Summary
Background
Polar bears (Ursus maritimus) are among those species most susceptible to the rapidly changing arctic climate, and their survival is of global concern. Despite this, little is known about polar bear species history. Future conservation strategies would significantly benefit from an understanding of basic evolutionary information, such as the timing and conditions of their initial divergence from brown bears (U. arctos) or their response to previous environmental change.
Results
We used a spatially explicit phylogeographic model to estimate the dynamics of 242 brown bear and polar bear matrilines sampled throughout the last 120,000 years and across their present and past geographic ranges. Our results show that the present distribution of these matrilines was shaped by a combination of regional stability and rapid, long-distance dispersal from ice-age refugia. In addition, hybridization between polar bears and brown bears may have occurred multiple times throughout the Late Pleistocene.
Conclusions
The reconstructed matrilineal history of brown and polar bears has two striking features. First, it is punctuated by dramatic and discrete climate-driven dispersal events. Second, opportunistic mating between these two species as their ranges overlapped has left a strong genetic imprint. In particular, a likely genetic exchange with extinct Irish brown bears forms the origin of the modern polar bear matriline. This suggests that interspecific hybridization not only may be more common than previously considered but may be a mechanism by which species deal with marginal habitats during periods of environmental deterioration.
Introduction
Recent and ongoing climatic changes across the northern latitudes, including elevated air temperatures, melting glacial ice, and rising sea levels, are reshaping the arctic ecosystem, with devastating consequences for arctic-adapted species. Among the most threatened of these is the polar bear, for which declining sea ice represents the greatest challenge to its survival [1]. Rapid declines in ice extent over the last 50 years have coincided with changes in the distribution, abundance, fecundity, and body size of polar bears [2]. Fewer sea-ice days mean longer open-water periods during summer, forcing polar bears onshore in search of food [1, 3] and leading to potentially dangerous interactions with humans [2]. An increase in terrestriality may also be a key driver for opportunistic interspecific breeding with brown bears, with several adult hybrid bears reported in the last five years [4].
Polar bears are proposed to have evolved from brown bears that became isolated on Siberian coastal enclaves as recently as the Middle Pleistocene, 800–150 thousand years ago (kya) [5]. Both morphological [6] and nuclear genetic data [7, 8] support a deep divergence between the two bear species, which show marked differences in dentition, integument, and physiology. The modern polar bear mitochondrial lineage, however, falls within the genetic diversity of brown bears; it is most closely related to those found in a population of brown bears from the Admiralty, Baranof, and Chichagof (ABC) islands of southeastern Alaska [9–13]. These data have been used to support the hypothesis that the morphological differences between the two species may be due to rapid adaptation of polar bears to their highly specialized arctic lifestyle [10, 14] and that the mitochondrial phylogeny reflects incomplete lineage sorting [14]. Alternatively, the sister relationship between ABC island brown bears and polar bears may be explained by recent hybridization between the two species and the introgression of a brown bear mitochondrial lineage into polar bears. Although both bears are strongly maternally philopatric, the geographic distribution of their matrilines is not stable through time [15], and lineage-wide extinctions may be frequent [16].
