Abstract
The most effective natural prevention against breast cancer is an early first full term pregnancy. Understanding how the protective effect is elicited will inform the development of new prevention strategies. To better understand the role of epigenetics in long-term protection, we investigated parity-induced DNA methylation in the mammary gland. FVB mice were bred or remained nulliparous and mammary glands harvested immediately after involution (early), or 6 months following involution (late), allowing identification of both transient and persistent changes. Targeted DNA methylation (109 Mb of Ensemble regulatory features) analysis was performed using the SureSelectXT Mouse Methy-seq assay and massively parallel sequencing. 269 genes were hypermethylated and 128 hypomethylated persistently at both the early and late time points. Pathway analysis of the persistently differentially methylated genes revealed Igf1r to be central to one of the top identified signaling networks, and Igf1r itself was one of the most significantly hypermethylated genes. Hypermethylation of Igf1r in the parous mammary gland was associated with a reduction of Igf1r mRNA expression. These data suggest that the IGF pathway is regulated at multiple levels during pregnancy, and that its modification might be critical in the protective role of pregnancy. This supports the approach of lowering IGF action for prevention of breast cancer, a concept which is currently being tested clinically.
Keywords: DNA Methylation, Pregnancy, IGF1R, Breast Cancer
Introduction
Pregnancy is the most effective method for breast cancer prevention. The very first report associating nulliparity with increased breast cancer risk was over 300 years ago. Bernardino Ramazzini, the father of industrial medicine, noticed that “tumors of the breast are found more often in nuns than any other women” and speculated that this was due to a life of celibacy (1). In 1970, this phenomenon was revisited in a land-mark case-control study finding that the risk of breast cancer in parous women who gave birth before the age of 20 is half that of nulliparous women (2). These observations were supported by further studies showing that women who gave birth at or before 20 or 25 years of age have a 50 and 38% reduction in lifetime risk of breast cancer respectively (3, 4); however, the detailed cellular and molecular mechanisms underlying this phenomenon in humans remain unclear. Studies have shown morphological changes and molecular alterations in the postpartum breast which are likely linked to the reduced breast cancer risk (5). In order for women to be protected from breast cancer for such a long period of time postpartum (30-40 years later), there must be a permanent alteration driving these molecular and morphological changes. We hypothesized that epigenetic alterations to the genome, which are inducible and long lasting, mediate pregnancy-induced protection against breast cancer.
Parity-induced protection from mammary cancer has been replicated in multiple rodent models including full-term pregnancy and pseudo-pregnancy using both carcinogen and spontaneous carcinoma models (6-8). Nulliparous animals treated with estradiol at a dose which results in circulating levels similar to pregnancy, leads to decreased carcinogen-induced mammary cancer, and administering estradiol plus progesterone enhances the protective effect presumably because these conditions better mimic a pregnant state (6, 7). The molecular and cellular mechanisms of pregnancy protection are likely multifold involving both changes in systemic hormones, and in cell differentiation, signaling, and survival in the mammary gland (9). An early full first term birth remains the single most effective natural method to prevent breast cancer, and the discovery of mechanisms driving this phenomenon will stimulate development of new prevention approaches.
Epigenetic changes are known to control mammary growth and development. During pregnancy and lactation, DNA is hypomethylated to allow expression of genes controlling remodeling of the gland and milk production (10). Additionally, changes in chromatin structure have been shown to be induced by parity. In nulliparous women, breast epithelial nuclei are large and euchromatic, in contrast to breast tissue from parous women which display small heterochromatic nuclei with strong methylation of histones at repressive marks (11). Epigenetic alterations are both stable and inducible, and thus can persistently alter gene expression and induce long-term phenotypic “memory-like” changes. Therefore it is likely that epigenetic regulations underlie the mechanisms responsible for the protective effect of pregnancy against breast cancer. We have utilized a new massively parallel targeted sequencing approach to analyze differentially methylated regions (DMRs) across the genome in parous and nulliparous mammary glands, with the goal of identifying pathways which are important in preventing breast cancer. The Igf1r, as well as other members of the IGF signaling pathway, displayed increased methylation with parity and may thus be involved in protecting parous individuals from breast cancer.
Materials and Methods
Animal Experiments
All animal experiments were approved by the IACUC of Baylor College of Medicine. FVB mice (Jackson Laboratories) at 50 days of age were bred to become parous or remained nulliparous (n=10 per group at each time point). Parous animals nursed for 21 days and were allowed to undergo involution for 28 days. Animals were euthanized immediately (day 120), and 6 months (day 300) post-involution, respectively. Nulliparous mice were euthanized at both time points. The experimental schema is shown in Supplementary Figure S1A. Tissues were harvested, flash frozen in liquid nitrogen, and stored at -80°C for further analysis.
Targeted Assessment of DNA Methylation in DMRs
Frozen mammary glands (n=6 per group at each time point) were cryo-homogenized and genomic DNA was isolated using the DNeasy Blood and Tissue reagents and protocol (Qiagen). Genomic DNA was sheared into 200 bp fragments using a Covaris S220 ultrasonicator. Sheared DNA underwent endrepair (XT Library Prepkit), polyA tail addition, and adapter ligation, verified by a 40 bp shift in size on an Agilent Bioanalyzer. Subsequently, the DNA was hybridized to SureSelectXT Mouse Methyl-Seq library probes overnight at 65°C, and captured with streptavidin magnetic beads. The eluted DNA was bisulfite converted (Zymo EZ DNA Meth Gold) and PCR amplified. Samples were then pooled and 3 samples per lane were analyzed by 100bp paired-end sequencing on an Illumina HiSeq 2500. The SureSelectXT Mouse Methyl-Seq design covers 109 Mb of Ensemble regulatory features (CpG shores and shelves, DNAse I hypersensitive sites, transcription factor binding sites, etc.), CpG islands, known tissue specific differentially methylated regions, and open regulatory elements.
Biostatistical Analysis of DNA Methylation in DMRs
Processing and Quality Control
The alignment reference genome used was mm9 assembly (12). The mm9 DNA reference genome was converted to a DNA methylation reference genome. Genome indexing was performed using Bismark genome preparation tools. FastQ files (23.4-146.4 million pairs of reads per sample, with an average 62 million pairs of reads) were aligned to the converted methylation reference using Bismark. The maximum number of mismatches permitted was 2. For valid paired-end alignments, the minimum insert size was 20 and the maximum insert size was 1,200. The remaining parameters from Bismark were used as default. Aligned reads outside of the targeted regions (Agilent SureSelectXT Mouse Kit) were removed. We also conducted hierarchical clustering on all samples from both time points (Supplementary Figure S2). One sample at the early time point (NP5) was removed from further analysis due to low mapping efficiency of 9%. One sample in each time point was removed due to non-conformity in the principal component analysis (PCA) (early – P12 is an outlier from all samples, late – NP4 is an outlier from all NP samples) (Supplementary Figure S3).
Differential Methylation Analysis
MethylKit software (a public R Package) was used to process sequencing data (13).
