Anthocyanin degradation by an intracellular laccase, mediated by epicatechin oxidation, causes the loss of red color in Lichi fruit peel.
Abstract
In contrast to the detailed molecular knowledge available on anthocyanin synthesis, little is known about its catabolism in plants. Litchi (Litchi chinensis) fruit lose their attractive red color soon after harvest. The mechanism leading to quick degradation of anthocyanins in the pericarp is not well understood. An anthocyanin degradation enzyme (ADE) was purified to homogeneity by sequential column chromatography, using partially purified anthocyanins from litchi pericarp as a substrate. The purified ADE, of 116 kD by urea SDS-PAGE, was identified as a laccase (ADE/LAC). The full-length complementary DNA encoding ADE/LAC was obtained, and a polyclonal antibody raised against a deduced peptide of the gene recognized the ADE protein. The anthocyanin degradation function of the gene was confirmed by its transient expression in tobacco (Nicotiana benthamiana) leaves. The highest ADE/LAC transcript abundance was in the pericarp in comparison with other tissues, and was about 1,000-fold higher than the polyphenol oxidase gene in the pericarp. Epicatechin was found to be the favorable substrate for the ADE/LAC. The dependence of anthocyanin degradation by the enzyme on the presence of epicatechin suggests an ADE/LAC epicatechin-coupled oxidation model. This model was supported by a dramatic decrease in epicatechin content in the pericarp parallel to anthocyanin degradation. Immunogold labeling transmission electron microscopy suggested that ADE/LAC is located mainly in the vacuole, with essential phenolic substances. ADE/LAC vacuolar localization, high expression levels in the pericarp, and high epicatechin-dependent anthocyanin degradation support its central role in pigment breakdown during pericarp browning.
Anthocyanins are one of the most abundant pigments in the plant kingdom, responsible for red, purple, and blue colors of flowers, fruits, and seeds. They play a central role in attracting pollinators and seed dispersers, as well as protecting plant tissues from biotic and environmental stress factors. Anthocyanin content in plants is dependent on the rate of their synthesis, their stability in the vacuoles, and the rate at which they are degraded. In contrast to the detailed molecular knowledge available on anthocyanin synthesis, there is little information on the catabolic pathway of anthocyanins in plants (Oren-Shamir, 2009).
Numerous color-changing phenomena in plants, which occur in different tissues and during diverse plant development processes, were found to be related to anthocyanin degradation (Oren-Shamir, 2009). Anthocyanins are often produced to protect young leaves, and are degraded as the leaves mature (Steyn et al., 2002). Anthocyanin accumulation is significantly induced in plants under stress conditions, such as high light intensity, low temperature, drought, or nutrition deficiency, and is degraded once the stress is eliminated (Winkel-Shirley, 2002; Rowan et al., 2009). Rapid anthocyanin degradation occurs in Brunfelsia calycina (yesterday-today-tomorrow) petals after flower opening (Zenner and Bopp, 1987; Vaknin et al., 2005). Anthocyanins are degraded during pear fruit maturity (Pyrus communis; Steyn et al., 2004) and throughout the senescence of litchi (Litchi chinensis; Zhang et al., 2001, 2005) and blood citrus fruit (Barbagallo et al., 2007). Additional examples of reduced anthocyanin concentrations occur during the development of grape berries (Mori et al., 2007) and rose flower buds (Dela et al., 2003) in response to high temperatures, probably due to reduced anthocyanin biosynthesis and induced anthocyanin degradation.
In vivo anthocyanin degradation was investigated mainly in fruits and ornamentals, in which preserving the pigmentation is crucial for maintaining high quality of the produce. These studies are based on the knowledge accumulated from pigment loss in fruit juices and wines (Cheynier et al., 1994; Kader et al., 1998). Polyphenol oxidases (PPOs) and peroxidase (POD) are the main enzymes that oxidize anthocyanins in fruit extracts, leading to the decrease in pigment content (Sarni et al., 1995; Kader et al., 1998). PPO and POD activities were detected during the change in color of litchi (Jiang and Fu, 1998; Zhang et al., 2005; Sun et al., 2008) and bayberry fruits (Myrica rubra; Fang et al., 2007) after harvest, and during the drying of plums (Raynal et al., 1989). The hydrolysis of anthocyanin catalyzed by β-glycosidases results in the breaking of the glycosidic bond and liberation of the corresponding anthocyanidin, which is then spontaneously converted to a colorless pseudobase (Huang, 1956), or subsequently oxidized by PPO and POD (Zhang et al., 2001, 2005). A β-glucosidase was partially purified from the fruit juice of Tarocco Sicilian blood oranges and found to be responsible for anthocyanin degradation both in the juice and the ripening fruit (Barbagallo et al., 2007).
Active enzymatic anthocyanin degradation dependent on novel mRNA and protein biosynthesis was demonstrated in B. calycina flower petals, which undergo a rapid color change from dark purple to white within 3 d after flower opening (Vaknin et al., 2005; Bar-Akiva et al., 2010). Recently, a basic peroxidase, BcPrx01, was found to be responsible for the in planta degradation of anthocyanins in B. calycina (Zipor et al., 2015). BcPrx01 has the ability to degrade complex anthocyanins. It colocalizes with these pigments in the vacuoles of petals, and both the mRNA and protein levels of BcPrx01 are greatly induced parallel to the degradation of anthocyanins.
Litchi is a tropical and subtropical fruit of high commercial value. The white translucent aril and attractive red color due to high content of anthocyanins in the pericarp contribute significantly to their value. However, litchi pericarp coloration changes after harvest from red to brown due to a 50% decrease in anthocyanin concentration, indicating pigment degradation (Fuchs et al., 1993; Holcroft and Mitcham, 1996; Zhang et al., 2001; Jiang et al., 2006; Mayer, 2006). Revealing the anthocyanin degradation pathway during pericarp browning may help preserve the original red coloration of the fruit pericarp. Postharvest browning of fruits and vegetables has long been considered a function of PPO, catalyzing the oxidation of an array of aromatic substrates resulting in brown colored by-products. The activity of PPO on exogenous substrates, such as catechol and 4-methylcatechol, was confirmed in litchi pericarp (Jiang and Fu, 1998; Sun et al., 2007). However, the purified PPO from litchi pericarp could oxidize anthocyanins only in the presence of exogenous catechol (Jiang, 2000; Zhang et al., 2005). This process is known as the anthocyanin-PPO-phenol model (Kader et al., 1998). The question still remains whether this is the process occurring in vivo in litchi pericarp.
Here, we describe the enzymatic process by which anthocyanins are degraded in litchi pericarp. The anthocyanin degradation enzyme (ADE) was purified to homogeneity and identified as a laccase (LAC), based on protein sequencing, gene cloning, and antibody verification. The ADE/LAC was found to be an intracellular enzyme prior to pericarp senescence, with epicatechin identified as one of its endogenous substrates involved in the coupled enzymatic anthocyanin degradation.
RESULTS
Detection of ADE Activity in the Pericarp Tissue
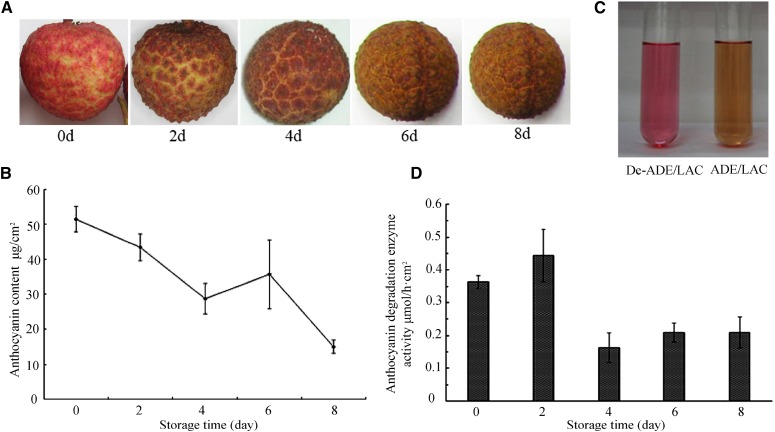
The anthocyanin concentration in the pericarp of red litchi fruit after harvest was around 53 μg cm−2. Pericarp browning began 2 d after harvest, and by 4 d, the characteristic red pigmentation disappeared. The anthocyanin content of the pericarp decreased to 53% of the initial content in 4 d and to around 32% in 8 d (Fig. 1, A and B). When partially purified pericarp anthocyanins were incubated with a crude pericarp protein extract, the pigments turned brown within 10 min. This decoloration was due to ADE activity, since the anthocyanins remained red when incubated with the denatured enzyme extract (Fig. 1C). The rate of ADE activity in the harvested pericarp was 0.36 μmol h−1 cm−2. The rate of anthocyanin degradation increased by about 20% 2 d after harvest, and then decreased to around 50% from the initial value 4 d after harvest (Fig. 1D).
Figure 1.
