Abstract
Many essential cellular processes including endocytosis and vesicle trafficking require alteration of membrane geometry. These changes are usually mediated by proteins that can sense and/or induce membrane curvature. Using spherical nanoparticle supported lipid bilayers (SSLBs), we characterize how SpoVM, a bacterial development factor, interacts with differently curved membranes by magic angle spinning solid-state NMR. Our results demonstrate that SSLBs are an effective system for structural and topological studies of membrane geometry-sensitive molecules.
Biological membranes exhibit diverse shapes, and there is a growing body of evidence that membrane topology plays key roles in many cellular processes.1,2 For example, cellular processes such as endocytosis, vesiculation, organelle synthesis, and cell division require transient membrane remodeling with a high degree of spatial and temporal accuracy. Further, entry of some bacteria and viruses into human cells also involves alteration of membrane structure.3 Recently, it was discovered that the curvature of the membrane bilayer can regulate the enzymatic activity (e.g., human ArfGAP1, tafazzin),4,5 and define the subcellular localization, of some proteins (e.g., bacterial development factors SpoVM and DivIVA).6–8 Increasingly, membrane geometry is viewed as an essential component of a microenvironment for membrane fusion and fission, protein localization, trafficking, and signaling.9,10 Membrane rearrangement is often governed by proteins that can sense and/or induce membrane curvature (e.g., BAR domain-containing proteins, dynamin, and epsins),11 but our understanding of the underlying molecular mechanisms of these processes remains rudimentary. Specifically, how do membrane geometry-sensitive molecules detect differences in curvature and how curvature affects their structures and membrane topology? In part, this gap in understanding is due to a lack of high resolution structural information in native-like membrane environments having a wide-range of stable and precisely controlled curvature.
To date, liposomes are the most common system for the structural analysis of membrane proteins. While relatively homogeneous small (~50 nm) and large unilamellar vesicles (~100 nm) can be prepared to mimic highly curved membranes, the preparation of giant unilamellar liposomes (GUVs) has been difficult. GUVs are not stable and are usually polydisperse in size and degree of curvature, limiting their use in structural studies.12 By contrast, spherical nanoparticle supported lipid bilayers (SSLBs),13 first introduced in 1990, preserve many characteristics of cell membranes such as lateral fluidity, impermeability to ionic species, and flexibility along the fatty acyl chain. The absorbed lipids form a single bilayer and are separated from the solid supports by a thin layer of water molecules (~20 Å, Figure 1). SSLBs can be prepared with well-defined curvatures (defined by the size of solid supports) and surface stiffness14,15 and have been used in biophysical studies of several bilayer-interacting peripheral membrane proteins.16,17 Here, we demonstrate the utility of SSLBs coupled with high-resolution magic angle spinning (HRMAS) solid-state NMR (ssNMR) for structural studies of membrane curvature-sensitive molecules. This system proves to be ideal for defining the role of membrane curvature in modulating protein–lipid interactions.
Figure 1.
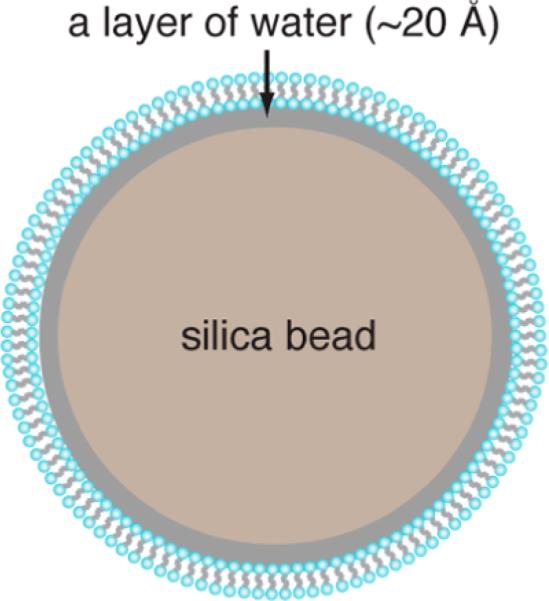
SSLB representation.
As an initial test system, we have examined the protein–lipid interaction of SpoVM. SpoVM is a small (26 residue) peripheral membrane protein from Bacillus subtilis.18 Under nutrient-limiting conditions, B. subtilis initiates a developmental pathway called sporulation in which a spherical internal organelle, termed the “forespore”, is formed that protects the cell's genetic material from environmental insults.19 SpoVM is among the first coat proteins that localize to the outer surface of the forespore to initiate coat assembly. In 2009, Ramamurthi et al. proposed that SpoVM exploits a novel mechanism for localization to the forespore.8 Specifically, SpoVM senses, and preferentially binds to, slightly convex curved outer surface of the forespore (~1 μm in diameter), determining its subcellular localization.8 Recently we dissected the molecular details of its curvature sensing using a SSLB-based in vitro fluorescence assay (Figure S1), solution NMR and molecular dynamics simulation.20 Here we report direct structural studies of SpoVM in SSLBs with native-like curvature.
