Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is a common form of muscular dystrophy in adults that is foremost characterized by progressive wasting of muscles in the upper body. FSHD is associated with contraction of D4Z4 macrosatellite repeats on chromosome 4q35 but this contraction is pathogenic only in certain “permissive” chromosomal backgrounds. Here we show that FSHD patients carry specific single nucleotide polymorphisms (SNPs) in the chromosomal region distal to the last D4Z4 repeat. This FSHD-predisposing configuration creates a canonical polyadenylation signal for transcripts derived from DUX4, a double homeobox gene of unknown function that straddles the last repeat unit and the adjacent sequence. Transfection studies revealed that DUX4 transcripts are efficiently polyadenylated and are more stable when expressed from permissive chromosomes. These findings suggest that FSHD arises through a toxic gain of function attributable to the stabilized distal DUX4 transcript.
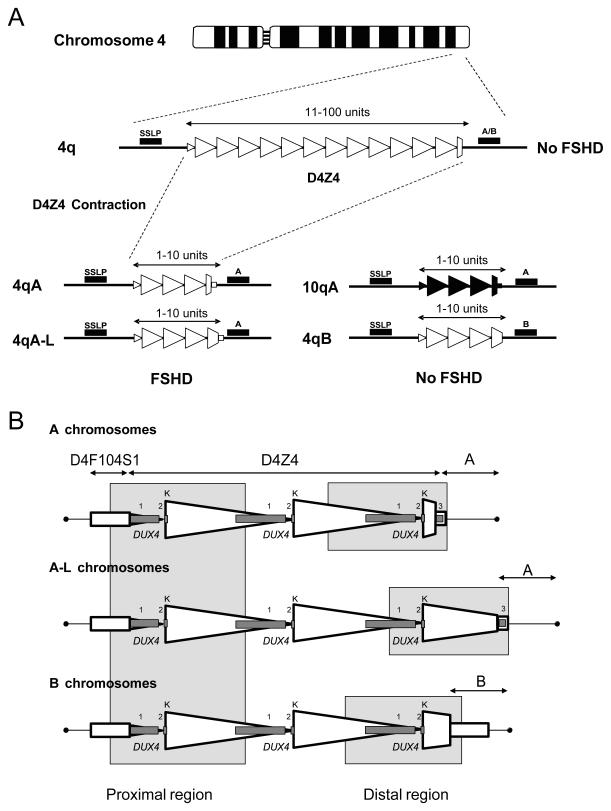
Autosomal dominant FSHD (FSHD1; OMIM 158900) is a common form of muscular dystrophy, affecting 1 in 20,000 people, that is characterized by progressive and often asymmetric weakness and wasting of facial, shoulder girdle and upper arm muscles (1). The disorder is most often caused by contraction of the D4Z4 macrosatellite repeat array in the subtelomeric region of chromosome 4q35 (2). This polymorphic macrosatellite repeat normally consists of 11-100 D4Z4 units, each 3.3 kb in size and ordered head-to-tail. Patients with FSHD1 have one repeat array of 1-10 units (Fig 1A). At least one unit of D4Z4 is required to develop FSHD (3).
Fig. 1.
Schematic overview showing the minimal genetic requirement for FSHD1. (A) The D4Z4 repeat array on chromosome 4q35 (open triangles) and its homologue on chromosome 10q26 (closed triangles), indicating the location of the simple sequence length polymorphism (SSLP) and 4qA/4qB polymorphisms that define the genetic background of the repeat. Patients with FSHD1 have a D4Z4 repeat array size of 1-10 units on 4qA but not on 4qB or 10q chromosomes. (B) Schematic of the D4Z4 repeat and flanking sequences on A, A-L (with an extended distal D4Z4 repeat unit) and B chromosomes. Each D4Z4 unit is defined by the KpnI restriction site (K). The proximal and distal regions that were sequenced are indicated. The exons of DUX4 are indicated as grey boxes numbered 1-3.
D4Z4 contraction needs to occur on a specific chromosomal background to cause FSHD. The chromosome 10q subtelomere contains an almost identical repeat array but contractions on this chromosome are non-pathogenic (Fig 1A). Translocated copies of the chromosome 4 and chromosome 10 repeat units are frequently encountered on either chromosome end (4). This complex genetic situation in which genetically almost identical repeat units can be exchanged between both chromosomes with apparently discordant pathological consequences has long hampered the identification of the disease mechanism.