Anthocyanin degradation and ADE activity during the browning of litchi fruit after harvest. Litchi fruit were stored, unpackaged, at 20°C ± 1°C and 70% relative humidity for 8 d. Change in fruit color (A) and anthocyanin contents (B) was recorded during fruit browning. C, Browning of litchi anthocyanin due to the ADE activity in the crude pericarp enzyme extract, with the denatured enzyme extract (De-ADE) as a reference. D, The time course of pericarp ADE activity during fruit browning. The values presented in C and D are means of three measurements from three individual extractions. Error bars indicate the sd of the values.
Purification of the Litchi Pericarip ADE
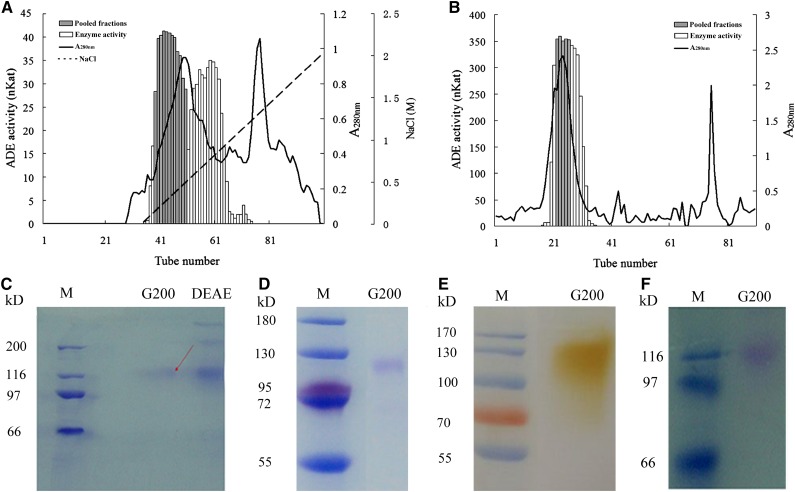
Purification of litchi pericarp ADE was achieved by ammonium sulfate fractionation, DEAE-Sepharose, and Sephadex G-200 column chromatography. After DEAE-Sepharose chromatography, two major ADE activity peaks partially overlapped the main protein peak (Fig. 2A). The eluted fractions with high ADE activity in the first activity peak (sample nos. 39–49) were pooled and further subjected to Sephadex G-200 chromatography, in which one major activity peak was detected overlapping the main protein peak (Fig. 2B). Fractions from the major activity peak with high activity (sample nos. 23–26) were pooled and found to contain a 73.9-fold higher active ADE concentration in comparison with the crude protein extract (Supplemental Table S1).
Figure 2.
Purification of the ADE from litchi pericarp. A, Total protein (280 nm) and ADE activity determined in DEAE-Sepharose chromatography fractions from pericarp crude enzyme extract, after ammonium sulfate fractionation. B, Total protein (280 nm) and ADE activity determined in Sephadex G-200 chromatography fractions from the fractions with ADE activity after the DEAE-Sepharose chromatography. Enzyme activities of pooled and nonpooled fractions, the A280, and the NaCl gradients (for A and B) are indicated. C, SDS-PAGE of samples from purification steps. D, Urea-SDS-PAGE of the purified ADE after Sephadex G-200 chromatography. E, In-gel activity assay of the purified ADE after Sephadex G-200 chromatography with partially purified litchi anthocyanins as substrates. F, Periodic acid-Schiff staining of the purified ADE. The unit of the ADE activity, Kat, is as described in Table II.
The protein profiles of ADE purification are presented in an SDS-PAGE (Fig. 2C). After the G-200, one major band at about 116 kD was observed by SDS-PAGE (Fig. 2C) or urea SDS-PAGE (Fig. 2D), indicating a high purity of the ADE protein, and that the protein is a single polypeptide enzyme. To further confirm that the purified protein contains ADE activity, the fraction was run on an activity gel. A single brown band of 116 kD was detected 4 min after incubating the gel in the litchi anthocyanin substrate, indicating ADE activity (Fig. 2E). A purple-magenta color was seen on this protein band after staining with periodic acid-Schiff, suggesting that this ADE protein is a glycoprotein (Fig. 2F).
The Litchi Pericarp ADE Is a Laccase
The major band seen in the final purified fraction with ADE activity was cut and sent for tandem mass spectrometry (MS/MS) protein sequencing. Four peptides with high similarity to published protein sequences were identified (Table I). The sequence of the first peptide, of 11 amino acids, showed 100% identity to the deduced protein sequence of a partial complementary DNA (cDNA) of a litchi Laccase (ACB22018.1). The sequences of the second and fourth peptides of 9 and 15 amino acids, respectively, were of 100% identity to the sequences of a laccase (AAB09228.1) of Acer pseudoplatanus. The third peptide, with 16 amino acids, is of 100% identity to a laccase (NP_196498.1) from Arabidopsis (Arabidopsis thaliana). Based on the fact that four peptides identified by MS/MS showed high identities to the protein sequences of laccases from different species, we assumed that the purified ADE enzyme is a laccase.
Table I. Fragment sequences of the litchi ADE detected by MS/MS and their identical hits from other plant species.
| Sequences Detected by MS/MS | Annotation | Gene Identified | Organisms | |
|---|---|---|---|---|
| 1 | PPEVNTIQLSK | Laccase | ACB22018.1 | L. chinensis |
| 2 | AVHYDFVVK | Laccase | AAB09228.1 | A. pseudoplatanus |
| 3 | ACPTPSDGPEYVTQCK | Laccase | NP_196498.1 | Arabidopsis |
| 4 | SMLTVNGSFPGLLLR | Laccase | AAB09228.1 | A. pseudoplatanus |
Substrate Specificity Analysis Further Confirmed Laccase Activity of ADE
The purified ADE, a putative laccase, showed the ability to oxidize a wide range of aromatic compounds (Table II). The purified ADE efficiently oxidized several o-diphenols, such as epicatechin, catechol, and 4-methyl catechol, as well as hydroquinone (a p-diphenol), two monolignols (coniferyl alcohol and sinapyl alcohol), and ABTS (a universal laccase substrate), strongly indicating the ADE is a laccase.
Table II. Substrate specificity of the litchi ADE/LAC.
—, Activity on these compounds was not detected.
| Substrate | Km | Va | V/Km | % V/Km |
|---|---|---|---|---|
| μM | nKat mg−1 | |||
| Epicatechin | 1.098×103 | 236.15 | 0.240 | 100 |
| Coniferyl alcohol | 74 | 3.1 | 0.041 | 17 |
| 2,2′-Azinobis-3-ethylbenzthiazolinesulfonic acid (ABTS) | 2.356×103 | 83.3 | 0.035 | 15 |
| 4-methylcatechol | 43.15×103 | 1.111×103 | 0.027 | 11 |
| Sinapyl alcohol | 239 | 5.4 | 0.023 | 10 |
| Catechin | 2.313×103 | 35.7 | 0.015 | 6 |
| Hydroquinone | 31.950×103 | 151.5 | 0.005 | 2 |
| Phenol (PhOH) | — | — | — | — |
| Phloroglucinol | — | — | — | — |
| p-Anisidine | — | — | — | — |
| Litchi anthocyaninsb | 72 | 1.25 | 0.017 | 7 |
| Litchi anthocyaninsc | — | — | — | — |
One Kat represents the enzyme activity that catalyzes the reaction of 1 mol substrate per second.
Litchi anthocyanins were partially purified by Amberlite XAD-7 column chromatography.
Litchi anthocyanins were highly purified by sequential Amberlite XAD-7 and Sephadex LH-20 column chromatography.
This is the first report in which epicatechin was included in the substrate specificity analysis for laccases or PPOs. The extinction coefficient of epicatechin was reported as ε 280 nm = 3,988 M−1 cm−1 (Kennedy and Jones, 2001). Overlapping absorbance spectra at 280 nm were found for epicatechin and its oxidation product, and an obvious increase at 380 nm was observed after the reaction was initiated by the addition of the purified ADE (Supplemental Fig. S1). Accordingly, the extinction coefficient for the epicatechin oxidation product was determined at 380 nm, and an ε 380 nm = 7,858 M−1 cm−1 was obtained (Supplemental Fig. S1). The Km (1.098 × 103 μm) for ADE on epicatechin was determined based on the ε 380 nm and not that of epicatechin at 280 nm 3,988 M−1 cm−1 (Kennedy and Jones, 2001). As judged by the ratio of V/Km, epicatechin appeared to be the favored substrate, whereas the second favored substrate of those tested was coniferyl alcohol, with a V/Km value only 17% of that for epicatechin. Hydroquinone was the least favored substrate (Table II). Both phloroglucinol and p-anisidine were not oxidized by the purified ADE. The V/Km value of the ADE-oxidizing partially purified litchi anthocyanins was 19.3% of that for epicatechin. However, ADE did not oxidize highly purified anthocyanins (Table II). The great difference in activity between the highly and partially purified anthocyanins suggests that certain endogenous substrates for the enzyme exist in the partially purified pigment necessary for the reaction to take place. To avoid confusion in this context, ADE was also denominated as ADE/LAC.
