Summary
A three-step procedure comprising (i) unnatural-amino-acid mutagenesis with 4-azido-phenylalanine, (ii) Staudinger-Bertozzi ligation with a probe-phosphine derivative, and (iii) in vitro reconstitution of RNA polymerase (RNAP) enables the efficient site-specific incorporation of a fluorescent probe, a spin label, a crosslinking agent, a cleaving agent, an affinity tag, or any other biochemical or biophysical probe, at any site of interest in RNAP. Straightforward extensions of the procedure enable the efficient site-specific incorporation of two or more different probes in two or more different subunits of RNAP. We present protocols for synthesis of probe-phosphine derivatives, preparation of RNAP subunits and the transcription initiation factor σ, unnatural amino acid mutagenesis of RNAP subunits and σ, Staudinger ligation with unnatural-amino-acid-containing RNAP subunits and σ, quantitation of labelling efficiency and labelling specificity, and reconstitution of RNAP.
Keywords: unnatural-amino-acid mutagenesis, Staudinger ligation, in vitro reconstitution, 4-azido-phenylalanine, fluorescent probes, Cy3B, Alexa647, phosphines, Cy3B-phosphine, Alexa647-phosphine, RNA polymerase, RNA polymerase α subunit, RNA polymerase β subunit, RNA polymerase β' subunit, RNA polymerase ω subunit, σ70
1. Introduction
Important biochemical and biophysical approaches for analysis of RNA polymerase (RNAP) structure and function--including affinity cleaving (1,2), photocrosslinking (3,4), and ensemble and single-molecule fluorescence resonance energy transfer (5–16)--require the ability to incorporate probes at specific sites within RNAP.
One approach to introduce a probe at a specific site within a protein is to perform site-directed mutagenesis to introduce a unique Cys residue at the site of interest followed by Cys-specific chemical modification to introduce the probe at the site of interest (1–17). This approach can be used to incorporate probes at any non-essential, solvent-accessible residue of Escherichia coli RNAP ω subunit (11), E. coli RNAP α subunit (after use of site-directed mutagenesis to substitute the pre-existing Cys residues at positions 54, 131, 176, and 269; 1), and E. coli transcription initiation factor σ70 (after use of site-directed mutagenesis to substitute the pre-existing Cys residues at positions 132, 291, and 295; 2, 5–16). The resulting probe-containing ω and α derivatives enable preparation of probe-containing RNAP derivatives by in vitro reconstitution of RNAP (1,11; see 19–23), and the resulting probe-containing σ70 derivatives enable preparation of probe-containing RNAP holoenzyme derivatives by addition to RNAP core (2, 5–16). However, this approach cannot be used to incorporate probes into the second-largest and largest RNAP subunits, β and β', since these very large polypeptides (1342 residues for E. coli β; 1407 residues for E. coli β') contain multiple pre-existing Cys residues that are essential and that therefore cannot be substituted without loss of function. Accordingly, this approach cannot be used for analysis of the principle determinants of RNAP conformation, RNAP catalytic activity, RNAP-DNA interactions, RNAP-RNA interaction, RNAP-substrate interactions, and RNAP-inhibitor interactions--all of which involve residues of β and β'.
Another approach to introduce a probe at a specific site within a protein is intein-mediated C-terminal labelling (8–10,12,24). This approach has been applied successfully to E. coli RNAP α, β, and β' subunits (8–10,12). However, since this approach is applicable only to protein C-termini, this approach, even when used in conjunction with "split-β" or "spilt-β’'" RNAP derivatives having non-native C-termini (9,12; see also 25,26), has enabled incorporation of probes at only a limited set of sites within α, β, and β' (αII residue 235; β residues 643 and 937; and β' residue 1377; 8–10,12).
In recent work, we have developed an approach that enables site-specific incorporation of probes at any site of interest in any RNAP subunit or σ70, and, moreover, that enables site-specific incorporation of probes at any two or more sites of interest in any two or more RNAP subunits or σ70 (16,27). The approach involves three steps: (i) unnatural-amino-acid mutagenesis to introduce 4-azido-L-phenylalanine at a site of interest in an RNAP subunit or σ70 (28,29), (ii) Staudinger-Bertozzi ligation with a probe-phosphine derivative to introduce a probe at the site of interest (30–32), and (iii) RNAP reconstitution (20–23). There are no constraints on the selection of the probe site, other than that the probe site should have a sidechain that is non-essential (i.e., that can be substituted without loss of function) and solvent-accessible (i.e., that can allow an appended probe to interact with solvent and not clash with RNAP, template, product, or substrate). In published work, we have used this approach to incorporate fluorescent probes at the tips of the E. coli RNAP β and β' pincers for single-molecule-FRET studies of opening and closing of the RNAP active-center cleft (β residue 106; β' residue 284; 16). In unpublished work, we have used this approach to incorporate probes at each of 17 additional sites in E. coli RNAP and σ70 (β residues 159, 164, 222, 357, 379, 482, 484, 643, and 937; β' residues 171, 405, 942, and 1183; σ70 residues 14, 36, 46, and 59), and we have shown that that the approach can be combined with Cys-specific modification to incorporate pairs of different probes at pairs of different sites within E. coli σ70. In other work, we have used the first two steps of the approach--unnatural amino acid mutagenesis and Staudinger-Bertozzi ligation--to enable site-specific incorporation of probes into the Rpo1' and Rpo2" subunits of archaeal RNAP and into an archaeal transcription factor (27).
In this report, we present detailed procedures for synthesis of phosphine derivatives of the fluorescent probes Cy3B and Alexa647, preparation of RNAP subunits and σ70, unnatural amino acid mutagenesis of RNAP subunits and σ70, Staudinger-Bertozzi ligation with unnatural-amino-acid-containing RNAP subunits and σ70, quantitation of labelling efficiency and labelling specificity, and reconstitution of RNAP.
2. Materials
2.1. Preparation of Probe Phosphines
Alexa Fluor 647 NHS ester (Molecular Probes) (see Notes 2–4)
NHS-Phosphine (Thermo Scientific) (see Note 3)
N-Hydroxysulfosuccinimide (NHSS) (Invitrogen)
Mono-trityl-ethylenediamine, acetic acid salt (Novabiochem)
N-Trityl-1,2-ethanediamine, hydrobromide salt (Sigma-Aldrich)
1-Methyl-2-diphenylphosphinoterephthalate (MDPT) (SynChem; see Note 5)
1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDAC) (Invitrogen)
N,N-Diisopropylethylamine (DIPEA) (Sigma-Aldrich)
Triethylamine (TEA) (Sigma-Aldrich)
Dimethylformamide, anhydrous (DMF) (Sigma-Aldrich)
Chloroform (Sigma-Aldrich)
Acetonitrile (HPLC grade) (Fisher Scientific)
Trifluoroacetic acid (TFA) (Sigma-Aldrich)
Methanol (Sigma-Aldrich)
α-Cyano-4-hydroxycinnamic acid (CHCA) (Sigma-Aldrich)
Drierite dessicant (VWR)
Argon (compressed)
Desiccator
Aluminum foil
Borosilicate glass vials with phenolic screw caps, 1.8 mL and 3.6 mL (VWR)
Micro magnetic stir bars, 2 × 7 mm (VWR)
Magnetic stirrer
Sub-micro quartz spectrophotometer cell, 10 mm path length (Starna Cells)
UV/Vis spectrophotometer (e.g., PerkinElmer Lambda 25)
Speedvac evaporator (Thermo Scientific)
Discovery BIO Wide Pore C18 HPLC Column, 25 cm × 10 mm, 10 µm (Sigma-Aldrich) (see Note 6)
Reversed-phase HPLC system equipped with diode array detector (e.g., Hitachi L-7455)
Fraction collector (e.g., ADVANTEC CHF122SC)
MALDI plate (e.g., Applied Biosystems Opti-TOF 384 Well Insert, 123 mm × 81 mm)
MALDI mass spectrometer (e.g., Applied Biosystems 4800)
2.2. Preparation of RNAP Subunits and σ70
2.2.1. Preparation of FLAG-αNTDI-GSGGSG-αNTDII
Plasmid encoding a fusion protein comprising an N-terminally FLAG-tagged first E. coli RNAP α subunit N-terminal domain (α residues 1–235; αNTDI), followed by a GlySerGlyGlySerGly linker, followed by a second E. coli RNAP α subunit N-terminal domain (α residues 1–235; αNTDII) (FLAG-αNTDI-GSGGSG-αNTDII; see Table 1)
Chemically competent E. coli strain BL21(DE3) (Invitrogen)
TYE agar containing 40 µg/mL kanamycin (10 g/L tryptone, 5 g/L yeast extract, 8 g/L NaCl, and 15 g/L agar; autoclave-sterilized without antibiotics; supplemented with antibiotics after cooling to 55°C; poured into sterile 100 × 15 mm Petri plates at ~25 mL/plate)
LB broth (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl; autoclave-sterilized)
40 mg/mL kanamycin (filter sterilized) (Sigma-Aldrich)
1 M IPTG (filter sterilized) (Gold Biotechnology)
Complete Protease inhibitor Cocktail Tablets, EDTA-Free (Roche Applied Sciences)
ANTI-FLAG M2 affinity gel column (Sigma-Aldrich)
FLAG peptide (Sigma-Aldrich)
0.1 M Glycine-HCl, pH 3.5
Ammonium sulfate (Sigma-Aldrich)
Buffer A (20 mM Tris-HCl, pH 7.9, 500 mM NaCl, and 10 mM EDTA)
Buffer B (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 5% glycerol)
Buffer C (50 mM Tris-HCl, pH 7.9, 200 mM KCl, 10 mM MgCl2, 1 mM EDTA, 5 mM 2-mercaptoethanol, and 20% glycerol)
6× SDS-loading buffer (190 mM Tris-HCl, pH 6.8, 6% SDS, 10% β-mercaptoethanol, 48% glycerol, and 0.3% bromophenol blue)
SDS-running buffer (25 mM Tris, pH 8.3, 250 mM glycine, and 0.1% SDS)
Destaining solution (10% acetic acid, 50% methanol, and 40% water)
Coomassie Brilliant Blue R-250 (Bio-Rad)
10% Polyacrylamide (37.5:1 acrylamide:bisacrylamide), 0.1% SDS, slab gel (10 × 7 × 0.075 cm)
Prestained protein molecular weight markers (7–210 kDa) (Bio-Rad)
Bradford Protein Assay Kit (Bio-Rad)
Spectra/Por 3 dialysis tubing (3.5 kDa molecular weight cutoff) (Spectrum Labs)
Dialysis membrane closures (VWR)
Econo-Pac 20 mL chromatography columns (Bio-Rad)
2.8 L Triple-baffled Fernbach flask (autoclave-sterilized) (Bellco Glass)
100 mL Pyrex beaker (VWR)
1 L Polypropylene copolymer centrifuge bottle with cap (VWR)
14 mL Polypropylene culture tubes with snap closures (autoclave-sterilized) (VWR)
1.7 mL Polypropylene microcentrifuge tubes (Denville Scientific)
15 mL Polypropylene centrifuge tube with cap (VWR)
50 mL Polypropylene centrifuge tube with cap (VWR)
Avestin EmulsiFlex-C5 cell disrupter (Avestin)
Microcentrifuge (e.g., Eppendorf 5417C)
Sorvall RC-3B Plus centrifuge (Thermo Scientific)
Sorvall RC-6 Plus centrifuge (Thermo Scientific)
Platform shaker
Table I.
