Abstract
Influenza A virus genome RNA segment 7 encodes three known mRNAs, two of which, M2 mRNA and M mRNA3, are derived by alternative splicing of the primary collinear mRNA transcript using alternative 5′ splice sites. The function of M mRNA3 is currently unknown, therefore we attempted to determine whether it is essential for virus replication. Recombinant viruses unable to produce M mRNA3 and/or M2 mRNA were created by mutating the shared 3′ splice site. Growth of the mutant viruses in M2-expressing MDCK cells was not significantly affected by the lack of M mRNA3. During the course of a wild-type virus infection, levels of M mRNA3 began to decrease while those of M2 mRNA increased, which may indicate a potential mechanism of alternative splicing control. These data suggest that neither M mRNA3 nor any potential protein product are essential for influenza virus replication in tissue culture.
Pre-mRNA splicing in eukaryotic cells is a highly efficient process catalysed by a protein–RNA complex, the spliceosome (Adams et al., 1996; Jurica & Moore, 2003; Zhou et al., 2002). A number of viruses, including human immunodeficiency virus type 1, adenovirus, human papillomavirus type 16 and murine leukemia virus, have evolved so that they utilize the host cell splicing machinery to their advantage (Bohne et al., 2007; Filippova et al., 2007; Kammler et al., 2006; Kraunus et al., 2006; Lutzelberger et al., 2006; Schaub et al., 2007; Tormanen et al., 2006). Alternative splicing is a method utilized by a number of viruses not only to encode a number of protein products on a single gene but also to control the levels of mRNA during infection. It can be accomplished by a number of mechanisms such as exon skipping, alternative 5′ or 3′ splicing and intron retention (Smith & Valcarcel, 2000). Characteristics common to alternatively spliced exons (Xing & Lee, 2006) are found in the influenza virus RNA segment 7 mRNA. Splicing of the primary mRNA transcript leads to the synthesis of three distinct mRNAs: the unspliced collinear transcript encoding the M1 protein and two spliced mRNAs from alternative 5′ splice sites, although they share the same 3′ splice site (Fig. 1a) (Inglis & Brown, 1981; Lamb & Choppin, 1981; Lamb et al., 1981). Cleavage at the distal 5′ splice site occurs after only 11 nt of viral specific mRNA and results in the synthesis of M mRNA3. The first AUG translation initiation codon within mRNA3 occurs downstream of the 3′ splice site and if utilized, would result in an as yet undiscovered 9 aa peptide that is identical in sequence to the nine C-terminal residues of the M1 protein. Cleavage at the proximal 5′ splice site occurs after nucleotide 51 and results in production of M2 mRNA. The M1 and M2 proteins share initiating methionine codons but as a result of splicing, they share only nine amino acids at the amino-terminus. The ratio of M2 : M1 mRNA within virus-infected cells increases during infection, and it appears that the regulation of mRNA levels occurs at the level of splicing (Valcarcel et al., 1991). Previous studies reported that RNA segment 7 splicing requires the host cell SF2/ASF splicing factor (Shih & Krug, 1996) and the synthesis of other viral proteins (Inglis & Brown, 1984), specifically the polymerase complex (Shih et al., 1995).
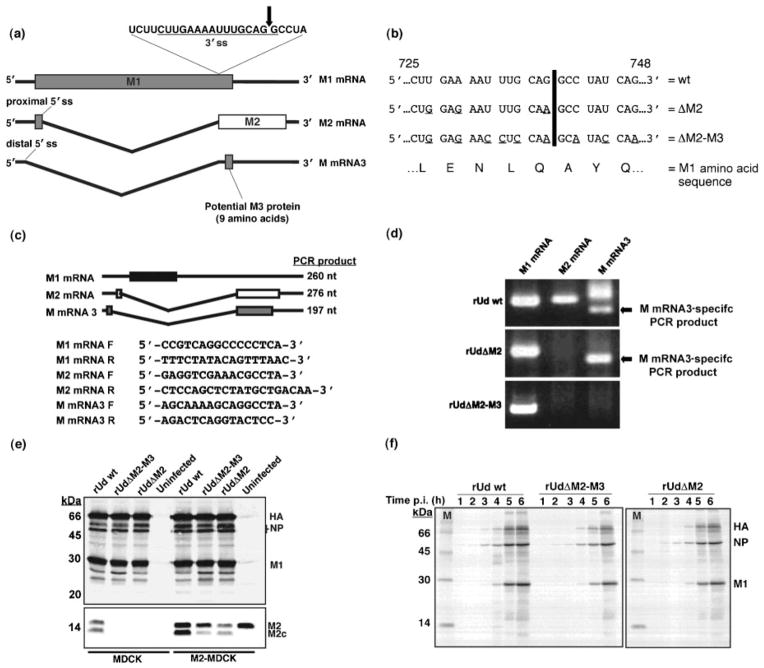
Fig. 1.