Cloning ADE/LAC Full-Length cDNA and Phylogenetic Analysis of the Deduced Protein Sequence
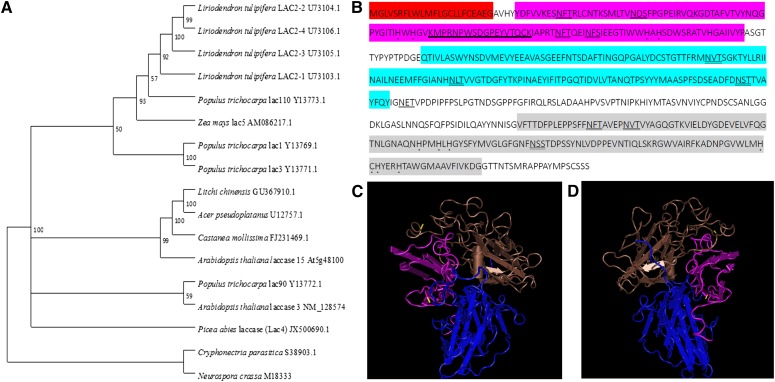
Based on the peptide sequences of the litchi ADE/LAC (Table I), degenerate primers were designed to amplify the cDNA fragments encoding the litchi ADE/LAC. An mRNA sequence of 1.770 kb with the complete coding sequence (CDS; 1.703 kb) was identified using the assembly of 3′ and 5′ ends of the fragment sequences. The sequence was uploaded to the National Center for Biotechnology Information (NCBI) as GU367910. The CDS encoded a protein of 567 amino acids and a molecular mass of around 62 kD. The difference in migration of the corresponding protein on SDS-PAGE may be due to the existence of carbohydrate moieties detected on the purified protein band (Fig. 2F). Several N-glycosylation sites are present in the deduced amino acid sequence (Fig. 3B). The sequence contains three cupredoxin domains that commonly appear in plant laccases, CuRO-1-LCC, CuRO-2-LCC and CuRO-3-LCC, in which two copper binding sites appear in the CuRO-1-LCC and CuRO-3-LCC domains, respectively. A secretion pathway protein signal was detected at the N terminus (Fig. 3B). Based on the 3D model of a fungal laccase (Protein Data Bank code no. 1GYC, Research Collaboratory for Structural Bioinformatics, Rutgers University, http://www.rcsb.org/), a putative 3D structure was built that contained three domains (Fig. 3, C and D). A phylogenetic tree including previously characterized plant laccases was constructed, indicating high homologies of the litchi ADE/LAC to a laccase of A. pseudoplatanus and Laccase 15 (TT10) of Arabidopsis (Fig. 3A).
Figure 3.
Sequence feature analysis of litchi ADE/LAC. A, A rooted neighbor-joining phylogenetic tree relating the litchi ADE/LAC (GU367910.1) with laccases from other plant species. The following sequences from NCBI were used: (1) highly identical laccases; (2) extensively studied laccases (A. pseudoplatanus Laccase [U12757.1], Populus euramericana Lac90 [Y13772.1], and Arabidopsis TRANSPARENT TESTA10 [TT10; At5g48100]); (3) four P. euramericana laccases and five Liriodendron tulipifera laccases, and two laccases from fungi. The gene bank accession numbers for the correspondent mRNA sequences coding the enzymes are presented. B, The deduced amino acid sequence. Putative domains include a signal peptide (red); three plant cupredoxin domains CuRO-1-LCC (purple), CuRO-2-LCC (blue), and CuRO-3-LCC (gray); putative copper binding sites of His-X-His (with a dot below). Potential N-glycosylation sites (Asn-Xaa-Ser/Thr) are doubly underlined. Peptides used for antibody production are underlined. A hypothetical three-dimensional (3D) structural model of ADE/LAC predicted by the SWISS-MODEL and viewed using Cn3D macromolecular structure viewer and Vector Alignment Search Tool is presented in two images (C and D). The arrangement of the domain structure is depicted with similar colors as in B.
The Anthocyanin Degradation Function of Litchi ADE/LAC Gene Was Confirmed by Transient Expression in Tobacco Leaves
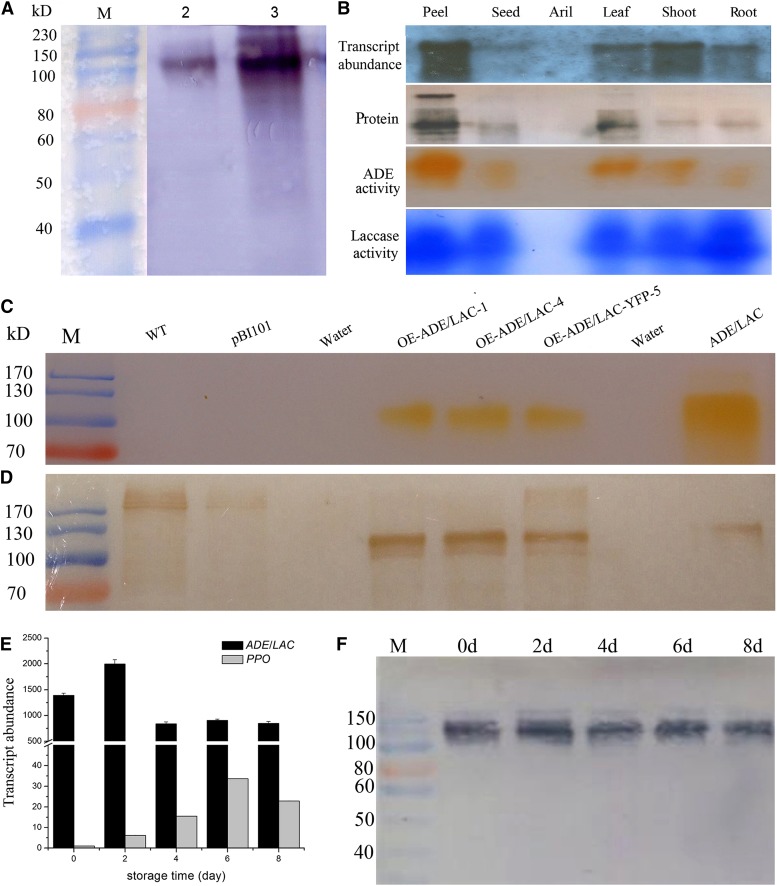
An anti-ADE/LAC antibody was raised against a deduced peptide located next to a trinuclear copper binding site in the CuRO-1-LCC domain at the N terminus of the laccase coded by the ADE/LAC gene (Fig. 3B) to avoid recognition by other copper-containing proteins. The antibody recognized the purified ADE protein displaying one band at 116 kD in SDS-PAGE (Fig. 2C; lane 1 in Fig. 4A), further confirming that the ADE protein was the laccase encoded by the identified ADE/LAC mRNA sequence. The anti-ADE/LAC antibody also recognized a protein of similar size in the total protein extract from the litchi pericarp, with a major band at 116 kD and some minor bands with higher molecular masses (lane 2 in Fig. 4A).
Figure 4.
ADE/LAC transcript, protein, and activity levels in fruit pericarp and transient expression of litchi ADE/LAC in tobacco. A, Immunodetection of ADE/LAC in the purified ADE (lane 2) and the crude pericarp enzyme extract (lane 3) by a polyclonal anti-ADE/LAC. B, Transcript abundance and protein activity levels of ADE/LAC in different litchi tissues. The transcript abundance was analyzed by northern hybridization with a digoxigenin (DIG)-labeled ADE/LAC-specific DNA probe. The protein levels were analyzed by immunodetection with the anti-ADE/LAC. The in-gel activities were assayed as described in Figure 2(D) with partially purified litchi anthocyanins for ADE substrate and 1,8-diaminonaphthalene (DAN) for laccase substrate. C, ADE activity assay in gel for the recombinant ADE/LAC transiently expressed in tobacco leaves. Protein extracts from the leaves of three lines, transiently overexpressed ADE/LAC or ADE/LAC-yellow fluorescent protein (YFP), were assayed for the activity in gel, with the wild-type (WT), empty vector control samples as negative controls and the native litchi ADE/LAC as a positive control. D, Immunodetection of the recombinant ADE/LAC transiently expressed in tobacco leaves. The recombinant ADE/LAC levels in the protein extracts in C were analyzed by immunodetection with the anti-ADE/LAC. E, The relative expression levels of ADE/LAC and PPO (JF926153) genes were analyzed by quantitative real-time (qRT)-PCR. F, ADE/LAC protein levels in the pericarp during fruit browning after harvest were analyzed as described in B. The details of the values presented in C are as described in Figure 1.
The open reading frame of ADE/LAC was transiently overexpressed by infiltration of Agrobacterium tumefaciens (containing pBI101-35S-ADE/LAC or 35S-YFP-ADE/LAC) into Nicotiana benthamiana leaves. ADE activity bands similar in size to the native litchi ADE/LAC were observed using an in-gel assay for the protein extracts from the two ADE/LAC and one ADE/LAC-YFP overexpression lines (Fig. 4C). The proteins from the overexpression lines with ADE activities were recognized by the above described anti-ADE/LAC. The molecular masses of these proteins were around 110 kD, slightly less in comparison with the native protein from litchi pericarp. No activity bands or proteins of the same mobility as litchi ADE/LAC were detected for wild-type or pBI101 empty vector control leaves, despite the recognition by the anti-ADE/LAC of several proteins above 170 kD (Fig. 4D). The ADE activities in the overexpression lines strongly confirmed the anthocyanin degradation function of the litchi ADE/LAC gene product.