Disease models were postulated in which D4Z4 repeat contractions cause chromatin remodeling and transcriptional deregulation of genes close to D4Z4. Indeed, contracted D4Z4 repeat arrays show partial loss of DNA methylation and of heterochromatic histone 3 lysine 9 trimethylation and heterochromatin protein 1γ markers consistent with a more open chromatin structure (5, 6). Transcriptional upregulation of genes proximal to D4Z4 was reported in FSHD1 patients (7), but could not be confirmed (8, 9).
Exchanges between repeat units of chromosomes 4 and 10 occur much less frequently than anticipated: most transclocated repeat units are relicts of ancient translocation events between chromosomes 4q and 10q (10). Of the two distal chromosome 4q configurations, 4qA and 4qB, only contractions of the 4qA form led to FSHD1 (11). Genetic follow-up studies unveiled consistent polymorphisms in the FSHD locus resulting in the recognition of at least 17 genetic variants of distal 4q (10). Contractions in the common variant 4A161 cause FSHD1, while contractions in many other variants such as the common 4B163 do not cause FSHD1 (Fig. 1A) (12). Thus it appears that chromosome 4A161 specific sequence variants are causally related to FSHD.
Because at least one D4Z4 unit is necessary to cause disease, we reasoned that the minimal pathogenic region might reside in the first or the last unit. The distal unit of the D4Z4 repeat was recently shown to have a transcriptional profile that differs from internal units (13, 14). While the major transcript in each unit is the DUX4 gene, which codes for a double homeobox protein, none of these transcripts seem to be stable, probably due to the absence of a polyadenylation signal in internal D4Z4 units. Spliced and unspliced transcripts of the DUX4 gene in the last unit, however, use a unique 3′ untranslated region (UTR) in the pLAM region (15) which is immediately distal to this last unit (Fig. 1B) and which contains a poly(A) signal that presumably stabilizes this distal transcript (13, 14). The DUX4 transcript of the distal D4Z4 unit encompasses two facultative introns in the 3′UTR. When expressed in C2C12 muscle cells, DUX4 causes a phenotype compatible with molecular observations in FSHD (16). This distal DUX4 transcript can be observed in FSHD1 myotubes but not in control myotubes (Fig. S1) (17).
To investigate why the 4A161 chromosome is permissive for disease, we compared the sequence of the 4A161 chromosome with that of common, non-permissive 4B163 and 10A166 chromosomes. We could not identify a sequence signature in the proximal D4Z4 unit of the repeat array that explained the permissiveness of the 4A161 chromosome (Fig. S2). However, immediately distal to D4Z4, in the adjacent pLAM sequence, we found a polymorphism potentially affecting polyadenylation of the distal DUX4 transcript. The DUX4 poly(A) signal ATTAAA, which is commonly used in humans (18), is present on the permissive 4A161 chromosome, while the corresponding ATCAAA sequence on chromosome 10q is not known to be a poly(A) signal (Fig. S2). Non-permissive 4qB chromosomes, like 4B163, lack pLAM altogether, including this poly(A) site (Fig. 1B). Another non-permissive 10qA chromosome (10A176T) (10) carries ATTTAA at this position which is also not known as a poly(A) signal (Figs. S2 and S3). In silico poly(A) signal prediction programs (19, 20) also recognized the DUX4 poly(A) signal in 4A161 but failed to identify potential poly(A) signals in non-permissive chromosomes 10A166, and 10A176T.
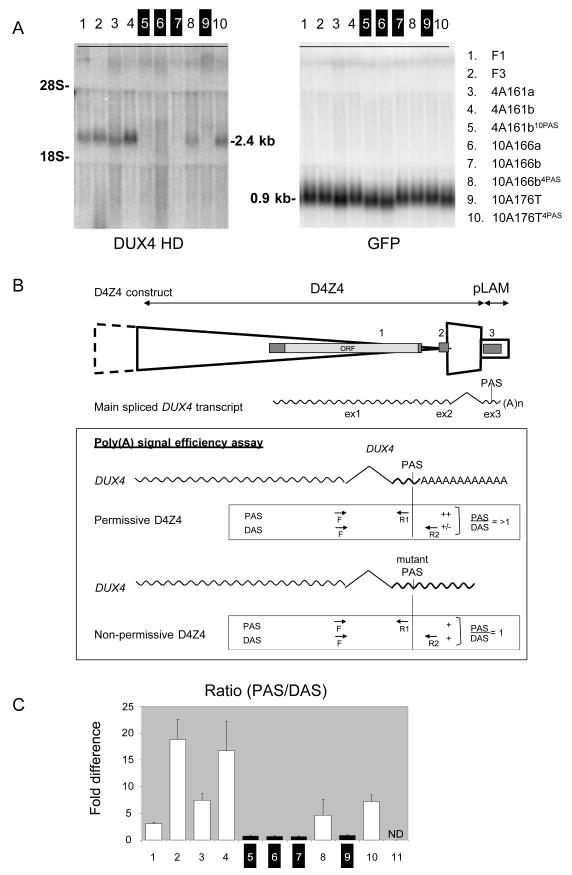