Generation and characterization of mutant viruses. (a) The splicing events of influenza A virus RNA segment 7. The arrow indicates the point of cleavage in the 3′ splice site (ss). (b) Nucleotide point mutations (underlined) introduced into the 3′ splice site of RNA segment 7, encoded on the pHH-M ‘rescue plasmid’, by site-directed mutagenesis. Numbers represent nucleotide position. The black line indicates the point of cleavage as marked in (a). (c) Schematic representation of segment 7-specific RT-PCR products. Primers were designed to allow the amplification of M1 mRNA- (black box), M2 mRNA- (white box) and M mRNA3- (grey box) specific PCR products (260, 276 and 197 nt, respectively). The primers for amplifying M2 mRNA and M mRNA3 products were designed to span the splice site junction and were based on those previously described by Cheung et al. (2005). Forward and reverse primers are indicated by F and R, respectively. (d) RT-PCR analysis of RNA segment 7-specific mRNAs in wt (rUd wt) and mutant (rUdΔM2 and rUdΔM2-M3) virus-infected MDCK cells. In the M mRNA3-specific lanes, the faster-migrating band is the M mRNA3-specific product and the slower migrating band is a product derived from M2 mRNA due to primer sequence similarities. (e, f) Analysis of viral protein synthesis in wt- and mutant virus-infected MDCK or M2-MDCK cells by Western blot (e) and radioimmunoprecipitation (1–6 h p.i.) (f) using an anti-Udorn antibody. Images were captured using Image Gauge version 3.3 software (Fuji Medical Systems).
Non-functional alternative exons are usually lost gradually through evolution and only exons with a useful function are retained over a longer time period (Xing & Lee, 2006). As M mRNA3 is evolutionarily conserved, this suggests that it is functionally relevant to the virus. In this report, we investigate whether M mRNA3 is essential for efficient viral replication. We demonstrate that influenza virus lacking M mRNA3 splicing capacity is viable.
It might be thought possible to separate the overlapping reading frames by creating a bicistronic or tricistronic RNA segment 7. However, such a construct eliminates alternative splicing constraints, thus the preliminary line of investigation would be obfuscated. Therefore, we chose to manipulate the existing splice sites within RNA segment 7. However, attempts to generate mutant viruses containing mutations in the distal 5′ splice site of RNA segment 7 were unsuccessful (data not shown), presumably due to the disruption of the viral promoter region. Although prevention of splicing at the distal 5′ splice site was successful in a previous study (Shih & Krug, 1996), it was not in the context of a viral RNA (vRNA) and such mutations may affect the vRNA promoter (Bourmakina & Garcia-Sastre, 2005; Lee & Seong, 1996, 1998).
Mutations were introduced into the 3′ splice site but as this is utilized for both M2 mRNA and M mRNA3 splicing, the generation of mutant viruses was performed in MDCK cells stably expressing the M2 protein (M2-MDCK cells) by reverse genetics, as described previously (Fodor et al., 1999; Neumann et al., 1999). Splicing at the wild type (wt) 3′ splice site occurs between two conserved guanines (Fig. 1a). As this region overlaps the M1 protein open reading frame, the mutagenesis strategy was to introduce a number of synonymous nucleotide mutations into the splice site (Fig. 1b). Two mutants were recovered and as subsequent analysis revealed one mutant lacked only M2 mRNA and not M mRNA3 (Fig. 1d) it was subsequently termed rUdΔM2, while the second mutant, which lacked both M2 mRNA and M mRNA3, was termed rUdΔM2-M3. MDCK cells were infected with wt (rUd wt) or mutant viruses at an m.o.i. of 1 for 6 h; cells were lysed and total cellular RNA was extracted using the RNEasy kit (Qiagen). mRNAs were reverse transcribed using an oligo(dT) primer and Super Reverse Transcriptase (Molecular Genetic Resources). Fragments of RNA segment 7-derived viral mRNAs (Fig. 1c) were amplified by PCR using Taq DNA polymerase (New England Biolabs). PCR samples were analysed on a 2 % agarose gel and each product was sequenced, using a 3100-Avant genetic analyser (Applied Biosystems), to confirm its identity. Analysis of RNA segment 7-specific mRNAs isolated from rUdΔM2 virus-infected MDCK cells, indicated that although M2 mRNA splicing was significantly reduced, there appeared to be no effect on M mRNA3 splicing (Fig. 1d). However, RT-PCR analysis of RNA segment 7-specific mRNAs isolated from rUdΔM2-M3 virus-infected cells confirmed that the mutations in the 3′ splice site prevented splicing of both M2 mRNA and M mRNA3 (Fig. 1d).
