Abstract
Coordinated replication and expression of mitochondrial genome is critical for metabolically active cells during various stages of development. However, it is not known whether replication and transcription can occur simultaneously without interfering with each other and whether mtDNA copy number can be regulated by the transcription machinery. We found that interaction of human transcription elongation factor, TEFM with mitochondrial RNA polymerase (mtRNAP) and nascent transcript prevents generation of replication primers and increases transcription processivity thereby serving as a molecular switch between replication and transcription, which appear to be mutually exclusive processes in mitochondria. TEFM may allow mitochondria to increase transcription rates and, as consequence, respiration and ATP production without the need to replicate mtDNA, which has been observed during spermatogenesis and early stages of embryogenesis.
The maternally-inherited circular mtDNA encodes subunits of complexes of the oxidative phosphorylation chain as well as tRNAs and ribosomal RNAs (1, 2). Transcription of human mtDNA is directed by two promoters, the LSP (light strand promoter) and the HSP1 (heavy strand promoter) located in opposing mtDNA strands, resulting in two almost genome-sized polycistronic transcripts that undergo extensive processing prior to polyadenylation and translation (3, 4). Intriguingly, transcription terminates prematurely about 120 bps downstream of LSP at a conserved in all vertebrates G-rich region, called conserved sequence block II (CSBII), as a result of formation of a hybrid G-quadruplex between nascent RNA and the non-template strand of DNA (5–7). This termination event occurs near the origin of replication of the heavy strand (oriH) (8) and generates a replication primer. According to the asymmetric model (9), replication then proceeds through about 2/3 of the mtDNA until the oriL sequence in the opposing strand becomes single stranded and forms a hairpin structure. The oriL hairpin is then recognized by mtRNAP which primes replication of the light strand (10). Since replication of mtDNA coincides with transcription in time and space, collisions between transcription and replication machineries are inevitable and, similar to bacterial and eukaryotic systems, likely have detrimental effects on mtDNA gene expression (11).
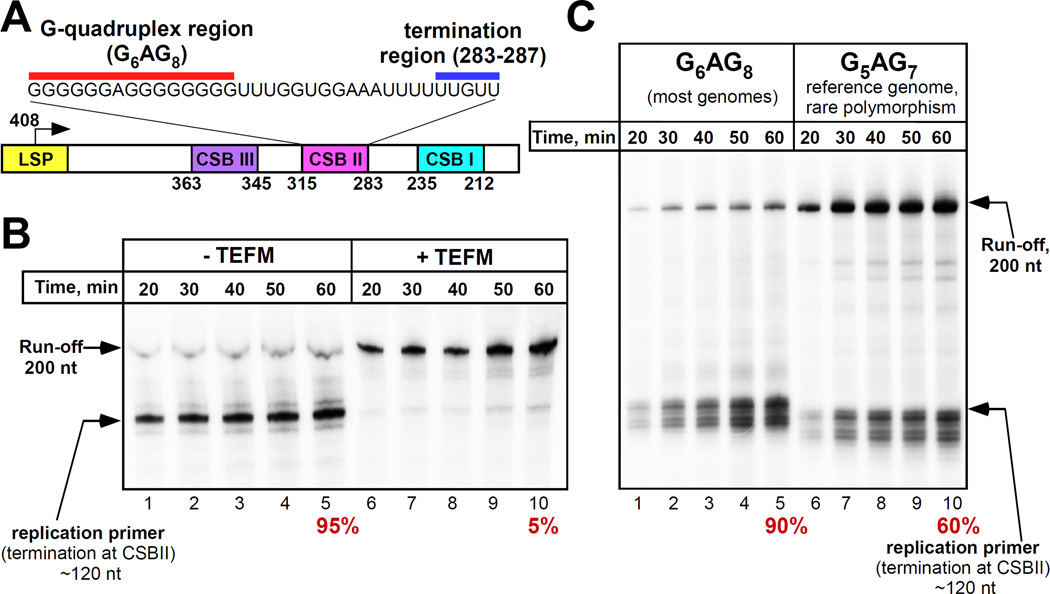
We analyzed the effects of a mitochondrial transcription elongation factor, TEFM, recently discovered by Minczuk and colleagues (12) on transcription of mtDNA. This protein was pulled down from mitochondrial lysates via mtRNAP and was found to stimulate non-specific transcription on promoter-less DNA; however its effect on promoter-driven transcription had not been determined (12). We found that in the presence of TEFM mtRNAP efficiently transcribes through CSBII (Fig. 1A,B). Thus, TEFM acts as an antitermination factor that prevents termination at CSBII and synthesis of a primer for mitochondrial DNA polymerase. We identified the exact location of the termination point in CSBII (fig. S1). MtRNAP terminates at the end of a U6 sequence (positions 287–283 in mtDNA) 16–18 nt downstream of the G-quadruplex (Fig. 1A). At this point, the 9 bp RNA-DNA hybrid in the elongation complex (EC) is extremely weak, as it is composed only of A-U / T-A pairs. This is reminiscent of intrinsic termination signals in prokaryotes where the formation of an RNA hairpin is thought to disrupt the upstream region of the RNA-DNA hybrid, and is followed by the run of 6–8 UMP residues that further destabilizes the complex (5, 13).