Differentially methylated regions were identified by creating 120bp windows (not overlapping) and filtering out any windows with less than 10 reads. All windows were normalized to adjust for bias across all samples. Windows which did not align in the targeted regions were discarded. β value was calculated as methylated CpG/Total CpGs. Logistic regression in methylkit was used with a sliding window analysis and a cutoff of q-value < 0.01 and methylation difference > 25% to identify differential methylation between parous and nulliparous samples at each time point. P values were adjusted to q values using the SLIM method to obtain false discovery rates (FDR) (14). We also assigned samples to parous and nulliparous groups in random combinations at each time point and permutated this procedure 100 times. Our results are highly significant compared to randomly assigning treatment groups (p<0.05 for all cases). A 25% difference in DNA methylation has been shown to induce a 2-fold reduction in gene expression (15). To eliminate any significance driven by outliers windows were further filtered out by a trimmed mean difference < 0.25 (mean value with 25% observation deleted at both ends). These data are presented in the first two tabs of the Supplemental Data 1 spreadsheet.
Meta-analysis
Once differentially methylated windows were identified, an adaptively weighted (AW) meta-analysis (16) was conducted to identify differentially methylation windows which persisted over time. The contribution of each window was marked with an AW weight (1: contribute, 0: not contribute). Persistently altered windows were defined as having weight (1, 1) with false discovery rate (FDR) <0.05, and effect size > 10% methylation difference for both time points. We again filtered out any significant windows driven by outliers using a trimmed mean difference < 0.10. Two samples were outliers on the PCA analysis and were therefore not included in the meta-analysis (late - P7, & P8). Windows were then annotated to genes using “BSgenome Mmusculus UCSC mm9” from bioconductor. The windows were classified as CpG island or CpG shore based on CpG category database by Wu et. al. (17). These windows were also annotated as promoter (TSS ± 1000 bp), exon, intron and intergenic based on genic parts category using TxDb.Mmusculus.UCSC.mm9.knownGene (R database) and IRange (R function) R package (18). These data are presented in the second two tabs of the Supplemental Data 1 spreadsheet.
Bisulfite Sequencing of Candidate Genes
Genomic DNA was bisulfite converted (Zymo EZ DNA Gold) and PCR amplified using primers designed by Meth Primer (Supplemental Table S1). PCR products were cloned into pCR 4.0 by a TOPO reaction and transformed into TOP10 competent cells (Invitrogen). After transformation cells were spread on ampicillin containing agar plates and 10-20 colonies were chosen for analysis. Colonies grew up overnight in ampicillin containing LB broth and DNA was isolated by Qiagen miniprep protocol. 1 ug of DNA was submitted for Sanger Sequencing at The University of Pittsburgh Genomics and Proteomics Core Facility. Sequences were processed using Sequencher 5.2.4 (Gene Codes) and BiQ Analyzer (Max-Planck-Institute for Informatics and Saarland University, Saarbrucken, German) (19). β value was calculated as methylated CpG/Total CpGs, and data was analyzed via logistic regression in R (glm function). The gene location of the Igf1r hypermethylation was drawn with FancyGene (20) using NCBI reference sequence number NM_010513.
Quantitative Real-time PCR
Frozen mammary glands were cryo-homogenized and RNA was isolated. Reverse transcription was conducted using iScript and qPCR was run on a Bio-Rad cfx 384 as previously described (21), using primers in Supplemental Table S1. Data was analyzed via students t-test using Prism 5, v5.40 (GraphPad).
Data Availability
The fastq files were submitted to SRA (SRP053781), which is linked to BioProject (Accession: PRJNA273963), and BioSample (Accessions:SAMN03324064-SAMN03324087).
Results
Experimental design
To discover parity-induced alterations in DNA methylation of the murine mammary gland, we bred FVB/NJ mice at approximately 50 days of age and euthanized them at two time points. One group (early) was euthanized immediately after involution, to identify changes directly induced by pregnancy, lactation and involution (n=8). The second group (late) was euthanized 6 months post-involution, to identify parity-induced changes which are persistent and remain long after involution. Age-matched virgins were included at both time points (Supplemental Fig S1A). In these studies whole mammary glands were utilized. To understand the distribution of cell types present in the gland we conducted quantitative RT-PCR for transcripts specific for particular cell types (1-Leukocyte (CD45); 2-Adipose (Adiponectin); 3-Luminal epithelial (ESR1 & CK18); 4-Myoepithelial (SMA & CK14); and 5-Fibroblast (FSP)). While some minor changes were detected, none were statistically significant, suggesting that no major changes in cell composition remain following pregnancy (Supplemental Fig S1B). In the case of CK14 the parous samples trended higher, but the two other stromal markers were not changed, and neither epithelial marker was changed. Immediately post-involution adiponectin tended to be lower in the parous samples, which was expected but was not significant. We are therefore confident that the cellular components of the mammary glands are not remarkably different comparing nulliparous and parous mice.
Targeted bisulfite sequencing of DMRs
Genomic DNA isolated from mammary tissues was used to conduct a targeted analysis of DMRs across the genome. DNA methylation was assessed using SureSelectXT Mouse Methyl-Seq Technology. On average 39 million pairs of reads per sample were aligned to the reference with a mapping efficiency of 50%∼68.1% and an average 71× sequencing depth was achieved (the coverage depth was 113× before alignment) (Supplemental Fig S3). The sequencing results were processed as described in the Materials and Methods, and quality control was conducted by Pearson correlation and principle component analysis (Supplemental Fig S3).
Differentially methylated genes in early and late parous mammary glands
To identify differentially methylated genes, logistic regression was used to determine significantly hyper/hypomethylated windows which were then annotated to genes. In the early time point, comparing parous and nulliparous mammary glands, 4385 windows representing 1880 genes were identified as differentially methylated, with 3507 windows (1535 genes) hypo-methylated, and 878 windows (446 genes) hyper-methylated (Supplemental Data 1). Additionally, 101 windows were both hyper and hypo-methylated. Of the 4385 differentially methylated windows, 2% were located in CpG islands (CpGi) and 17% in CpGi shores as annotated by UCSC genome browser (Fig 1A). For those windows in genomic regions, 9% were located in promoter regions, 9% in exons, 48% in introns, and 35% in intergenic regions (Fig 1A). At the late time point a total of 8884 windows (2888 genes) including 6561 windows (2260 genes) hypermethylated and 2323 windows (872 genes) hypomethylated. 244 genes were both hyper and hypomethylated. The distribution of the methylated sites in CpG islands and shores and genomic regions was almost identical in both early and late samples (Fig 1A). In the SureSelectXT Mouse Methy-Seq library 7% of the probes are located in CpG islands and 11% in shores, while we detected only 2% in CpG islands and 16% in shores. Additionally, 35% of the library probes are located in introns, 12% in exons, 8% in Promoters, and 45% in intergenic regions. Our data is substantially enriched for intronic regions and CpG shores, with fewer differentially methylated windows in intergenic regions and CpG islands. This difference is highly significant by Chi Square analysis (genomic region – early p=4.75E-51, late p=4.91E-148, CpGi/shores – early p=1.19E-63, late p=3.15E-124). It is possible that DNA methylation in introns and CpG shores plays a greater role than previously thought.
