Abstract
We developed a loop-mediated isothermal amplification (LAMP) method to rapidly diagnose Wheat streak mosaic virus (WSMV) during quarantine inspections of imported wheat, corn, oats, and millet. The LAMP method was developed as a plant quarantine inspection method for the first time, and its simplicity, quickness, specificity and sensitivity were verified compared to current reverse transcription-polymerase chain reaction (RT-PCR) and nested PCR quarantine methods. We were able to quickly screen for WSMV at quarantine sites with many test samples; thus, this method is expected to contribute to plant quarantine inspections.
Keywords: LAMP, loop-mediated isothermal amplification, quarantine, wheat streak mosaic virus, WSMV
Wheat streak mosaic virus (WSMV) is an unreported virus in Korea (Animal, Plant and Fisheries Quarantine and Inspection Agency, 2013) and quarantine has raised the possibility that WSMV can cause serious food loss and economic damage (French and Robertson, 1994) by infecting wheat, corn, oats, and millet (Lee et al., 2013a). Quarantine inspections have used reverse transcription-polymerase chain reaction (RT-PCR) and nested PCR to detect pathogens since 2012 in Korea (Lee, 2013; Lee et al., 2013b; Lee et al., 2013c). However, these methods require 10 hrs for results, so they are inconvenient for field use where rapid inspection and subsequent quarantine are needed. Therefore, in this study, we developed a LAMP assay as an inspection method to detect WSMV in a one-step quick screen during quarantine inspection.
Samples of WSMV and reference viruses [Cereal chlorotic mottle virus (CCMV) and Cucumber mosaic virus (CMV)] were collected through import approval of prohibited goods. Nucleic acids were extracted and cDNA was synthesized using a method reported previously (Lee and Shin, 2014; Lee et al., 2014; Lee et al., 2015; Shin and Rho, 2014). RNA of 170–200 ng/μl was extracted from the samples, and 100 μl cDNA was synthesized from the RNA.
The WSMA complete genome (NC_001886), 15 WSMV strains and their base sequences, and the base sequences of 20 reference virus strains that could infect the same host were collected from the National Center for Biotechnology Information. Two sets of WSMV-specific LAMP primers were designed based on the base sequences collected using the LAMP primer designing software PrimerExplorer (Table 1).
Table 1.
Loop-mediated isothermal amplification primer sequence of the amplified poly-protein coding gene to rapidly detect Wheat streak mosaic virus
| Set | Primer | Sequence (5′→3′) | Length (nt) | G+C (%) | Tm (°C) |
|---|---|---|---|---|---|
| WSMV_1 | F3 | GAGTTGGAGGAAAGGAAA | 18 | 44.4 | 49.0 |
| B3 | TCCGAAAACGGTAGAAAG | 18 | 44.4 | 49.3 | |
| FIP | GGACCAGAATATGTGAGTCGTATCTTTCACACGCAGAGT | 39 | - | - | |
| BIP | AGGAAGGTTACTCACCTTTGCGGAGTATAACTTGCAAGACG | 41 | - | - | |
|
| |||||
| WSMV_2 | F3 | TACGACTCACATATTCTGGT | 20 | 40.0 | 50.2 |
| B3 | GCCTATGAGTATAACTTGCA | 20 | 40.0 | 49.4 | |
| FIP | TCGGTCCTCGGAGATAGCGGAAGAAAGGTTGGAGAA | 36 | - | - | |
| BIP | ACAAAGGGAGTTCCTTGAGGATTTTCCGCAAAGGTGAGTAA | 41 | - | - | |
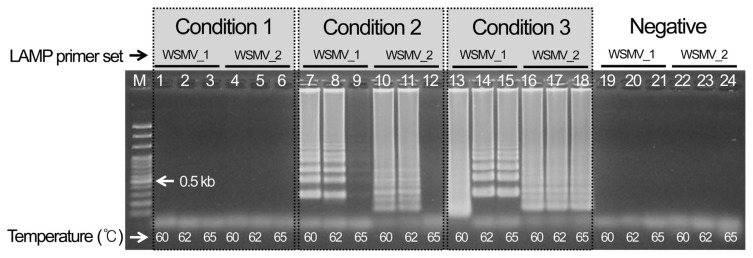
Three WSMV template cDNAs were chosen and reacted for 1 hr at three different temperatures (60, 62, and 65°C) to select the best LAMP conditions for detecting WSMV after 10 min at 95°C and 1 min at 4°C. As a result, condition 3 using an 11 μl total volume and no distilled water showed the most specific reaction. The analysis indicated that the specific reaction occurred at all temperatures, so the most stable conditions at 62°C were selected (Fig. 1). In addition, set 1 of the cDNAs for the CCMV and CMV reference viruses indicated nonspecific amplification for both the reference virus and the negative control, so it finally selected as the LAMP primer combination for detecting WSMV (Supplementary Fig. 1).