We used a spatially explicit Bayesian inference approach to examine different phylogeographic histories of brown bear and polar bear matrilines sampled over at least the last 100 kya. We incorporated data from every geographic location known to have been occupied by brown bears during the time since the estimated most recent common ancestor (MRCA) of modern polar bear matrilines (circa 45 kya; [10, 17]), including a previously unsampled Late Pleistocene population of brown bears from the vicinity of modern-day Britain and Ireland. We also included two recently published ancient polar bear matrilines, both believed to come from bones older than 110 kya [10, 14], and 51 mitochondrial sequences from modern and Holocene (up to 8000 years old) polar bears, resulting in a data set that spans the present geographic distributions of both bear species.
Results and Discussion
Late Pleistocene/Holocene Phylogeography of Brown and Polar Bears
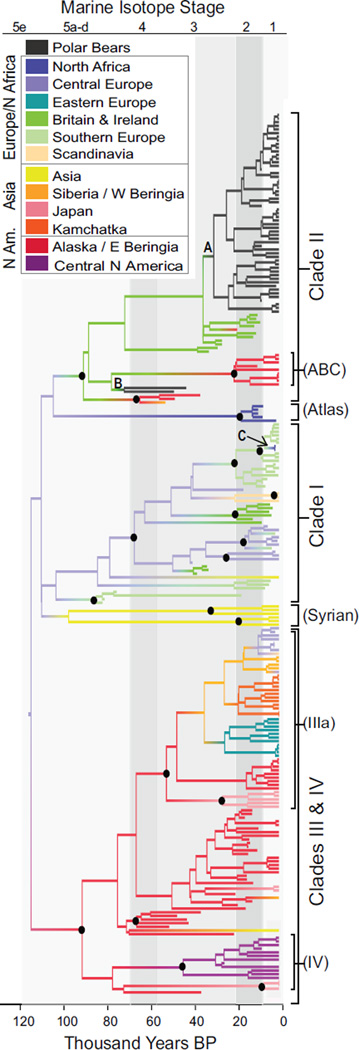
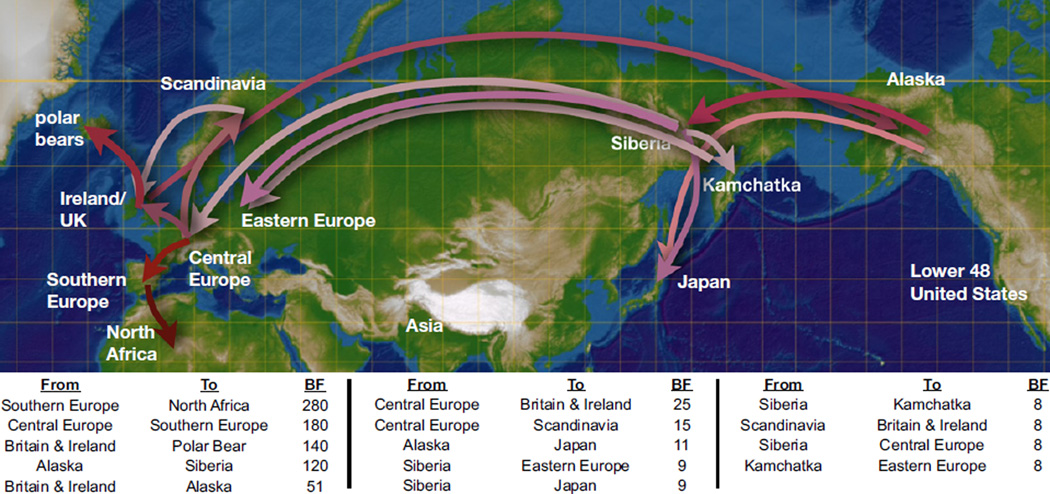
The recent history of polar bear and brown bear matrilines was characterized by long periods of geographic stability punctuated by episodes of rapid matrilineal dispersal, often over considerable geographic distances (Figure 1; Figure 2; see also Figure S1 available online). This is best illustrated by the inferred radiation of brown bears out of Alaska during the marine isotope stage (MIS) 3 interstadial (Bayes factor, BF [18], support for Alaska in comparison to any other location = 45), which was a period of peak environmental conditions for ice-age megafauna [19]. This rapid, long-distance radiation (clade IIIa, Figure 1) established clusters of closely related matrilines in Japan, Kamchatka, Siberia, and both Eastern and Central Europe by 14 kya.
Figure 1. Maximum Clade Credibility Genealogy Describing the Estimated Evolutionary History of Sampled Brown and Polar Bear Matrilines.
Maximum clade credibility (MCC) genealogy resulting from a phylogeographic BEAST [39] analysis of 242 brown bears and polar bears ranging in age from 120 thousand years ago (kya) to modern. Colors along the branches describe the most probable geographic location of each lineage. Black circles indicate major nodes with >85% posterior support, summarized from the combined output of three Markov chain Monte Carlo chains run for 150 million iterations each and sampled every 10,000 iterations. Letters A–C highlight nodes discussed in the main text. Background shades of gray indicate warm (light gray) and cool (dark gray) marine isotope stages. As noted previously [15, 16], the branching order among the earliest diverging branches is not well resolved, with the exception of very high support (99.97%) for monophyly of the clade III/IV lineage. See also Figure S1. Table S1 contains detailed information about the specimens used in this analysis.
Figure 2. Reconstructed Spatial Diffusion Pathways of Brown and Polar Bear Matrilines over the Last 120,000 Years.
Map indicating the 13 locations to which each matriline was assigned and the 14 significant diffusion pathways that describe the maternal phylogeographic history of brown bears and polar bears over the last circa 120 kya. For the phylogeographic analyses, we assigned all polar bears to a single geographic location, depicted here as Svalbard. Nonreversible diffusion rates are estimated across the entire distribution of posterior trees and therefore reflect average rates of diffusion over time. Rates are considered to be significantly different from zero with Bayes factor (BF) > 8. The significant diffusion pathways are shown in pink, with increasing significance indicated by darker shades and arrows indicating the direction of diffusion. An interactive visualization of the diffusion process of the ancestral bear matrilines over time is available at http://www.phylogeography.org/BEARS.html. Figure S2 describes the results of two sensitivity analyses performed to assess the phylogeographic results depicted here and in Figure 1.
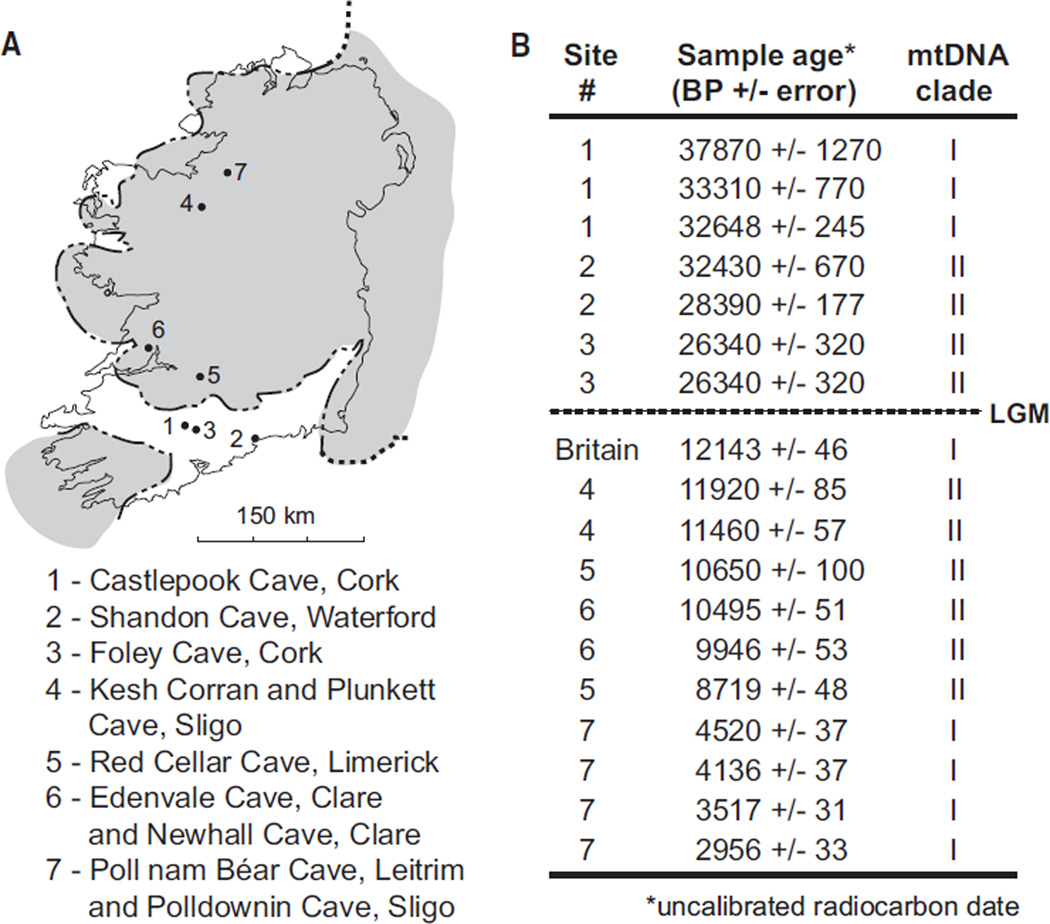