To determine whether the lack of M2 mRNA or M mRNA3 splicing had an effect on viral protein synthesis, MDCK or M2-MDCK cells were infected with wt, rUdΔM2 or rUdΔM2-M3 virus at an m.o.i. of 1 for 16 h. Cells were lysed in RIPA buffer [1 % deoxycholic acid, 1 % Triton X-100, 0.1 % SDS, 10 mM Tris (pH 7.4), 0.15 M NaCl], lysates were separated on a 15 % polyacrylamide gel and polypeptides were analysed by immunoblotting using either a goat–anti-Udorn antibody or the anti-M2 monoclonal antibody 14C2. The levels of viral protein expression were similar for all three viruses regardless of the cell type (Fig. 1e). As rUdΔM2 and rUdΔM2-M3 viruses result in little or no M2 mRNA production, M2 protein was absent from MDCK cells infected with either virus. In virus-infected M2-MDCK cells, M2 protein migrated as two bands (Fig. 1e), the faster migrating M2c is derived from cleavage of the C-terminal region of M2 (Zhirnov et al., 2002).
To determine whether the lack of RNA segment 7-specific mRNA splicing affected the kinetics of viral protein synthesis, MDCK cells were infected with either rUd wt, rUdΔM2 or rUdΔM2-M3 virus at an m.o.i. of 3; at hourly intervals, proteins were radiolabelled with 50 μCi (1.85 MBq) Redivue Promix L-[35S] (Amersham Biosciences) in Dulbecco’s modified Eagle’s medium (DMEM) deficient in methionine and cysteine for 30 min. Viral proteins were immunoprecipitated with a goat–anti-Udorn antibody and analysed by SDS-PAGE. Radioactivity was detected using a Fuji BioImager 1000 (Fuji Medical Systems). The levels of viral protein synthesis in all virus-infected cells were virtually indistinguishable at each time post-infection (p.i.) (Fig. 1f).
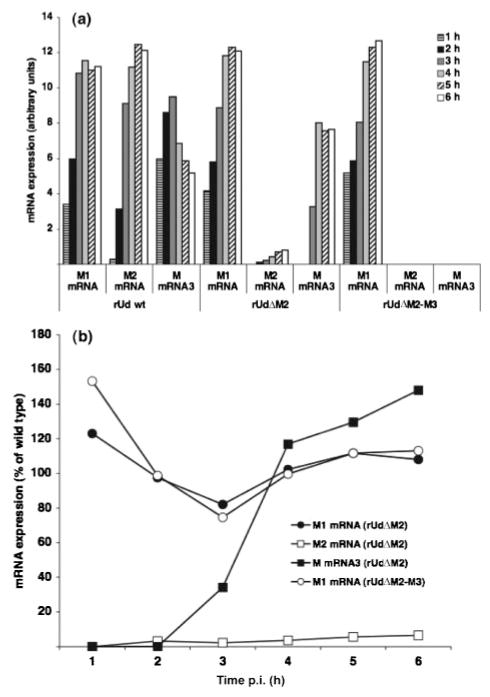
The relative quantities of RNA segment 7 mRNAs in wt and mutant virus-infected cells were determined. Due to the large degree of sequence similarity between the various species of mRNA, it proved difficult to design a probe for use in quantitative PCR experiments that could distinguish accurately between the different mRNAs. Therefore, a semiquantitative RT-PCR was performed. MDCK cells were infected with wt, rUdΔM2 or rUdΔM2-M3 virus at an m.o.i. of 1 and RNA was extracted every hour from 1 to 6 h p.i. mRNAs were subjected to RT-PCR analysis as described above and individual bands were quantified by densitometry using Image Gauge version 3.3 software. M1 mRNA was detected 1 h p.i. for all three viruses and gradually increased in concentration over time (Fig. 2a). rUdΔM2-M3 virus-infected cells did not contain M2 mRNA; however, rUdΔM2 virus-infected cells did contain very small amounts of M2 mRNA from 2 h p.i., which increased gradually with time. These data suggest that the introduced mutations did not abolish M2 mRNA splicing but reduced its efficiency. Presumably due to the reduction in splicing efficiency, the onset of M mRNA3 splicing was delayed by approximately 2 h in rUdΔM2 virus-infected cells compared with that of wt virus. Fig. 2(b) shows the expression levels of mRNAs from mutant virus-infected cells expressed as a percentage of that in wt virus-infected cells. Although M1 mRNA levels were slightly higher in mutant virus-infected cells up to 2 h p.i. (which may be due to a reduction in splicing events), levels were similar to wt virus-infected cells from 4 h p.i. onwards. M2 mRNA within rUdΔM2 virus-infected cells was above the background level from 2 h p.i. and the levels remained at approximately 5–10 % of that of wt virus throughout infection. Four hours after inoculation, levels of M mRNA3 in rUdΔM2 virus-infected cells were slightly higher than those of wt virus. It is possible that this was due to either the reduction in M mRNA3 splicing in wt virus-infected cells at later times after infection or the reduction in M2 mRNA splicing in rUdΔM2 virus-infected cells, thereby allowing an increase in M mRNA3 splicing events.