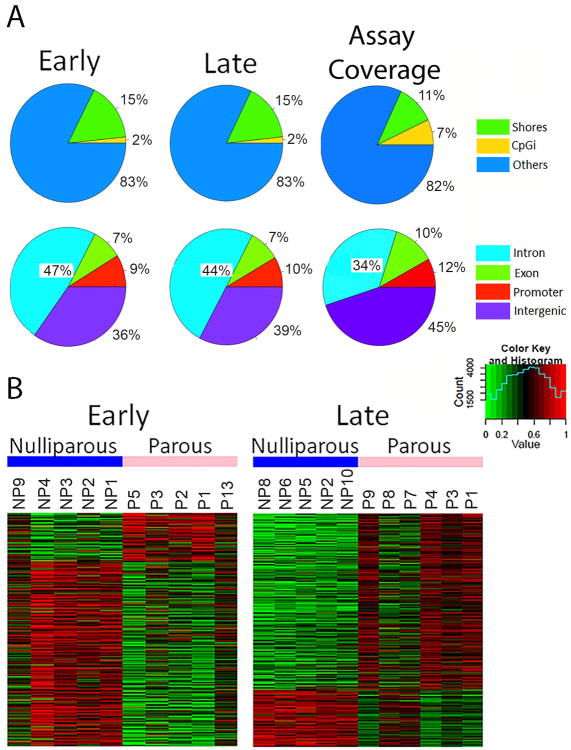
Figure 1. Parity-induced DNA methylation in the mouse mammary gland.
(A) Pie charts depicting areas of the genome which are differentially methylated at the early and late time points as well as the distribution of library probes in the SureSelectXT Methyl-Seq assay. Upper panel shows the percent of CpG islands and shores, while lower panel shows the genomic region (B) A heat map of differentially methylated windows after hierarchical clustering in mammary glands from parous mice and age-matched nulliparous mice is depicted at each time point. Windows which had at least a coverage of 10, a q value < 0.01, and a difference in methylation (between nulliparous and parous) of at least 25%. We additionally filtered out windows with an absolute trimmed mean difference of < 0.25. Methylation is represented as β value with green as hypomethylated and red as hypermethylated. At the early time point 4385 windows were differentially methylated (3507 hypomethylated, and 878 hypermethyalted). At the late time point 8884 windows were differentially methylated (2323 hypomethylated, and 6561 hypermethylated).
Recently, mammary specific differentially methylated regions have been identified (15, 22). 21 breast specific differentially methylated genes were identified in these two publications and 13 are represented in the SureSelectXT Methyl-Seq assay. Although these were identified in human samples we found two genes to be significantly differentially methylated with parity in our mouse study, Mgmt and Hlf. Mgmt is hyperhethylated with parity at the late time point while hypomethylated at the early time point, and Hlf is hypomethylated at the late time point (Table 1). The significantly differentially methylated windows are presented by heat map (Fig 1B). Windows shown have a significance of at least q < 0.01, and a methylation difference between nulliparous and parous greater than 25%.
Table 1.
Mammary specific differentially methylated genes.
| Genes | SureSelectXT Coverage | Publication |
|---|---|---|
| LOC100271722 | FALSE | Zhang 2013 |
| LOC150381 | FALSE | Zhang 2013 |
| MIRLET7 | TRUE | Zhang 2013 |
| MIRLET7A3 | FALSE | Zhang 2013 |
| MIRLET7B | FALSE | Zhang 2013 |
| MIRLET7BGH | FALSE | Zhang 2013 |
| ALX4 | TRUE | Avraham 2014 |
| FEV | TRUE | Avraham 2014 |
| HLF | TRUE | Avraham 2014 |
| HOXA11 | TRUE | Avraham 2014 |
| LYL1 | TRUE | Avraham 2014 |
| Neurog | TRUE | Avraham 2014 |
| PAX9 | TRUE | Avraham 2014 |
| GATA5 | TRUE | Avraham 2014 |
| MGMT | TRUE | Avraham 2014 |
| SOX10 | TRUE | Avraham 2014 |
| SREBF1 | TRUE | Avraham 2014 |
| ST18 | TRUE | Avraham 2014 |
| TP73 | FALSE | Avraham 2014 |
| TRIM20 | FALSE | Avraham 2014 |
| ZNF436 | FALSE | Avraham 2014 |
Validation of the targeted methyl-sequencing of DMRs
In order to first validate our assay using an orthogonal approach, we employed bisulfite sequencing on the same DNA that was utilized in the Methyl-Seq assay. Genes with the largest difference in beta-value and the highest coverage were chosen for validation, specifically Slc9a1, Pstpip2, St6gal1, Irf2, Ece1, and Lck (Supplemental Data 1 and Fig. 2A). Pstpip2 and St6gal1 were confirmed as hypomethylated and Slc9a1, and Irf2 were confirmed as hypermethylated by bisulfite sanger sequencing, while a non-significant trend was noted for Ece1 and Lck (Fig. 2B and Supplemental Fig S4). As an additional control, we further interrogated the Methyl-Seq data by investigating regions of the genome known to be maternally imprinted (Grb10, Cdkn1c, Igf1r/Airn, Htr2, Xist & H19/Igf1) and found that as expected, approximately 50% of the CpGs were methylated in each of these regions (Supplemental Fig. S5).
Figure 2. Confirmation of Methyl-Seq DNA methylation by bisulfite sequencing.
(A) β values for the top differentially methylated genes with the highest coverage in the Methyl-Seq assay are depicted. (B) DNA methylation after bisulfite sequencing is represented as β values. Logistic regression was used to compare nulliparous and parous samples for each gene (*** = p<0.001, ** = p<0.01).
Persistent DNA methylation changes in the parous mammary gland
To determine which genes were persistently differentially methylated in the parous mammary gland 6 months post-involution (late), we performed meta-analysis using 493,473 windows from the early time point, and 473,714 windows from the late time point (432,946 windows overlapped). This analysis identified 624 genes (660 windows) and 322 genes (330 windows) that were hyper- and hypomethylated in the early group, and remained altered in the late group. After filtering by trimmed means > 0.10, 276 windows (269 genes) were hypermethylated, and 131 windows (128 genes) hypomethylated (Fig. 3, Supplemental Data 1). A supervised hierarchical clustering analysis was also performed and is displayed in Supplemental Fig S6. Ingenuity Pathway Analysis (Ingenuity.com) on the persistently hypermethylated, hypomethylated, and both sets of altered genes simultaneously showed several signaling pathways to be altered including P70S6K, Rho GTPases, Protein Kinase A, and others. The full data set from the pathway analysis is presented in the Supplemental Data 1 spreadsheet. Among the top persistently hypermethylated genes identified in the Methyl-Seq assay (Table 2) which was also central to one of the top signaling networks in the pathway analysis (Supplemental Fig. S7) was Igf1r.
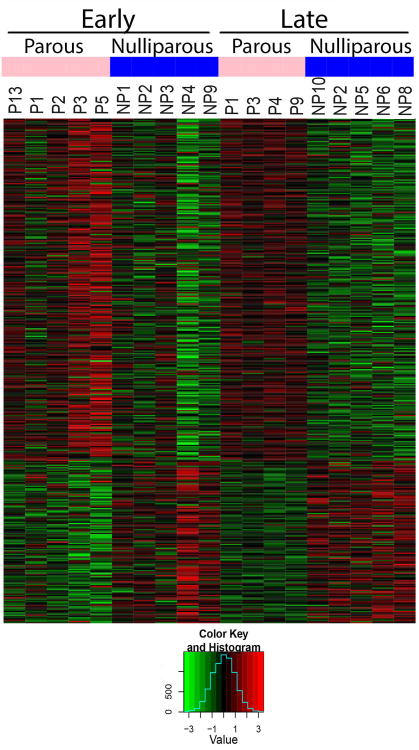
Figure 3. Identification of regions with persistently altered DNA methylation.
