A 26-mer DNA aptamer AS1411 that is capable of targeting nucleolin was conjugated to satellite assemblies for targeted imaging of the nucleolin-overexpressed breast cancer cell line 4T1. As shown in Figure 4, the cells treated with the aptamer-functionalized satellite assemblies showed both strong upconversion fluorescence and light scattering signals. The near complete overlap of the two imaging signals further confirms the cellular uptake of the hetero-assemblies. In comparison, the satellite assemblies modified with control DNA (A27) showed much less binding to the 4T1 cells, confirming the targeting role of the specific aptamer.
Figure 4.
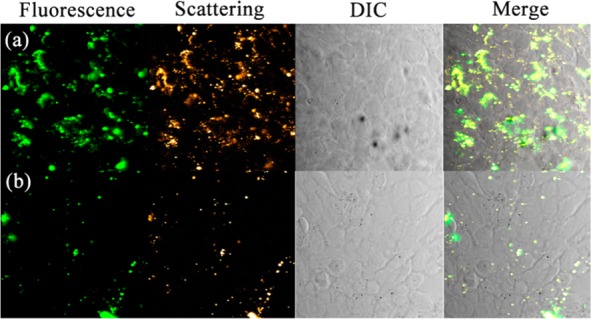
Targeted dual-modality imaging of cancer cells with the hetero-assemblies. Confocal microscopy images of 4T1 cells treated with (a) DNA aptamer and (b) control DNA functionalized satellite assemblies. Fluorescence emission was collected in the range of 520–560 nm with the excitation of NIR light (980 nm).
In conclusion, we have demonstrated regiospecific assembly of plasmonic upconversion hetero-superstructures based on DNA–inorganic interfacial interactions. Such multifunctional superassemblies not only offer plasmonic and upconversion optical properties but also enable a polyvalent DNA surface, which may find wide applications in bioimaging, nanomedicine, and photovoltaics.
Acknowledgments
This work was supported by the U.S. National Institute of Health (Grant ES016865).
Supporting Information Available
Detailed synthesis and characterization data as well as supplementary results. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- a Wang L.; Xu L.; Kuang H.; Xu C.; Kotov N. A. Acc. Chem. Res. 2012, 45, 1916. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wang Y.; Chen G.; Yang M.; Silber G.; Xing S.; Tan L. H.; Wang F.; Feng Y.; Liu X.; Li S.; Chen H. Nat. Commun. 2010, 1, 87. [DOI] [PubMed] [Google Scholar]; c Nie Z.; Petukhova1 A.; Kumacheva E. Nat. Nanotechnol. 2010, 5, 15. [DOI] [PubMed] [Google Scholar]; d Glotzer S. C.; Solomon M. J. Nat. Mater. 2007, 6, 557. [DOI] [PubMed] [Google Scholar]
- a Mirkin C. A.; Letsinger R. L.; Mucic R. C.; Storhoff J. J. Nature 1996, 382, 607. [DOI] [PubMed] [Google Scholar]; b Seeman N. C. Nature 2003, 421, 427. [DOI] [PubMed] [Google Scholar]; c Rothemund P. W. K. Nature 2006, 440, 297. [DOI] [PubMed] [Google Scholar]; d Tan L. H.; Xing H.; Lu Y. Acc. Chem. Res. 2014, 47, 1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Ding B. Q.; Deng Z. T.; Yan H.; Cabrini S.; Zuckermann R. N.; Bokor J. J. Am. Chem. Soc. 2010, 132, 3248. [DOI] [PubMed] [Google Scholar]; b Maye M. M.; Nykypanchuk D.; Cuisinier M.; van der Lelie D.; Gang O. Nat. Mater. 