Fig. 2.
(a) Analysis of RNA segment 7-specific mRNAs in wt- and mutant virus-infected MDCK cells, from 1–6 h p.i. by semiquantitative RT-PCR. (b) mRNA expression in mutant virus-infected cells expressed as a percentage of that of wt virus-infected cells.
M mRNA3 has the capacity to encode a 9 aa peptide, however, this has not been identified to date. Various attempts to identify the M3 peptide by immunoblotting, immunoprecipitation and immunofluorescence experiments using M3-specific antisera or expression of epi-tope-tagged peptides were unsuccessful (data not shown). However, it is difficult to interpret negative data and it is conceivable that more sensitive techniques are required, especially if the peptide were rapidly degraded.
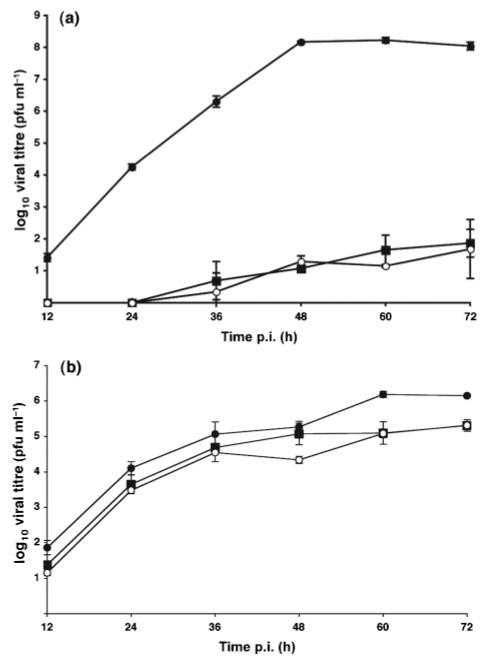
As the rUdΔM2-M3 virus does not encode its own M2 protein, growth curves were performed in both MDCK and M2-MDCK cells. Cells were infected with rUd wt, rUdΔM2 or rUdΔM2-M3 virus at an m.o.i. of 0.001 and overlaid with serum-free DMEM containing 2.5 μg N-acetyl-trypsin ml−1. Cell supernatants were harvested every 12 h until 72 h p.i., cell debris was removed by centrifugation at 1125 g for 5 min and samples were titrated by plaque assay. The growth of both mutant viruses was significantly attenuated in MDCK cells (Fig. 3a). Wild type virus reached its peak titre at 48 h p.i., whereas both mutants were attenuated 107-fold at this time point. The most plausible explanation for the defect in growth is that it results from a lack of the M2 protein in mutant virus-infected cells. However, it cannot be ruled out that lesser amounts of mRNA3 result in a reduction of viral fitness in ways that differ from loss of M2 protein function. When the growth analysis was performed in M2-MDCK cells, both mutant viruses were attenuated 5- to 10-fold compared with wt virus (Fig. 3b). It is clear that the M2-MDCK cells did not completely restore the growth of the mutant viruses to a level similar to the wt virus. However, despite the lack of M mRNA3 in rUdΔM2-M3 virus-infected M2-MDCK cells, both mutant viruses replicated to similar levels, which suggests that M mRNA3 is not essential for the growth of influenza virus in tissue culture. It is interesting to note that the maximum titre of wt virus in M2-MDCK cells was 100-fold lower than in MDCK cells. This is a characteristic of virus growth in M2-MDCK cells, which may be a result of an overexpression of the M2 ion channel protein. Although it would be interesting to examine the mutant viruses in an animal model system, the requirement for M2-transcomplementation and the toxicity of M2 expression means that experiments cannot be done readily. In this report, it was found that although splicing of M2 mRNA in rUdΔM2 virus-infected cells was greatly reduced, the splicing of M mRNA3, although slightly delayed, still reached levels that were found in wt virus-infected cells. As both mRNAs utilize the same mutated 3′ splice site, the difference seems most likely to be due to the ‘stronger’ 5′ splice site of M mRNA3 (Shih & Krug, 1996). Previous studies showed that this ‘stronger’ distal 5′ splice site must be blocked by the viral polymerase to allow initiation of M2 mRNA splicing (Shih & Krug, 1996; Shih et al., 1995). If the alternative splicing of RNA segment 7 results in a ‘switch’ from M mRNA3 splicing to that of M2 mRNA, it is conceivable that levels of M mRNA3 synthesis would decrease as those of M2 mRNA increase. This was apparent from 3 h p.i. (Fig. 2a), however, to conclusively prove this, a more quantitative approach would be required.