High Protein and Transcript Abundance of the ADE/LAC Detected in the Pericarp, with Increasing Levels after Harvest
ADE/LAC protein levels in different litchi organs were detected by the anti-ADE/LAC antibody. The highest ADE/LAC level was found in the pericarp with significantly lower levels in the leaf, stem, root, and seed, and no detected ADE/LAC protein in the aril (Fig. 4B; Supplemental Fig. S2). The protein level in different organs correlated with its transcript abundance, analyzed by a DIG-labeled probe generated at the 3′ untranslated ration region of ADE/LAC mRNA (GU367910; Fig. 4B). Furthermore, the in-gel activities using partially purified litchi anthocyanins as substrate also correlated with the above protein and transcript levels (Fig. 4B; Supplemental Fig. S2). When using DAN, a universal substrate for phenol oxidase, higher activity levels were detected in the root and pericarp in comparison with other tissues (Fig. 4B; Supplemental Fig. S2). The correlated patterns of the transcript, protein, and activity levels confirm that the litchi ADE is indeed a laccase, with high expression levels in the pericarp. High expression levels of this enzyme in the pericarp are in correlation with the rapid anthocyanin degradation and browning occurring in this tissue after harvest.
Litchi fruit were harvested at the mature red stage and stored at 20°C and 70% relative humidity without packaging. The expression levels of the ADE/LAC gene peaked 2 d after harvest, correlating with the dramatic color change occurring during this time (Fig. 4E). The transcript abundances of ADE/LAC during the eight postharvest days were about 1,000-fold higher than those of the PPO gene, whose transcripts was found in the pericarp (Wang et al., 2014). The transcript abundance of ADE/LAC after harvest correlated with the ADE/LAC protein levels, with highest levels at day 2 (Fig. 4F).
The effect of several inhibitors for PPOs and laccases was tested on the ADE activity in the crude litchi pericarp protein extract. ADE activity was found to be sensitive to laccase inhibitors such as CTAB or SDS, but not to PPO inhibitors such as p-coumaric acid, cinnamic acid, ferulic acid, and polyvinylpyrrolidone (Supplemental Table S2). These results, in addition to the dramatically higher transcript abundance of ADE/LAC in comparison with PPO, suggest that ADE/LAC may play a major role in anthocyanin degradation during fruit browning.
Laccase Activity Is Localized Mainly in the Mesocarp Zone of the Pericarp
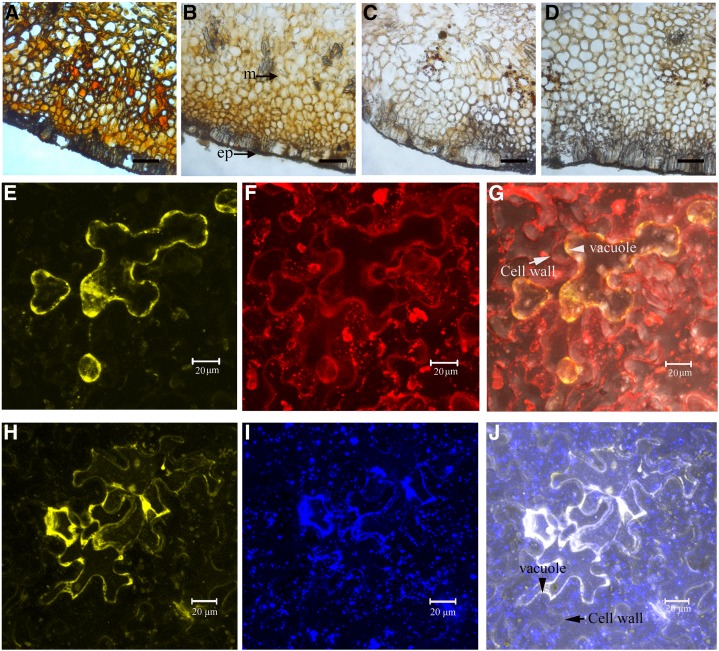
When litchi fruit pericarp sections were incubated with artificial laccase substrates, the cells in the mesocarp closest to the epicarp acquired a distinct red-brown or brown color (Fig. 5, A and B), suggesting laccase activity. No color change was observed in controls that did not contain a substrate (Fig. 5C) or in the boiled section controls (Fig. 5D). In most cells, the brown coloration was present in both the intracellular and extracellular spaces.
Figure 5.
Histochemical localization of ADE/LAC in litchi pericarp and subcellular location of ADE/LAC-YFP in tobacco leaf epidermal cells. A, Cross-sections of litchi pericarp incubated with 4-methylcatechol resulted in a red-brown coloration in the mesocarp. B, Incubation with ABTS resulted in brown coloration in the mesocarp. C, Control sections without incubation of 4-methylcatechol or ABTS. D, Boiled control gave no coloration. M, Mesocarp; ep, epicarp. Bars = 25 mm. N. benthamiana leaves were infiltrated with A. tumefaciens containing pBI101.1-ADE/LAC-YFP, were incubated in the growth chamber for 2 d, and inspected using confocal laser scanning microscopy. Some YFP fluorescence was detected at the boundary of Lysotracker red-stained vacuoles, and some was inside the vacuoles of a plasmolyed epidermal cell (E and F). The YFP partially overlapped with red fluorescence (G). Note that the cell wall was also lightly stained with Lysotracker red and was easily distinguished from the vacuole/protoplast after plasmolysis of the cell, as indicated by the arrows. No overlapping with the YFP was observed at the cell wall. Most of the YFP fluorescence was overlapped with the blue florescence in lines or in clusters, but not in dots (H and I), resulting in white fluorescence (J). The lines and clusters of the blue fluorescence stained by ER-Tracker Blue-White DPX indicated the location of the endoplasmic reticulum network (I). The cell wall was indicated in the overlapping image in J.
Litchi ADE/LAC, Located in the Vacuoles, Is Secreted to the Extracellular Space after Pericarp Browning
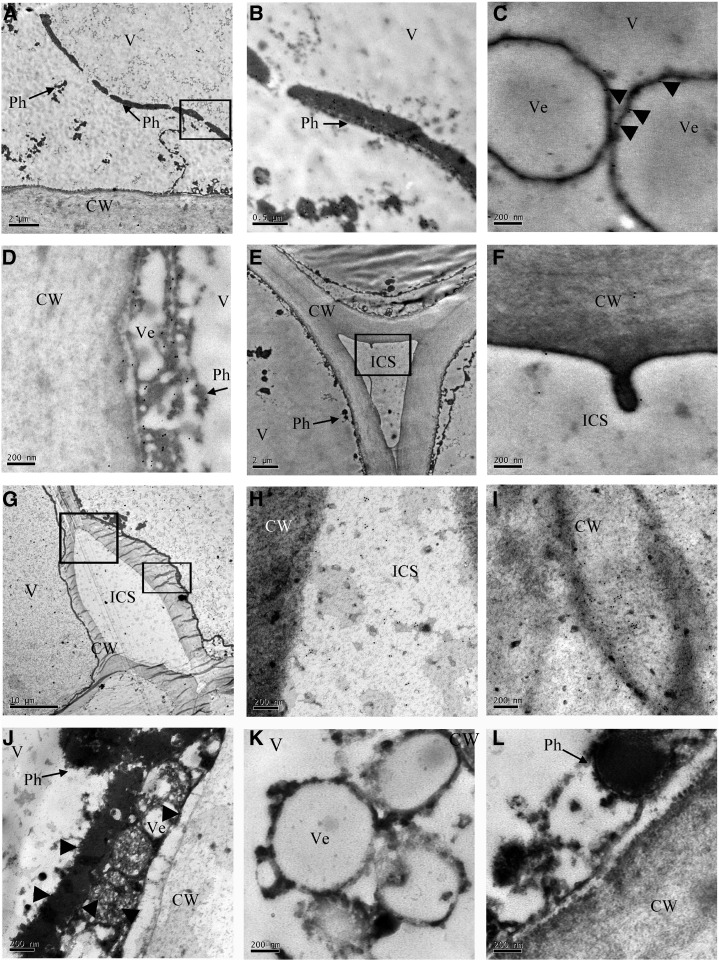
Pericarp tissue of litchi fruit, 0 to 4 d postharvest, was ultrasectioned, labeled by anti-ADE/LAC immunogold, and analyzed by transmission electron microscopy (TEM). The ultrastructure of the pericarp cells revealed a large vacuole and the cytoplasm compressed toward the cell wall (Fig. 6, A, E, and G; Supplemental Fig. S3). Numerous phenolic grains (Phs) were distributed in the vacuoles (Fig. 6, A, E, and G), and some accumulated in small vesicles inside the vacuoles (Fig. 6, C and K; Supplemental Fig. S3). In cells from day 0 fruit, numerous gold particles colocalized with these Phs inside the vacuole (Fig. 6, B–D), and few gold particles were distributed at the cell wall and intercellular space (Fig. 6, E and F). An increase in gold particles along the cell wall in comparison with the intercellular space was observed at day 4 when the fruit completely lost their red color (Fig. 1A) and the cell wall disintegrated (Fig. 6, G–I). The labeling proved to be specific for ADE/LAC, since no gold particles were observed in cells without treatment with anti-ADE/LAC (Fig. 6, K and L).