To explore whether these polymorphisms affect the distal DUX4 transcript, we transfected the last D4Z4 unit and flanking pLAM sequence of permissive and non-permissive chromosomes in C2C12 cells and assessed the stability of the distal DUX4 transcript by Northern blot analysis (Fig. 2A). We also examined the relative potency of the poly(A) signals on the permissive and non-permissive chromosomes in directing polyadenylation of the distal DUX4 transcript. We studied polyadenylation site usage indirectly by using a Q-RT-PCR assay (21) in which we compared DUX4 transcript levels proximal and distal of the poly(A) site (Fig. 2B). The use of the predicted poly(A) signal was verified by 3′RACE (Fig. S4). We also transfected constructs in which the poly(A) signal of permissive chromosomes was replaced by those of non-permissive chromosomes, and vice versa. We found that DUX4 transcripts were stable (Fig. 2A) and efficiently polyadenylated (Fig. 2C) when we used constructs from permissive chromosomes or when the poly(A) signal of a permissive chromosome was introduced on constructs derived from non-permissive chromosomes. Consistently, when constructs derived from non-permissive chromosomes were transfected, no DUX4 transcripts could be detected on Northern blot and polyadenylation was inefficient. DUX4 stability and polyadenylation efficiency decreased when the poly(A) signal of permissive constructs was replaced by non-permissive sequences. Altogether, constructs with a bona fide poly(A) signal produced stable transcripts and showed 4-16 fold higher polyadenylation efficiency than constructs with a mutation in the poly(A) signal. This suggests that increased polyadenylation, and hence stability, of the distal DUX4 transcript may be centrally involved in FSHD pathogenesis.
Fig. 2.
DUX4 expression analysis after transfection of the distal D4Z4 unit and flanking pLAM sequence into C2C12 mouse myoblast cells. (A) Northern blot analysis of C2C12 mouse myoblast cells transfected with genomic constructs (see fig 2B) derived from permissive (lanes 1-4) or non-permissive chromosomes (lanes 6, 7 and 9) or constructs in which the poly (A) signals from non-permissive chromosomes are replaced by those from permissive chromosomes (4PAS; lanes 8 and 10) and vice versa (10mPAS; lane 5). Only constructs with canonical poly (A) signals show a DUX4 transcript. Co-transfected EGFP gene was used as a control. (B) Schematic of the transfected sequence including the distal D4Z4 unit and flanking pLAM sequence, The DUX4 gene and the poly(A) signal are part of the transfected sequence. The main spliced DUX4 transcript and its open reading frame (ORF) are indicated. The primers used to quantify polyadenylation efficiency are indicated. The boxed area shows the design of the assay to infer the polyadenylation efficiency of the DUX4 poly(A) signal. The forward primer straddles exons 2 and 3 and is therefore specific for the spliced DUX4 transcript. The primer set Proximal to the poly(A) Signal (PAS) and a primer set using the same forward primer but a reverse primer Distal to the poly(A) Signal (DAS) are indicated as well as the expected PAS/DAS ratios. C) Bar diagram with the observed PAS/DAS ratios of permissive chromosomes (lanes 3 and 4), non-permissive chromosomes (lanes 6, 7 and 9), and permissive chromosomes in which the poly(A) signal is replaced for a sequence derived from a non-permissive chromosome (10mPAS; lane 5) or vice versa (4PAS; lanes 8 and 10), or of pathogenic chromosomes derived from families F1 and F3 (lanes 1 and 2). Lane 11 is negative control. ND = not detected Bars represent values of quadruple experiments with standard errors of the mean.