Fig. 3.
Multi-step growth analysis of wt (●) and mutant viruses (■, rUdΔM2-M3; ○, rUdΔM2) at an m.o.i. of 0.001 in MDCK (a) or M2-MDCK (b) cells. Results represent means±SD of three independent experiments.
We have shown that M mRNA3 is not required for efficient growth of influenza virus in tissue culture, albeit in an M2-deficient background. Although a peptide could not be detected, it cannot be ruled out that the putative M3 protein is synthesized in virus-infected cells at a level that is not easily detectable. It is possible that the M mRNA3 5′ splice site is retained as it overlaps the viral promoter sequence; therefore, M mRNA3 is a byproduct of this feature. However, it is also possible that M mRNA3 itself is dispensable but its splicing is required to regulate the splicing of M2 mRNA. To prove this hypothesis conclusively, it would be necessary to remove the distal 5′ splice site but unfortunately, this does not allow the recovery of viable virus.
Acknowledgments
This work was supported by Research Grant R01 AI-20201 from the National Institute of Allergy and Infectious Diseases. D. J. was an Associate and R. A. L. is an Investigator of the Howard Hughes Medical Institute.
References
- Adams MD, Rudner DZ, Rio DC. Biochemistry and regulation of pre-mRNA splicing. Curr Opin Cell Biol. 1996;8:331–339. doi: 10.1016/s0955-0674(96)80006-8. [DOI] [PubMed] [Google Scholar]
- Bohne J, Schambach A, Zychlinski D. New way of regulating alternative splicing in retroviruses: the promoter makes a difference. J Virol. 2007;81:3652–3656. doi: 10.1128/JVI.02105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourmakina SV, Garcia-Sastre A. The morphology and composition of influenza A virus particles are not affected by low levels of M1 and M2 proteins in infected cells. J Virol. 2005;79:7926–7932. doi: 10.1128/JVI.79.12.7926-7932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TK, Guan Y, Ng SS, Chen H, Wong CH, Peiris JS, Poon LL. Generation of recombinant influenza A virus without M2 ion-channel protein by introduction of a point mutation at the 5′ end of the viral intron. J Gen Virol. 2005;86:1447–1454. doi: 10.1099/vir.0.80727-0. [DOI] [PubMed] [Google Scholar]
- Filippova M, Johnson MM, Bautista M, Filippov V, Fodor N, Tungteakkhun SS, Williams K, Duerksen-Hughes PJ. The large and small isoforms of human papillomavirus type 16 E6 bind to and differentially affect procaspase 8 stability and activity. J Virol. 2007;81:4116–4129. doi: 10.1128/JVI.01924-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, Garcia-Sastre A. Rescue of influenza A virus from recombinant DNA. J Virol. 1999;73:9679–9682. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis SC, Brown CM. Spliced and unspliced RNAs encoded by virion RNA segment 7 of influenza virus. Nucleic Acids Res. 1981;9:2727–2740. doi: 10.1093/nar/9.12.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis SC, Brown CM. Differences in the control of virus mRNA splicing during permissive or abortive infection with influenza A (fowl plague) virus. J Gen Virol. 