A heat map of persistently differentially methylated (significantly differentially methylated immediately post-involution and remaining differentially methylated 6 months post-involution) windows after an adaptive weighted meta-analysis in mammary glands from parous mice and age-matched nulliparous mice is depicted. Windows displayed have an FDR < 0.05, and a difference in methylation (between nulliparous and parous) greater than 10%. Methylation is represented as β value with green as hypomethylated and red as hypermethylated.
Table 2. IGF1R is among the top 20 persistently altered genes.
Differentially methylated genes were identified by logistic regression and those that were altered early and remained altered later were considered persistently altered. A meta-analysis was used to compare the early and late time points to determine which changes we altered by parity and remained after 6 months. The differential methylation remaining at the late time point (Labeled “Trimmed_Means. 2 in the Supplemental Data File) and FDR from the meta-analysis are listed as well as the genomic feature the differential methylation occurs in (Ex=exon, In=intron, Sh=shore, Prom=promoter).
| Hypermethylated | Hypomethylated | ||||||
|---|---|---|---|---|---|---|---|
| Gene | Diff Meth | FDR | Feature | Gene | Diff Meth | FDR | Feature |
| Trp53bp2 | 37.90 | 3.86E-20 | Intron | Gprc5b | -10.16 | 1.17E-04 | Intron |
| Nnat | 34.63 | 3.86E-20 | Ex/In/Sh | Sim1 | -10.17 | 1.57E-07 | Ex/In |
| Frmd4a | 34.20 | 3.86E-20 | Intron | Dlg5 | -10.24 | 4.69E-03 | Intron |
| Clybl | 33.12 | 3.86E-20 | Intron | Nup88 | -10.43 | 3.86E-20 | In/Sh |
| Lhfpl2 | 32.66 | 3.86E-20 | Intron | Mtor | -10.66 | 9.21E-04 | Intron |
| Rabep1 | 30.75 | 3.86E-20 | Intron | Ehhadh | -10.70 | 3.86E-20 | Intron |
| Igf1r | 30.63 | 3.86E-20 | Intron | Ptgis | -10.73 | 2.5E-02 | Intron |
| Fez1 | 29.99 | 3.86E-20 | Intron | Asap3 | -10.79 | 5.86E-06 | Intron |
| Cdh4 | 29.97 | 3.86E-20 | Intron | Eif4e | -10.91 | 2.37E-05 | Intron |
| Rbm20 | 29.87 | 3.86E-20 | Intron | Slc27a2 | -10.94 | 9.61E-04 | Intron |
| Cldn14 | 29.84 | 3.86E-20 | Intron | Itga5 | -10.95 | 1.14E-05 | In/Sh |
| Unc5c | 29.43 | 1.57E-07 | Intron | Asb2 | -10.95 | 1.26E-06 | Intron |
| Mapk4 | 28.81 | 1.57E-07 | Intron | Gm8066 | -11.00 | 0.000143 | Shore |
| Rap1gap | 28.06 | 3.86E-20 | Ex/In | Arhgap10 | -11.04 | 3.86E-20 | Intron |
| Vasp | 27.55 | 3.86E-20 | Ex/In | Usp6nl | -11.07 | 6.12E-07 | In/Sh |
| Myog | 27.10 | 3.86E-20 | Exon | A930024E 05Rik | -11.13 | 2.96E-06 | In/Sh |
| Tspan7 | 27.03 | 3.86E-20 | Intron | Slmo1 | -11.23 | 3.86E-20 | Intron |
| Zmym6 | 25.87 | 3.86E-20 | Prom/In/Sh | Ncoa3 | -11.41 | 3.87E-07 | Intron |
| Gm9618 | 25.78 | 3.86E-20 | Intergenic | AI662270 | -11.44 | 5.53E-04 | Prom/In |
| Nr2f2 | 25.64 | 3.86E-20 | In/Sh | Myom2 | -11.58 | 1.57E-07 | intron |
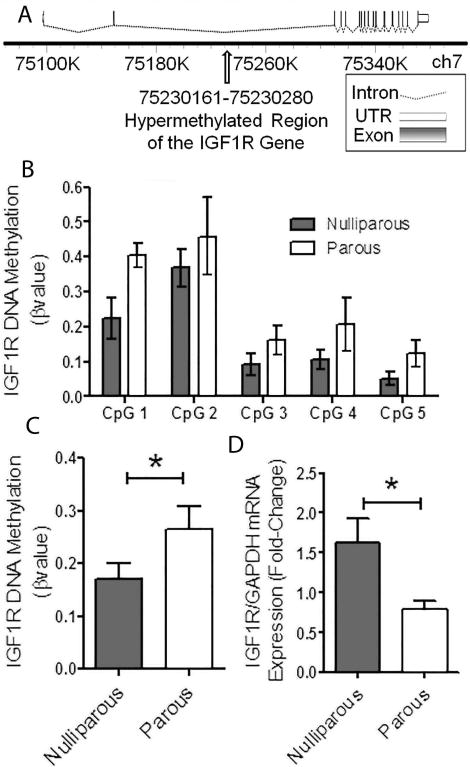
Igf1r is persistently methylated in the parous mammary gland
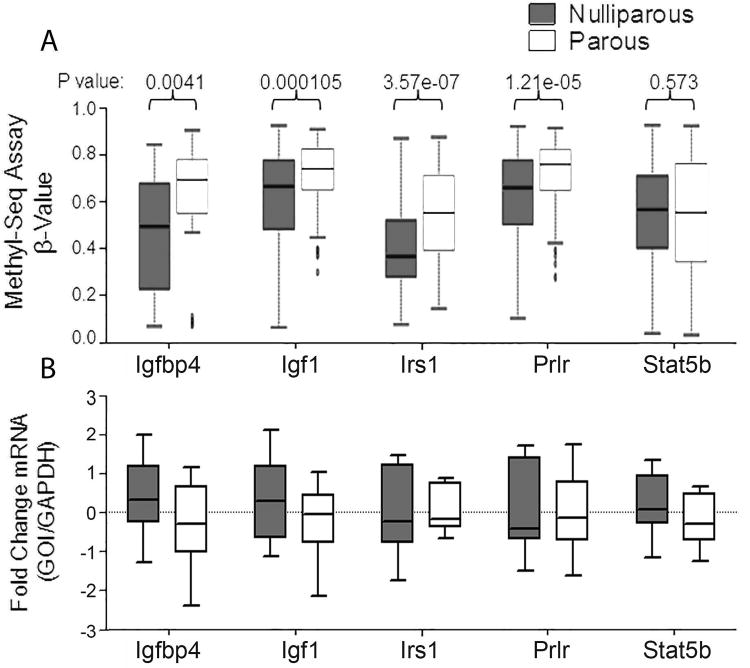
Genes which are persistently altered at both the early and late time points are of great biological significance and serve to address our hypothesis that long term epigenetic alterations underlie the protective effect of pregnancy. We have previously reported that parity results in a significant decrease in circulating GH in rats that altered GH/IGF signaling in the mammary gland, and others have shown alterations on the GH/IGF axis in humans (23, 24). Interestingly, in the current study we found that Igf1r was seventh among the top hypermethylated genes after meta-analysis, indicating it is methylated in the mammary gland post-partum and remains methylated long after involution (Table 2). Since the GH/IGF pathway has previously been implicated in the protective effect of pregnancy and it is among the top persistently hypermethylated genes we further investigated this family. This hypermethylation occurs in the largest intron of the Igf1r gene between exons 2 & 3 (Fig. 4A). This intron also contains other epigenetic modifications including histone 3 lysine 4 mono and tri methylation as displayed in the UCSC Genome Browser, indicating that this region is likely involved in gene expression regulation (Supplemental Fig. S8A). We confirmed these findings by bisulfite sequencing and found a significant increase in Igf1r methylation at the same above mentioned intron in parous animals 6 months post-involution (Fig. 4B & C, Supplemental Fig. S8B). This increase in intron methylation was associated with a decrease in Igf1r mRNA expression in the mammary gland of parous animals (Fig. 4D). We interrogated other IGF pathway genes in the Methyl-Seq data analysis and found several to have regions which displayed significantly altered DNA methylation in the Methyl-Seq analysis (Table 3). The 5 genes with the highest ratio (number of significantly altered windows/the total number of windows), Irs1, Igf1, Igfbp4, Prlr, and Stat5b, all display increased methylation with parity in the Methyl-Seq assay (Fig. 5A), and almost all showed a trend towards decreased mRNA expression in the parous mammary gland (Fig. 5B). The differentially methylated windows in the top 3 IGF pathway genes, Irs1, Igf1, and Igfbp4, overlap with other epigenetic modification, indicating that they are likely involved in gene regulation (Supplemental Fig. S9).