Figure 1. TEFM prevents termination at CSBII.
(A) Schematic illustration of the human mtDNA region downstream of LSP promoter. The numbers below the scheme corresponds to the reference human mtDNA; the transcribed sequence of CSBII is shown at the top. (B) MtRNAP does not terminate at CSBII in the presence of TEFM. Termination efficiency (%) is indicated below the panel. (C) A rare polymorphism in a reference mtDNA results in a decreased efficiency of transcription termination at CSBII.
Human mtDNA is highly polymorphic in the CSBII region; coincidently, the reference mitochondrial genome (Cambridge) contains a rare polymorphism in the G-quadruplex - G5AG7, whereas the majority of mtDNA from various haplogroups have two additional G-residues (G6AG8) (14). We found that the termination efficiency of mtRNAP was significantly lower at G5AG7-CSBII (Fig. 1C) suggesting an effect of G-run length on quadruplex formation, and underscoring the importance of further studies of various polymorphisms in this region.
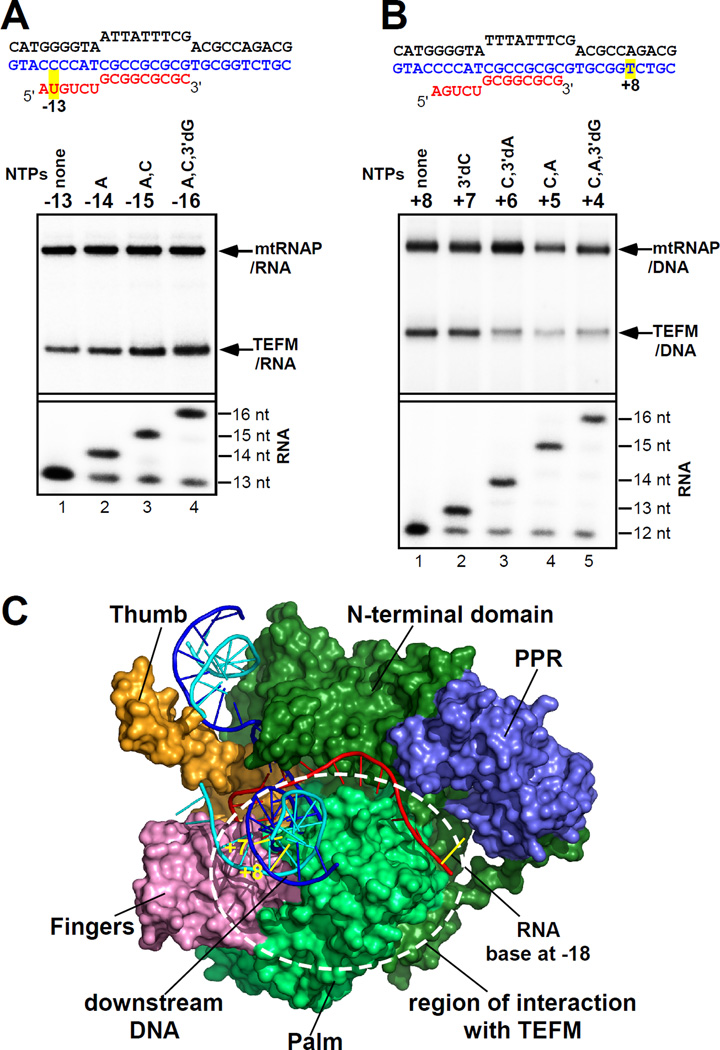
In considering a putative mechanism of TEFM antitermination activity, we investigated whether it can interact with the nascent transcript and thus interfere with the formation of the quadruplex structure. We assembled ECs on a nucleic acid scaffold containing a photo reactive analog of uridine, 4-thio-uridine, 13 nt downstream from the 3’ end of RNA, and walked mtRNAP along the template by incorporation of appropriate substrate NTPs (Fig. 2A). We observed efficient cross-linking between TEFM and RNA when the photo-reactive base was 15–16 bps away from the 3’end of RNA. Additionally, using a template DNA containing the LSP promoter and CSBII, we probed interactions of TEFM with RNA having a photo-reactive probe (6-thio GMP) 9–16 or 18–25 nt away from the 3’ end of RNA (fig. S2). We found that TEFM efficiently cross-linked to the G-quadruplex region of RNA, further confirming its interactions with the nascent transcript (fig. S2).