Plasmids encoding RNAP subunits and σ70
| Plasmid | Relevant Characteristics | Source |
|---|---|---|
| pET28a-NF-αNTDI-αNTDII | KmR; ori-pBR322; PT7-FLAG-rpoA(1–235)-rpoA(1–235) | (16) |
| pET21d-rpoB-CH6 | ApR; ori-pBR322; PT7-rpoB-His6 | (16) |
| pET21a-rpoC-CH6 | ApR; ori-pBR322; PT7-rpoC-His6 | (16) |
| pT7ω | ApR; ori-pBR322; ori-fl; PT7-rpoZ | (9) |
| pGEMD | ApR; ori-pBR322; PT7-rpoD | (18) |
2.2.2 Preparation of β, β' and ω
Plasmids encoding E. coli RNAP β, β, and ω subunits (see Table 1)
Chemically competent E. coli strain BL21(DE3) (Invitrogen)
TYE agar containing 40 µg/mL kanamycin (10 g/L tryptone, 5 g/L yeast extract, 8 g/L NaCl, and 15 g/L agar; autoclave-sterilized without antibiotic; supplemented with antibiotic after cooling to 55°C; poured into sterile 100 × 15 mm Petri plates at ~25 mL/plate)
LB broth (10 g/L tryptone, 5 g/L yeast extract, and 10g/L NaCl; autoclave-sterilized)
100 mg/mL Ampicillin (filter-sterilized)
1 M IPTG (filter-sterilized) (Gold Biotechnology)
10% Sodium desoxycholate (Sigma-Aldrich)
10% n-Octyl-β-D-glucopyranoside (Sigma-Aldrich)
Complete Protease inhibitor Cocktail Tablets, EDTA-Free (Roche Applied Sciences)
Glycerol (Fisher Scientific)
Buffer D (40 mM Tris-HCl, pH 7.9, 300 mM KCl, 10 mM EDTA, 1 mM DTT)
2.8 L Triple-baffled Fernbach flask (autoclave-sterilized) (Bellco Glass)
50 mL Erlenmeyer flask (autoclave-sterilized) (VWR)
100 mL Pyrex beaker (VWR)
1 L Polypropylene copolymer centrifuge bottle with cap (VWR)
14 mL Polypropylene culture tubes with snap closures (autoclave-sterilized) (VWR)
1.7 mL Polypropylene microcentrifuge tubes (Denville Scientific)
50 mL Polypropylene centrifuge tube with cap (VWR)
Avestin EmulsiFlex-C5 cell disrupter (Avestin)
Branson Sonifier 450 Sonicator (VWR)
Microcentrifuge (e.g., Eppendorf 5417C)
Sorvall RC-3B centrifuge (Thermo Scientific)
Sorvall RC-5B centrifuge (Thermo Scientific)
Platform shaker
2.2.3 Preparation of σ70
Plasmid encoding E. coli σ70 subunit (pGEMD; Table 1)
Chemically competent E. coli strain BL21(DE3) (Invitrogen)
TYE agar containing 200 µg/mL ampicillin (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl, and 15 g/L agar; autoclave-sterilized without antibiotic; supplemented with antibiotic after cooling to 55°C; poured into 100 × 15 mm Petri plates at ~25 mL/plate)
LB broth (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl; autoclave-sterilized)
100 mg/mL Ampicillin (filter-sterilized) (Sigma-Aldrich)
1 M IPTG (filter-sterilized) (Gold Biotechnology)
Complete Protease inhibitor Cocktail Tablets, EDTA-Free (Roche Applied Sciences)
2% Lysozyme (~50,000 U/mg) (Sigma-Aldrich)
Triton X-100 (Sigma-Aldrich)
10% Sodium desoxycholate (Sigma-Aldrich)
10% n-Octyl-β-D-glucopyranoside (Sigma-Aldrich)
Glycerol (Fisher Scientific)
Buffer E (40 mM Tris-HCl of pH7.9, 300 mM KCl, 10 mM EDTA)
Buffer F (50 mM Tris–HCl of pH 7.9, 6 M guanidine hydrochloride, 10 mM MgCl2, 10 µM ZnCl2, 1 mM EDTA, 10 mM DTT, and 10% glycerol)
Buffer G (20 mM Tris-HCl, pH 7.9, 200 mM NaCl, 0.1 mM EDTA, 1 mM DTT, and 50% glycerol)
Buffer TGEβ (20 mM Tris-HCl, pH 7.9, 0.1 mM EDTA, 5 mM 2-mercaptoethanol, and 5% glycerol)
Buffer TGED (20 mM Tris-HCl, pH 7.9, 0.1 mM EDTA, 1 mM DTT, and 5% glycerol)
6× SDS-loading buffer (190 mM Tris-HCl, pH 6.8, 6% SDS, 10% β-mercaptoethanol, 48% glycerol, and 0.3% bromophenol blue)
SDS-running buffer (25 mM Tris-HCl, pH 8.3, 250 mM glycine, and 0.1% SDS)
Destaining solution (10% acetic acid, 50% methanol, and 40% water)
Coomassie Brilliant Blue R-250 (Bio-Rad)
10% Polyacrylamide (37.5:1 acrylamide:bisacrylamide), 0.1% SDS, slab gel (10 × 7 × 0.075 cm)
Prestained protein molecular weight markers (7–210 kDa) (Bio-Rad)
Bradford Protein Assay Kit (Bio-Rad)
Spectra/Por 3 dialysis tubing (3.5 kDa molecular weight cutoff) (Spectrum Labs)
Dialysis membrane closures (VWR)
Amicon Ultra-15 centrifugal filter unit (30 kDa molecular-weight cutoff) (EMD Millipore)
2.8 L Triple-baffled Fernbach flask (autoclave-sterilized) (Bellco Glass)
50 mL Erlenmeyer flask (VWR)
100 mL Pyrex beaker (VWR)
1 L Polypropylene copolymer centrifuge bottle with cap (VWR)
1.7 mL Polypropylene microcentrifuge tubes (Denville Scientific)
14 mL Polypropylene culture tubes with snap closures (autoclave-sterilized) (VWR)
50 mL Polypropylene centrifuge tube with cap (VWR)
Mono-Q HR 10/10 chromatography column (GE healthcare)
ÄKTA Purifier chromatography system (GE Healthcare)
Avestin EmulsiFlex-C5 cell disrupter (Avestin)
Branson Sonifier 450 Sonicator (VWR)
Microcentrifuge (e.g., Eppendorf 5417C)
Sorvall RC-3B Plus centrifuge (Thermo Scientific)
Sorvall RC-6 Plus centrifuge (Thermo Scientific)
Platform shaker
2.3. Unnatural-Amino-Acid Mutagenesis of RNAP Subunits and σ70
2.3.1. Unnatural-Amino-Acid Mutagenesis of FLAG-αNTDI-GSGGSG-αNTDII, β, β' or ω
Plasmid encoding FLAG-αNTDI-GSGGSG-αNTDII, β, β’, or ω (Table 1)
Plasmid pEVOL-pAzF (29; provided by P. Schultz, The Scripps Research Institute, La Jolla CA)
Chemically competent E. coli strain BL21(DE3) (Invitrogen)
QuikChange II Site-Directed Mutagenesis Kit (Agilent)
LB broth (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl; autoclave-sterilized)
TYE agar containing 50 µg/mL chloramphenicol, or 50 µg/mL chloramphenicol and 40 µg/mL kanamycin, or 50 µg/mL chloramphenicol and 100 µg/mL ampicillin (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl, and 15 g/L agar; autoclave-sterilized without antibiotics; supplemented with antibiotic after cooling to 55°C; poured into sterile 100 × 15 mm Petri plates; ~25 mL/plate)
50 mg/mL Chloramphenicol (filter-sterilized)
40 mg/mL Kanamycin (filter-sterilized)
1000 mg/mL Ampicillin (filter-sterilized)
20% Arabinose (filter-sterilized)
1 M IPTG (filter-sterilized) (Gold Biotechnology)
4-Azido-L-phenylalanine (ChemImpex)
Buffer D (40 mM Tris-HCl, pH 7.9, 300 mM KCl, 10 mM EDTA, 1 mM DTT)
Buffer H (10 mM potassium acetate, pH 7.0, 10 mM KCl, 50 mM CaCl2, and 10% glycerol)
2.8 L Triple-baffled Fernbach flask (autoclave-sterilized) (Bellco Glass)
50 mL Erlenmeyer flask (VWR)
1.7 mL Polypropylene microcentrifuge tubes (Denville Scientific)
14 mL Polypropylene culture tubes with snap closures (autoclave-sterilized) (VWR)
Microcentrifuge (e.g., Eppendorf 5417C)
Platform shaker
2.3.2. Unnatural-Amino-Acid Mutagenesis of σ70
Plasmid encoding σ70 (Table 1)
Plasmid pEVOL-pAzF (29; provided by P. Schultz, The Scripps Research Institute, La Jolla CA)
Chemically competent E. coli strain BL21(DE3) (Invitrogen)
QuikChange II Site-Directed Mutagenesis Kit (Agilent)
LB broth (10 g/L tryptone, 5 g/L yeast extract, and 10g/L NaCl; autoclave-sterilized)
TYE agar containing 50 µg/mL chloramphenicol or containing 35 µg/mL chloramphenicol and 200 µg/mL ampicillin (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl, and 15 g/L agar; autoclave-sterilized without antibiotics; supplemented with antibiotic after cooling to 55°C; poured into sterile 100 × 15 mm Petri plates; ~25 mL/plate)
M9+ medium (1 × M9 minimal salts, 0.4% D-glucose, 2 mM MgSO4, 0.1 mM CaCl2, 0.1 mM FeSO4, 3 nM (NH4)6Mo7O24, 400 nM H3BO3, 30 nM CoCl2, 10 nM CuSO4, 80 nM MnCl2, 20 nM ZnSO4, 0.4 mg/L choline chloride, 0.5 mg/L folic acid, 0.5 mg/L nicotinamide, 1 mg/L myo-inositol, 1 mg/L pyridoxal HCl, 2 mg/L thiamine HCl, 0.05 mg/L riboflavin, 1 mg/L biotin; prepared freshly before use from 10× M9 minimal salts, 20% glucose, 1 M MgSO4, 1 M CaCl2, 0.2 M FeSO4, 1000× vitamins, and autoclaved water)
50 mg/mL Chloramphenicol (filter-sterilized)
100 mg/mL Ampicillin (filter-sterilized)
20% L-Arabinose (filter-sterilized)
1 M IPTG (filter-sterilized) (Gold Biotechnology)
4-Azido-L-phenylalanine (ChemImpex)
10× M9 minimal salts (67.8 g/L Na2HPO4-7H2O, 30 g/L KH2PO4, 10 g/L NH4Cl, 5 g/L NaCl; autoclave-sterilized)
20% D-Glucose (prepared in autoclaved water, filter-sterilized)
1 M MgSO4 (prepared in autoclaved water, filter-sterilized)
1 M CaCl2 (prepared in autoclaved water, filter-sterilized)
0.2 M FeSO4
1000× Vitamins (0.4 g/L choline chloride, 0.5 g/L folic acid, 0.5 g/L nicotinamide, 1g/L myo-inositol, 1 g/L pyridoxal HCl, 2 g thiamine HCl, 0.05 g/L riboflavin, and 1 g/L biotin) (Sigma-Aldrich)
Buffer H (10 mM potassium acetate, pH 7.0, 10 mM KCl, 50 mM CaCl2, and 10% glycerol)
Buffer I (40 mM Tris-HCl, pH 7.9, 300 mM KCl, 10 mM EDTA, 10% glycerol)
2.8 L Triple-baffled Fernbach flask (autoclave-sterilized) (Bellco Glass)
250 mL Erlenmeyer flask (VWR)
1.7 mL Polypropylene microcentrifuge tubes (Denville Scientific)
14 mL Polypropylene culture tubes with snap closures (autoclave-sterilized) (VWR)
Microcentrifuge (e.g., Eppendorf 5417C)
Platform shaker
2.4. Staudinger-Bertozzi Ligation of Unnatural-Amino-Acid-Containing RNAP Subunits and σ70
2.3.1. Staudinger-Bertozzi Ligation of Unnatural-Amino-Acid-Containing FLAG-αNTDI-GSGGSG-αNTDII, β, β' or ω
4-azidophenylalanine-containing FLAG-αNTDI-GSGGSG-αNTDII, β, β, or ω (from Subheading 3.3.1.)