Clade II was much more geographically widespread during MIS 3 than it is today and comprised brown bears from Siberia, Alaska, and modern-day Britain and Ireland, as well as the two ancient polar bear matrilines (Figure 1). After the last glacial maximum (LGM), clade II was isolated to small regions of Alaska (ABC islands) and the vicinity of modern-day Ireland (Figure 1). Modern brown bears living on the ABC islands share a matrilineal common ancestor 37–10 kya (median 24 kya), a timing coincident with climatic changes surrounding the LGM that may have led to their isolation from brown bears living on the mainland. In Ireland, brown bears carrying the clade II matriline are present prior to 26 kya and again after 12 kya, but no bear fossils of any kind have been dated to the LGM (Table S2; Figure 3), suggesting that suitable habitat may not have persisted in Ireland through the peak of the last ice age.
Figure 3. Ages and Geographic Locations of the Sampled Fossil British and Irish Brown Bears.
Locations of origin (A) and uncalibrated accelerator mass spectrometry radiocarbon ages and mtDNA clade assignments (B) for British and Irish bears included in study. The shaded area in (A) indicates the traditional limit of the last glacial maximum (LGM) in Ireland. However, recent work suggests that the shaded area probably represents a subsequent ice sheet readvance occurring 20.9–14.7 kya [32] and that Ireland was completely covered in ice from circa 27–23 kya [29, 31]. Details of the Irish bear samples and ancient DNA sequencing are provided in Table S2.
The Origin and Evolution of the Polar Bear Matriline
As has been shown previously, all modern polar bear matrilines cluster within clade II (Figure 1). However, in contrast to previous analyses, we found no evidence for reciprocal monophyly between brown and polar bears within this clade, nor do our results support a sister relationship of polar bears with the ABC island brown bears. Instead, the inferred common matrilineal ancestor of modern polar bears falls within the genetic diversity of Irish brown bears (BF = 53). The low posterior support for modern polar bear monophyly (Figure 1, node A) is due to modern polar bear lineages frequently clustering into several clades within the diversity of clade II Irish brown bears in the posterior distribution of trees.
The two older polar bear matrilines, one isolated from a jawbone from Svalbard [10] associated with a stratigraphic layer dated to 130–110 kya [20] and another from a rib bone from northern Norway believed to date to circa 115 kya [14], also fall within clade II (Figure 1, node B). However, although these two sequences are closely related to each other, they are not directly ancestral to the modern polar bear matriline: modern polar bear matrilines cluster with the ancient polar bear matrilines with less than 5% posterior probability. If incomplete lineage sorting were to explain these results, as suggested previously [14], the coalescent event of the two lineages in question would have to have taken place prior to speciation. Because the coalescence of all modern polar bear matrilines postdates the age of the two ancient bears (and therefore the speciation date), hybridization followed by matrilineal introgression is much more likely than incomplete lineage sorting to explain the observed data.
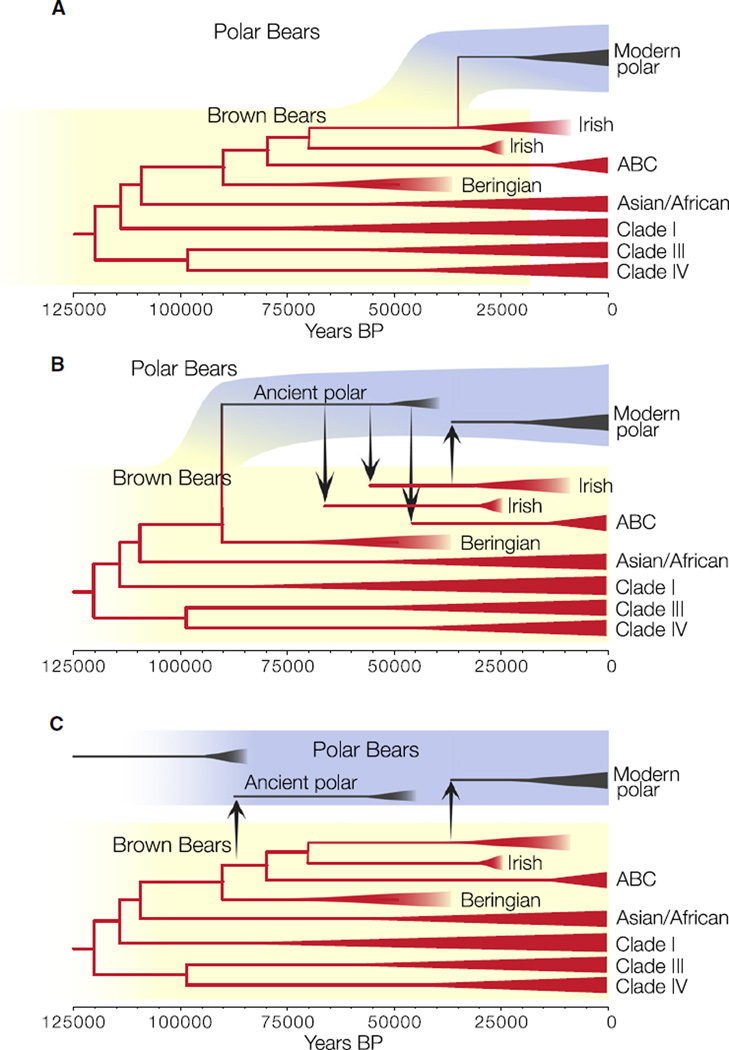
Figure 4 describes three hypothetical scenarios that may explain the phylogeographic relationships presented here. In the scenario depicted in Figure 4A, the timing of the initial divergence between brown bear and polar bear matrilines is the same as our estimated MRCA of modern polar bear matrilines. This scenario describes most closely the prevailing hypothesis explaining the mitochondrial data: a sister relationship between polar bear matrilines and those of ABC island brown bears. However, although it is consistent with conditions during the LGM favoring the emergence of a cold-adapted species, the timeline of this scenario conflicts with both the existence of the two ancient polar bears and the high degree of morphological and behavioral dissimilarity between brown bears and polar bears. This hypothesis could only be tenable if the ancient polar bear sequences contain errors that force them to fall erroneously outside of the diversity of the modern polar bears and the ancient Irish and ABC island brown bears.