2009, 8, 388. [DOI] [PubMed] [Google Scholar]; c Macfarlane R. J.; Lee B.; Jones M. R.; Harris N.; Schatz G. C.; Mirkin C. A. Science 2011, 334, 204. [DOI] [PubMed] [Google Scholar]; d He Y.; Ye T.; Ribbe A. E.; Mao C. J. Am. Chem. Soc. 2011, 133, 1742. [DOI] [PubMed] [Google Scholar]; e Xu L.; Kuang H.; Xu C.; Ma W.; Wang L.; Kotov N. A. J. Am. Chem. Soc. 2012, 134, 1699. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Sharma J.; Chhabra R.; Cheng A.; Brownell J.; Liu Y.; Yan H. Science 2009, 323, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Zhao W. A.; Gao Y.; Kandadai S. A.; Brook M. A.; Li Y. F. Angew. Chem., Int. Ed. 2006, 45, 2409. [DOI] [PubMed] [Google Scholar]; h Lee J. H.; Wernette D. P.; Yigit M. V.; Liu J.; Wang Z.; Lu Y. Angew. Chem., Int. Ed. 2007, 46, 9006. [DOI] [PubMed] [Google Scholar]; i Tan S. J.; Campolongo M. J.; Luo D.; Cheng W. Nat. Nanotechnol. 2011, 6, 268. [DOI] [PubMed] [Google Scholar]; j Pal S.; Deng Z.; Wang H.; Zou S.; Liu Y.; Yan H. J. Am. Chem. Soc. 2011, 133, 17606. [DOI] [PubMed] [Google Scholar]
- a Sharma J.; Ke Y. G.; Lin C. X.; Chhabra R.; Wang Q. B.; Nangreave J.; Liu Y.; Yan H. Angew. Chem., Int. Ed. 2008, 47, 5157. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Tikhomirov G.; Hoogland S.; Lee P. E.; Fischer A.; Sargent E. H.; Kelley S. O. Nat. Nanotechnol. 2011, 6, 485. [DOI] [PubMed] [Google Scholar]; c Deng Z.; Samanta A.; Nangreave J.; Yan H.; Liu Y. J. Am. Chem. Soc. 2012, 134, 17424. [DOI] [PubMed] [Google Scholar]; d Liu Y. Nat. Nanotechnol. 2011, 6, 463. [DOI] [PubMed] [Google Scholar]
- a Maune H. T.; Han S. P.; Barish R. D.; Bockrath M.; Goddard W. A.; Rothemund P. W. K.; Winfree E. Nat. Nanotechnol. 2010, 5, 61. [DOI] [PubMed] [Google Scholar]; b Zhao W.; Gao Y.; Brook M. A.; Li Y. F. Chem. Commun. 2006, 3582. [DOI] [PubMed] [Google Scholar]
- a Schreiber R.; Do J.; Roller E.-M.; Zhang T.; Schüller V. J.; Nickels P. C.; Feldmann J.; Liedl T. Nat. Nanotechnol. 2014, 9, 74. [DOI] [PubMed] [Google Scholar]; b Zhang Y.; Lu F.; Yager K. G.; van der Lelie D.; Gang O. Nat. Nanotechnol. 2013, 8, 865. [DOI] [PubMed] [Google Scholar]; c Zhang C.; Macfarlane R. J.; Young K. L.; Choi C. H. J.; Hao L.; Auyeung E.; Liu G.; Zhou X.; Mirkin C. A. Nat. Mater. 2013, 12, 741. [DOI] [PubMed] [Google Scholar]; d Wang R. S.; Nuckolls C.; Wind S. J. Angew. Chem., Int. Ed. 2012, 51, 11325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Wang F.; Liu X. Chem. Soc. Rev. 2009, 38, 976. [DOI] [PubMed] [Google Scholar]; b Haase M.; Schäfer H. Angew. Chem., Int. Ed. 2011, 50, 5808. [DOI] [PubMed] [Google Scholar]; c Mader H. S.; Kele P.; Saleh S. M.; Wolfbeis O. S. Curr. Opin. Chem. Biol. 2010, 14, 582. [DOI] [PubMed] [Google Scholar]; d Feng W.; Sun L.; Zhang Y.; Yan C. Coord. Chem. Rev. 2010, 254, 1038. [Google Scholar]; e Zhou J.; Liu Z.; Li F. Chem. Soc. Rev. 2012, 41, 1323. [DOI] [PubMed] [Google Scholar]; f Li Z.; Zhang Y.; Jiang S. Adv. Mater. 