Probe-phosphine (from Subheading 3.1.)
Buffer J (50 mM Tris-HCl, pH 7.9, 6 M guanidine-HCl, and 5% glycerol)
Dimethylformamide (DMF) (Sigma-Aldrich)
Bradford Protein Assay kit (BioRad)
Bio-Gel P30 (BioRad)
Econo-Pac 20 mL chromatography column (BioRad)
1.7 mL Polypropylene microcentrifuge tubes (Denville Scientific)
Sub-micro quartz spectrophotometer cell, 10 mm path length (Starna Cells)
UV/Vis spectrophotometer (e.g., PerkinElmer Lambda 25)
Temperature-controlled rocker (e.g., Lab Line)
2.3.1 Staudinger-Bertozzi Ligation of Unnatural-Amino-Acid-Containing σ70
4-Azidophenylalanine-containing σ70 (from Subheading 3.3.2.)
Probe-phosphine (from Subheading 3.1.)
Buffer G (20 mM Tris-HCl, pH 7.9, 200 mM NaCl, 0.1 mM EDTA, 1 mM DTT, and 50% glycerol)
Buffer J (50 mM Tris-HCl, pH 7.9, 6 M guanidine-HCl, and 5% glycerol)
Buffer TGEβ (20 mM Tris-HCl, pH 7.9, 0.1 mM EDTA, 5 mM 2-mercaptoethanol, and 5% glycerol)
Buffer TGED (20 mM Tris-HCl, pH 7.9, 0.1 mM EDTA, 1 mM DTT, and 5% glycerol)
Dimethylformamide (DMF) (Sigma-Aldrich)
Bradford Protein Assay kit (BioRad)
Spectra/Por 3 dialysis tubing (3.5 kDa molecular weight cutoff) (Spectrum Labs)
Dialysis membrane closures (VWR)
Bio-Gel P30 (BioRad)
Econo-Pac 20 mL chromatography column (BioRad)
Mono-Q HR 10/10 chromatography column (GE healthcare)
ÄKTA Purifier chromatography system (GE Healthcare)
1.7 mL Polypropylene microcentrifuge tubes (Denville Scientific)
Sub-micro quartz spectrophotometer cell, 10 mm path length (Starna Cells)
UV/Vis spectrophotometer (e.g., PerkinElmer Lambda 25)
temperature-controlled rocker (Lab Line)
2.5 Quantitation of Labelling Efficiency and Specificity
Labelled FLAG-αNTDI-GSGGSG-αNTDI I, β, β, or σ70 (from Subheading 3.4.)
Ni:NTA agarose (Qiagen)
Imidazole (Sigma-Aldrich)
Buffer J (50 mM Tris-HCl, pH 7.9, 6 M guanidine-HCl, and 5% glycerol)
Dialysis bags (3.5 kDa molecular-weight cutoff; regenerated cellulose) (Spectrum Labs)
Sub-micro quartz spectrophotometer cell, 10 mm path length (Starna Cells)
UV/Vis spectrophotometer (e.g., Perkin Elmer Lambda 25)
2.6 Reconstitution of RNAP core and RNAP holoenzyme
Labelled and/or unlabelled FLAG-αNTDI-GSGGSG-αNTDI, β, β, and ω (for reconstitution of RNAP core; from Subheading 3.2. and/or Subheading 3.4.), or labelled and/or unlabelled FLAG-αNTDI-GSGGSG-αNTDII, β, β, ω, and σ70 (for reconstitution of RNAP holoenzyme; from Subheading 3.2. and/or Subheading 3.4.)
ANTI-FLAG M2 affinity gel (Sigma-Aldrich)
FLAG peptide (Sigma-Aldrich)
0.1 M Glycine-HCl, pH 3.5
2-Mercaptoethanol (Sigma-Aldrich)
Glycerol (Fisher Scientific)
Buffer F (50 mM Tris-HCl, pH 7.9, 6M guanidine-HCl, 10 mM MgCl2, 10µM ZnCl2, 1 mM EDTA, 10 mM DTT, and 10% glycerol)
Buffer K (50 mM Tris-HCl, pH 7.9, 200 mM KCl, 10 mM MgCl2, 10µM ZnCl2, 1 mM EDTA, 5 mM β-mercaptoethanol, and 20% glycerol)
Buffer B (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 5% glycerol)
Buffer L (30 mM Tris-HCl, pH 7.9, 0.1 mM EDTA, 200 mM NaCl, 1 mM 2-mercaptoethanol, and 5% glycerol)
Buffer M (25 mM Tris-HCl, pH 7.9, 300 mM NaCl, 0.1 mM EDTA, 1 mM 2-mercaptoethanol, and 5% glycerol)
6× SDS-loading buffer (190 mM Tris-HCl, pH 6.8, 6% SDS, 10% β-mercaptoethanol, 48% glycerol, and 0.3% bromophenol blue)
SDS-running buffer (25 mM Tris-HCl, pH 8.3, 250 mM glycine, and 0.1% SDS)
Destaining solution (10% acetic acid, 50% methanol, and 40% water)
Coomassie Brilliant Blue R-250 (Bio-Rad)
10% Polyacrylamide (37.5:1 acrylamide:bisacrylamide), 0.1% SDS, slab gel (10 × 7 × 0.075 cm)
Prestained protein molecular weight markers (7–210 kDa) (Bio-Rad)
Bradford Protein Assay Kit (BioRad)
Dialysis bags (3.5 kDa molecular-weight cutoff; regenerated cellulose) (Spectrum Labs)
Amicon Ultra-4 centrifugal filter unit (100 kDa molecular-weight cutoff) (EMD Millipore)
Econo-Pac 20 mL chromatography column (BioRad)
1.7 mL Polypropylene microcentrifuge tubes (Denville Scientific)
50 mL Polypropylene centrifuge tubes (VWR)
100 mL Polypropylene centrifuge tubes (VWR)
Microcentrifuge (e.g., Eppendorf 5417C)
Mono-Q HR 10/10 chromatography column (GE healthcare)
ÄKTA Purifier chromatography system (GE Healthcare)
3. Methods
3.1. Preparation of Probe Phosphines
3.1.1. Preparation of Cy3B-Phosphine
3.1.1.1. 2-(Tritylamino)ethyl-amido-Cy3B
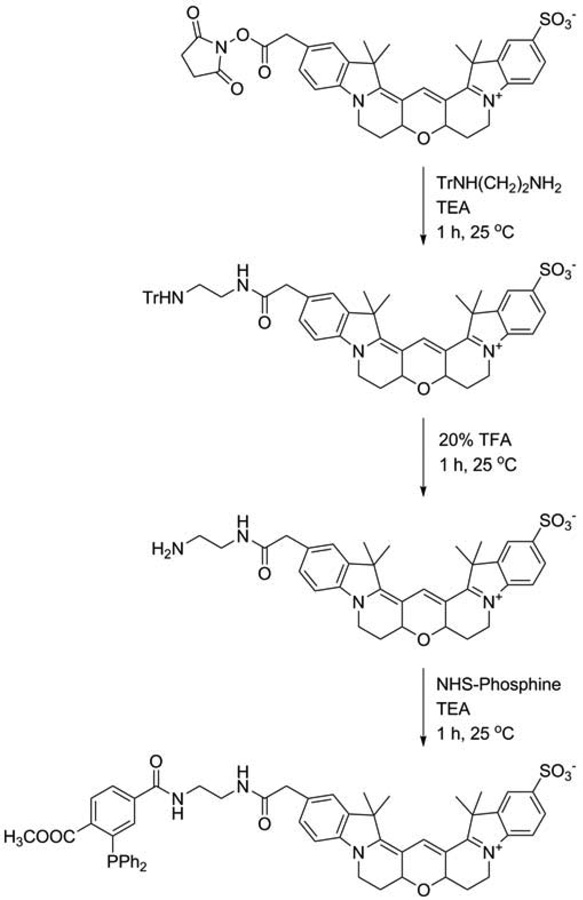
Dissolve 23.5 mg (65 µmol) mono-trityl-ethylenediamine, acetic acid salt, in 200 µL anhydrous DMF in a 1.8 mL glass vial containing a micro stir bar. Add 5.0 mg (6.5 µmol) Cy3B NHS ester followed by 60 µL (430 µmol) TEA. Cap vial and wrap with aluminum foil to protect from light. Stir reaction mixture on magnetic stirrer for 1 h at room temperature. (See Notes 2–4.)