Figure 4. Three Hypothetical Scenarios Describing the Divergence between Brown Bear and Polar Bear Matrilines.
(A) Recent speciation.
(B) Medium speciation plus multiple hybridization events.
(C) Ancient speciation plus more recent hybridization.
The branching order of the bear matrilines is based on the MCC tree in Figure 1. Colored background shading indicates the hypothetical polar bear (blue) and brown bear (yellow) autosomal lineages.
The scenario depicted in Figure 4B accommodates the two ancient polar bears by allowing brown bear and polar bear matrilines to diverge earlier than depicted in Figure 4A. In this scenario, the maternal MRCA of polar bears, Irish brown bears, and ABC island brown bears diverged from ancestral brown bears carrying the clade II matriline during the Pleistocene. Because of uncertainty surrounding the age of the two ancient polar bear fossils, we allow the age of both mitochondrial line-ages to be random variables drawn from a distribution ranging from the oldest bound on their stratigraphic dates (130 kya) to the most recent bound on their radiocarbon dates (40 kya) using a tip-sampling method [21]. The estimated age of the MRCA of clade II matrilines is 135–111 kya, roughly similar to but slightly younger than that estimated by either Lindqvist et al. [10] (152 kya) or Davison et al. [14] (160 kya). This most likely reflects differences between methods used to calibrate the molecular clock, because the incorporation of an external (fossil) calibration will result in a slower overall evolutionary rate [14]. In our analyses, the age of this particular node is highly influenced by the age of the ancient polar bears. When the ages of the two ancient polar bear sequences are sampled between 130–100 kya, rather than 130–40 kya, the estimated age of the MRCA of this node is 166–120 kya, overlapping with both our estimates using the less constrained age and the previous estimates.
If the Irish bears carrying clade II matrilines, or at least those Irish bears associated with radiocarbon ages postdating the LGM, were actually polar bears and not brown bears, then fewer hybridization events would be required to explain the observed data (in Figure 4B, only the transfer of the polar bear matriline to the ABC island brown bears would be required). However, bone isotopic data indicates that all Irish bears had a terrestrial diet similar to that of late Pleistocene brown bears from Alaska [15] and dissimilar to the markedly marine diet of polar bears (Supplemental Experimental Procedures). The scenario depicted in Figure 4B therefore predicts the following: an older polar bear matriline, represented in our analysis by the two ancient polar bears, became widespread across the Arctic. These polar bears hybridized opportunistically with small, coastal populations of brown bears, leading in at least two instances to fixation of the polar bear matriline in brown bear populations (Ireland and the ABC islands). More recently, additional hybridization occurred in Ireland, followed by fixation of the Irish brown bear matriline in polar bears. This marks the MRCA of the modern polar bear matriline.
A single bear from Fairbanks, Alaska whose mtDNA sequence was isolated as part of a previous study [15] also falls within the cluster of Irish brown bear and modern polar bear matrilines. The fossil from which this sequence was isolated is part of a large, geographically extensive collection of bones from across Alaska made by Otto Geist in the late 1930s that is now housed at the American Museum of Natural History in New York. Because the fossil was an ulna belonging to a juvenile, it could only be identified morphologically as Ursus. However, unlike the Irish brown bears, the isotopic signature of this Alaskan bear is decidedly marine [15], leading the authors of the original publication to conclude that the specimen was incorrectly provenanced and likely came from the coast of Alaska rather than from Fairbanks. Even if the provenance of this specimen is wrong, its radiocarbon age (19,360 ± 140 uncalibrated radiocarbon years before present; lab accession number OxA-10036) suggests that polar bears with clade II matrilines were present in Alaska throughout the LGM, providing additional support for the multiple-hybridization scenario depicted in Figure 4B and making it possible that more than one Irish matriline was captured by polar bears prior to the Holarctic fixation of the modern matriline.