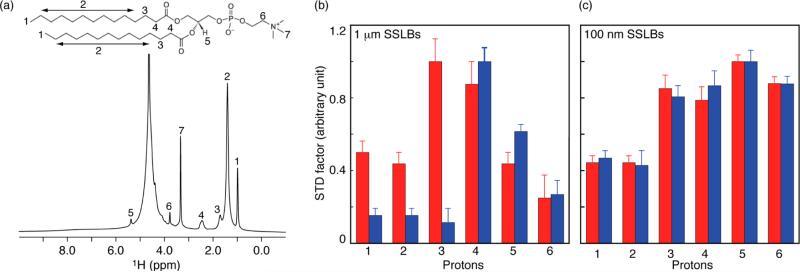
In solution the C-terminal 11 to 23 residues of SpoVM adopts an amphipathic α-helix that is deeply embedded into the membrane.20 We rationalized that this is a result of its atypical helical structure that presents a large hydrophobic surface with only one positively charged residue, Arg17, on its polar face when bound to the membrane. As an initial test of this hypothesis, we replaced Arg17 with Asp (SpoVMR17D) and determined that this variant mislocalized in vivo.20 Structurally, SpoVMR17D adopts a similar α-helical structure to the wild-type protein in the presence of liposomes, as judged by circular dichroism (Figure S2). The loss of in vivo activity of SpoVMR17D could be due to perturbations in its membrane insertion depth, as a consequence of losing the “snorkeling” effect of the Arg upon replacement with the Asp.21 To address this, we compared the insertion depths of wild-type and R17D SpoVM variant using HRMAS spectroscopy and 1 μm SSLBs with native forespore-like curvature. Figure 2a shows a representative 1D 1H HRMAS spectrum of DMPC lipids coating 1 μm silicon beads. Despite a relatively small volume fraction of lipid bilayers of these large SSLBs (~0.2 mg lipids on 20 mg beads in a Bruker 4 mm rotor insert), proton direct detection provided sufficient sensitivity to observe the lipid resonances. Furthermore, the spectrum displayed high resolution, which is a result of the flexibility of fluid lipids of these SSLBs. Multiple protons from the lipid headgroup, glycerol backbone, and fatty acid chains were well-resolved at the moderate sample spinning speed of 10 kHz. Since these proton resonances reflect different membrane depths, they are ideal for monitoring the localization of specific protein–lipid interactions as a function of membrane curvature.
Figure 2.
Curvature-dependent membrane insertion depth. (a) 1D 500 MHz 1H HRMAS spectrum of DMPC-coated 1 μm SSLBs recorded at 30 °C, 10 kHz spinning speed. STD factors of SpoVM (red) and SpoVMR17D (blue) bound to DMPC-coated (b) 1 μm SSLBs at a molar ratio of 1:100. (c) 100 nm SSLBs at a molar ratio of 1:280.
To define the membrane insertion depth of SpoVM and SpoVMR17D, we exploited the saturation transferred difference (STD) NMR experiment. In our protein–lipid preparations the spin-diffusion process is very fast among protein protons, but it is slow between lipid protons that reside in different regions of the bilayer such as the headgroup, glycerol backbone, and acyl chains since it is mainly an intermolecular process.22–24 Under these conditions, selectively saturating protein aromatic resonances rapidly equilibrates protein protons, and the degree of transfers between protein and lipid protons reflects their averaged proximity. The STD effects for SpoVM and SpoVMR17D peptides bound to 1 μm DMPC-coated SSLBs are shown in Figure 2b. For SpoVM, the maximum STD factor was on lipid H3 protons with substantial effects on the methyl and methylene protons of acyl chains and smaller effects on the headgroups. Conversely, the largest effect for SpoVMR17D was on lipid H4 protons and on protons of the glycerol backbone and headgroups, but only minimal transfer to the methyl and methylene protons of acyl chains. Thus, these data supported a model in which SpoVM inserts deeply into the membrane with its hydrophobic side chains interacting with lipid acyl chains, while SpoVMR17D insertion is shallower, due to the shorter and negatively charged Asp side-chain, mainly interacting with lipid head groups and glycerol backbones.