1984;65:153–164. doi: 10.1099/0022-1317-65-1-153. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kammler S, Otte M, Hauber I, Kjems J, Hauber J, Schaal H. The strength of the HIV-1 3′ splice sites affects Rev function. Retrovirology. 2006;3:89. doi: 10.1186/1742-4690-3-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraunus J, Zychlinski D, Heise T, Galla M, Bohne J, Baum C. Murine leukemia virus regulates alternative splicing through sequences upstream of the 5′ splice site. J Biol Chem. 2006;281:37381–37390. doi: 10.1074/jbc.M601537200. [DOI] [PubMed] [Google Scholar]
- Lamb RA, Choppin PW. Identification of a second protein (M2) encoded by RNA segment 7 of influenza virus. Virology. 1981;112:729–737. doi: 10.1016/0042-6822(81)90317-2. [DOI] [PubMed] [Google Scholar]
- Lamb RA, Lai CJ, Choppin PW. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: colinear and interrupted mRNAs code for overlapping proteins. Proc Natl Acad Sci U S A. 1981;78:4170–4174. doi: 10.1073/pnas.78.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Seong BL. Mutational analysis of influenza B virus RNA transcription in vitro. J Virol. 1996;70:1232–1236. doi: 10.1128/jvi.70.2.1232-1236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Seong BL. Nucleotides in the panhandle structure of the influenza B virus virion RNA are involved in the specificity between influenza A and B viruses. J Gen Virol. 1998;79:673–681. doi: 10.1099/0022-1317-79-4-673. [DOI] [PubMed] [Google Scholar]
- Lutzelberger M, Reinert LS, Das AT, Berkhout B, Kjems J. A novel splice donor site in the gag-pol gene is required for HIV-1 RNA stability. J Biol Chem. 2006;281:18644–18651. doi: 10.1074/jbc.M513698200. [DOI] [PubMed] [Google Scholar]
- Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub MC, Lopez SR, Caputi M. Members of the heterogeneous nuclear ribonucleoprotein H family activate splicing of an HIV-1 splicing substrate by promoting formation of ATP-dependent spliceosomal complexes. J Biol Chem. 2007;282:13617–13626. doi: 10.1074/jbc.M700774200. [DOI] [PubMed] [Google Scholar]
- Shih SR, Krug RM. Novel exploitation of a nuclear function by influenza virus: the cellular SF2/ASF splicing factor controls the amount of the essential viral M2 ion channel protein in infected cells. EMBO J. 1996;15:5415–5427. [PMC free article] [PubMed] [Google Scholar]
- Shih SR, Nemeroff ME, Krug RM. The choice of alternative 5′ splice sites in influenza virus M1 mRNA is regulated by the viral polymerase complex. Proc Natl Acad Sci U S A. 1995;92:6324–6328. doi: 10.1073/pnas.92.14.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- Tormanen H, Backstrom E, Carlsson A, Akusjarvi G. L4-33K, an adenovirus-encoded alternative RNA splicing factor. J Biol Chem. 2006;281:36510–36517. doi: 10.1074/jbc.M607601200. [DOI] [PubMed] [Google Scholar]
- Valcarcel J, Portela A, Ortin J. Regulated M1 mRNA splicing in influenza virus-infected cells. J Gen Virol. 1991;72:1301–1308. doi: 10.1099/0022-1317-72-6-1301. [DOI] [PubMed] [Google Scholar]
- Xing Y, Lee C. Alternative splicing and RNA selection pressure – evolutionary consequences for eukaryotic genomes. Nat Rev Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- Zhirnov OP, Konakova TE, Wolff T, Klenk HD. NS1 protein of influenza A virus down-regulates apoptosis. J Virol. 2002;76:1617–1625. doi: 10.1128/JVI.76.4.1617-1625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]