Remove stir bar (e.g., using a magnet), and dry reaction mixture under vacuum in Speedvac (see Note 7).
Re-dissolve dried materials in 1 mL 50% acetonitrile in water. Purify product by reversed-phase HPLC on a Discovery BIO Wide Pore C18 HPLC column (solvent A, water; solvent B, acetonitrile; gradient, 30–100% B in A; 30 min at 2 mL/min; see Note 6). Inject 200 µL for each run. Collect 1 mL fractions. Identify product-containing fractions using diode-array detector. Product exhibits an absorbance maximum at 559 nm and elutes at ~22 min.
Dry product-containing fractions under vacuum in Speedvac (see Note 7).
Re-dissolve in 200 µL methanol.
Mix a 4 µL aliquot with 76 µL water, and measure OD559 using spectrophotometer. Concentration of product is calculated as OD559/130000 M.
Mix 1 µL aliquot with 1 µL saturated CHCA in 90% methanol in water, spot mixture on MALDI plate, dry in air, and analyze using MALDI mass spectrometer in positive-ion reflector mode (calculated m/z, 845.37; see Note 8).
Dry product in open vial under vacuum in Speedvac (see Note 7).
Cap vial, wrap with aluminum foil, and store dry in desiccator at −20°C.
3.1.1.2. 2-(Amino)ethyl-amido-Cy3B
Dissolve 4.2 mg (5.0 µmol) 2-(tritylamino)ethyl-amido-Cy3B from Subheading 3.1.1.1. in 200 µL chloroform in a 1.8 mL glass vial containing a micro stir-bar. Add 50 µL (650 µmol) TFA. Cap vial and wrap with aluminum foil to protect from light. Stir reaction mixture on magnetic stirrer for 1 h at room temperature. (See Note 4.)
Remove stir bar (e.g., using a magnet), and dry reaction mixture under vacuum in Speedvac (see Note 7).
Re-dissolve in 1 mL 50% acetonitrile and 0.1% TFA in water. Purify product by reversed-phase HPLC on Discovery BIO Wide Pore C18 HPLC column (solvent A, 0.1% TFA in water; solvent B, 0.1% TFA in acetonitrile; gradient, 20–80% B in A; 30 min at 2 mL/min; see Note 6). Inject 200 µL for each run. Collect 1 mL fractions. Product-containing fractions are identified using diode-array detector. Product exhibits an absorbance maximum at 559 nm and elutes at ~8 min.
Dry product-containing fractions under vacuum in Speedvac (see Note 7).
Re-dissolve product in 200 µL methanol.
Mix a 4 µL aliquot with 76 µL water, and measure OD559 using spectrophotometer. Concentration of product is calculated as OD559/130000 M.
Mix 1 µL aliquot with 1 µL saturated CHCA in 90% methanol in water, spot mixture on MALDI plate, dry in air, and analyze using MALDI mass spectrometer in positive-ion reflector mode (calculated m/z, 603.26).
Dry product solution in open vial under vacuum in Speedvac (see Note 7).
Cap vial, wrap with aluminum foil, and store dry in desiccator at −20°C.
3.1.1.3. 2-(Phosphinylamido)ethyl-amido-Cy3B (Cy3B-Phosphine)
Dissolve 2.4 mg (4.0 µmol) 2-(amino)ethyl-amido-Cy3B from Subheading 3.1.1.2. in 100 µL anhydrous DMF in a 1.8 mL glass vial containing a micro stir bar. Add 9.2 mg (20 µmol) NHS-phosphine, and 40 µL (290 µmol) TEA. Remove air in vial by blowing argon above reaction mixture. Cap vial and wrap with aluminum foil to protect from light. Stir reaction mixture on magnetic stirrer for 1 h at room temperature. (See Notes 3–4.)
Remove stir bar (e.g., using a magnet), and dry reaction mixture under vacuum in Speedvac (see Note 7).
Re-dissolve in 1 mL 50% acetonitrile and 0.1% TFA in water. Purify product by reversed-phase HPLC on Discovery BIO Wide Pore C18 HPLC column (solvent A, 0.1% TFA in water; solvent B, 0.1% TFA in acetonitrile; gradient, 30–100% B in A; 30 min at 2 mL/min; see Note 6). Inject 200 µL sample for each run. Collect 1 mL fractions. Product-containing fractions are identified using a diode-array detector. Product exhibits an absorbance maximum at 559 nm and elutes at ~17 min.
Dry product-containing fractions under vacuum in Speedvac (see Note 7).
Re-dissolve product in 200 µL methanol.
Mix a 4 µL aliquot with 76 µL water, and measure OD559 using spectrophotometer. Concentration of product is calculated as OD559/130000 M.
Mix 1 µL aliquot with 1 µL saturated CHCA in 90% methanol in water, spot mixture on MALDI plate, dry in air, and analyze using MALDI mass spectrometer in positive-ion reflector mode (calculated m/z, 949.34).
Dry product in open vial under vacuum in Speedvac (see Note 7).
Cap vial, wrap with aluminum foil, and store dry in desiccator at −20°C.
3.1.2. Preparation of Alexa647-Phosphine
3.1.2.1. 2-(Tritylamino)ethyl-amido-Alexa647
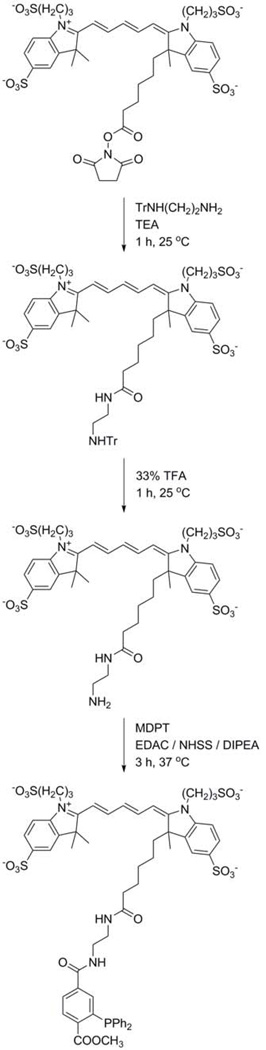
Dissolve 23 mg (60 µmol) N-trityl-1,2-ethanediamine hydrobromide in 1 mL anhydrous DMF in 3.6 mL glass vial containing a micro stir bar. Add 5.0 mg (5.0 µmol) Alexa Fluor 647 NHS ester and 10 µL (71 µmol) TEA to DMF solution. Cap vial and wrap with aluminum foil to protect from light. Stir reaction mixture on magnetic stirrer for 1 h at room temperature. (See Notes 2–4.)
Remove stir bar (e.g., using a magnet), and dry reaction mixture under vacuum in Speedvac (see Note 7).
Re-dissolve in 1 mL of 50% acetonitrile in water. Purify product by reversed-phase HPLC on Discovery BIO Wide Pore C18 HPLC column (solvent A, water; solvent B, acetonitrile; gradient, 10–75% B in A; 30 min at 2 mL/min; see Note 6). Inject 200 µL sample for each run. Collect 1 mL fractions. Product-containing fractions are identified using diode-array detector. Product exhibits an absorbance maximum at 650 nm and elutes at ~20 min.
Dry product-containing fractions under vacuum in Speedvac (see Note 7).
Re-dissolve in 200 µL methanol.
Mix 4 µL aliquot with 76 µL water, and measure OD650 using spectrophotometer. Concentration of product is calculated as OD650/240000 M.
Mix 1 µL aliquot with 1 µL saturated CHCA in 90% methanol in water, spot mixture on MALDI plate, dry in air, and analyze using MALDI mass spectrometer in positive-ion reflector mode (calculated m/z, 1143.36; see Note 8).
Dry product solution in open vial under vacuum in Speedvac (see Note 7).
Cap vial, wrap with aluminum foil, and store dry in desiccator at −20°C.
3.1.2.2. 2-(Amino)ethyl-amido-Alexa647
Dissolve 5.0 mg 2-(tritylamino)ethyl-amido-Alexa647 from Subheading 3.1.2.1. (4.4 µmol) in 200 µL chloroform in 1.8 mL glass vial containing a micro stir bar. Add 100 µL TFA (1.3 mmol) to the solution. Cap vial and wrap with aluminum foil to protect from light. Stir reaction mixture on magnetic stirrer for 1 h at room temperature. (See Note 4.)
Remove stir bar (e.g., using a magnet), and dry reaction mixture under vacuum in Speedvac (see Note 7).
Re-dissolve in 1 mL 50% acetonitrile and 0.1% TFA in water. Purify product by reversed-phase HPLC on Discovery BIO Wide Pore C18 HPLC column (solvent A, 0.1% TFA in water; solvent B, 0.1% TFA in acetonitrile; gradient, 10–90% B in A; 30 min at 2 mL/min; see Note 6). Inject 200 µL sample for each run. Collect 1 mL fractions. Product-containing fractions are identified
using diode-array detector. Product exhibits an absorbance maximum at 650 nm and elutes at ~7.5 min.
Dry product-containing fractions under vacuum in Speedvac (see Note 7).
Re-dissolve in 200 µL methanol.
Mix a 4 µL aliquot with 76 µL water, and measure OD650 using spectrophotometer. Concentration of product is calculated as OD650/240000 M.
Mix 1 µL aliquot with 1 µL saturated CHCA in 90% methanol in water, spot mixture on MALDI plate, dry in air, and analyze using MALDI mass spectrometer in positive-ion reflector mode (calculated m/z, 901.25).
Dry product in open vial under vacuum in Speedvac (see Note 7).
Cap vial, wrap with aluminum foil, and store dry in desiccator at −20°C.