Figure 4. IGF1R is significantly hypermethylated in parous animals 6 months after involution and this is translated to gene expression.
(A) A depiction of the mouse Igf1r gene as drawn by FancyGene with the site of hypermethylation in the second intron highlighted by an arrow. The number line represents the genome location. (B) Bisulfite sequencing of Igf1r DNA is depicted as β values for each CpG site (n=8). (C) Total methylation of the Igf1r as β values is represented. Logistic regression was used to analyze the methylation level (n=8, *=p<0.05). (D) mRNA expression of the IGF1R. Students t-test was used to analyze mRNA expression (n=9, *=p<0.05).
Table 3. IGF pathway members also display altered DNA methylation.
The total number of 120bp windows available for each gene was compared to the number of windows which were significantly differentially methylated with parity to obtain the ratio.
| Early | Late | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Total Windows | Significant Windows | Ratio | Hyper | Hypo | Total Windows | Significant Windows | Ratio | Hyper | Hypo |
| Irs1 | 36 | 2 | 0.0556 | 0 | 2 | 35 | 19 | 0.5429 | 19 | 0 |
| Igf1 | 26 | 3 | 0.1154 | 0 | 3 | 25 | 13 | 0.5200 | 12 | 1 |
| Igfbp4 | 28 | 5 | 0.1786 | 1 | 4 | 20 | 9 | 0.4500 | 9 | 0 |
| Stat5b | 81 | 7 | 0.0864 | 3 | 4 | 85 | 36 | 0.4235 | 18 | 18 |
| Prlr | 29 | 3 | 0.1034 | 0 | 3 | 29 | 12 | 0.4138 | 12 | 0 |
| Shc | 21 | 1 | 0.0476 | 0 | 1 | 21 | 8 | 0.3810 | 8 | 0 |
| Igfbp3 | 11 | 0 | 0.0000 | 0 | 0 | 8 | 3 | 0.3750 | 3 | 0 |
| Igf2r | 32 | 4 | 0.1250 | 1 | 3 | 28 | 10 | 0.3571 | 7 | 3 |
| Igfbp6 | 3 | 0 | 0.0000 | 0 | 0 | 3 | 1 | 0.3333 | 1 | 0 |
| Ghr | 45 | 5 | 0.1111 | 0 | 5 | 45 | 14 | 0.3111 | 13 | 1 |
| Akt1 | 22 | 0 | 0.0000 | 0 | 0 | 20 | 6 | 0.3000 | 6 | 0 |
| Akt2 | 29 | 2 | 0.0690 | 0 | 2 | 29 | 8 | 0.2759 | 8 | 0 |
| Irs2 | 46 | 10 | 0.2174 | 0 | 10 | 45 | 12 | 0.2667 | 12 | 0 |
| Sos | 27 | 1 | 0.0370 | 1 | 0 | 34 | 9 | 0.2647 | 3 | 6 |
| Stat5a | 16 | 0 | 0.0000 | 0 | 0 | 23 | 6 | 0.2609 | 4 | 2 |
| Src | 50 | 1 | 0.0200 | 1 | 0 | 48 | 10 | 0.2083 | 10 | 0 |
| Stat3 | 51 | 6 | 0.1176 | 3 | 3 | 54 | 10 | 0.1852 | 5 | 5 |
| Erk | 12 | 0 | 0.0000 | 0 | 0 | 13 | 2 | 0.1538 | 2 | 0 |
| Igfbp2 | 14 | 0 | 0.0000 | 0 | 0 | 12 | 1 | 0.0833 | 0 | 1 |
| Irs4 | 19 | 0 | 0.0000 | 0 | 0 | 20 | 1 | 0.0500 | 1 | 0 |
| Igfbp5 | 3 | 0 | 0.0000 | 0 | 0 | 3 | 0 | 0.0000 | 0 | 0 |
| PI3K | 2 | 0 | 0.0000 | 0 | 0 | 2 | 0 | 0.0000 | 0 | 0 |
| Igfbp1 | 2 | 0 | 0.0000 | 0 | 0 | 2 | 0 | 0.0000 | 0 | 0 |
Figure 5. IGF pathway members are hypermethylated with parity.
(A) Methyl-Seq analysis of IGF pathway members at the late time point. Logistic regression was implemented to compare nulliparous and parous. (B) mRNA expression of the IGF pathway members analyzed by quantitative RT-PCR.
Discussion
The most significant modifiable factor affecting a woman's risk for developing breast cancer is an early age at first full term birth (FFTB). Although the protective effect of an early FFTB was identified decades ago, the mechanisms underlying this effect remain to be elucidated. Age at FFTB is increasing in women in the United States and likely represents a contributing factor to the increase in breast cancer incidence (25). Understanding how an early FFTB prevents breast cancer may identify promising new avenues to develop novel breast cancer prevention strategies. In this study we have characterized epigenetic changes following pregnancy, and found that Igf1r and other IGF family members are hypermethylated and downregulated. This study provides support to the concept of inhibiting the GH/IGF pathway for breast cancer prevention.