Figure 2. Interactions of TEFM with the components of the elongation complex.
(A) TEFM interacts with the RNA in a scaffold EC. Top panel: cross-linking of labeled RNA to the proteins indicated; bottom panel, RNA extension. (B) TEFM interacts with the downstream DNA. (C) Putative region of interaction with TEFM. Structure of human mtRNAP EC (surface representation, PDB ID 3BOC) is shown with the major domains indicated (fingers - pink, palm - light green, N-terminal domain - dark green, thumb - orange, DNA template strand - blue, non-template strand - cyan, RNA - red). The 9 nt long RNA in the EC is extended by 9 nt to model the trajectory of the nascent RNA.
The catalytic domains of mtRNAP and the single-subunit RNAP from bacteriophage T7 share high sequence and structural homology (15). When we compared the structural organization of human mtRNAP EC (15) and T7 RNAP EC (16), one striking difference became apparent – the former polymerase makes very few contacts with the downstream DNA duplex (fig. S3). In contrast, in T7 RNAP EC, the N-terminus of the polymerase makes long-range electrostatic interactions with the entire downstream duplex. These extensive interactions with the downstream DNA may explain the high processivity of T7 RNAP (16). Since TEFM binds the C-terminus of mtRNAP (12) we hypothesized that it may also bind the downstream DNA to compensate for the lack of mtRNAP-DNA interactions. To check this, we incorporated 4-thio dTMP into the DNA template in the downstream region and assembled the EC using RNA-DNA scaffolds. We found that TEFM efficiently cross-linked to the +7 and +8 bases in the DNA template strand (Fig. 2B and fig. S4A). The DNA-TEFM cross-linking was mtRNAP-specific, as no efficient cross-linking was detected when yeast mtRNAP was used (fig. S4B).
The binding site of TEFM on mtRNAP has been previously identified in the C-terminal region, between residues 768–1230 (12). These data, taken together with the RNA and DNA cross-linking data suggest that TEFM likely binds to the palm subdomain of mtRNAP in proximity to the PPR domain and interacts with the emerging RNA transcript (Fig. 2C). It is therefore likely that TEFM antitermination activity is due to direct interference with the formation of the bulky G-quadruplex structure between the palm subdomain and the PPR domain of mtRNAP or due to the allosteric effect of quadruplex formation on complex stability. Similar mechanisms of antitermination have been suggested for lambda phage antiterminators Q and N and a number of bacterial antiterminators which were shown to interact with both RNAP and its nascent transcript that would otherwise form a hairpin structure (17, 18).