In Figure 4C, brown bear and polar bear matrilines diverged prior to the mitochondrial diversification within brown bears. This allows more time for the evolution of each species’ distinctive phenotype and is consistent with hypotheses of at least a Middle Pleistocene origin for polar bears [5] but requires multiple hybridizations with brown bears to explain both the Svalbard polar bear matriline and the modern polar bear matriline. If this hypothesis is correct, autosomal markers should support a divergence between polar bears and brown bears prior to the divergence of all extant brown bears. To test this, we analyzed 20 nuclear loci isolated from all eight extant species within the Ursidae [7, 8, 22]. The small evolutionary distance between brown bears and polar bears prohibits precise estimates of the brown/polar divergence; however, the 95% highest posterior density interval of estimated coalescence dates spans the interval 2 Mya–400 kya. Although there are a number of reasons unrelated to hybridization why the mitochondrial MRCA would be more recent than most autosomal common ancestor dates, such as a smaller mitochondrial effective population size, this date range predates by a considerable margin the estimated MRCA of polar bear and brown bear matrilines.
Regardless of which scenario prevails, modern polar bears across their Holarctic distribution all share a common matrilineal ancestor within the last 51–20 kya. If polar bears were already widely distributed at this time, this suggests a complete replacement of the previous mitochondrial lineage within a remarkably short time frame. Such a recent coalescence is feasible given current estimates of their effective population size (Ne): if the mitochondrial haplotypes were selectively equivalent and assuming a female Ne of 2,025 based on an estimated 18% of females reproducing in a total census size of 10,000–12,500 females and a female generation time of 9.7 years [23], our sample of 51 modern mitochondrial alleles would coalesce with 95% probability in 93.2–12.9 kya (median 33.4 kya; Supplemental Experimental Procedures).
The timescale we present is derived from evolutionary rates calibrated using dated ancient DNA sequences; previous work has suggested that this may overestimate rates and therefore underestimate time since divergence [24]. However, recent evidence suggests that time dependency may not significantly affect rate estimates over relatively short time frames, such as those spanned by our genealogies [25]. Crucially, our phylogeographic estimates of the timing of establishment of four different brown bear mtDNA lineages coincide with paleontological evidence of the first bears in those locations, providing independent support for the accuracy of the tip-calibrated mtDNA evolutionary rates. Specifically, this includes the establishment of the clade III/IV lineage in Alaska (BF = 27) 105–70 kya [26], the first appearance of clade IIIa in eastern Europe by 24–5 kya (median 12 kya) [27], the recolonization of Scandinavia by bears 28–9 kya (median 11 kya) [27, 28], and the movement of bears carrying clade I from Spain into North Africa 8.5–1.6 kya (median 3.2 kya) (Figure 1, node C, which likely reflects translocation of bears by the Romans or Carthaginians for organized wild animal combat).
The Role of Hybridization in Polar Bear Evolution
Hybridization between brown bears and polar bears occurred during a period of rapid climate change, when fluctuations in the amount and distribution of habitat in the North Atlantic would have provided ample opportunity for their ranges to overlap, and therefore optimal conditions for opportunistic mating. These same rapid changes may have led to population size fluctuations, which in turn could contribute to the rapid fixation of introgressing mitochondrial lineages. The British-Irish ice sheet (BIIS) reached its maximum extent 22–20 kya, with major tidewater glaciers on the western shelf and down the Irish Sea basin into the Celtic Sea [29–31], providing an ideal habitat for polar bears. Pockets of ice-free refugia comprising open, treeless steppe tundra similar to that of Beringia may have remained in southern Ireland during the LGM [32, 33], or at least up until 26 kya, when our radiocarbon dates indicate that bears were still present in Ireland, and again after the initial deglaciation of the BIIS 24–23 kya [29, 34]. Because most of Ireland would have been uninhabitable even during the period leading up to the LGM (Figure 3), brown bears may have been forced to use transient, suitable habitat such as sea ice shelves, nunataks, or land exposed by lower sea levels, increasing the potential for contact with polar bears.
The modern distribution of bear matrilines likely arose as a consequence of long-term local stability punctuated by rapid, climate-driven dispersals and opportunistic mating between polar bears and brown bears. Today, the arctic climate is again changing rapidly, and the habitat of brown and polar bears is once again beginning to overlap, providing the opportunity for the two species to hybridize. Although the evolutionary role of hybridization is not yet completely understood, the recent proliferation of molecular genetic data is revealing a growing number of examples from both plants and animals [35]. Hybridization is known to occur more frequently in regions where population density is low, or where species are near the edge of their ecological range [36], and in some circumstances may provide the means to replace damaged alleles or to transfer novel traits between species, providing a fitness advantage to hybrid offspring [35].
Whereas vulnerable populations of both bear species are currently protected, the protection status of hybrids is less clear, with a 1996 proposed US policy to protect hybrids [37] yet to be finalized. Although the extent of any fitness differential between hybrid brown/polar bears and their parents remains unclear, given the increasing evidence of hybridization among many threatened arctic taxa [4], it may be appropriate to reconsider protection of hybrids, because they may play an underappreciated role in the survival of species. Our results suggest that, although the genetic mixing observed in bears today may be an important component of the long-term evolution of the polar bear, brown and polar bears have remained evolutionarily distinct lineages over geological time, suggesting that they are likely to remain as such in the future.
Experimental Procedures
Data Collection
We extracted DNA from 23 ancient Irish bears, 30 historic polar bears, four early Holocene (circa 8 kya) polar bears, and 17 modern polar bears (Table S1; Table S2). Fragments of the mtDNA hypervariable region (HVR) were selected and amplified so as to overlap with previously published ancient and modern matrilines (Supplemental Experimental Procedures). Matrilineal clade assignments for ancient Irish bears were verified by amplifying an additional fragment from the mitochondrial cytochrome b (cytb) gene. Protocols to control for contamination and validate ancient results were followed at each step. Mitochondrial sequence data have been deposited at GenBank with the accession numbers JF900098–JF900175. Additional HVR sequences were collected from GenBank to compile a data set of 242 brown and polar bear matrilines representing their entire geographic distribution over the last 120,000 years. Twenty nuclear loci were also collected from GenBank for each of the eight extant species of Ursidae.