2008, 20, 4765. [Google Scholar]; g Ju Q.; Tu D.; Liu Y.; Li R.; Zhu H.; Chen J.; Chen Z.; Huang M.; Chen X. J. Am. Chem. Soc. 2012, 134, 1323. [DOI] [PubMed] [Google Scholar]; h Nam S. H.; Bae Y. M.; Il Park Y.; Kim J. H.; Kim H. M.; Choi J. S.; Lee K. T.; Hyeon T.; Suh Y. D. Angew. Chem., Int. Ed. 2011, 50, 6093. [DOI] [PubMed] [Google Scholar]; i Abel K. A.; Boyer J. C.; van Veggel F. C. J. M. J. Am. Chem. Soc. 2009, 131, 14644. [DOI] [PubMed] [Google Scholar]; j Li L.; Zhang R.; Yin L.; Zheng K.; Qin W.; Selvin P. R.; Lu Y. Angew. Chem., Int. Ed. 2012, 51, 6121. [DOI] [PMC free article] [PubMed] [Google Scholar]; k Li L.-L.; Wu P.; Hwang K.; Lu Y. J. Am. Chem. Soc. 2013, 135, 2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Wang F.; Deng R.; Wang J.; Wang Q.; Han Y.; Zhu H.; Chen X.; Liu X. Nat. Mater. 2011, 10, 968. [DOI] [PubMed] [Google Scholar]; b Liu Y.; Tu D.; Zhu H.; Li R.; Luo W.; Chen X. Adv. Mater. 2010, 22, 3266. [DOI] [PubMed] [Google Scholar]; c Zhang Y.; Zhang L.; Deng R.; Tian J.; Zong Y.; Jin D.; Liu X. J. Am. Chem. Soc. 2014, 136, 4893. [DOI] [PubMed] [Google Scholar]
- Liu J.; Lu Y. Nat. Protoc. 2006, 1, 246. [DOI] [PubMed] [Google Scholar]
- a Shi F.; Wang J.; Zhang D.; Qin G.; Qin W. J. Mater. Chem. 2011, 21, 13413. [Google Scholar]; b Li Z.; Zhang Y.; Jiang S. Adv. Mater. 2008, 20, 4765. [Google Scholar]
- a Yin Y.; Alivisatos A. P. Nature 2005, 437, 664. [DOI] [PubMed] [Google Scholar]; b Tao A. R.; Habas S.; Yang P. D. Small 2008, 4, 310. [Google Scholar]
- a Gugliotti L. A.; Feldheim D. L.; Eaton B. E. Science 2004, 304, 850. [DOI] [PubMed] [Google Scholar]; b Chiu C. Y.; Li Y. J.; Ruan L. Y.; Ye X. C.; Murray C. B.; Huang Y. Nat. Chem. 2011, 3, 393. [DOI] [PubMed] [Google Scholar]; c Wang Z.; Tang L.; Tan L. H.; Li J.; Lu Y. Angew. Chem., Int. Ed. 2012, 51, 9078. [DOI] [PubMed] [Google Scholar]
- Whaley S. R.; English D. S.; Hu E. L.; Barbara P. F.; Belcher A. M. Nature 2000, 405, 665. [DOI] [PubMed] [Google Scholar]
- Costa D.; Burrows H. D.; da Graça Miguel M. Langmuir 2005, 21, 10492. [DOI] [PubMed] [Google Scholar]
- a Li F.; Gao D.; Zhai X.; Chen Y.; Fu T.; Wu D.; Zhang Z.-P.; Zhang X.- E.; Wang Q. Angew. Chem., Int. Ed. 2011, 50, 4202. [DOI] [PubMed] [Google Scholar]; b Jin Y.; Gao X. Nat. Nanotechnol. 2009, 4, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