3.1.2.3 2-(Phosphinylamido)ethyl-amido-Alexa647 (Alexa647-Phosphine)
Dissolve 21 mg (110 µmol) EDAC in 250 µL degassed water, 21 mg (78 µmol) NHSS in 250 µL degassed water, 5.0 mg (5.5 µmol) 2-(amino)ethyl-amido-Alexa64 from Subheading 3.1.2.2 in 200 µL DMF, and 30 mg (75 µmol) MDPT in 250 µL DMF. Combine solutions in a 3.6 mL glass vial containing a micro stir bar. Add more DMF until the precipitate dissolves (~700 µL more DMF). Add 28 µL (160 µmol) DIPEA. Remove air in vial by blowing argon above reaction mixture. Cap vial and wrap with aluminum foil to protect from light. Stir reaction mixture on magnetic stirrer for 3 h at 37°C. (See Notes 4–5.)
Remove stir bar (e.g., using a magnet), and dry reaction mixture under vacuum in Speedvac (see Note 7).
Re-dissolve in 1 mL of 50% acetonitrile and 0.1% TFA in water. Purify product by reversed-phase HPLC equipped on Discovery BIO Wide Pore C18 HPLC column (solvent A, 0.1% TFA in water; solvent B, 0.1% TFA in acetonitrile; gradient, 30–100% B in A; 30 min at 2 mL/min; see Note 6). Inject 200 µL sample for each run. Collect 1 mL fractions. Product-containing fractions are identified using a diode-array detector. Product exhibits an absorbance maximum at 650 nm and elutes at ~10 min.
Dry product-containing fractions under vacuum in Speedvac (see Note 7).
Re-dissolve in 200 µL methanol.
Mix a 4 µL aliquot with 76 µL water, and measure OD650 using spectrophotometer. Concentration of product is calculated as OD650/240000 M.
Mix 1 µL aliquot with 1 µL saturated CHCA in 90% methanol in water, spot mixture on MALDI plate, dry in air, and analyze using MALDI mass spectrometer in positive-ion reflector mode (calculated m/z, 1247.33).
Dry product in open vial under vacuum in Speedvac (see Note 7).
Cap vial, wrap with aluminum foil, and store dry in desiccator at −20°C.
3.2 Preparation of RNAP Subunits and σ70
3.2.1 Preparation of FLAG-αNTDI-GSGGSG-αNTDII
Transform chemically competent E. coli strain BL21(DE3) per instructions of vendor with plasmid pET28a-NF-αNTDI-αNTDII. Plate transformants to TYE agar containing 40 µg/mL kanamycin and incubate 16 h at 37°C. (See Note 9.)
Inoculate single colony into 10 mL LB containing 40 µg/mL kanamycin in autoclave-sterilized 50 mL Erlenmeyer flask, and shake vigorously for 12 h at 37°C.
Transfer to 15 mL polypropylene centrifuge tube. Centrifuge 5 min at 3000 × g at room temperature, and discard supernatant. Wash cell pellet twice with 5 mL LB.
Re-suspend cell pellet in 5 mL LB, inoculate into 1 L LB containing 40 µg/mL kanamycin in 2.8 L Fernbach flask, and shake vigorously at 37°C until OD600 = 0.6. Add 1 mL 1 M IPTG, and shake vigorously for an additional 3 h at 37°C.
Transfer culture to 1 L polypropylene copolymer centrifuge bottle, and collect cells by centrifugation 20 min at 5000 × g at 4°C. Discard supernatant.
Re-suspend cell pellet in 50 mL buffer A containing one protease inhibitor cocktail tablet at 4°C.
Lyse cells using Avestin EmulsiFlex-C5 cell disrupter, passing suspension through cell disruptor at pressure of 10,000 psi, collecting lysate in 100 mL glass beaker on ice. Repeat three times.
Transfer lysate to two 50 mL polypropylene centrifuge tubes. Centrifuge 20 min at 20000 × g at 4°C. Collect supernatant.
Transfer supernatant to 100 mL glass beaker. Add 35 g ammonium sulfate per 100 mL supernatant, and stir 20 min on ice.
Transfer suspension to two 50 mL polypropylene centrifuge tubes. Centrifuge 10 min at 20000 × g at 4°C. Discard supernatant.
Re-suspend pellet in 10 mL buffer B. Load supernatant onto two 5 mL ANTI-FLAG M2 affinity gel columns pre-equilibrated with 20 mL buffer B (prepared by pouring 10 mL ANTI-FLAG M2 suspension into 20 mL Econo-Pac column, removing snap-off tip at bottom of column, allowing liquid to drain, and activating column with 15 mL 0.1 M glycine-HCl, pH 3.5, prior to equilibration with buffer B; see Note 10). Collect flow-through, and re-load onto column twice. Wash column with 50 mL buffer B. Elute column with fifteen 2 mL fractions of buffer B containing 0.1 mg/mL FLAG peptide.
Transfer 30 µL aliquots of each fraction into 1.7 mL polypropylene microcentrifuge tubes, and add 6 µL 6× SDS-loading buffer, heat 5 min at 100°C, and apply to 10% polyacrylamide (37.5:1 acrylamide:bisacrylamide), 0.1% SDS, slab gel (10 × 7 × 0.075 cm). As marker, load into adjacent lane 5 µl prestained protein molecular-weight markers. Electrophorese in SDS-running buffer at 25 V/cm until bromophenol blue reaches bottom of gel. Stain gel by gently shaking for 10 min in 50 mL 0.2% Coomassie Brilliant Blue R-250 in destaining solution. Destain by gently shaking for 1 h in 100 mL destaining solution.
Pool fractions containing FLAG-αNTDI-GSGGSG-αNTDII. Dialyze using 3.5-kDa molecular-weight-cutoff dialysis tubing against two 1 L changes of Buffer C for 12 h at 4°C.
Transfer to 50 mL polypropylene centrifuge tube. Add ammonium sulfate to final concentration of 0.3 g/mL, and rock gently 20 min at 4°C. Centrifuge 10 min at 20000 × g at 4°C. Remove all except ~5 mL of supernatant Re-suspend pellet in remaining ~ 5 mL of supernatant.
Divide into 1.5 mL aliquots, and transfer to 1.7 mL polypropylene microcentrifuge tubes. Store at −80°C (stable for at least 1 y). Expected yield: 15–20 mg (4–6 mg/aliquot). Expected purity: >99%.
3.2.2 Preparation of β, β' and ω
Transform chemically competent E. coli strain BL21(DE3) per instructions of vendor with plasmid pET21d-rpoB-CH6, pET21a-rpoC-CH6, or pT7ω. Plate transformants to TYE agar containing 100 µg/mL ampicillin and incubate 16 h at 37°C.
Inoculate single colony into 10 mL LB containing 100 µg/mL ampicillin in 50 mL autoclave-sterilized Erlenmeyer flask, and shake vigorously for 12 h at 37°C.
Inoculate culture from step 2 into 1 L LB containing 100 µg/mL ampicillin in 2.8 L Fernbach flask, and shake vigorously at 37°C until OD600 = 0.6. Add 1 mL 1 M IPTG, and shake vigorously for 3 h at 37°C.
Transfer culture to 1 L polypropylene copolymer centrifuge bottle. Harvest cells by centrifugation 30 min at 4000 × g at 4°C.
Re-suspend cell pellet in 50 mL Buffer D containing 0.2% sodium desoxycholate and one protease inhibitor cocktail tablet at 4°C. Transfer suspension into 50 ml polypropylene centrifuge tube, and place tube on ice.
Lyse cells in using Avestin EmulsiFlex-C5 cell disrupter, passing suspension through cell disruptor at pressure of 10,000 psi, collecting lysate in 100 mL glass beaker on ice. Repeat three times.
Transfer lysate to 50 ml polypropylene centrifuge tube. Centrifuge 20 min at 15000 × g at 4°C. Discard supernatant.
Re-suspend pellet in 10 mL Buffer D containing 0.2 % n-octyl-β-D-glucopyranoside at 4°C. Sonicate with five 30 s sonication pulses at 25% maximum sonicator output (2 min pause between each pulse). Centrifuge 20 min at 15000 × g at 4°C. Discard supernatant.
Re-suspend pellet in 10 mL Buffer D containing 0.2 % n-octyl-β-D-glucopyranoside at 4°C. Sonicate as in step 8. Centrifuge 20 min at 15000 × g at 4°C. Discard supernatant.
Re-suspend pellet in 6 mL Buffer D at 4°C. Place tube on ice, and sonicate 10 s at 25% maximum sonicator output. Divide into 1.5 ml aliquots, and transfer to 1.7 mL polypropylene microcentrifuge tubes. Centrifuge 5 min at 13000 × g at 4°C. Discard supernatant.
Add 100 µl ice-cold Buffer D containing 10% glycerol. Store at −80°C (stable for at least 1 y). Expected yield: 50–100 mg (12.5–25 mg/aliquot). Expected purity: 50–90%.
3.2.3 Preparation of σ70
Transform chemically competent E. coli strain BL21(DE3) per instructions of vendor with plasmid pGEMD. Plate transformants to TYE agar containing 200 µg/mL ampicillin and incubate 16 h at 37°C.
Inoculate single colony into 10 mL LB containing 200 µg/mL ampicillin in 50 mL Erlenmeyer flask, and shake vigorously for 12 h at 37°C.
Inoculate culture from step 2 into 1 L LB containing 200 µg/mL ampicillin in 2.8 L Fernbach flask, and shake vigorously at 37°C until OD600 = 0.6. Add 1 mL 1 M IPTG, and shake vigorously for an additional 3 h at 37°C.
Transfer culture to 1 L polypropylene copolymer centrifuge bottle, and collect cells by centrifugation 20 min at 5000 × g at 4°C. Discard supernatant.
Re-suspend cell pellet in 40 mL Buffer E containing 0.2% sodium desoxycholate and one protease inhibitor cocktail tablet at 4°C.
Lyse cells in using Avestin EmulsiFlex-C5 cell disrupter, passing suspension through cell disruptor at pressure of 10,000 psi, collecting lysate in 100 mL glass beaker on ice. Repeat three times.
Transfer lysate to 50 mL polypropylene centrifuge tube. Centrifuge 20 min at 15000 × g at 4°C. Discard supernatant.
Re-suspend pellet in 20 mL Buffer E containing 0.2% sodium desoxycholate, 0.2% n-octyl-β-D-glucopyranoside, and 0.02% lysozyme, and sonicate 2 min at 40% duty cycle and 40% maximum output. Centrifuge 20 min at 15000 × g at 4°C. Discard supernatant.