Model Development
We extended a recent approach to infer the spatial dynamics of measurably evolving populations [38] that now allows for different rates of diffusion between locations depending on the direction traveled, thereby providing a more realistic approximation of the geographic diffusion process of large mammals through time. Our novel approach employs a continuous-time Markov chain (CTMC) model in which diffusion between n locations is characterized by an n × n rate matrix, Λ, that contains n(n − 1) off-diagonal, nonnegative rate parameters λij for i, j = 1,…,n. Previously, phylogenetic modeling (e.g., [38]) assumed that λij = λji, or at least that Λ is similar to a symmetric matrix, as in, for example, the HKY and GTR models.
For our general case, it is possible to decompose Λ = V D V−1, where matrix V is a set of real eigenvectors and matrix D is of block diagonal form with 1 × 1 submatrices of real eigenvalues and 2 × 2 submatrices of complex conjugate pairs of eigenvalues. To draw inference under this extended model, we devised new algorithms to compute the likelihood of the observed locations at the tips of the tree by determining the CTMC finite-time transition probabilities exp(Λt) for real times t along each branch. Because the amount of phylogeographic data is small, we adopted a Bayesian stochastic search variable selection (BSSVS) procedure to select among all possible migration pathway scenarios [38]. We provide details of this model and its implementation in BEAST [39] with BEAGLE [40] (Supplemental Experimental Procedures).
Phylogenetic Analyses
For the phylogeographic analyses, we ran two independent Markov chain Monte Carlo (MCMC) simulations of 50 million iterations using the model described above for two different data sets: (1) using just the HVR and (2) incorporating complete mtDNA genomes for ten bears. We assumed the HKY+G model of nucleotide substitution and either the flexible Bayesian skyride [41] or Bayesian skyline [42] coalescent priors [41] (Supplemental Experimental Procedures). Mean accelerator mass spectrometry radiocarbon dates for ancient specimens calibrated the molecular clock. Six ancient specimens originating from underrepresented geographic regions were included: one from Siberia [15], three from Valdegoba Cave, Spain [43], and two ancient polar bears from Norway [10, 14]. For each of these, the age of the sequences was assumed a priori to arise from a lognormal distribution reflecting the most likely age range of that specimen. Tip ages were numerically integrated via sampling in the MCMC chain for each of these six specimens simultaneously with evolutionary and demographic parameters [21]. Initially, we performed analyses with and without the post-mortem damage (PMD) model [44] to assess the influence of potentially damaged nucleotides sites on the phylogenetic analysis. A BF comparison [18] indicated that the PMD model did not improve the model fit, and we excluded the PMD model from further analyses.
In all MCMC simulations, we subsampled the posterior realizations every 10,000 iterations to decrease autocorrelation and discarded the first 10% as chain burn-in. For a given data set, we combined remaining samples from all chains to draw inference. Effective sample size estimates and parameter trace plots supported MCMC convergence. We computed the summary maximum clade credibility (MCC) tree (Figure 1) using TreeAnnotator and developed a BF test [18] to identify which diffusion links were statistically significant (BF > 8; Figure 2).
We performed two different sensitivity analyses that are described in detail in Supplemental Experimental Procedures. First, to investigate the impact of unsampled populations on the Irish-polar bear phylogeographic link, we simulated unknown (cryptic) lineages in clade II and measured relevant BF support for the conclusion of Irish ancestry of modern polar bear sequences (Figure S2A). Second, we explored the effect of sampling heterogeneity on root state probabilities using location state randomization (Figure S2B).
To provide a spatial projection, we converted the MCC tree, inferred posterior modal node locations, and median node heights into a keyhole markup language (KML) file. The resulting interactive visualization is available from http://www.phylogeography.org/BEARS.html.
To compare nuclear and mitochondrial species topologies and to further test the hypothesis that polar bear matrilines are recently derived from a subset of those in brown bears, we performed additional analyses using up to 20 nuclear loci (Supplemental Experimental Procedures). For these analyses, we assumed a Yule prior to inform the speciation rate. We assumed both strict and relaxed clocks and calibrated the phylogenies by drawing the age of the radiation of the Ursinae from a lognormal distribution with 95% of the probability density between 5 and 3 Mya [45].
Supplementary Material
Acknowledgments
We thank E. Regehr, T. Evans, the US Fish and Wildlife Service, and the Marine Mammals Tissue Bank for providing modern polar bear samples for analysis and G. Zazula for providing ancient samples. C.J.E. was supported by the Irish Research Council for Science Engineering and Technology (SC/2002/510 and 04/BR/B0390). M.A.S. is partially supported by National Institutes of Health (NIH) grant R01 GM086887 and a John Simon Guggenheim Memorial Fellowship. P.L. is supported by the Fund for Scientific Research (FWO) Flanders. B.S. and T.L.F. are supported by NIH R01 GM083603-01 and National Science Foundation (NSF) ARC-0909456. The research leading to these results received funding from the European Research Council under the Seventh Framework Programme (FP7/2007- 2013)/ERC grant agreement number 260864. Collaboration between B.S., M.A.S., A.R., and P.L. was supported by the National Evolutionary Synthesis Center (NESCent) grant NSF EF-0423641.
Footnotes
Accession Numbers
New brown and polar bear mitochondrial sequence data have been deposited at GenBank with the accession numbers JF900098–JF900175.