To examine whether membrane curvature affects insertion depth of these proteins, we collected STD data on highly curved 100 nm DMPC-coated SSLBs (Figure 2c). On this membrane, both SpoVM and SpoVMR17D peptides showed similar large STD effects on protons in the lipid glycerol backbone and headgroup, suggesting shallow penetration. Further, at higher protein/lipid ratios, substantial reductions in resonance intensity as a result of increased line broadening were consistently seen on choline methyl protons upon peptide binding (Figure S3). Together, the results indicated that SpoVM and SpoVMR17D only shallowly insert into highly curved bilayers. Thus, our data provided direct experimental support that curvature can regulate insertion depth of membrane proteins, which in turn regulate protein activities.25–29
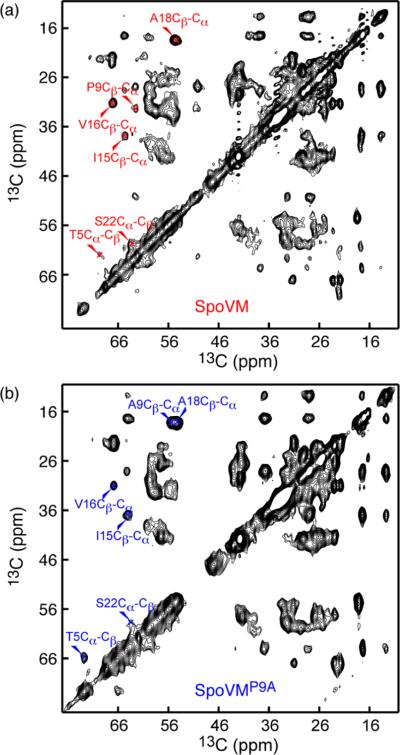
We further sought to validate our solution NMR results, which were performed in planar bicelles, in the more native-like lipid environments of SSLBs. Figure 3 shows 2D high-resolution 13C–13C correlation spectra of SpoVM and SpoVMP9A, a functionally impaired variant that does not specifically recognize membrane curvature in vivo and in vitro. Both peptides can bind to the highly curved 50 nm DMPC-coated SSLBs. Based on their characteristic chemical shifts, several resonances of SpoVM and SpoVMP9A were readily assigned (Figure 3a,b); their Cα secondary shifts (red in Figure S5) are in agreement with their secondary shifts observed in isotropic bicelles (black in Figure S5). In addition, Ala9 Cα and Cβ chemical shifts at 55.5 and 18.2 ppm and the change of Thr5 resonance positions from 62.2 and 69.5 ppm for SpoVM to 65.9 and 72.8 ppm for SpoVMP9A were consistent with a structural rearrangement of N-terminal residues from a loop in SpoVM to an α-helical conformation in SpoVMP9A. These data support our earlier conclusion that disruption of SpoVM helix by Pro9 is necessary for SpoVM curvature sensing.20
Figure 3.
Structure validations of SpoVM and SpoVMP9A in SSLBs. 2D MAS 13C–13C correlation spectra for 13C-,15N-labeled SpoVM (a) and SpoVMP9A (b) bound to DMPC-coated 50 nm SSLBs. The data were collected on an 800 MHz Bruker NMR instrument with 35 ms mixing time,10 kHz spinning speed, 1.5 s recycle delay at 15 °C (sample temperature) for increased spectral sensitivity. The total acquisition time was about 36 h. Temperature-dependent static 31P NMR spectra indicate no lipids are detached from these SSLBs to form small discoidal structures in the gel phase (Figure S4).
An overlay of SpoVM spectra bound to SSLBs and liposomes showed remarkably similarity and comparable resolution (Figure S6). These observations indicate that the presence of a support does not affect the protein structure. The stability of SSLBs during sample spinning is validated by their typical chemical shift powder patterns of the static 31P spectra collected before and after MAS experiments (Figure S7). Furthermore, SpoVM binding does not alter the bilayer structure (Figure S8), since SpoVM only functions as a curvature-sensing molecule and does not tabulate or vesiculate membranes. Together, these observations indicate that SSLBs are suitable for structural studies by MAS ssNMR.
In summary, our study demonstrates that SSLBs are an effective model system for structural and topological characterization of membrane geometry-sensitive molecules by MAS ssNMR. The spectral resolution is comparable to that from liposomes, and spectral sensitivity is adequate for studying protein–lipid and lipid–lipid interactions. Furthermore, we provide direct experimental evidence that bilayer curvature can affect the insertion depth of peripheral membrane proteins.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. T. A. Cross and K. C. Huang for helpful discussions, and Nissan Chemical Industries, Ltd. for 50 and 100 nm silicon beads. The MAS ssNMR data were performed at the National High Magnetic Field Laboratory, which is supported by NSF Cooperative Agreement no. DMR-1157490, the State of Florida, and the U.S. Department of Energy. This work was supported by the National Institutes of Heath NIGMS (R01GM105963 to F.T. and R01GM094526 to J.M.F.) and the NIH Intramural Research Program, National Cancer Institute, Center for Cancer Research (K.S.R.)