Figure 6.
Subcellular localization of ADE/LAC in litchi pericarp. TEM immunolocalization of the ADE/LAC in the pericarp cells of fruit at day 0 (A–F) and day 4 (G–J) after harvest. A, Ultrastructure of litchi pericarp cells at 0 d exhibited the cytoplasm pressed toward the cell wall by the central large vacuole. In addition, several long Phs were detected in the vacuole. B, A magnified image of the area indicated by the square in A. Numerous gold particles appeared in the long Phs in the vacuole (V). C, A magnified image showing numerous gold particles in the Phs located on the surface of the vesicle (Ve) in the vacuole. D, An additional magnified image showing numerous gold particles in the Phs on the surface of the vesicles close to the cell wall. E, Ultrastructure of litchi pericarp cell at 0 d revealed the cell wall (CW) and intercellular space (ICS). F, Magnified image of the square in E. Few gold particles appeared in the cell wall and intercellular space (ICS). G, Ultrastructure of litchi pericarp cells of 4 d fruit after pericarp browning displayed disintegration of the cell wall. H, A magnified image of the upper square in G. Numerous gold particles in ICS. I, A magnified image of the lower square in G. Numerous gold particles were detected in the disintegrated cell wall. J, Fewer gold particles were detected in Phs after pericarp browning at 4 d than in those at 0 d. K and L, Control images without anti-ADE/LAC incubation showed no gold particles.
For further subcellular localization of ADE/LAC, the enzyme was expressed in tobacco leaves (N. benthamiana) as a fusion to yellow fluorescent protein (YFP; Fig. 5). In the epidermal tobacco leaf cells, the yellow fluorescence of the ADE/LAC-YFP fusion proteins was observed in protoplasts of the plasmolyzed cells, but not in the cell wall and intercellular space (Fig. 5, E and H). The fluorescent fused protein accumulated mainly in the vacuoles and ER (Fig. 5, G and J). The endoplasmic reticulum (ER) localization of the ADE/LAC protein in tobacco cells is in agreement with the gene sequence analysis (SignalP 4.1 server, http://www.cbs.dtu.dk/services/SignalP/), predicting a secretion protein signal via the ER. The destination of the secretion might be either the vacuole or the extracellular space, as observed by immunogold labeling TEM of cells before and after senescence.
The results suggest that ADE/LAC is an intracellular enzyme colocalizing with Phs before the disintegration of the pericarp cells.
Epicatechin Was Identified as an Endogenous Substrate from Litchi Pericarp for ADE/LAC
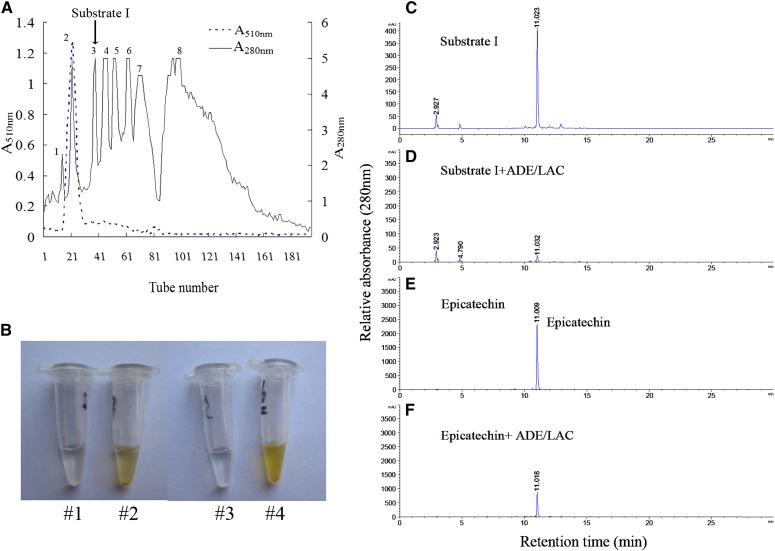
Litchi ADE/LAC degraded anthocyanins only in the partially purified litchi anthocyanin extract and not in the pure pigment preparation (Table II). This may indicate that certain ADE/LAC substrates exist in the partially purified extract that are essential for anthocyanin degradation activity. To identify the essential substrates required for ADE/LAC activity, compounds in the partially purified extract were separated and tested for ADE/LAC activity. Eight major fractions with peaks at 280 nm were detected. The second peak, which was of high absorbance at 510 nm (A510) as well, was identified as the major anthocyanin (cyanindin-3-rutinoside) in litchi pericarp, and was chosen as the highly purified pigment in the following reactions (Fig. 7A). Of the seven remaining fractions, only two were oxidized by the purified ADE/LAC and turned brown due to this reaction (third and eighth peaks; Fig. 7B). The fraction correlating with the third peak was named substrate I.
Figure 7.
Purification and identification of the endogenous substrate for ADE/LAC in the litchi pericarp. A, Two hundred eighty- and 510-nm absorbance determined in the fractions of Sephadex LH-20 chromatography for the partially purified litchi anthocyanins. B, The color change of the reactions of substrate I (#2) or epicatechin (#4) by ADE/LAC. The reactions with the denatured ADE/LAC served as references (#1 and #3). C to F, HPLC analysis of the reaction products after reactions #1 to 4 in B.
The activity of the ADE/LAC on substrate I was analyzed by HPLC, with epicatechin, the major phenol in litchi pericarp (Zhang et al., 2000), as a standard reference. When substrate I was incubated with the denatured enzyme, a major peak and several minor peaks were detected. The major peak at 11.0 min showed the same retention time as the epicatechin standard (Fig. 7, C and E). After incubation with the native enzyme, the peak at 11.0 min decreased dramatically, indicating that the compound in this fraction was catalyzed by the enzyme (Fig. 7, C and D). The peak of the standard epicatechin also decreased when incubated with the ADE/LAC enzyme (Fig. 7, E and F). The fraction from substrate I that eluted at 11.0 min was collected and further analyzed by liquid chromatography (LC)-mass spectrometry (MS). A negative ion of Mr 287.2 was of the highest abundance in comparison with the other ions (Supplemental Fig. S4). This ion showed the same molecular mass and similar MS/MS profile as epicatechin when subjected to the same analysis (Supplemental Fig. S4). These results suggest that the major active compound in substrate I is epicatechin.
Epicatechin-Mediated ADE/LAC Anthocyanin Degradation May Be Responsible for the Pigment Loss during Litchi Fruit Browning
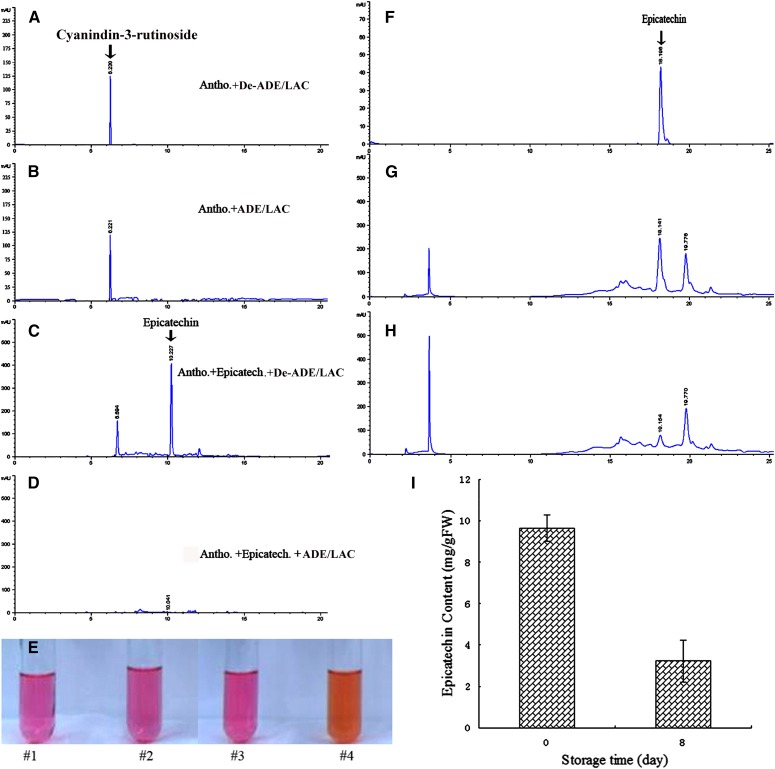
In vitro anthocyanin degradation by the ADE/LAC was analyzed by HPLC. No obvious decrease in anthocyanin content was detected when the pigment alone was incubated with either the active or boiled ADE/LAC enzyme (Fig. 8, A and B). However, when in the presence of epicatechin, a dramatic decrease in both anthocyanin and epicatechin content was observed after addition of the active ADE/LAC enzyme (Fig. 8D). The boiled enzyme had no effect on either anthocyanins or epicatechin levels (Fig. 8C). The degradation of anthocyanins, detected by HPLC, correlated with the color change of the reactions from bright red to brown (Fig. 8E).