Epigenetic changes are known to play integral roles in the mammary gland (10). Since epigenetic modifications are both inducible and stable, they can persistently alter gene expression in a long lasting ‘memory-like’ fashion, likely playing a role in the protective effect of pregnancy against breast cancer, as speculated by Jerry et. al. (26). Ghosh et al. investigated DNA methylation in breast tissue of 30 women (19 parous, 16 nulliparous) (27). Using a genome-wide methyl binding protein pull down to isolate all methylated DNA (a non-targeted approach), their analysis identified FOXA1 as hypermethylated and possibly silenced with parity (27). They speculated that this could attenuate the effects of ERα leading to a reduced susceptibility to breast cancer. They also found an IGF family member altered in their analysis. The IGF acid labile subunit (IGFALS) was hypomethylated in their analysis. The IGFALS is responsible for carrying IGF1 in the circulation and altering the levels of IGFALS may reduce IGF1 activity in target tissues. Additionally, histone modifications are likely important. Tri methylation of lysine 27 on histone 3 (H3K27me3) was identified in a genome wide histone methylation screen to increase during pregnancy in rodents (28). Our in silico data shows that histone modifications, including but not limited to H3K27, overlap with parity-induced DNA methylation changes in IGF pathway genes (Supplemental Figure S9).
Parity-induced genome-wide gene expression changes have been investigated in both rodents and women. The first study to characterize changes in expression across the genome identified a gene signature indicative of parity in multiple rat strains and mice (29). In women a genome-wide change in expression is associated with parity and parous women who never develop breast cancer display a different signature than parous women who do develop breast cancer (30), and an 11 gene signature can identify parity in women (31). Of note, in our data at the early time point the vast majority of differentially methylated genes were hypomethylated, while 6 months after involution there is a dramatic shift to hypermethylation. This hypomethylation at the early time point could signify an immense induction of expression of genes involved in the remodeling process of involution which is multifaceted and would likely produce a complex gene signature.
Our unbiased screen identified Igf1r as one of the most hypermethylated and persistently epigenetically altered genes in parous mammary gland. The Igf1r and other members of the GH/IGF pathway are critical for mammary development and also play a role in pregnancy, lactation, and involution (32-35). The Igf1r has been shown to be important during lactation in the mouse mammary gland as overexpressing a dominant/negative IGF1R reduced alveolar development and milk protein production (36). Consistent with hypermethylation of the Igf1r intron, we found that Igf1r mRNA was decreased in parous mammary gland. Several other IGF family members also showed epigenetic alterations and gene expression changes. Other genome-wide expression studies have identified components of the GH/IGF pathway to be downregulated in the parous mammary gland (29, 37). IGF1 was found to be significantly decreased in parous mammary gland, while IGFBP5 was increased (29). IGF1 was also found to be decreased in the breast of parous women (37). In addition to mammary specific changes, we have previously demonstrated that circulating GH is decreased in parous animals leading to a reduction in protein activation of key IGF/GH pathway signaling molecules in the mammary gland (23). These data, in conjunction with previous gene expression studies, and our data reported here strongly support the concept that downregulation of the GH/IGF axis (both endocrine and intrinsic to the mammary gland) may be involved in the protective effect of pregnancy against breast cancer.
Epigenetic modifications have been linked to breast cancer risk in multiple studies. At least three dietary compounds (folate, choline, and sulforaphane) can alter the epigenome and reduce cancer risk (38-44). Folate, a water soluble vitamin, had been shown to alter DNA methylation by modulating both DNA methyltransferases and methylbinding proteins (38). In women, folate or choline (a methyl donor food) intake is inversely correlated with breast cancer risk in multiple studies (39, 42, 43, 45). Sulforaphane is a modulator of histone acetylation and inhibits growth in multiple breast cancer cells, and also reduces susceptibility to DMBA induced tumorigenesis growth in animals (40, 41, 44). If pregnancy is indeed modulating disease risk through epigenetic mechanisms which reduce tumorigenic pathways such as the IGF/GH pathway, it might be possible that modulating these epigenetic events in young women by dietary or therapeutic interventions will ultimately reduce risk of breast cancer.
The current landscape of evidence-based prevention therapies include inhibitors of estrogen regulation namely, tamoxifen, raloxifene, anastrozole, and exemestane (46-49). Interestingly, parity has been associated with a reduction in ER/PR+ breast cancers indicating a role for hormonal preventative therapies (50, 51). Unfortunately, while these therapies are effective, they also have side-effects and thus these prevention therapies are only recommended to women of high breast cancer risk (49, 52). One major avenue of investigation for chemoprevention, currently under clinical investigation in high risk women, is inhibition of the IGF pathway (53, 54). For example, a ten day treatment of a somatostatin analog (SOM230, Signifor, Novartis) decreased proliferation and increased apoptosis in ductal carcinoma in situ. The treatment also reduced IGF-IR, pAKT and pERK (55).
Our study utilized a novel DNA methylation screen by conducting next generation sequencing on bisulfite converted DNA, which has been enriched for areas of the genome expected to be differentially methylated. This targeted approach enhanced our analysis by providing over 100× coverage of specific regions important in DNA methylation. Although highly novel and innovative, there were some limitations in our design. For example, the mouse estrous cycles were not synchronized and therefore the mice may have been in different stages of the cycle at euthanasia. This is unlikely to skew our results since DNA methylation is relatively stable, and the estrus stage of the cycle, when the greatest changes would likely occur, is relatively short therefore the majority of glands were not likely harvested during estrus. Additionally, we analyzed the entire mammary gland including stroma and epithelium; therefore it is unclear in which compartments these epigenetic changes occur.
In conclusion, we have shown that members of the GH/IGF pathway undergo DNA methylation in response to pregnancy which may contribute to a reduction in their expression. This pathway has been implicated in breast cancer and also been shown to be reduced with parity in multiple previous studies. These data together indicate that the GH/IGF pathway maybe critical in pregnancy protection from breast cancer, and prevention strategies targeting this pathway should undergo further research.
Supplementary Material
Acknowledgments
I would like to thank Drs. Kevin Poon and Alex Siebold with Agilent Technologies for their outstanding support and early access to the SureSelectXT Mouse Methyl-Seq kit, as well as Ora L. Britton for mouse husbandry and experimentation. This project used the UPCI Tissue and Research Pathology Services shared facilities that are supported in part by award P30CA047904.
Financial Support: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the University of Pittsburgh. This work was supported in part by the National Cancer Institute of the National Institutes of Health under award number R01CA94118 (A. V. Lee) and T32CA186873 (T. A. Katz). A. V. Lee is a recipient of a Scientific Advisory Council award from Susan G. Komen for the Cure, and a Hillman Foundation Fellow. The authors acknowledge support from the University of Pittsburgh Cancer Institute, UPMC, and by the University of Pittsburgh Center for Simulation and Modeling through the supercomputing resources provided.