Fig. 1.
Optimal selection of LAMP conditions for detecting Wheat streak mosaic virus.
The LAMP method developed here can detect WSMV in imported samples within 1.5 hrs (30 min for cDNA synthesis and 60 min for LAMP) after RNA extraction. Thus, this method reduces the time to detect WSMV from the previous 10 hrs using the RT-PCR, electrophoresis, nested PCR, and additional electrophoresis steps. Furthermore, it is much simpler and easier to use as the standard quarantine method. Additionally, specificity improved compared to that of the PCR method.
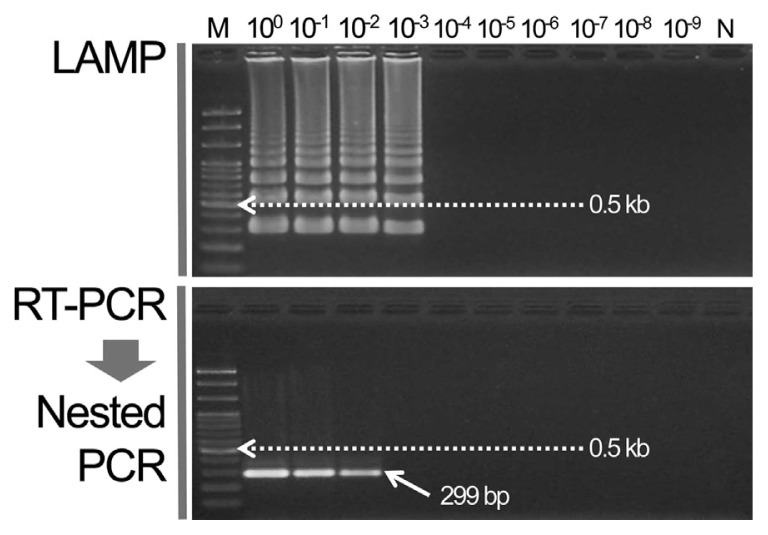
The WSMV cDNA template for LAMP was diluted up to 10−9. The RT-PCR primer combination [forward: 5′-TGG CGA TGA AGA TGT CAG-3′, reverse: 5′-CCA TTT CTG TGA AGG CTT T-3′ (834 bp)] was used previously to diagnose WSMV. The PCR template result was used for the nested PCR reaction to amplify the specific 299 bp gene (Lee et al., 2013a). The detection sensitivities of the two methods were compared. As a result, a specific band was formed at up to a 10−2 dilution in the nested PCR but the specific ladder from the LAMP assay was analyzed at up to a 10−3 dilution (Fig. 2).
Fig. 2.
Compare sensitivity of LAMP and nested PCR assay for detecting Wheat streak mosaic virus.
After LAMP assay, the prepared 10 μl of LAMP products were digested with 10 U of restriction enzyme BfaI (New England Biolabs, US) at 37°C for 2 hrs, then they were subjected to electrophoresis in a 1.5% agarose gel and visualized (digestion fragments [194 + 156 bp]) (Supplementary Fig. 2).
The LAMP assay was applied to quickly screen for WSMV during quarantine inspection of imported wheat, corn, oats, and millet. Instead of using Taq DNA polymerase, LAMP amplifies denatures, anneals, and extends at the same temperature by using Bst polymerase, resulting in quicker detection than that of the existing PCR method. In addition, the 5′→3′ exonuclease feature provides high detection sensitivity and a quick result (Ahn et al., 2010; Cho et al., 2013). The LAMP developed in this study is simple and more specific rather than the current two-step quarantine methods using RT-PCR and nested PCR and reduced processing time by approximately 8 hrs. Furthermore, the LAMP method had 10 fold higher detection sensitivity than the current two-step PCR method. We expect that this LAMP method will be applied for use during quarantine inspections in the future.