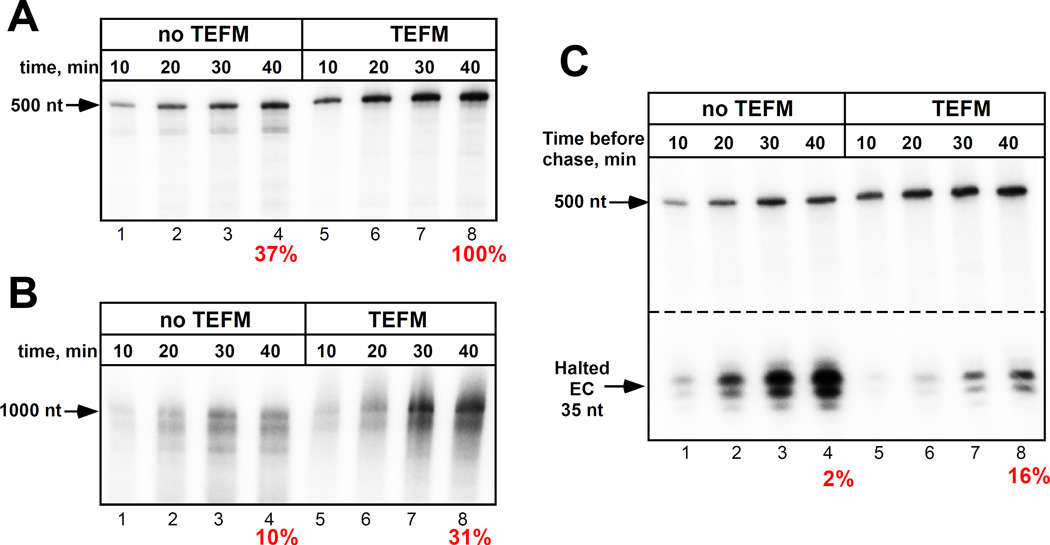
Because TEFM interacts with the downstream DNA (which may stabilize the EC), we decided to investigate its effect on mtRNAP processivity. We found that the efficiency of run-off synthesis by mtRNAP was low if the size of the product exceeded 500 nt (Fig. 3A,B). Indeed, the processivity of mtRNAP was substantially lower as compared to a related T7 RNAP (which is known to transcribe large fragments of DNA) and was apparently not sufficient to transcribe a 16,000 bps long mtDNA. In the presence of TEFM, the efficiency of synthesis of long RNA was significantly increased (Fig. 3A,B), suggesting that TEFM indeed acts as a processivity factor, as has been originally proposed (12).
Figure 3. TEFM increases processivity and stability of mtRNAP EC.
(A,B) TEFM increases the EC processivity. Transcription was performed using LSP templates lacking CSBII sequence to generate 500 nt (A) or 1000 nt (B) run-off products. Efficiency of synthesis of 500 nt RNA in the presence of TEFM is taken as 100%. (C) TEFM increases the EC stability. Complexes halted at +35 in the presence or absence of TEFM were incubated for the times indicated and chased with CTP. Numbers below indicate efficiency of a halted EC extension (%).
We next probed if TEFM contributes to the stability of the elongating mtRNAP and, as consequence, to its antitermination activity. To demonstrate this, we halted the EC 35 bps downstream of the promoter start site by omitting one of the substrate NTPs (CTP, Fig. 3C and fig.S5), in the presence or absence of TEFM. After 10–40 min the complexes were chased to allow synthesis of a run-off product. In the absence of TEFM, accumulation of a 35-mer RNA product predominated, with little production of runoff, indicating that the halted EC is unstable, rapidly dissociates and reinitiates, but is inefficiently chased (Fig. 3C and fig.S5). In contrast, in the presence of TEFM little, if any, of 35-mer RNA product accumulates, but more efficient synthesis of full length products is observed, indicating that under these conditions the EC is stable and may subsequently be extended.
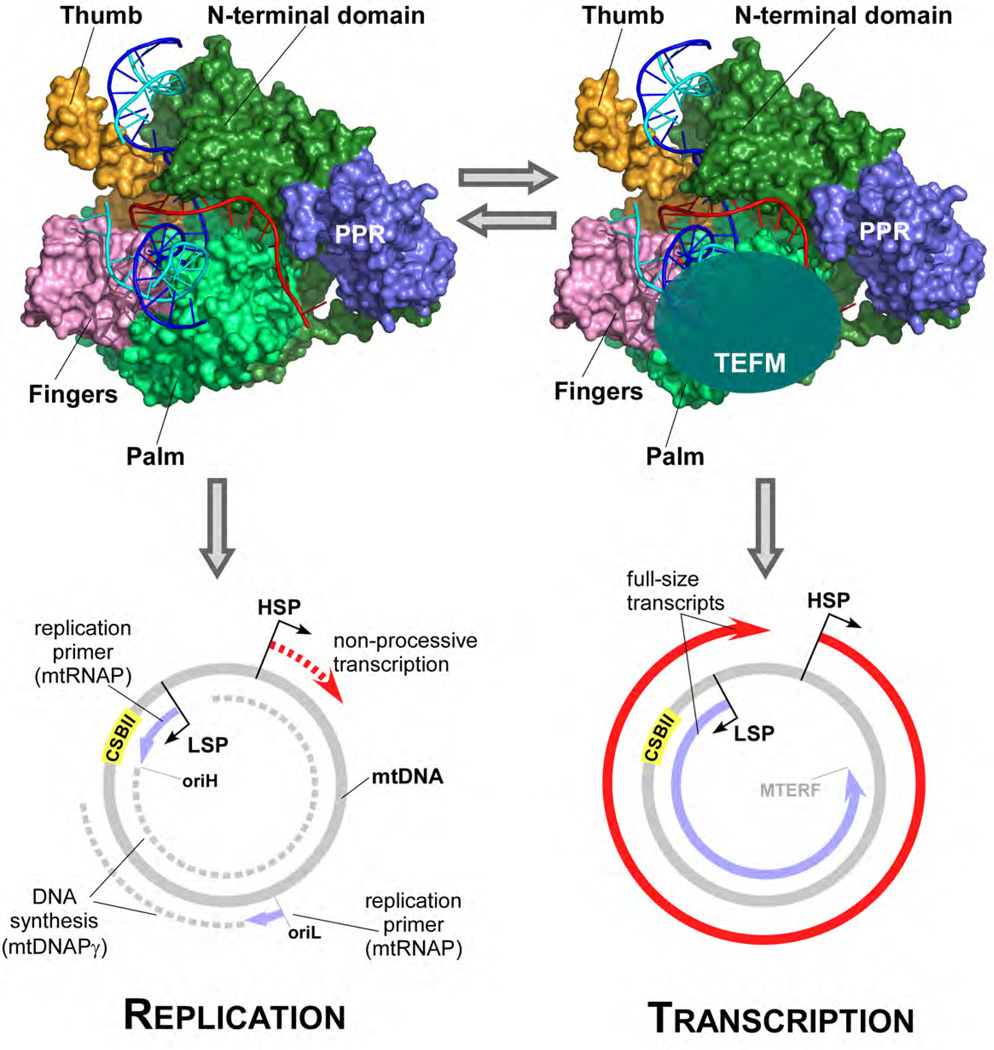