Footnotes
Conflict of Interest: There are no conflicts of interest to report
References
- 1.Mustacchi P. Ramazzini and Rigoni-Stern on parity and breast cancer. Clinical impression and statistical corroboration. Archives of internal medicine. 1961;108:639–42. doi: 10.1001/archinte.1961.03620100131018. [DOI] [PubMed] [Google Scholar]
- 2.MacMahon B, Cole P, Lin TM, Lowe CR, Mirra AP, Ravnihar B, et al. Age at first birth and breast cancer risk. Bulletin of the World Health Organization. 1970;43:209–21. [PMC free article] [PubMed] [Google Scholar]
- 3.Li CI, Malone KE, Daling JR, Potter JD, Bernstein L, Marchbanks PA, et al. Timing of menarche and first full-term birth in relation to breast cancer risk. American journal of epidemiology. 2008;167:230–9. doi: 10.1093/aje/kwm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schonfeld SJ, Pfeiffer RM, Lacey JV, Jr, Berrington de Gonzalez A, Doody MM, Greenlee RT, et al. Hormone-related risk factors and postmenopausal breast cancer among nulliparous versus parous women: An aggregated study. American journal of epidemiology. 2011;173:509–17. doi: 10.1093/aje/kwq404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russo IH, Russo J. Pregnancy-induced changes in breast cancer risk. Journal of mammary gland biology and neoplasia. 2011;16:221–33. doi: 10.1007/s10911-011-9228-y. [DOI] [PubMed] [Google Scholar]
- 6.Guzman RC, Yang J, Rajkumar L, Thordarson G, Chen X, Nandi S. Hormonal prevention of breast cancer: mimicking the protective effect of pregnancy. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2520–5. doi: 10.1073/pnas.96.5.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajkumar L, Guzman RC, Yang J, Thordarson G, Talamantes F, Nandi S. Short-term exposure to pregnancy levels of estrogen prevents mammary carcinogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11755–9. doi: 10.1073/pnas.201393798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raafat A, Strizzi L, Lashin K, Ginsburg E, McCurdy D, Salomon D, et al. Effects of age and parity on mammary gland lesions and progenitor cells in the FVB/N-RC mice. PloS one. 2012;7:e43624. doi: 10.1371/journal.pone.0043624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barton M, Santucci-Pereira J, Russo J. Molecular pathways involved in pregnancy-induced prevention against breast cancer. Frontiers in endocrinology. 2014;5:213. doi: 10.3389/fendo.2014.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rijnkels M, Kabotyanski E, Montazer-Torbati MB, Hue Beauvais C, Vassetzky Y, Rosen JM, et al. The epigenetic landscape of mammary gland development and functional differentiation. Journal of mammary gland biology and neoplasia. 2010;15:85–100. doi: 10.1007/s10911-010-9170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russo J, Santucci-Pereira J, de Cicco RL, Sheriff F, Russo PA, Peri S, et al. Pregnancy-induced chromatin remodeling in the breast of postmenopausal women. International journal of cancer Journal international du cancer. 2012;131:1059–70. doi: 10.1002/ijc.27323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kent WJSC, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome biology. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang HQ, Tuominen LK, Tsai CJ. SLIM: a sliding linear model for estimating the proportion of true null hypotheses in datasets with dependence structures. Bioinformatics. 2011;27:225–31. doi: 10.1093/bioinformatics/btq650. [DOI] [PubMed] [Google Scholar]
- 15.Avraham A, Cho SS, Uhlmann R, Polak ML, Sandbank J, Karni T, et al. Tissue specific DNA methylation in normal human breast epithelium and in breast cancer. PloS one. 2014;9:e91805. doi: 10.1371/journal.pone.0091805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Tseng G. An adaptively weighted statistic for detecting differential gene expression when combining multiple transcriptomic studies. The Annals of Applied Statistics. 2011;5:994–1019. [Google Scholar]
- 17.Wu H, Caffo B, Jaffee HA, Irizarry RA, Feinberg AP. Redefining CpG islands using hidden Markov models. Biostatistics. 2010;11:499–514. doi: 10.1093/biostatistics/kxq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, et al. Software for computing and annotating genomic ranges. PLoS computational biology. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–8. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 20.Rambaldi D, Ciccarelli FD. FancyGene: dynamic visualization of gene structures and protein domain architectures on genomic loci. Bioinformatics. 2009;25:2281–2. doi: 10.1093/bioinformatics/btp381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sikora MJ, Cooper KL, Bahreini A, Luthra S, Wang G, Chandran UR, et al. Invasive lobular carcinoma cell lines are characterized by unique estrogen-mediated gene expression patterns and altered tamoxifen response. Cancer research. 2014;74:1463–74. doi: 10.1158/0008-5472.CAN-13-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang B, Zhou Y, Lin N, Lowdon RF, Hong C, Nagarajan RP, et al. Functional DNA methylation differences between tissues, cell types, and across individuals discovered using the M&M algorithm. Genome research. 2013;23:1522–40. doi: 10.1101/gr.156539.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dearth RK, Delgado DA, Hiney JK, Pathiraja T, Oesterreich S, Medina D, et al. Parity-induced decrease in systemic growth hormone alters mammary gland signaling: a potential role in pregnancy protection from breast cancer. Cancer Prev Res (Phila) 2010;3:312–21. doi: 10.1158/1940-6207.CAPR-09-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmes MD, Pollak MN, Hankinson SE. Lifestyle correlates of plasma insulin-like growth factor I and insulin-like growth factor binding protein 3 concentrations. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2002;11:862–7. [PubMed] [Google Scholar]
- 25.M TJ, H BE. First births to older women continue to rise. National Center for Health Statistics Data Brief. 2014;152 [Google Scholar]
- 26.Jerry DJ, Makari-Judson G, Crisi GM, Dunphy KA. Pregnancy offers new insights into mechanisms of breast cancer risk and resistance. Breast cancer research: BCR. 2013;15:312. doi: 10.1186/bcr3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh S, Gu F, Wang CM, Lin CL, Liu J, Wang H, et al. Genome-wide DNA methylation profiling reveals parity-associated hypermethylation of FOXA1. Breast cancer research and treatment. 2014;147:653–9. doi: 10.1007/s10549-014-3132-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pal B, Bouras T, Shi W, Vaillant F, Sheridan JM, Fu N, et al. Global changes in the mammary epigenome are induced by hormonal cues and coordinated by Ezh2. Cell reports. 2013;3:411–26. doi: 10.1016/j.celrep.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 29.Blakely CM, Stoddard AJ, Belka GK, Dugan KD, Notarfrancesco KL, Moody SE, et al. Hormone-induced protection against mammary tumorigenesis is conserved in multiple rat strains and identifies a core gene expression signature induced by pregnancy. Cancer research. 2006;66:6421–31. doi: 10.1158/0008-5472.CAN-05-4235. [DOI] [PubMed] [Google Scholar]
- 30.Russo J, Balogh GA, Russo IH. Full-term pregnancy induces a specific genomic signature in the human breast. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;17:51–66. doi: 10.1158/1055-9965.EPI-07-0678. [DOI] [PubMed] [Google Scholar]