Re-suspend the pellet in 20 mL Buffer E containing 0.2% sodium desoxycholate and 0.5% Triton X-100, and sonicate as in step 7. Centrifuge 20 min at 15000 × g at 4°C. Discard supernatant.
Solubilize pellet in 40 mL Buffer F. Break pellet by pipetting gently. Incubate 20 min at 4°C.
Centrifuge 20 min at 15000 × g at 4°C. Transfer supernatant to new 50 mL polypropylene centrifuge tube.
Dialyze using 3.5 kDa molecular-weight-cutoff dialysis tubing against two 2 L changes of Buffer TGEβ containing 0.2 M NaCl for 16 h at 4°C.
Remove particulates by centrifugation for 20 min at 15000 × g at 4°C. Apply supernatant at 1 mL/min flow-rate to Mono-Q HR 10/10 column pre-equilibrated in 40 mL Buffer TGED containing 0.2 M NaCl. Wash column with 16 mL Buffer TGED containing 0.2 M NaCl. Elute column at 1 mL/min flow-rate with 160 mL linear gradient of 0.2–0.6 M NaCl in Buffer TGED. Collect 2 mL fractions.
Transfer 10 µL aliquot of each fraction to 1.7 mL polypropylene microcentrifuge tube, and add 2 µL 6× SDS-loading buffer, heat 5 min at 100°C, and apply to 10% polyacrylamide (37.5:1 acrylamide:bisacrylamide), 0.1% SDS, slab gel (10 × 7 × 0.075 cm). As marker, load into adjacent lane 5 µl prestained protein molecular-weight markers. Electrophorese in SDS-running buffer at 25 V/cm until bromophenol blue reaches bottom of gel. Stain gel by gently shaking for 10 min in 50 mL 0.2% Coomassie Brilliant Blue R-250 in destaining solution. Destain by gently shaking for 1 h in 100 mL destaining solution.
Pool fractions containing σ70 (typically fractions near 0.36 M NaCl in Buffer TGED), and concentrate ~6-fold using 30 kDa molecular-weight-cutoff Amicon Ultra centrifugal filter unit.
Determine protein concentration using Bradford Protein Assay Kit per procedure of vendor.
Dialyze pooled fractions using 3.5 kDa molecular-weight-cutoff dialysis tubing against 500 mL buffer G for 8 h at 4°C (or, as simpler alternative, mix pooled fractions with equal volume of glycerol), and store in 1 mL aliquots at −80°C (stable for at least 1 y). Expected yield: 50–80 mg (5–8 mg/aliquot). Expected purity: >95%.
3.3 Unnatural-amino-acid Mutagenesis of RNAP Subunits and σ70
3.3.1 Unnatural-Amino-Acid Mutagenesis of FLAG-αNTDI-GSGGSG-αNTDII, β, β' and ω
Introduce amber codon (TAG) at site of interest in plasmid pET28a-NF-αNTDI-αNTDII, pET21d-rpoB-CH6, pET21a-rpoC-CH6, or pT7ω by site-directed mutagenesis using QuikChange II Site-Directed Mutagenesis Kit per instructions of vendor.
Transform chemically competent E. coli strain BL21(DE3) per instructions of vendor with plasmid pEVOL-pAzF. Plate transformants to TYE agar plates containing 50 µg/mL chloramphenicol and incubate 16 h at 37°C.
Pick single colony and inoculate into 5 mL LB broth containing 50 µg/mL chloramphenicol in 14 mL polypropylene culture tube. Shake vigorously 16 h at 37°C.
Inoculate 50 µl into 5 mL LB broth containing 50 µg/mL chloramphenicol in 14 mL polypropylene culture tube. Shake vigorously at 37°C until OD600 = 0.3–0.4.
Transfer 1 mL aliquot to 1.7 mL microcentrifuge tube. Centrifuge 10 min at 1000 × g at 4°C, and discard supernatant.
Re-suspend pellet gently in 300 µl of Buffer H. Incubate 20 min on ice. Centrifuge 10 min at 1000 × g at 4°C, and discard supernatant.
Re-suspend pellet gently in 100 µl Buffer H. Freeze in dry-ice/ethanol bath and store at −70°C.
Transform chemically competent BL21(DE3) pEVOL-pAzF cells from step 7 with amber-codon-containing plasmid from step 1. Plate transformants onto TYE agar containing 50 µg/mL chloramphenicol and either 40 µg/mL kanamycin (for pET28a-NF-αNTDI-αNTDII derivatives) or 100 µg/mL ampicillin (for pET21d-rpoB-CH6, pET21a-rpoC-CH6, or pT7ω derivatives). Incubate 16 h at 37°C.
Pick single colony and inoculate into 10 mL LB broth containing 50 µg/mL chloramphenicol and either 40 µg/mL kanamycin (for pET28a-NF-αNTDI-αNTDII derivatives) or 100 µg/mL ampicillin (for pET21d-rpoB-CH6, pET21a-rpoC-CH6, or pT7ω derivatives) in 50 mL autoclave-sterilized Erlenmeyer flask, and shake vigorously for 12 h at 37°C.
Inoculate culture from step 10 into 1 L LB broth containing 50 µg/mL chloramphenicol and either 40 µg/mL kanamycin (for pET28a-NF-αNTDI-αNTDII derivatives) or 100 µg/mL ampicillin (for pET21d-rpoB-CH6, pET21a-rpoC-CH6, or pT7ω derivatives) in 2.8 L Fernbach flask, and shake vigorously at 37°C until OD600 = 0.6. Add 4-azido-L-phenylalanine to final concentration of 1 mM, add 1 mL 20% arabinose and 1 mL 1 M IPTG, and shake vigorously for 4 h at 37°C in the dark.
Follow steps 5–15 from Subheading 3.2.1 (for pET28a-NF-αNTDI-αNTDII derivatives) or steps 4–11 from Subheading 3.2.2 (for pET21d-rpoB-CH6, pET21a-rpoC-CH6, or pT7ω derivatives) to purify the resulting 4-azidophenylalanine-labelled proteins.
3.3.2 Unnatural-Amino-Acid Mutagenesis of σ70
Introduce amber codon (TAG) at site of interest in plasmid pGEMD by site-directed mutagenesis using QuikChange II Site-Directed Mutagenesis Kit per instructions of vendor.
Perform steps 2–7 from Subheading 3.3.1.
Transform chemically competent BL21(DE3) pEVOL-pAzF cells from step 2 with amber-codon-containing plasmid from step 1. Plate transformants onto TYE agar containing 35 µg/mL chloramphenicol and 200 µg/mL ampicillin. Incubate 16 h at 37°C.
Pick single colony and inoculate into 50 mL LB broth containing 35 µg/mL chloramphenicol and 200 µg/mL ampicillin in 250 mL autoclave-sterilized Erlenmeyer flask, and shake vigorously for 16 h at 37°C.
Transfer to 50 mL polypropylene tube, and centrifuge 5 min at 3000 × g at room temperature. Discard supernatant.
Re-suspend pellet in 10 mL M9+ medium, inoculate into 1 L M9+ medium containing 35 µg/mL chloramphenicol, 200 µg/mL ampicillin, and 1 mM 4-azido-L-phenylalanine in 2.8 L Fernbach flask, and shake vigorously at 37°C in the dark until OD600 = 0.5. Add 1 mL 20% L-arabinose, and shake vigorously at 37°C in the dark until OD600 = 0.6. Add 1 mL 1 M IPTG, and shake vigorously for 3 h at 37°C in the dark.
Follow steps 4–9 from Subheading 3.2.3. to purify the resulting 4-azidophenylalanine-labelled protein.
Re-suspend pellet in 10 mL Buffer E containing 10% glycerol. Store in 1.5 mL aliquots at −80°C (stable for at least 1 y). Expected yield: 30–50 mg (4.5–7.5 mg/aliquot).
3.4 Staudinger-Bertozzi Ligation of Unnatural-Amino-Acid-Containing RNAP Subunits and σ70
3.4.1 Staudinger-Bertozzi Ligation of Unnatural-Amino-Acid-Containing FLAG-αNTDI-GSGGSG-αNTDII, β, β', or ω
Thaw aliquot of 4-azidophenylalanine-labelled FLAG-αNTDI-GSGGSG-αNTDII, β, β’, or ω from Subheading 3.3.1 by placing on ice for 10 min. Centrifuge 1 min at 13000 × g. Discard supernatant.
Solubilize pellet in 1 mL Buffer J. Mix with pipette. Centrifuge 2 min at 13,000 × g. Transfer supernatant to new 1.7 mL polypropylene microcentrifuge tube. Determine protein concentration using Bradford Protein Assay Kit per procedures of vendor (typically 9–12 mg/mL).
Prepare 2.55 mL solution containing 30 µM solubilized 4-azidophenylalanine-labelled protein in Buffer J in 15 mL polypropylene centrifuge tube.
Prepare 0.45 mL solution containing 2 mM probe-phosphine from Subheading 3.1 in DMF.
Add solution of step 4 to solution of step 3. Incubate 16 h at 37°C, rocking gently.
Apply 1 mL aliquot of reaction mixture of step 5 to each of three 10 mL Bio-Gel P30 columns (prepared by adding 20 mL BioGel P30 to 20 mL Econo Pac column, removing snap-off tip at bottom, and allowing liquid to drain; pre-equilibrated in Buffer J by application of 30 mL Buffer J to top of column bed). Wash each column with 3 mL Buffer J. Elute each column with 4 mL buffer J. Collect 1 mL fractions.
Identify fractions containing probe-labelled protein by removing 20 µl aliquots, diluting with 80 µl Buffer J, and measuring UV/Vis absorbance using spectrophotometer (559 nm for Cy3B-labelled protein; 650 nm for Alexa647-labelled protein).
Pool fractions containing probe-labelled protein, add glycerol to a final concentration of 5%, and store in 1 mL aliquots at −80°C (not stable to prolonged storage; use within 48 h). Expected yield: 0.5–10 mg (0.15–3 mg per aliquot).
3.4.2 Staudinger-Bertozzi Ligation of Unnatural-Amino-Acid-Containing σ70
Thaw aliquot of 4-azidophenylalanine-labelled σ70 from Subheading 3.3.2 by placing on ice for 10 min.
Perform steps 2–8 from Subheading 3.4.1.
Pool fractions containing probe-labelled protein. Centrifuge 20 min at 15000 × g at 4°C. Transfer supernatant to 15 mL polypropylene centrifuge tube.