Supplementary Material
Table 2.
Optimal loop-mediated isothermal amplification conditions
| Components | Condition (μl) | ||
|---|---|---|---|
|
| |||
| 1 | 2 | 3 | |
| Template | 1.5 | 1.5 | 1.5 |
| Bst. 10× reaction buffer | 2 | 2 | 2 |
| dNTP (10 mM) | 2 | 2 | 2 |
| Primers | |||
| F3 (10 pmol) | 0.6 | 0.6 | 0.6 |
| B3 (10 pmol) | 0.6 | 0.6 | 0.6 |
| FIP (10 pmol) | 1.4 | 1.4 | 1.4 |
| BIP (10 pmol) | 1.4 | 1.4 | 1.4 |
| Bst. polymerase (12 U) | 1.5 | 1.5 | 1.5 |
| ddH2O | 9 | 4 | 0 |
|
| |||
| Total (μl) | 20 | 15 | 11 |
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant number: 2009-0093829).
References
- Ahn YC, Nam YH, Park SN, Cho MH, Seo JW, Yoon IK, Park YH, Jang WC. Detection of Mycobacterium tuberculosis by loop-mediated isothermal amplification assay. J Korean Chem Soc. 2008;52:273–280. doi: 10.5012/jkcs.2008.52.3.273. [DOI] [Google Scholar]
- Animal, Plant and Fisheries Quarantine and Inspection Agency. List of plant quarantine viruses in Korea newly revised in 2013. Res Plant Dis. 2013;19:67–75. [Google Scholar]
- Cho MH, Jang WC, Choi JG. Detection for methicillin resistant Staphylococcus aureus in using bio-chip based loop mediated isothermal amplification assay. J Korean Chem Soc. 2013;57:81–87. doi: 10.5012/jkcs.2013.57.1.81. [DOI] [Google Scholar]
- French R, Robertson NL. An RT-PCR method for the detection of the wheat streak mosaic virus. J Virol Methods. 1994;49:93–100. doi: 10.1016/0166-0934(94)90059-0. [DOI] [PubMed] [Google Scholar]
- Lee S. PhD thesis. Dankook University; Cheonan, Chungcheongnam-do, Korea: 2013. A study of molecular biological detection methods for seed-transmitted viruses in quarantine. [Google Scholar]
- Lee S, Cha M, Kim SM, Heo NY, Shin YG, Lee SH. Development of nucleotide primers for diagnostic RT-PCR and nested PCR detection of three seed-transmitted viruses (CRLV, SpLV and WClMV) in quarantine. J Agric Life Sci. 2014;48:75–83. doi: 10.14397/jals.2014.48.3.75. [DOI] [Google Scholar]
- Lee S, Kang EH, Chu YM, Shin YG, Ahn TY. Development of PCR diagnosis system for plant quarantine seed-borne Wheat streak mosaic virus. Korean J Microbiol. 2013a;49:112–117. doi: 10.7845/kjm.2013.3013. [DOI] [Google Scholar]
- Lee S, Kang EH, Heo NY, Kim SM, Kim YJ, Shin YG. Detection of Carnation necrotic fleck vrus and Carnation ringspot virus using RT-PCR. Res Plant Dis. 2013b;19:36–44. doi: 10.5423/RPD.2013.19.1.036. [DOI] [Google Scholar]
- Lee S, Kang EH, Shin YG, Lee SH. Development of RT-PCR and nested PCR for detection of four quarantine plant viruses belonging to Nepovirus. Res Plant Dis. 2013c;19:220–225. doi: 10.5423/RPD.2013.19.3.220. [DOI] [Google Scholar]
- Lee S, Lee JY, Shin YG, Lee SH, Ahn TY. Development and verification of nested PCR assay for detection of Tobacco rattle virus in plant quarantine. J Bacteriol Virol. 2015;45:54–61. doi: 10.4167/jbv.2015.45.1.54. [DOI] [Google Scholar]
- Lee S, Shin YG. Development and practical use of RTPCR for seed-transmitted Prune dwarf virus in quarantine. Plant Pathol J. 2014;30:178–182. doi: 10.5423/PPJ.NT.10.2013.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YG, Rho JY. Development of a PCR diagnostic system for Iris yellow spot tospovirus in quarantine. Plant Pathol J. 2014;30:440–444. doi: 10.5423/PPJ.NT.06.2014.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.