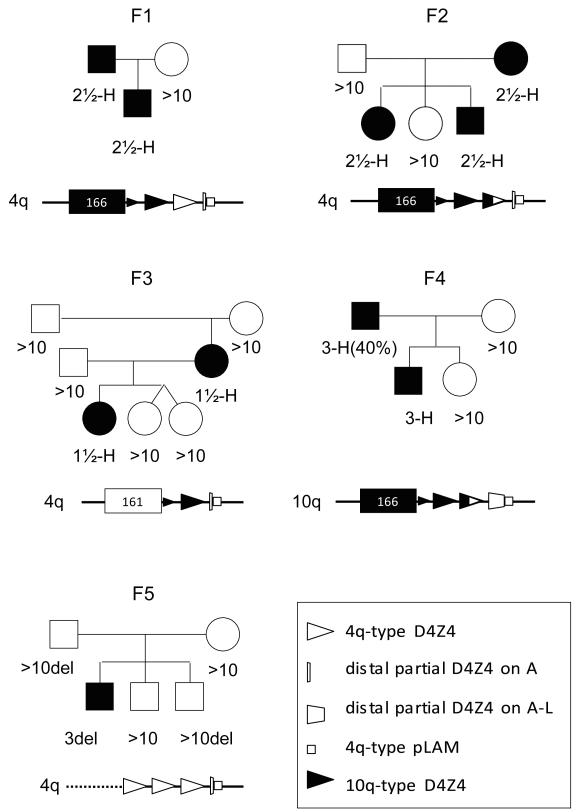
We next studied FSHD1 patients with unusual hybrid D4Z4 repeat array structures that contain mixtures of 4-type and 10-type units. We identified four families (F1-F4) with one or more individuals with FSHD1, carrying a contracted D4Z4 repeat array that commences with 10-type units, and ends with 4-type units (Fig. 3). In family F3 we identified a patient with a de novo meiotic rearrangement between chromosomes 4q and 10q leaving one and a half 10-type repeat unit on a permissive 4A161 chromosome. In family F4 the mildly affected father is a mosaic FSHD1 patient (22) due to a mitotic contraction of such hybrid repeat array. The mosaic pathogenic repeat starts with two and a half 10-type D4Z4 unit and ends with one and a half 4-type repeat unit. This repeat array in the father was transmitted to his affected son demonstrating its pathogenicity and, surprisingly, it was found to reside on chromosome 10 (Fig. S6). Only the distal end of the D4Z4 repeat array was transferred to chromosome 10q so that none of the FSHD candidate genes located proximal to the D4Z4 repeat array were co-transferred to chromosome 10 (Fig. S6). This report of a FSHD1 family linked to chromosome 10 apparently precludes a key role for proximal 4q genes in the pathogenesis of FSHD. Altogether, all unusual FSHD1-causing repeat arrays reported here thus share the commonality of a terminal 4qA repeat unit with a directly adjacent pLAM sequence.
Fig. 3.
Pedigrees of FSHD1 families with complex pathogenic chromosomes. Families F1-F4 all carry a hybrid D4Z4 repeat that commences with chromosome 10-type repeat units (closed triangles), but end with 4-type repeat units (open triangles). In family F3 a meiotic rearrangement between chromosomes 4 and 10 generated a short hybrid repeat structure on 4A161. In family F4, this pathogenic repeat is located on chromosome 10 and originates from a mitotic D4Z4 contraction in the mildly affected father which is transmitted to his affected son. Family F5 represents a disease chromosome in which in addition to partial deletion of the D4Z4 repeat, the region proximal to the D4Z4 repeat is also deleted.
We also analyzed other disease permissive chromosome 4 variants (Fig. S7): 4A161L was previously described (10, 15), while 4A159 and 4A168 are newly discovered uncommon permissive variants from a survey of >300 independent patients with FSHD. In addition, we studied >2,000 control individuals and identified additional non-permissive chromosome variants: 4B168, 10A164 and 10B161T (Fig. S3). Thus D4Z4 contractions on 4A161, 4A161L, 4A159 and 4A168 chromosomes are pathogenic, while D4Z4 contractions on 4B163, 4B168, 10A166, 10A164, 10B161T and 10A176T chromosomes are non-pathogenic.
We sequenced the first and last D4Z4 units and flanking sequences in these newly identified permissive and non-permissive chromosomes. (Figs. 1B and S2). In support of our earlier data, there is no common sequence in the proximal D4Z4 region that unifies FSHD permissive chromosomes. At the distal end all permissive chromosomes differed very little in sequence and all contained a canonical DUX4 poly(A) signal, while non-permissive chromosomes, showed much more sequence variation relative to the permissive chromosomes. The only exception, 4B163, has a D4Z4 sequence highly identical to 4A161 but, importantly, lacks the pLAM sequence (Fig. 1). The permissive 4A161L chromosome is identical to 4A161 but carries an extended D4Z4 sequence preceding an identical pLAM sequence (Figs. 1B and S2). Sequence analysis of the distal D4Z4-pLAM region of the pathogenic chromosome in our four families with complex repeat array structures showed a sequence identical to the permissive 4A161 sequence. Transfection experiments with D4Z4-pLAM sequences derived from the disease chromosomes of families F1 and F3 showed transcript stabilities and polyadenylation efficiencies of the distal DUX4 transcript comparable to 4A161 chromosomes (Fig. 2B). This demonstrates that DUX4 can also be efficiently produced from these chromosomes. Altogether, our study demonstrates that all patients with FSHD1 that came to our attention have an identical sequence in the last D4Z4 unit and immediately flanking pLAM sequence and it shows that specific sequence variants unique to the permissive haplotypes confer pathogenicity to the repeat irrespective of its chromosomal localization (Fig. S8).