Figure 8.
Degradation of anthocyanin by ADE/LAC in the presence of epicatechin. HPLC analysis of the reactions with the highly purified litchi anthocyanins (cyanidin-3-rutinoside) and ADE/LAC in the absence (B) or presence (D) of epicatechin, with no-enzyme controls for the relevant reactions (A and C). E, Color change of anthocyanins after the above described reactions (A–D) as indicated by #1 to 4, respectively. HPLC analysis of epicatechin in the pericarp extracts of the fruit at 0 d (G) and 8 d (H), with an epicatechin standard (F) as reference. I, The epicatechin contents detected by HPLC (E–G) are presented.
Epicatechin was detected in the total phenol extract of fruit pericarps, being a major phenol component in this tissue (Fig. 8, F and G; Zhang et al., 2000). The epicatechin content decreased to about 25% of its original concentration after 8 d (Fig. 8, G–I), parallel to the change in fruit color to brown.
DISCUSSION
Anthocyanins are important plant pigments both as attractive colors and as phyto-protectors in a variety of stress situations. They are finely regulated via both anthocyanin biosynthesis and degradation at certain development stages and in response to environment conditions. Although much is known about the biosynthesis of anthocyanins, very little is known about their in planta degradation mechanisms (Mol et al., 1998; Griesbach, 2005; Oren-Shamir, 2009). In the current study, the enzyme responsible for anthocyanin degradation during litchi pericarp browning after harvest was identified and characterized.
By using partially purified litchi anthocyanins from litchi fruit pericarp, we detected ADE activity in the crude pericarp protein extract and purified the enzyme to homogeneity. The purified ADE was suggested to be a laccase based on its sequence. The gene encoding the ADE/LAC was cloned and shown to actively degrade anthocyanins when transiently expressed in tobacco leaves. The ADE/LAC is located in the vacuole before pericarp browning. Pericarp anthocyanin degradation by the LAC/ADE was shown to be coupled to epicatechin oxidation.
Laccases: Versatile Enzymes in Plants
Laccases (benzenediol:oxygen oxidoreductase, EC 1.10.3.2) are blue multicopper oxidases that catalyze the oxidation of a range of aromatic compounds, such as mono-, di-, and polyphenols, aromatic amines, and methoxyphenols under reduction of oxygen to water (Messerschmidt, 1997; Burke and Cairney, 2002). They are ubiquitous enzymes that are found in plants, fungi, bacteria, and insects (Giardina et al., 2010). In plants, laccases participate in lignin biosynthesis, carrying out the oxidative polymerization of monolignols (Sterjiades et al., 1992, 1993; Ranocha et al., 1999; Schuetz et al., 2014). These enzymes were also found to be involved in additional physiological processes, such as cytokinin homeostasis (Galuszka et al., 2005), resistance to phenolic pollutants (Wang et al., 2004), flavonoid polymerization in seed coats (Pourcel et al., 2005), and iron metabolism (Hoopes and Dean, 2004). To our knowledge, this is the first report of a plant laccase involved in anthocyanin degradation.
Litchi ADE/LAC Was Glycosylated and Showed High Activity to Epicatechin
Laccase activity was first discovered in the sap of the Japanese lacquer tree Rhus vernicifera (Yoshida, 1883). To date, only a small number of plant laccases were purified and characterized biochemically (Sterjiades et al., 1992; Ranocha et al., 1999; Otto and Schlosser, 2014). A laccase was first purified to homogeneity from the medium of sycamore maple (A. pseudoplatanus) suspension cultures, and found to be 97.4 kD (Sterjiades et al., 1992). A 90-kD poplar (P. euramericana) laccase was purified to homogeneity from a lignifying tissue, and the full-length cDNA sequence coding the protein was achieved by PCR based on the peptide sequences of the enzyme (Ranocha et al., 1999). Recently, a hetero-oligomer of laccases of around 220 kD was purified from the culture supernatants of the green soil alga (Tetracystis aeria; Otto and Schlosser, 2014). Here, we purified a 116-kD laccase (ADE/LAC) responsible for anthocyanin degradation during litchi pericarp browning. The litchi 116-kD ADE/LAC was found to be a single polypeptide, as revealed by urea SDS-PAGE analysis, and larger than the predicted size based on the full-length amino acid sequences deduced by the cDNA sequences, suggesting posttranslation modification. Similar results were found with a poplar laccase, which was found to be 90 kD in size according to a western blot due to high glycosylation, in contrast to the predicted 60 kD based on sequence prediction (Ranocha et al., 1999). Glycan moieties were also detected in the litchi ADE/LAC by the Schiff reagent for sugar moiety detection (Fig. 2E). Similar to the amino acid sequence of the poplar laccase (Ranocha et al., 1999), multiple putative N-glycosylation sites were also inferred in litchi ADE/LAC (Fig. 3B).
The A. pseudoplatanus and P. euramericana laccases showed high activity to oxidize the monolignols, sinapyl, coniferyl, and p-coumaryl alcohols to form water-insoluble polymers (dehydrogenation polymers; Sterjiades et al., 1992; Ranocha et al., 1999). The litchi ADE laccase also oxidized coniferyl and sinapyl alcohol, but had the highest activity to epicatechin as a substrate. Activity with epicatechin was also predicted for an Arabidopsis laccase responsible for the brown color development of seed coats (Pourcel et al., 2005). To the best of our knowledge, this study is the first to present epicatechin as a laccase substrate in vitro.
The Litchi ADE/LAC Is Located in the Vacuole of the Pericarp Cells before Initiation of Senescence
Laccases have often been considered to be located in the extracellular cell wall space since they were purified from culture supernatants of sycamore (A. pseudoplatanus; Driouich et al., 1992; Sterjiades et al., 1992). Most of the laccases identified in plants have a predicted N-terminal signal peptide that could direct proteins into the secretory pathway (Cai et al., 2006). The extracellular localization of the A. pseudoplatanus laccases was well correlated with its function in the lignification of the secondary cell wall (Driouich et al., 1992; Sterjiades et al., 1992). However, it is not clear whether the laccases with other functions are also located in the extracellular space. For example, a laccase in ryegrass (Lolium perenne; LpLAC3) did not possess a signal peptide, suggesting that LpLAC3 may be located inside the cell (Gavnholt et al., 2002). Here, we confirmed the intracellular location of the ADE/LAC enzyme based on immunogold labeling in the pericarp cells of day 0 fruit, as well as fluorescence images of the YFP-fused ADE/LAC in tobacco epidermal cells. The ADE/LAC sequence was predicted to contain a secretion pathway signal peptide, directing the protein to the secretion pathway via the ER, similar to many proteins containing a secretion pathway signal peptide (Nothwehr and Gordon, 1990). This prediction was correlated with the location of the ADE/LAC-YFP fusion protein observed in tobacco epidermal cells (Fig. 5, H–J). In general, the protein secretory pathway in plants begins in the ER, passes through the Golgi complex, and finally reaches its final destination, which may be the vacuole, other compartments, or the extracellular space. The movement between the components in the pathway is mainly by vesicle trafficking (Xiang et al., 2013). In the mature litchi pericarp cells, most of the organelles were disintegrated, and ER, Golgi complex, and cytoplasm were hardly observed. However, some vesicles were found in the vacuole and close to the cell wall of the mature pericarp tissue. By immunogold labeling and TEM, the ADE/LAC protein was shown to be located in the vacuole of the pericarp cells of freshly red fruit; some colocalized with vesicles, indicating that the vacuoles may be the destination of the secretion pathway before pericarp browning. Interestingly, the enzyme colocalized with Phs distributed in the large vacuoles of the litchi pericarp cells (Fig. 6, B and C). Phs were easily observed in the vacuoles of plant cells, such as in banana (Musa acuminata) root cells (Beckman and Mueller, 1970), the mesocarp and endocarp parenchyma cells of walnut (Juglans regia) fruit pericarp, in seed coat cells (Wu et al., 2009), and in pericarp cells of apple (Malus domestica) fruit buds (Konarska, 2014). The colocalization of the ADE/LAC with the Phs indicates that ADE/LAC may be involved in the deposit of phenolic compounds in the vacuoles via oxidation and polymerization of the Phs. Anthocyanins and other flavonoids, such as epicatechin, are located in vacuoles (Kitamura et al., 2004), and contact with these compounds with ADE/LAC may lead to anthocyanin degradation. The distribution of ADE/LACs in the cell walls and intercellular space was observed only after pericarp browning (Fig. 6, H and I), suggesting that the secretion of the enzymes into extracellular space may be correlated with cell senescence.