Our in vitro data demonstrate that TEFM increases the general processivity of ECs, and that in the absence of TEFM mtRNAP is unable to efficiently produce long transcripts and instead terminates at CSBII, thereby priming replication of the heavy strand of mtDNA (Fig. 4). Consistent with this, a knockdown of TEFM in human cells results in a decrease of promoter-distal RNA transcripts (12). We therefore propose that replication and transcription are mutually exclusive processes in human mitochondria allowing the corresponding machineries to avoid the detrimental consequences of head-on collisions (Fig. 4). As such, TEFM serves as a molecular switch that would allow the organelles to either replicate the mtDNA and regulate its copy number or to elevate its transcription rates. This switch can be important for a number of developmental processes, such as embryogenesis and spermatogenesis, during which an active transcription of mtDNA is observed without replication (19–25). While TEFM may not be the only factor affecting mtDNA copy number (26), the proposed mechanism provides solution to these important biological processes. Future experiments should address the question as to how this switch is regulated in mitochondria of actively metabolizing cells.
Figure 4. Model for a replication-transcription switch in mitochondria.
In the absence of TEFM, mtRNAP initiates transcription at the LSP promoter but terminates at CSBII and produces a 120 nt replication primer at the replication origin oriH. The primer is used by replisome for replication of the heavy strand of mtDNA during which a separate initiation event involving mtRNAP generates a replication primer at oriL. Due to low processivity, transcription from the HSP promoter does not produce a full-size transcript (bottom left). When TEFM is bound, mtRNAP cannot terminate at CSBII and generates full-size transcripts from both LSP and HSP promoters (bottom right). TEFM antitermination activity and its effects on EC processivity ensures that replication events do interfere with the expression of mitochondrial genome.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kathrin Schwinghammer for help in cloning and isolation of TEFM and Bill McAllister and Max Gottesman for the critical discussion and reading of the manuscript. This work was supported in part by NIH RO1GM104231.