Dialyze using 3.5 kDa molecular-weight-cutoff dialysis tubing against two 1 L changes of Buffer TGEβ containing 0.2 M NaCl for 16 h at 4°C.
Remove particulates by centrifugation for 20 min at 15000 × g at 4°C. Apply supernatant at 1 mL/min flow-rate to Mono-Q HR 10/10 column pre-equilibrated in 40 mL Buffer TGED containing 0.2 M NaCl. Wash column with 16 mL Buffer TGED containing 0.2 M NaCl. Elute column at 1 mL/min flow-rate with 160 mL linear gradient of 0.2–0.6 M NaCl in Buffer TGED. Collect 2 mL fractions.
Identify fractions containing probe-labelled protein by removing 20 µl aliquots, diluting with 80 µl Buffer TGED, and measuring UV/Vis absorbance using spectrophotometer (559 nm for Cy3B-labelled protein; 650 nm for Alexa647-labelled protein).
Mix pooled fractions with an equal volume of glycerol, and store in 1 mL aliquots at −80°C (stable for at least 1 y). Expected yield: 0.5–1 mg (50–100 µg per aliquot).
3.5 Quantitation of Labelling Efficiency and Specificity
3.5.1 Quantitation of labelling efficiency and specificity for FLAG-αNTDI-GSGGSG-αNTDII, β, or β'
Apply 0.5–1.0 mg of probe-labelled FLAG-αNTDI-GSGGSG-αNTDII, β, or β' from Subheading 3.4.1 to 1 mL Ni:NTA agarose column pre-equilibrated in Buffer J containing 5 mM imidazole. Wash column with 5 mL buffer J containing 20 mM imidazole. Elute column with 3 mL buffer J containing 300 mM imidazole. Collect 0.5 mL fractions.
Identify fractions containing probe-labelled protein by removing 20 µl aliquots, diluting with 80 µl Buffer J, and measuring UV/Vis absorbance using spectrophotometer.
Pool fractions containing probe-labelled protein, and dialyze using 3.5 kDa molecular-weight-cutoff dialysis tubing against 500 mL buffer D for 16 h at 4°C.
Remove 20 µl aliquot, dilute with 80 µl Buffer J, and measure UV/Vis absorbance using spectrophotometer.
- Calculate the concentration of probe-labelled protein and the labelling efficiency, as :
where A280 is the measured absorbance at 280 nm, Amax is the measured absorbance at the long-wavelength absorbance maximum of fluorescent probe F (559 nm and 652 nm for Cy3B and Alexa647, respectively), εP,280 is the molar extinction coefficient of protein P at 280 nm (13410 M−1 cm−1, 86070 M−1 cm−1, and 100060 M−1 cm−1 for FLAG-αNTDI-GSGGSG-αNTDII, β, and β', respectively), εF,280 is the molar extinction coefficient of fluorescent probe F at 280 nm (10400 M−1 cm−1 and 7350 M−1 cm−1 for Cy3B and Alexa647, respectively), and εF,max is the extinction coefficient of fluorescent probe F at its long-wavelength absorbance maximum (130000 M−1 cm−1 and 240000 M−1 cm−1 for Cy3B and Alexa647, respectively). - In order to quantify labelling specificity for probe-labelled FLAG-αNTDI-GSGGSG-αNTDII, β, or β', perform parallel Staudinger-Bertozzi ligation using non-4-azido-phenylalanine-labelled FLAG-αNTDI-GSGGSG-αNTDII, β, or β', respectively, analyze product as in steps 1–6 above, and calculate labelling specificity as:
where efficiencyP-azide is the labelling efficiency with the 4-azidophenylalanine-containing protein and efficiencyP is the labelling efficiency with the corresponding non-4-azidophenylalanine-containing protein.
3.5.1 Quantitation of labelling efficiency and specificity for σ70
Thaw aliquot of probe-labelled from Subheading 3.4.2 by placing on ice for 10 min.
Remove 20 µl aliquot, dilute with 80 µl Buffer TGED, and measure UV/Vis absorbance using spectrophotometer.
- Calculate the concentration of probe-labelled protein and the labelling efficiency, as:
where A280 is the measured absorbance at 280 nm, Amax is the measured absorbance at the long-wavelength absorbance maximum of fluorescent probe F (559 nm and 652 nm for Cy3B and Alexa647, respectively), εP,280 is the molar extinction coefficient of protein P at 280 nm (41370 M−1 cm−1 for σ70), εF,280 is the molar extinction coefficient of fluorescent probe F at 280 nm (10400 M−1 cm−1 and 7350 M−1 cm−1 for Cy3B and Alexa647, respectively), and εF,max is the extinction coefficient of fluorescent probe F at its long-wavelength absorbance maximum (130000 M−1 cm−1 and 240000 M−1 cm−1 for Cy3B and Alexa647, respectively). - In order to quantify the labelling specificity of probe-labelled σ70, perform parallel Staudinger-Bertozzi ligation using non-4-azido-phenylalanine-labelled σ70, respectively, analyze product as in steps 1–3 above, and calculate labelling specificity as:
where efficiencyP-azide is the labelling efficiency with the 4-azidophenylalanine-containing protein and efficiencyP is the labelling efficiency with the corresponding non-4-azidophenylalanine-containing protein.
3.6 Reconstitution of labelled RNAP core and labelled RNAP holoenzyme
For RNAP derivatives containing unlabelled FLAG-αNTDI-GSGGSG-αNTDII, unlabelled β, unlabelled β', and/or unlabelled ω, thaw aliquots of unlabelled FLAG-αNTDI-GSGGSG-αNTDII, unlabelled β or β’, and/or unlabelled ω by placing on ice for 10 min. Centrifuge each aliquot 1 min at 13000×g. Discard each supernatant. Solubilize each pellet in 1 mL buffer J. If protein concentrations have not been determined previously, determine protein concentration using Bradford Protein Assay Kit per procedure of vendor.
For RNAP derivatives containing labelled FLAG-αNTDI-GSGGSG-αNTDII, labelled β, labelled β', labelled ω, and/or unlabelled or labelled σ70, thaw aliquots of labelled FLAG-αNTDI-GSGGSG-αNTDII, labelled β, labelled β', labelled ω, and/or unlabelled or labelled σ70 by placing on ice for 10 min. If protein concentrations have not been determined previously, determine protein concentration using Bradford Protein Assay Kit per procedure of vendor.
Prepare RNAP core reconstitution mixture by combining 4.2 mg (80 nmol) FLAG-α-NTDI-GSGGGSG-NTDII, 3 mg (20 nmol) β, 7.8 mg (50 nmol) β’, and 200 mg (200 nmol) ω, in 60 mL buffer F in a 100 mL polypropylene tube.
Dialyze RNAP core reconstitution mixture in regenerated-cellulose dialysis bag against two 2 L changes of buffer K for 16 h at 4°C.
Transfer RNAP core reconstitution mixture to 50 mL polypropylene centrifuge tubes. Centrifuge 30 min at 20000 × g at 4°C.
Transfer supernatant to new 50 mL polypropylene centrifuge tubes. For preparation of RNAP core, incubate tubes 45 min at 30°C. For preparation of RNAP holoenzyme, supplement each tube with 0.35 mg (5 nmol) σ70, and incubate 45 min at 30°C.
Centrifuge 30 min at 20000 × g. Transfer supernatant to new 100 mL polypropylene centrifuge tubes at 4°C.
During incubation in step 6, prepare 3 mL ANTI-FLAG M2 column by pouring 6 mL ANTI-FLAG M2 suspension into 20 mL Econo-Pac column, removing snap-off tip at bottom of column, and allowing liquid to drain; wash column with 9 mL 0.1 M glycine-HCl, pH 3.5; and wash column with 15 mL buffer B (see Note 10).
Apply supernatant from step 7 to ANTI-FLAG M2 column. Collect and re-load flow-through. Wash column with 30 mL buffer B. Elute column with buffer B containing 0.1 mg/mL FLAG peptide. Collect 1 mL fractions.
Identify fractions containing protein using Bradford Protein Assay Kit.
Dialyze pooled protein-containing fractions in regenerated-cellulose dialysis bag against 1L buffer L for 16 h at 4°C.
Centrifuge 20 min at 20000 × g. Transfer supernatant to new 50 mL polypropylene centrifuge tube.
Apply sample to Mono-Q HR 10/10 column pre-equilibrated in buffer M. Wash column with 24 mL buffer M. Elute column with 160 mL linear gradient of 300–500 mM NaCl. Collect 2 mL fractions.
Identify fractions by SDS-PAGE and Coomassie staining. Pool fractions containing RNAP.
Concentrate pooled fractions to ~125 µL using 100 kDa molecular-weight-cutoff Amicon Ultra-4 centrifugal filter unit.
Transfer sample into 1.7 mL polypropylene microcentrifuge tubes. Add 2-mercaptoethanol to final concentration of 1 mM and glycerol to final concentration of 50%. Mix and store at −20°C. Typical yields are ~0.2 mg per 60 mL reconstitution mixture.
Figure 1.
Synthesis of Cy3B-phosphine.
Figure 2.
Synthesis of Alexa647-phosphine.
Acknowledgements
We thank Peter Schultz and Ryan Mehl for plasmids. This work was supported by National Institutes of Health grant GM041376 and a Howard Hughes Medical Institute Investigatorship to R.H.E.
Footnotes
We have listed suppliers for all reagents in our protocol. Sufficiently pure reagents from alternative suppliers likely will suffice.
Cy3B has excitation and emission maxima of 559 nm and 570 nm, respectively. Alexa647 has excitation and emission maxima of 650 nm and 668 nm, respectively. Cy3B and Alexa647 have been used as a donor-acceptor pair for determination of distances by measurement of fluorescence resonance energy transfer (FRET; 16). When Cy3B and Alexa647 are used as a donor-acceptor pair, Ro, the distance at which donor-acceptor FRET efficiency is half-maximal is ~60 Å (16).
NHS esters are moisture-sensitive. Store NHS-ester containing compounds dry in desiccator containing Drierite dessicant at −20°C. Equilibrate vials to room temperature before opening. Prepare stock solutions immediately before use.
Fluorescent compounds are light-sensitive. Minimize exposure to light.
MDPT can be synthesized by procedures in 31.
Solvents for HPLC should be de-gassed by bubbling argon for 15 min. A Pasteur pipette is used to direct argon to bottom of solvent container, allowing bubbling from bottom of solvent container. De-gassing is particularly important for solvents for HPLC of phosphine derivatives, since dissolved oxygen in non-de-gassed solvents oxidizes phosphines to non-reactive phosphine oxides.