Litchi ADE Is a Laccase and Not a PPO
The existence of enzymatic activity on catechol or 4-methylcatechol has long been documented for litchi pericarp (Fuchs et al., 1993; Holcroft and Mitcham, 1996; Jiang et al., 2006). Since these two diphenols are the favorite substrates for catechol oxidase or o-diphenol:oxygen oxidoreductase (EC 1.10.3.1), a subclass of PPOs, PPOs have long been considered as the key enzymes functioning in litchi pericarp browning (Jiang and Fu, 1998; Jiang et al., 2006). Using 4-methylcatechol as a substrate, PPO was partially purified from litchi pericarp, and the anthocyanin degradation activity of PPO was demonstrated in vitro in the presence of 4-methylcatechol (Jiang and Fu, 1998). However, due to the lack of protein sequencing in previous studies and the overlap of substrates between PPOs and laccases, it was difficult to ascribe the above-described activity to PPOs or laccases (Jiang and Fu, 1998; Sun et al., 2007). In some studies laccases were classified as part of the PPO superfamily based on its activity on some di- and polyphenols. However laccases belong to a blue multicopper oxidase superfamily whose members have a broader range of substrate specificity than PPOs, which include tyrosinases (EC 1.14.18.1) and catechol oxidase (EC 1.10.3.1), but not laccases (Mayer, 2006; Sun et al., 2006).
The transcript, protein, and activity levels of ADE/LAC were higher in litchi fruit pericarp in comparison with other tissues, such as the fruit aril, correlating to the browning effect occurring in the pericarp but not in the aril. Recently, a litchi PPO gene was cloned using genomic DNA as a template, and its full-length cDNA sequence was obtained (Wang et al., 2014). In the current study, we compared the expression levels of PPO and ADE/LAC in the pericarp tissue during fruit browning and found that ADE/LAC expression levels were about 1,000-fold higher than those of the PPO. Screening the effect of PPO or laccase selective inhibitors, it was found that anthocyanin degradation activity in the crude enzyme extract was dependant on laccase and not on PPO activity (Supplemental Table S2).
Anthocyanin Degradation in Litchi Pericarp Is Coupled to Epicatechin Oxidation
Here, we demonstrate that the highly purified litchi ADE/LAC could not catalyze the oxidation of pure anthocyanin directly, probably due to the presence of sugar moieties in the anthocyanin molecule. The presence of phenolic substrates is crucial for this activity (Fig. 8A). Anthocyanin degradation by ADE/LAC is reminiscent of the model for PPOs, in which PPOs first oxidize phenols to quinones, and then the latter are consequently oxidative polymerized with anthocyanins, the so-called anthocyanin-PPO-phenol system (Sarni et al., 1995; Kader et al., 1998; Jiang, 2000; Zhang et al., 2005). The requirement of additional substrates for oxidative polymerization of anthocyanins explains the reason that ADE/LAC can degrade anthocyanins only from the partially purified extract containing the endogenous phenolic substrates, and not from the homogeneous pigment preparation.
Using the pure ADE laccase enzyme as a tool, we isolated the natural substrate in the partially purified anthocyanin mixture and showed it to be epicatechin. The fact that ADE laccase showed high activity to epicatechin and litchi pericarp contained high levels of epicatechin (Zhang et al., 2000, 2001; Sun et al., 2008) suggests that the anthocyanin-ADE/LAC-epicatechin reaction is the major reaction ascribed to the litchi pericarp anthocyanin degradation. The decrease in epicatechin content after pericarp browning further supports this inference. An Arabidopsis laccase (TT10) also showed activity to epicatechin, but not with quercetin-3-O-rhamnoside, the main glycosylated flavonoids in the seed coat. The fact that the oxidative polymerization of quercetin-3-O-rhamnoside was blocked in a laccase dysfunction mutant (tt10) leading to higher amounts of epicatechin and quercetin-3-O-rhamnoside remaining in the seed coat during maturation indicated that epicatechin is also required for the oxidative polymerization of quercetin-3-O-rhamnoside by TT10 laccase during the browning of the Arabidopsis seed coat (Pourcel et al., 2005).
CONCLUSION
This is the first report, to the best of our knowledge, in which a specific laccase was identified as the enzyme responsible for in planta epicatechin-mediated anthocyanin degradation. Similar enzymes may be identified in the future as responsible for anthocyanin degradation in other fruits after harvest.
MATERIALS AND METHODS
Plant Materials
Litchi fruit (Litchi chinensis ‘Huaizhi’) were harvested from a commercial orchard in the suburb of Guangzhou City, South China, at the fully mature red stage, and transported to the laboratory within 2 h. The fruit were selected for uniformity of shape and color, washed with 0.1% (w/v) hypochlorite in tap water for 2 min, and divided into two groups, each weighing approximately 50 kg. One group was peeled and its pericarp frozen in liquid N2 and stored at −80°C for further analysis. The second group was divided into 15 plastic baskets, with 20 fruit in each, and stored in a controlled environment room at 20°C and about 70% relative humidity for 8 d. On days 0, 2, 4, 6, and 8, three baskets of fruit were taken out for pericarp sampling for three biological replicates. The pericarp was quickly cut into discs 1 cm in diameter, frozen in liquid N2, and stored at −80°C prior to measurement.
Anthocyanin Extraction and Quantification
Litchi pericarp samples (10 discs each) from fruit stored for different lengths of time were blanched with 50 mL of 0.1 m HCl in water. Anthocyanin concentration of the extracts was determined by the pH differential method (Wrolstad et al., 1982). Five milliliters of pericarp extract was diluted either in 25 mL of 0.4 m KCl-HCl buffer (pH 1.0) or in 25 mL of 0.4 m citric acid-Na2HPO4 buffer (pH 4.5), respectively. The absorbance of the dilutions was measured by a spectrophotometer (Shimadzu UV- 2450) at 510 nm. Anthocyanin concentration was calculated as cyanidin-3-glucoside by the method of Wrolstad et al. (1982).
Litchi ADE Substrate Preparation and Activity Assay
Litchi pericarp anthocyanins were extracted in 0.1 m HCl solution and partially purified by Amberlite XAD-7 resin (Sigma-Aldrich) column chromatography, according to Zhang et al. (2004). The red fractions were collected and concentrated by a rotary evaporator (Heidolph). The concentration of the condensed anthocyanins was determined using the pH differential method as described above. Litchi ADE substrate was prepared by dilution of the condensed anthocyanin extract to a concentration of about 0.05 m in 0.1 m sodium acetate buffer (pH 4.0).
Thirty discs of frozen litchi pericarp was ground in liquid N2 to a fine powder, and ADE was extracted by homogenizing the powder with 10 mL of 0.05 m phosphate buffer (pH 7.0) with Polyclar AT (insoluble polyvinylpyrrolidone; 10% of peel by weight) and 1% (w/v) protein inhibitors (Sigma-Aldrich). The homogenate was centrifuged for 20 min at 12,532g and 4°C, and then the supernatant was collected as the crude enzyme extract. ADE activity assay was carried out according to the method of Zhang et al., (2001) with minor modifications. Crude enzyme extract (0.1 mL) was added to the 2 mL of ADE substrate. The mixture was incubated for 20 min at 40°C. The reaction was terminated by adding 2 mL of 0.1 m HCl in methanol. The rates of anthocyanin degradation were determined by the decrease in A510 when compared with the reaction set up in parallel with denatured enzyme that had been boiled for 10 min. Enzyme activity was expressed as the degradation of 1 μmol anthocyanins per hour at 40°C.
ADE Purification
One hundred grams of frozen litchi pericarp was subjected to crude enzyme extraction as described above. The extract was fractionated further by precipitation in 30% to 70% saturation of ammonium sulfate and centrifugation at 12,532g for 20 min. The precipitate was resuspended in 0.05 m phosphate buffer (pH 7.0) and dialyzed overnight against the same buffer. The dialyzed solution was applied to a DEAE-Sepharose (Sigma-Aldrich) column (1.5 × 50 cm) previously equilibrated with 0.05 m phosphate buffer (pH 7.0). ADE was eluted from the column with an NaCl linear gradient (0–0.3 m) in the phosphate buffer at a flow rate of 0.5 mL min−1. Fractions of 2 mL were collected and assayed both for ADE activity and protein concentration (A280). Two major peaks with ADE activity were observed (Fig. 2), and the fractions with high enzymatic activity in the first peak, were pooled and concentrated in an ultrafiltration centrifuge tube (Vivaspin 15; Sartorius). The ADE-enriched fraction was loaded onto a Sephadex G-200 column (1.5 × 50 cm; Sigma-Aldrich), preequilibrated with 0.05 m phosphate buffer (pH 7.0), and eluted with 0.05 m phosphate buffer (pH 7.0) at a flow rate of 0.2 mL min−1. Fractions of 1 mL were collected and assayed for ADE activity and A280, and the fractions with the highest activity and the highest ratio of activity to protein were pooled.