Supplemental Information
Supplemental Information includes two figures, two tables, Supplemental Experimental Procedures, and Supplemental Acknowledgments and can be found with this article online at doi:10.1016/j.cub.2011.05.058.
References
- 1.Amstrup SC, Marcot BG, Davis DC. Forecasting the Range-wide Status of Polar Bears at Selected Times in the 21st Century. Reston, VA: US Geological Survey Administrative Report; 2007. [Google Scholar]
- 2.Derocher AE, Lunn NJ, Stirling I. Polar bears in a warming climate. Integr. Comp. Biol. 2004;44:163–176. doi: 10.1093/icb/44.2.163. [DOI] [PubMed] [Google Scholar]
- 3.Stirling I, Richardson E, Thiemann GW, Derocher AE. Unusual predation attempts of polar bears on ringed seals in the southern Beaufort Sea: Possible significance of changing spring ice conditions. Arctic. 2008;61:14–22. [Google Scholar]
- 4.Kelly BP, Whiteley A, Tallmon D. The Arctic melting pot. Nature. 2010;468:891. doi: 10.1038/468891a. [DOI] [PubMed] [Google Scholar]
- 5.Kurtén B. The evolution of the polar bear, Ursus maritimus (Phipps) Acta Zool. Fenn. 1964;108:1–30. [Google Scholar]
- 6.Slater GJ, Figueirido B, Louis L, Yang P, Van Valkenburgh B. Biomechanical consequences of rapid evolution in the polar bear lineage. PLoS ONE. 2010;5:e13870. doi: 10.1371/journal.pone.0013870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pagès M, Calvignac S, Klein C, Paris M, Hughes S, Hänni C. Combined analysis of fourteen nuclear genes refines the Ursidae phylogeny. Mol. Phylogenet. Evol. 2008;47:73–83. doi: 10.1016/j.ympev.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 8.Yu L, Li QW, Ryder OA, Zhang YP. Phylogeny of the bears (Ursidae) based on nuclear and mitochondrial genes. Mol. Phylogenet. Evol. 2004;32:480–494. doi: 10.1016/j.ympev.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Cronin MA, Amstrup KS, Garner GW, Vyse ER. Interspecific and intraspecific mitochondrial DNA variation in North American bears (Ursus) Can. J. Zool. 1991;69:2985–2992. [Google Scholar]
- 10.Lindqvist C, Schuster SC, Sun Y, Talbot SL, Qi J, Ratan A, Tomsho LP, Kasson L, Zeyl E, Aars J, et al. Complete mitochondrial genome of a Pleistocene jawbone unveils the origin of polar bear. Proc. Natl. Acad. Sci. USA. 2010;107:5053–5057. doi: 10.1073/pnas.0914266107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shields GF, Adams D, Garner G, Labelle M, Pietsch J, Ramsay M, Schwartz C, Titus K, Williamson S. Phylogeography of mitochondrial DNA variation in brown bears and polar bears. Mol. Phylogenet. Evol. 2000;15:319–326. doi: 10.1006/mpev.1999.0730. [DOI] [PubMed] [Google Scholar]
- 12.Shields GF, Kocher TD. Phylogenetic relationships of North American ursids based on analysis of mitochondrial DNA. Evolution. 1991;45:218–221. doi: 10.1111/j.1558-5646.1991.tb05279.x. [DOI] [PubMed] [Google Scholar]
- 13.Talbot SL, Shields GF. Phylogeography of brown bears (Ursus arctos) of Alaska and paraphyly within the Ursidae. Mol. Phylogenet. Evol. 1996;5:477–494. doi: 10.1006/mpev.1996.0044. [DOI] [PubMed] [Google Scholar]
- 14.Davison J, Ho SYW, Bray S, Korsten M, Vulla E, Hindrikson M, Østbye K, Østbye E, Lauritzen S-E, Austin J, et al. Late-Quaternary biogeographic scenarios for a wild mammal model species, the brown bear (Ursus arctos) Quat. Sci. Rev. 2011;30:418–430. [Google Scholar]
- 15.Barnes I, Matheus P, Shapiro B, Jensen D, Cooper A. Dynamics of Pleistocene population extinctions in Beringian brown bears. Science. 2002;295:2267–2270. doi: 10.1126/science.1067814. [DOI] [PubMed] [Google Scholar]
- 16.Calvignac S, Hughes S, Tougard C, Michaux J, Thevenot M, Philippe M, Hamdine W, Hänni C. Ancient DNA evidence for the loss of a highly divergent brown bear clade during historical times. Mol. Ecol. 2008;17:1962–1970. doi: 10.1111/j.1365-294X.2008.03631.x. [DOI] [PubMed] [Google Scholar]
- 17.Ho SY, Saarma U, Barnett R, Haile J, Shapiro B. The effect of inappropriate calibration: Three case studies in molecular ecology. PLoS ONE. 2008;3:e1615. doi: 10.1371/journal.pone.0001615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suchard MA, Weiss RE, Sinsheimer JS. Bayesian selection of continuous-time Markov chain evolutionary models. Mol. Biol. Evol. 2001;18:1001–1013. doi: 10.1093/oxfordjournals.molbev.a003872. [DOI] [PubMed] [Google Scholar]
- 19.Zazula GD, Froese DG, Schweger CE, Mathewes RW, Beaudoin AB, Telka AM, Harington CR, Westgate JA. Palaeobotany: Ice-age steppe vegetation in east Beringia. Nature. 2003;423:603. doi: 10.1038/423603a. [DOI] [PubMed] [Google Scholar]
- 20.Ingólfsson Ó, Wiig Ø. Late Pleistocene fossil find in Svalbard: The oldest remains of a polar bear (Ursus maritimus Phipps, 1744) ever discovered. Polar Res. 2009;28:455–462. [Google Scholar]