Reaction mixtures and HPLC fractions for preparation are dried under vacuum in a Speedvac without heating. It typically takes several hours to dry microliter-scale reaction mixtures and ≥12 h to dry milliliter-scale HPLC fractions.
When analyzing trityl-containing compounds by mass spectrometry with CHCA as MALDI matrix, acid-catalyzed detritylation may occur, resulting in non-detection of the m/z signal for the trityl-containing compound and detection instead of the m/z signal for the corresponding detritylated compound.
FLAG-αNTDI-GSGGSG-αNTDII is a fusion protein comprising an N-terminally FLAG-tagged first E. coli RNAP α subunit N-terminal domain (α residues 1–235; αNTDI), followed by a GlySerGlyGlySerGly linker, followed by a second E. coli RNAP α subunit N-terminal domain (α residues 1–235; αNTDII) (16). The use in this procedure of FLAG-αNTDI-GSGGSG-αNTDII instead of N-terminally FLAG-tagged wild-type RNAP α subunit results in higher yields and equal or higher specific activities (16). RNAP derivatives containing FLAG-αNTDI-GSGGSG-αNTDII behave indistinguishably from RNAP derivatives containing wild-type α subunit in transcription initiation and elongation (16).
ANTI-FLAG M2 columns should be activated with glycine-HCl no more than 20 min before application of samples.
References
- 1.Miyake R, Murakami K, Owens J, Greiner D, Ozoline O, Ishihama A, Meares C. Dimeric association of Escherichia coli RNA polymerase α subunits, studied by cleavage of single-cysteine alpha subunits conjugated to iron-(S)-1-[p-(bromoacetamido)benzyl]ethylenediaminetetraacetate. Biochem. 1998;37:1344–1349. doi: 10.1021/bi9723313. [DOI] [PubMed] [Google Scholar]
- 2.Owens J, Chmura A, Murakami K, Fujita N, Ishihama A. Mapping the promoter DNA sites proximal to conserved regions of σ70 in an Escherichia coli RNA polymerase-lac UV5 open promoter complex. Biochem. 1998;37:7670–7675. doi: 10.1021/bi980188n. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Ebright Y, Ebright RH. Identification of the target of a transcription activator protein by protein-protein photocrosslinking. Science. 1994;265:90–92. doi: 10.1126/science.8016656. [DOI] [PubMed] [Google Scholar]
- 4.Miller A, Wood D, Ebright RH, Rothman-Denes L. RNA polymerase beta' subunit: a target of DNA binding-independent activation. Science. 2004;75:1655–1657. doi: 10.1126/science.275.5306.1655. [DOI] [PubMed] [Google Scholar]
- 5.Callaci S, Heyduk E, Heyduk T. Conformational changes of Escherichia coli RNA polymerase σ70 factor induced by binding to the core enzyme. J. Biol. Chem. 1998;273:32995–33001. doi: 10.1074/jbc.273.49.32995. [DOI] [PubMed] [Google Scholar]
- 6.Callaci S, Heyduk E, Heyduk T. Core RNA polymerase from E. coli induces a major change in the domain arrangement of the σ70 subunit. Mol. Cell. 1999;3:229–238. doi: 10.1016/s1097-2765(00)80313-5. [DOI] [PubMed] [Google Scholar]
- 7.Heyduk E, Heyduk T. Architecture of a complex between the σ70 subunit of Escherichia coli RNA polymerase and the nontemplate strand oligonucleotide. J. Biol. Chem. 1999;274:3315–3322. doi: 10.1074/jbc.274.6.3315. [DOI] [PubMed] [Google Scholar]
- 8.Mukhopadhyay J, Kapanidis A, Mekler V, Kortkhonjia E, Ebright YW, Ebright RH. Translocation of σ70 with RNA polymerase during transcription: fluorescence resonance energy transfer assay for movement relative to DNA. Cell. 2001;106:453–463. doi: 10.1016/s0092-8674(01)00464-0. [DOI] [PubMed] [Google Scholar]
- 9.Mekler V, Kortkhonjia E, Mukhopadhyay J, Knight J, Revyakin A, Kapanidis A, Niu W, Ebright YW, Levy R, Ebright RH. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. [DOI] [PubMed] [Google Scholar]
- 10.Mukhopadhyay J, Mekler V, Kortkhonjia E, Kapanidis A, Ebright YW, Ebright RH. Fluorescence resonance energy transfer (FRET) in analysis of transcription-complex structure and function. Meths. Enzymol. 2003;371:144–159. doi: 10.1016/S0076-6879(03)71010-6. [DOI] [PubMed] [Google Scholar]
- 11.Mukhopadhyay J, Sineva E, Knight J, Levy R, Ebright RH. Antibacterial peptide microcin J25 inhibits transcription by binding within, and obstructing, the RNA polymerase secondary channel. Mol. Cell. 2004;14:739–751. doi: 10.1016/j.molcel.2004.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knight J, Mekler V, Mukhopadhyay J, Ebright RH, Levy R. Distance-restrained docking of rifampicin and rifamycin SV to RNA polymerase using systematic FRET measurements: developing benchmarks of model quality and reliability. Biophys. J. 2005;88:925–938. doi: 10.1529/biophysj.104.050187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapanidis A, Margeat E, Laurence T, Doose S, Ho S, Mukhopadhyay J, Kortkhonjia E, Mekler V, Ebright RH, Weiss S. Retention of transcription Initiation factor σ70 in transcription elongation: single-molecule analysis. Mol. Cell. 2005;20:347–356. doi: 10.1016/j.molcel.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Margeat E, Kapanidis A, Tinnefeld P, Wang Y, Mukhopadhyay J, Ebright RH, Weiss S. Direct observation of abortive initiation and promoter escape within single immobilized transcription complexes. Biophys. J. 2006;90:1419–1431. doi: 10.1529/biophysj.105.069252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapanidis A, Margeat E, Ho S, Kortkhonjia E, Weiss S, Ebright RH. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakraborty A, Wang D, Ebright YW, Korlann Y, Kortkhonjia E, Kim T, Chowdhury S, Wigneshweraraj S, Irschik H, Jansen R, Nixon BT, Knight J, Weiss S, Ebright RH. Opening and closing of the bacterial RNA polymerase clamp. Science. 2012;337:591–595. doi: 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebright Y, Chen Y, Pendergrast PS, Ebright R. Incorporation of an EDTA-metal complex at a rationally selected site within a protein: application to EDTA-iron DNA affinity cleaving with catabolite gene activator protein (CAP) and Cro. Biochem. 1992;31:10664–10670. doi: 10.1021/bi00159a004. [DOI] [PubMed] [Google Scholar]
- 18.Igarashi K, Ishihama A. Bipartite functional map of the E. coli RNA polymerase α subunit: Involvement of the C-terminal region in transcription activation by cAMP-CRP. Cell. 1991;65:1015–1022. doi: 10.1016/0092-8674(91)90553-b. [DOI] [PubMed] [Google Scholar]
- 19.Kashlev M, Martin E, Polyakov A, Severinov K, Nikiforov V, Goldfarb A. Histidine-tagged RNA polymerase: dissection of the transcription cycle using immobilized enzyme. Gene. 1993;130:9–14. doi: 10.1016/0378-1119(93)90340-9. [DOI] [PubMed] [Google Scholar]
- 20.Tang H, Severinov K, Goldfarb A, Ebright RH. Rapid RNA polymerase genetics: one-day, no-column preparation of reconstituted recombinant Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. USA. 1995;92:4902–4906. doi: 10.1073/pnas.92.11.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang H, Kim Y, Severinov K, Goldfarb A, Ebright RH. Escherichia coli RNA polymerase holoenzyme: rapid reconstitution from recombinant α, β, β', and σ subunits. Meths. Enzymol. 1996;273:130–134. doi: 10.1016/s0076-6879(96)73012-4. [DOI] [PubMed] [Google Scholar]
- 22.Naryshkin N, Kim Y, Dong Q, Ebright RH. Site-specific protein-DNA photocrosslinking: analysis of bacterial transcription initiation complexes. Meths. Mol. Biol. 2001;148:337–361. doi: 10.1385/1-59259-208-2:337. [DOI] [PubMed] [Google Scholar]
- 23.Naryshkin N, Druzhinin S, Revyakin A, Kim Y, Mekler V, Ebright RH. Static and kinetic site-specific protein-DNA photocrosslinking: analysis of bacterial transcription initiation complexes. Methods Mol. Biol. 2009;543:403–437. doi: 10.1007/978-1-60327-015-1_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Severinov K, Muir T. Expressed protein ligation, a novel method for studying protein-protein interactions in transcription. J. Biol. Chem. 1998;273:16205–16209. doi: 10.1074/jbc.273.26.16205. [DOI] [PubMed] [Google Scholar]
- 25.Severinov K, Mustaev A, Severinova E, Bass I, Kashlev M, Landick R, Nikiforov V, Goldfarb A, Darst S. Assembly of functional Escherichia coli RNA polymerase containing β subunit fragments. Proc. Natl. Acad. Sci. USA. 1995;92:4591–4595. doi: 10.1073/pnas.92.10.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Severinov K, Mustaev A, Kukarin A, Muzzin O, Bass I, Darst S, Goldfarb A. Structural modules of the large subunits of RNA polymerase. Introducing archaebacterial and chloroplast split sites in the β and β' subunits of Escherichia coli RNA polymerase. J. Biol. Chem. 1996;271:27969–27974. doi: 10.1074/jbc.271.44.27969. [DOI] [PubMed] [Google Scholar]
- 27.Grohmann D, Nagy J, Chakraborty A, Klose D, Fielden D, Ebright RH, Michaelis J, Werner F. The Initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Mol. Cell. 2011;43:263–274. doi: 10.1016/j.molcel.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chin J, Santoro S, Martin A, King D, Wang L, Schultz P. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J. Am. Chem. Soc. 2002;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 29.Young T, Ahmad I, Yin J, Schultz P. An enhanced system for unnatural amino acid mutagenesis in E. coli . J. Mol. Biol. 2009;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 30.Saxon E, Bertozzi C. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 31.Kiick K, Saxon E, Tirrell D, Bertozzi C. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc. Natl. Acad. Sci. USA. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakraborty A, Wang D, Ebright Y, Ebright RH. Azide-specific labeling of biomolecules by Staudinger-Bertozzi ligation: phosphine derivatives of fluorescent probes suitable for single-molecule fluorescence spectroscopy. Meths Enzymol. 2010;472:19–30. doi: 10.1016/S0076-6879(10)72018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]