Finally, this distal pLAM region is also preserved in individuals with FSHD1 in whom the deleted region extends proximally to the D4Z4 repeat array (F5 in Fig. 3) as well as in FSHD2 patients, who have a classical FSHD phenotype but show a similar local chromatin relaxation on a 4A161 chromosome independent of D4Z4 repeat array contraction (6, 23).
Our study puts forward a plausible, genetic model for FSHD. In this model, two polymorphisms create a polyadenylation site for the distal DUX4 transcript, located in the pLAM sequence. In combination with the chromatin relaxation of the repeat, this leads to increased DUX4 transcript levels. FSHD may arise through a toxic gain of function attributable to the stabilized distal DUX4 transcript. Our study thus not only explains the striking chromosome-specificity of the disorder, but also provides a genetic mechanism that may unify the genetic observations in patients with FSHD.
Supplementary Material
Acknowledgments
We thank all patients and family members for their participation. This study was supported by the Fields Center for FSHD and Neuromuscular Research; the Netherlands Organization for Scientific Research NWO 917.56.338; Breakthrough Project Grant by the Netherlands Genomics Initiative NWO 93.51.8001; the National Institutes of Health P01NS069539, the Muscular Dystrophy Association; the Shaw Family Foundation, a Marjorie Bronfman Fellowship grant from the FSH Society, the Pacific Northwest Friends of FSH Research, Centro Investigación Biomédica en Red para Enfermedades Neurodegenerativas (CIBERNED), Basque Government (Fellowship grant, N° 2008111011), and Instituto Carlos III, ILUNDAIN Fundazioa.
Footnotes
Materials and Methods
Figs. S1, S2, S3, S4, S5, S6, S7
References and Notes
- 1.Tawil R, van der Maarel SM. Muscle Nerve. 2006;34:1. doi: 10.1002/mus.20522. [DOI] [PubMed] [Google Scholar]
- 2.Wijmenga C, et al. Nat. Genet. 1992;2:26. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 3.Tupler R, et al. J. Med. Genet. 1996;33:366. doi: 10.1136/jmg.33.5.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Greef JC, Frants RR, van der Maarel SM. Mutat. Res. 2008;647:94. doi: 10.1016/j.mrfmmm.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Overveld PG, et al. Nat. Genet. 2003;35:315. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- 6.Zeng W, et al. PLoS. Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabellini D, Green M, Tupler R. Cell. 2002;110:339. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 8.Klooster R, et al. Eur. J. Hum. Genet. 2009;17:1615. doi: 10.1038/ejhg.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masny PS, et al. Eur. J. Hum. Genet. 2010;18:448. doi: 10.1038/ejhg.2009.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemmers RJ, et al. Am. J. Hum. Genet. 2010;86:364. doi: 10.1016/j.ajhg.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemmers RJ, et al. Nat. Genet. 2002;32:235. doi: 10.1038/ng999. [DOI] [PubMed] [Google Scholar]
- 12.Lemmers RJ, et al. Am. J. Hum. Genet. 2007;81:884. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixit M, et al. Proc. Natl. Acad. Sci. U.S.A. 2007;104:18157. [Google Scholar]
- 14.Snider L, et al. Hum. Mol. Genet. 2009;18:2414. doi: 10.1093/hmg/ddp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Deutekom JC, et al. Hum. Mol. Genet. 1993;2:2037. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 16.Bosnakovski D, et al. EMBO J. 2008;27:2766. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Materials and methods are available as supporting material on Science Online. 2010.
- 18.Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Genome Res. 2000;10:1001. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmed F, Kumar M, Raghava GP. In Silico Biol. 2009;9:13. [PubMed] [Google Scholar]
- 20.Liu H, Han H, Li J, Wong L. Bioinformatics. 2005;21:671. doi: 10.1093/bioinformatics/bth437. [DOI] [PubMed] [Google Scholar]
- 21.Ahn SH, Kim M, Buratowski S. Mol. Cell. 2004;13:67. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 22.van der Maarel SM, et al. Am. J. Hum. Genet. 2000;66:26. doi: 10.1086/302730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Greef JC, et al. Hum. Mutat. 2009;30:1449. doi: 10.1002/humu.21091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.