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b08303.
Detailed experimental procedures, fluorescence, CD, 1H HRMAS, static 31P, and 13C–13C correlation spectra for SpoVM and variants (PDF)
REFERENCES
- 1.McMahon HT, Gallop JL. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 2.Groves JT, Kuriyan J. Nat. Struct. Mol. Biol. 2010;17:659–665. doi: 10.1038/nsmb.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melikyan GB. Curr. Opin. Virol. 2014;4:1–7. doi: 10.1016/j.coviro.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlame M, Acehan D, Berno B, Xu Y, Valvo S, Ren M, Stokes DL, Epand RM. Nat. Chem. Biol. 2012;8:862–869. doi: 10.1038/nchembio.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bigay J, Gounon P, Robineau S, Antonny B. Nature. 2003;426:563–566. doi: 10.1038/nature02108. [DOI] [PubMed] [Google Scholar]
- 6.Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, Marenduzzo D, Hamoen LW. EMBO J. 2009;28:2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramamurthi KS, Losick R. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13541–13545. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Science. 2009;323:1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rangamani P, Lipshtat A, Azeloglu EU, Calizo RC, Hu MF, Ghassemi S, Hone J, Scarlata S, Neves SR, Iyengar R. Cell. 2013;154:1356–1369. doi: 10.1016/j.cell.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmick M, Bastiaens PIH. Cell. 2014;156:1132–1138. doi: 10.1016/j.cell.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Shen HY, Pirruccello M, De Camilli P. Cell. 2012;150:1300–U226. doi: 10.1016/j.cell.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walde P, Cosentino K, Engel H, Stano P. ChemBioChem. 2010;11:848–865. doi: 10.1002/cbic.201000010. [DOI] [PubMed] [Google Scholar]
- 13.Bayerl TM, Bloom M. Biophys. J. 1990;58:357–362. doi: 10.1016/S0006-3495(90)82382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buranda T, Huang J, Ramarao GV, Ista LK, Larson RS, Ward TL, Sklar LA, Lopez GP. Langmuir. 2003;19:1654–1663. [Google Scholar]
- 15.Savarala S, Ahmed S, Ilies MA, Wunder SL. Langmuir. 2010;26:12081–12088. doi: 10.1021/la101304v. [DOI] [PubMed] [Google Scholar]
- 16.Galneder R, Kahl V, Arbuzova A, Rebecchi M, Radler JO, McLaughlin S. Biophys. J. 2001;80:2298–2309. doi: 10.1016/S0006-3495(01)76201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Picard F, Paquet MJ, Dufourc EJ, Auger M. Biophys. J. 1998;74:857–868. doi: 10.1016/S0006-3495(98)74009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin PA, Fan N, Ricca E, Driks A, Losick R, Cutting S. Mol. Microbiol. 1993;9:761–771. doi: 10.1111/j.1365-2958.1993.tb01736.x. [DOI] [PubMed] [Google Scholar]
- 19.Tan IS, Ramamurthi KS. Environ. Microbiol. Rep. 2014;6:212–225. doi: 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gill RL, Castaing JP, Hsin J, Tan IS, Wang X, Huang KC, Tian F, Ramamurthi KS. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E1908–E1915. doi: 10.1073/pnas.1423868112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strandberg E, Killian JA. FEBS Lett. 2003;544:69–73. doi: 10.1016/s0014-5793(03)00475-7. [DOI] [PubMed] [Google Scholar]
- 22.Yau WM, Gawrisch K. J. Am. Chem. Soc. 2000;22:3971–3972. [Google Scholar]
- 23.Soubias O, Gawrisch K. J. Am. Chem. Soc. 2005;127:13110–13111. doi: 10.1021/ja0538942. [DOI] [PubMed] [Google Scholar]
- 24.Aucoin D, Camenares D, Zhao X, Jung J, Sato T, Smith SO. J. Magn. Reson. 2009;197:77–86. doi: 10.1016/j.jmr.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiang W, Sun Y, Weliky DP. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15314–15319. doi: 10.1073/pnas.0907360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambroso MR, Hegde BG, Langen R. Proc. Natl. Acad. Sci. U. S. A. 2014;111:6982–6987. doi: 10.1073/pnas.1402233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strandberg E, Zerweck J, Wadhwani P, Ulrich AS. Biophys. J. 2013;104:L09–L11. doi: 10.1016/j.bpj.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T, Cady SD, Hong M. Biophys. J. 2012;102:787–794. doi: 10.1016/j.bpj.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soubias O, Teague WE, Hines KG, Gawrisch K. Biochimie. 2014;107:28–32. doi: 10.1016/j.biochi.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.