Footnotes
REFERENCES AND NOTES
- 1.Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 2.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Molecular cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 3.Hallberg BM, Larsson NG. Making Proteins in the Powerhouse. Cell Metab. 2014;20:226–240. doi: 10.1016/j.cmet.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Morozov YI, Agaronyan K, Cheung AC, Anikin M, Cramer P, Temiakov D. A novel intermediate in transcription initiation by human mitochondrial RNA polymerase. Nucleic Acids Res. 2014;42:3884–3893. doi: 10.1093/nar/gkt1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wanrooij PH, Uhler JP, Shi Y, Westerlund F, Falkenberg M, Gustafsson CM. A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 2012;40:10334–10344. doi: 10.1093/nar/gks802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wanrooij PH, Uhler JP, Simonsson T, Falkenberg M, Gustafsson CM. G-quadruplex structures in RNA stimulate mitochondrial transcription termination and primer formation. Proc Natl Acad Sci U S A. 2010;107:16072–16077. doi: 10.1073/pnas.1006026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci U S A. 1985;82:351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang D, Miyako K, Kai Y, Irie T, Takeshige K. In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J Biol Chem. 1997;272:15275–15279. doi: 10.1074/jbc.272.24.15275. [DOI] [PubMed] [Google Scholar]
- 9.Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 10.Fuste JM, Wanrooij S, Jemt E, Granycome CE, Cluett TJ, Shi Y, Atanassova N, Holt IJ, Gustafsson CM, Falkenberg M. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol Cell. 2010;37:67–78. doi: 10.1016/j.molcel.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 11.Pomerantz RT, O'Donnell M. What happens when replication and transcription complexes collide? Cell Cycle. 2010;9:2537–2543. doi: 10.4161/cc.9.13.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minczuk M, He J, Duch AM, Ettema TJ, Chlebowski A, Dzionek K, Nijtmans LG, Huynen MA, Holt IJ. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic acids research. 2011;39:4284–4299. doi: 10.1093/nar/gkq1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An allosteric path to transcription termination. Mol Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 15.Schwinghammer K, Cheung A, Morozov AI, Agaronyan K, Temiakov D, Cramer P. Structure of mitochondrial RNA polymerase elongation complex. Nat Struct Mol Biol. 2013;20:1298–1303. doi: 10.1038/nsmb.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahirov TH, Temiakov D, Anikin M, Patlan V, McAllister WT, Vassylyev DG, Yokoyama S. Structure of a T7 RNA polymerase elongation complex at 2.9 A resolution. Nature. 2002;420:43–50. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- 17.Gusarov I, Nudler E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell. 2001;107:437–449. doi: 10.1016/s0092-8674(01)00582-7. [DOI] [PubMed] [Google Scholar]
- 18.Shankar S, Hatoum A, Roberts JW. A transcription antiterminator constructs a NusA-dependent shield to the emerging transcript. Mol Cell. 2007;27:914–927. doi: 10.1016/j.molcel.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rantanen A, Larsson NG. Regulation of mitochondrial DNA copy number during spermatogenesis. Human reproduction. 2000;15(Suppl 2):86–91. doi: 10.1093/humrep/15.suppl_2.86. [DOI] [PubMed] [Google Scholar]
- 20.Thundathil J, Filion F, Smith LC. Molecular control of mitochondrial function in preimplantation mouse embryos. Molecular reproduction and development. 2005;71:405–413. doi: 10.1002/mrd.20260. [DOI] [PubMed] [Google Scholar]
- 21.Wai T, Ao A, Zhang X, Cyr D, Dufort D, Shoubridge EA. The role of mitochondrial DNA copy number in mammalian fertility. Biology of reproduction. 2010;83:52–62. doi: 10.1095/biolreprod.109.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carling PJ, Cree LM, Chinnery PF. The implications of mitochondrial DNA copy number regulation during embryogenesis. Mitochondrion. 2011;11:686–692. doi: 10.1016/j.mito.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Van Blerkom J. Mitochondrial function in the human oocyte and embryo and their role in developmental competence. Mitochondrion. 2011;11:797–813. doi: 10.1016/j.mito.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Monnot S, Samuels DC, Hesters L, Frydman N, Gigarel N, Burlet P, Kerbrat V, Lamazou F, Frydman R, Benachi A, Feingold J, Rotig A, Munnich A, Bonnefont JP, Steffann J. Mutation dependance of the mitochondrial DNA copy number in the first stages of human embryogenesis. Hum Mol Genet. 2013;22:1867–1872. doi: 10.1093/hmg/ddt040. [DOI] [PubMed] [Google Scholar]
- 25.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.