- 31.Asztalos S, Gann PH, Hayes MK, Nonn L, Beam CA, Dai Y, et al. Gene expression patterns in the human breast after pregnancy. Cancer Prev Res (Phila) 2010;3:301–11. doi: 10.1158/1940-6207.CAPR-09-0069. [DOI] [PubMed] [Google Scholar]
- 32.Kleinberg DL, Feldman M, Ruan W. IGF-I: an essential factor in terminal end bud formation and ductal morphogenesis. J Mammary Gland Biol Neoplasia. 2000;5:7–17. doi: 10.1023/a:1009507030633. [DOI] [PubMed] [Google Scholar]
- 33.Ruan W, Kleinberg DL. Insulin-like growth factor I is essential for terminal end bud formation and ductal morphogenesis during mammary development. Endocrinology. 1999;140:5075–81. doi: 10.1210/endo.140.11.7095. [DOI] [PubMed] [Google Scholar]
- 34.Ruan W, Monaco ME, Kleinberg DL. Progesterone stimulates mammary gland ductal morphogenesis by synergizing with and enhancing insulin-like growth factor-I action. Endocrinology. 2005;146:1170–8. doi: 10.1210/en.2004-1360. [DOI] [PubMed] [Google Scholar]
- 35.Richards RG, Klotz DM, Walker MP, Diaugustine RP. Mammary gland branching morphogenesis is diminished in mice with a deficiency of insulin-like growth factor-I (IGF-I), but not in mice with a liver-specific deletion of IGF-I. Endocrinology. 2004;145:3106–10. doi: 10.1210/en.2003-1112. [DOI] [PubMed] [Google Scholar]
- 36.Sun Z, Shushanov S, LeRoith D, Wood TL. Decreased IGF type 1 receptor signaling in mammary epithelium during pregnancy leads to reduced proliferation, alveolar differentiation, and expression of insulin receptor substrate (IRS)-1 and IRS-2. Endocrinology. 2011;152:3233–45. doi: 10.1210/en.2010-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peri S, de Cicco RL, Santucci-Pereira J, Slifker M, Ross EA, Russo IH, et al. Defining the genomic signature of the parous breast. BMC Med Genomics. 2012;5:46. doi: 10.1186/1755-8794-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghoshal K, Li X, Datta J, Bai S, Pogribny I, Pogribny M, et al. A folate- and methyl-deficient diet alters the expression of DNA methyltransferases and methyl CpG binding proteins involved in epigenetic gene silencing in livers of F344 rats. The Journal of nutrition. 2006;136:1522–7. doi: 10.1093/jn/136.6.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong Z, Ambrosone CB, McCann SE, Zirpoli G, Chandran U, Hong CC, et al. Associations of dietary folate, Vitamins B6 and B12 and methionine intake with risk of breast cancer among African American and European American women. International journal of cancer Journal international du cancer. 2014;134:1422–35. doi: 10.1002/ijc.28466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katz TA, Huang Y, Davidson NE, Jankowitz RC. Epigenetic reprogramming in breast cancer: from new targets to new therapies. Annals of medicine. 2014;46:397–408. doi: 10.3109/07853890.2014.923740. [DOI] [PubMed] [Google Scholar]
- 41.Pledgie-Tracy A, Sobolewski MD, Davidson NE. Sulforaphane induces cell type-specific apoptosis in human breast cancer cell lines. Molecular cancer therapeutics. 2007;6:1013–21. doi: 10.1158/1535-7163.MCT-06-0494. [DOI] [PubMed] [Google Scholar]
- 42.Xu X, Gammon MD, Zeisel SH, Lee YL, Wetmur JG, Teitelbaum SL, et al. Choline metabolism and risk of breast cancer in a population-based study. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2008;22:2045–52. doi: 10.1096/fj.07-101279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang D, Baumgartner RN, Slattery ML, Wang C, Giuliano AR, Murtaugh MA, et al. Dietary intake of folate, B-vitamins and methionine and breast cancer risk among Hispanic and non-Hispanic white women. PloS one. 2013;8:e54495. doi: 10.1371/journal.pone.0054495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Kensler TW, Cho CG, Posner GH, Talalay P. Anticarcinogenic activities of sulforaphane and structurally related synthetic norbornyl isothiocyanates. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:3147–50. doi: 10.1073/pnas.91.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bassett JK, Baglietto L, Hodge AM, Severi G, Hopper JL, English DR, et al. Dietary intake of B vitamins and methionine and breast cancer risk. Cancer causes & control: CCC. 2013;24:1555–63. doi: 10.1007/s10552-013-0232-y. [DOI] [PubMed] [Google Scholar]
- 46.Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. Journal of the National Cancer Institute. 1998;90:1371–88. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 47.Fisher BJ, Perera FE, Cooke AL, Opeitum A, Stitt L. Long-term follow-up of axillary node-positive breast cancer patients receiving adjuvant tamoxifen alone: patterns of recurrence. Int J Radiat Oncol Biol Phys. 1998;42:117–23. doi: 10.1016/s0360-3016(98)00177-1. [DOI] [PubMed] [Google Scholar]
- 48.Vogel VG, Costantino JP, Wickerham DL, Cronin WM, Cecchini RS, Atkins JN, et al. Update of the National Surgical Adjuvant Breast and Bowel Project Study of Tamoxifen and Raloxifene (STAR) P-2 Trial: Preventing breast cancer. Cancer Prev Res (Phila) 2010;3:696–706. doi: 10.1158/1940-6207.CAPR-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waters EA, McNeel TS, Stevens WM, Freedman AN. Use of tamoxifen and raloxifene for breast cancer chemoprevention in 2010. Breast cancer research and treatment. 2012;134:875–80. doi: 10.1007/s10549-012-2089-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma H, Bernstein L, Pike MC, Ursin G. Reproductive factors and breast cancer risk according to joint estrogen and progesterone receptor status: a meta-analysis of epidemiological studies. Breast cancer research: BCR. 2006;8:R43. doi: 10.1186/bcr1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pike MC, Pearce CL, Wu AH. Prevention of cancers of the breast, endometrium and ovary. Oncogene. 2004;23:6379–91. doi: 10.1038/sj.onc.1207899. [DOI] [PubMed] [Google Scholar]
- 52.Freedman AN, Yu B, Gail MH, Costantino JP, Graubard BI, Vogel VG, et al. Benefit/risk assessment for breast cancer chemoprevention with raloxifene or tamoxifen for women age 50 years or older. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:2327–33. doi: 10.1200/JCO.2010.33.0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kleinberg DL, Ameri P, Singh B. Pasireotide, an IGF-I action inhibitor, prevents growth hormone and estradiol-induced mammary hyperplasia. Pituitary. 2011;14:44–52. doi: 10.1007/s11102-010-0257-0. [DOI] [PubMed] [Google Scholar]
- 54.Ruan W, Fahlbusch F, Clemmons DR, Monaco ME, Walden PD, Silva AP, et al. SOM230 inhibits insulin-like growth factor-I action in mammary gland development by pituitary independent mechanism: mediated through somatostatin subtype receptor 3? Molecular endocrinology. 2006;20:426–36. doi: 10.1210/me.2005-0283. [DOI] [PubMed] [Google Scholar]
- 55.Singh B, Smith JA, Axelrod DM, Ameri P, Levitt H, Danoff A, et al. Insulin-like growth factor-I inhibition with pasireotide decreases cell proliferation and increases apoptosis in pre-malignant lesions of the breast: a phase 1 proof of principle trial. Breast cancer research: BCR. 2014;16:463. doi: 10.1186/s13058-014-0463-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The fastq files were submitted to SRA (SRP053781), which is linked to BioProject (Accession: PRJNA273963), and BioSample (Accessions:SAMN03324064-SAMN03324087).