- 21.Shapiro B, Ho SY, Drummond AJ, Suchard MA, Pybus OG, Rambaut A. A Bayesian phylogenetic method to estimate unknown sequence ages. Mol. Biol. Evol. 2011;28:879–887. doi: 10.1093/molbev/msq262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakagome S, Pecon-Slattery J, Masuda R. Unequal rates of Y chromosome gene divergence during speciation of the family Ursidae. Mol. Biol. Evol. 2008;25:1344–1356. doi: 10.1093/molbev/msn086. [DOI] [PubMed] [Google Scholar]
- 23.Cronin MA, Amstrup SC, Talbot SL, Sage GK, Amstrup KS. Genetic variation, relatedness, and effective population size of polar bears (Ursus maritimus) in the southern Beaufort Sea Alaska. J. Hered. 2009;100:681–690. doi: 10.1093/jhered/esp061. [DOI] [PubMed] [Google Scholar]
- 24.Ho SY, Kolokotronis SO, Allaby RG. Elevated substitution rates estimated from ancient DNA sequences. Biol. Lett. 2007;3:702–705. doi: 10.1098/rsbl.2007.0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burridge CP, Craw D, Fletcher D, Waters JM. Geological dates and molecular rates: Fish DNA sheds light on time dependency. Mol. Biol. Evol. 2008;25:624–633. doi: 10.1093/molbev/msm271. [DOI] [PubMed] [Google Scholar]
- 26.Kurtén B, Anderson E. Pleistocene Mammals of North America. New York: Columbia University Press; 1980. [Google Scholar]
- 27.Sommer R, Benecke N. The recolonisation of Europe by brown bears Ursus arctos Linnaeus 1758 after the last glacial maximum. Mammal Rev. 2005;35:156–164. [Google Scholar]
- 28.Rinterknecht VR, Clark PU, Raisbeck GM, Yiou F, Bitinas A, Brook EJ, Marks L, Zelcs V, Lunkka JP, Pavlovskaya IE, et al. The last deglaciation of the southeastern sector of the Scandinavian ice sheet. Science. 2006;311:1449–1452. doi: 10.1126/science.1120702. [DOI] [PubMed] [Google Scholar]
- 29.Clark CD, Hughes ALC, Greenwood SL, Jordan C, Sejrup HP. Pattern and timing of retreat of the last British-Irish Ice Sheet. Quat. Sci. Rev. 2010 in press. Published online October 28, 2010. [Google Scholar]
- 30.Coxon P, MacCarron SG. In: The Geology of Ireland. Second Edition. Holland H, Sanders IA, editors. Dunedin, UK: Dunedin Academic Press; 2009. pp. 355–396. [Google Scholar]
- 31.O’Cofaigh C, Telfer MW, Bailey RM, Evans DJA. Late Pleistocene chronostratigraphy and ice sheet limits, southern Ireland. Quat. Sci. Rev. 2010 in press. Published online April 8, 2010. [Google Scholar]
- 32.Clark CD, Gibbard PL, Rose J. Pleistocene glacial limits in England, Scotland and Wales. In: Ehlers J, Gibbard PL, editors. Quaternary Glaciations: Extent and Chronology, Part 1: Europe. Amsterdam: Elsevier; 2004. pp. 47–82. [Google Scholar]
- 33.Stewart JR, Lister AM, Barnes I, Dalén L. Refugia revisited: Individualistic responses of species in space and time. Proc. Biol. Sci. 2010;277:661–671. doi: 10.1098/rspb.2009.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiverrell RC, Thomas GSP. Extent and timing of the last glacial maximum (LGM) in Britain and Ireland: A review. J. Quat. Sci. 2010;25:535–549. [Google Scholar]
- 35.Rieseberg LH. Evolution: Replacing genes and traits through hybridization. Curr. Biol. 2009;19:R119–R122. doi: 10.1016/j.cub.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 36.Arnold ML. Natural Hybridization and Evolution. Oxford: Oxford University Press; 1997. [Google Scholar]
- 37.United States Fish and Wildlife Service and National Oceanic and Atmospheric Administration. Endangered and threatened wildlife and plants; Proposed policy and proposed rule on the treatment of intercrosses and intercross progeny (“hybridization”) Fed. Regist. 1996;61:4710–4713. [Google Scholar]
- 38.Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suchard MA, Rambaut A. Many-core algorithms for statistical phylogenetics. Bioinformatics. 2009;25:1370–1376. doi: 10.1093/bioinformatics/btp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minin VN, Bloomquist EW, Suchard MA. Smooth skyride through a rough skyline: Bayesian coalescent-based inference of population dynamics. Mol. Biol. Evol. 2008;25:1459–1471. doi: 10.1093/molbev/msn090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- 43.Valdiosera CE, García N, Anderung C, Dalén L, Crégut-Bonnoure E, Kahlke RD, Stiller M, Brandström M, Thomas MG, Arsuaga JL, et al. Staying out in the cold: Glacial refugia and mitochondrial DNA phylogeography in ancient European brown bears. Mol. Ecol. 2007;16:5140–5148. doi: 10.1111/j.1365-294X.2007.03590.x. [DOI] [PubMed] [Google Scholar]
- 44.Rambaut A, Ho SY, Drummond AJ, Shapiro B. Accommodating the effect of ancient DNA damage on inferences of demographic histories. Mol. Biol. Evol. 2009;26:245–248. doi: 10.1093/molbev/msn256. [DOI] [PubMed] [Google Scholar]
- 45.Wayne RK, Van Valkenburgh B, O’Brien SJ. Molecular distance and divergence time in carnivores and primates. Mol. Biol. Evol. 1991;8:297–319. doi: 10.1093/oxfordjournals.molbev.a040651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.