ADE SDS-PAGE/Urea SDS-PAGE Separation, Immunodetection, and Sequencing
The purified protein was denatured by 10 min of boiling in Laemmli’s sample buffer prior to separation using 10% (w/v) SDS-PAGE with or without 8 m urea following standard conditions (Sambrook et al., 1989). The gel was stained with Coomassie Brilliant Blue R-250 (Sigma-Aldrich), and the band of 116 kD in the 10% (w/v) SDS-PAGE was excised and sent for protein sequencing by Sangon Biotech via Biolynx peptide sequencing.
A polyclonal anti-ADE/LAC antibody was generated by Invitrogen using a peptide that was synthesized based on the amino acid sequence at the N terminus of ADE/LAC by Invitrogen. The antibody was purified by antigen affinity chromatography and proved to strongly cross react with the purified ADE/LAC by ELISA. No cross reaction was found for the preimmune serum. For immunodetection, the purified protein or equal amounts of total proteins from different tissues or the pericarp at different time points after fruit harvest were resolved by 10% (w/v) SDS-PAGE and electroblotted onto a nitrocellulose membrane (GE Healthcare) following standard conditions (Sambrook et al., 1989). The membranes were then incubated with the polyclonal anti-ADE/LAC antibody and visualized by the chromogenic technique using the Enhanced HRP-DAB Chromogenic Substrate Kit (Pierce).
In-Gel Activity Assay and Sugar Moiety Detection
Seminative electrophoresis of either the purified or the crude extract proteins was separated on a 10% (w/v) SDS-PAGE without protein sample boiling. The gel was rinsed twice with 50 mm citric acid buffer (pH 3.0) for 5 min to remove SDS, and immersed in the same buffer containing 50 mm partially purified litchi anthocyanins as the substrate until formation of brown pigmentation was seen. Laccase activity was assayed by immersion of the seminative gels in 50 mm DAN in 50 mm citric acid buffer (pH 5.0) according to the methods of Hoopes and Dean (2004). The gels were stained with Schiff reagent for sugar moiety detection according to the method of Gilks et al. (1988).
Substrate Specificity and Behavior toward Effectors
Substrate specificity for a large number of aromatic compounds was tested with the purified ADE/LAC enzyme. Reactions were carried out in 20 mm phosphate buffer (pH 7.0) at 40°C. The compounds tested were epicatechin, 4-methyl catechol, ABTS, catechin, p-anisidine, hydroquinone, phenol, phloroglucinol, coniferyl and sinapyl alcohols, partially purified litchi anthocyanins as described above, and highly purified litchi anthocyanin prepared by Sephadex LH-20 (see the following). Extinction coefficients and concentrations of the substrates, except for epicatechin, were taken from Sterjiades et al. (1993), and the Km and Vmax values for each substrate were determined by extrapolation of Lineweaver and Burk plots. The spectra of epicatechin before and after oxidation by ADE were scanned by spectrophotometer (Shimadzu UV-2450) from 200 to 600 nm. Because epicatechin and its product had overlapping absorbance spectra at the extinction maximum (280 nm; Kennedy and Jones, 2001), and an obvious increase in A380 was recorded in spectra of epicatechin oxidation products, the extinction coefficient of the epicatechin oxidation product was determined at 380 nm. Concentrations of 0.01, 0.02, 0.04, 0.06, 0.08, and 0.1 g L−1 epicatechin were incubated with 0.5 μg of purified ADE for 20 h until the absorbance was stable and the substrate was assumed to be completely converted to product. By plotting the A380 against the epicatechin concentration, the extinction coefficient of the epicatechin oxidation product was estimated.
Several known effectors of PPO and laccases, including p-coumaric acid (1 mm), cinnamic acid (1 mm), ferulic acid (1 mm), polyvinylpyrrolidone (1 mm), SDS (5 mm), and CTAB (5 mm), were tested for their effect on the ADE activity in the crude enzyme extract from litchi pericarp. The partially purified litchi anthocyanins prepared as described above were used as substrate.
Litchi ADE/LAC cDNA Cloning, Sequence Features, and Potential 3D Structure Prediction of the Deduced Protein Sequence
Total RNA was extracted from the pericarps using a modified CTAB-LiCl protocol (Chang et al., 1993). First-strand cDNA was synthesized by PrimeScript (Takara) reverse transcriptase and used as a template to amplify the fragment of the litchi ADE/LAC with degenerate primers designed according to the peptide sequences (Table II; Supplemental Table S2). A 250-bp fragment was amplified and used to design primers (Supplemental Table S3) for 3′ RACE and 5′ RACE to obtain the full-length cDNA sequence of the litchi ADE/LAC.
The deduced protein sequence encoded by a full-length cDNA sequence of the litchi ADE/LAC was obtained by the open reading frame finder software in NCBI (http://www.ncbi.nlm.nih.gov). The putative signal peptide (N-terminal propeptide) in the sequence was identified using the SIGNALP software (Petersen et al., 2011; http://www.cbs.dtu.dk/services/SignalP/). The domain structure was inferred by BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi). A hypothetical 3D structural model and the putative copper binding sites of ADE/LAC were predicted by SWISS-MODEL (http://www.swissmodel.expasy.org/) based on a homolog of a fungal laccase (Protein Data Bank no. 1GYC, Research Collaboratory for Structural Bioinformatics, Rutgers University, http://www.rcsb.org/). The hypothetical 3D structural model was viewed using Cn3D macromolecular structure viewer (http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml) and Vector Alignment Search Tool (http://www.ncbi.nlm.nih.gov/Structure/VAST/vastsearch.html). Putative N-glycosylation sites were predicted by NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/).
Molecular Phylogenetic Analyses of Laccases
Alignment with litchi ADE/LAC and 16 Laccase sequences was performed by ClustalX (Thompson et al., 1997). Heuristic maximum parsimony searches were conducted in PAUP version 4.0b10 (Swofford, 2002) using tree-bisection-reconnection branch swapping and 100 random addition replicates, with unordered (Fitch) parsimony. Bootstrap support values (Felsenstein, 1985) were calculated from 1,000 replicate analyses, using a single random addition replication per bootstrap resampling, and with the maximal number of saved trees per replicate set to 1,000.
Transient Expression of Litchi ADE/LAC, ADE/LAC-YFP in Tobacco Leaves
Nicotiana benthamiana (Nb) plants were grown in a chamber maintained at 23°C to 25°C under a 16-h light/8-h dark cycle. The preparation of Agrobacterium tumefaciens suspension and infiltrations were carried out as described by Sainsbury et al., (2009). A. tumefaciens strain LBA4404 containing pBI101.1-ADE/LAC or pBI101.1-ADE/LAC-YFP vectors was grown at 28°C to stationary phase in Luria-Bertani medium with both kanamycin (50 mg L−1) and rifampicin (50 mg L−1). The cells were pelleted by centrifugation at 2,000g, then resuspended in MMA buffer (10 mm MES [pH 5.6], 10 mm MgCl2, 100 μm acetosyringone) to an absorbance at 600 nm as 1.2 and incubated at ambient temperature for 2 to 4 h. The suspensions were pressure infiltrated into 4-week-old Nb leaves using a syringe. The tissue was harvested 3 d after infiltration for protein extraction. The plants were incubated under normal growing conditions for 2 to 3 d. The sequences of the primers used in the pBI101.1-ADE/LAC or pBI101.1-ADE/LAC-YFP vector construction are listed in Supplemental Table S4.
Histochemical Staining and Subcellular Localization of ADE/LAC by Immunolgold Labeling and Fluorescence Imaging of ADE/LAC-YFP in Tobacco Leaves
Eighty-millimeter-thick litchi pericarp sections were hand cut using a razor blade, rinsed in cold sodium acetate buffer (50 mm, pH 5.0), and incubated for 20 min at 25°C with catalase (100 mg mL−1; Sigma-Aldrich) to eliminate endogenous hydrogen peroxide. Samples were then rinsed again and incubated overnight at 37°C in the dark with 2.3 mm ABTS or 11 mm 4-methylcatechol in sodium acetate buffer (pH 5.0) containing catalase (100 mg mL−1). The two controls were pericarp sections incubated in buffer without substrate, and sections were boiled for 15 min to destroy enzyme activity.
For subcellular immunogold localization, 2- × 2-mm discs of litchi pericarp were fixed in 2% (v/v) glutaraldehyde, 4% (w/v) paraformaldehyde, and 100 mm phosphate-buffered saline (PBS; pH 7.2) for 12 h, washed three times for 30 min in 100 mm PBS (pH 7.2), and dehydrated in 30%, 50%, 70%, 80%, 85%, 90%, 95%, and 100% (v/v) of ethanol at 4°C. The samples were infiltrated with LRWhite (Electron Microscopy Sciences) via three intermediate steps at 2:1, 1:1, and 1:2 (v/v) mixture of ethanol:LRWhite (12 h each time). Finally, the mixture was replaced by pure LRWhite, kept for 12 h at −20°C, and then changed once with fresh LRWhite and kept for 1 d at −20°C. Samples were then transferred to capsules with fresh LRWhite and cured under two 15-W ultraviolet lamps (360 nm) for at least 24 h at −20°C, and then continued curing for 2 d at room temperature. Ultrathin sections (60 nm) were cut using a Sorvall MT-6000 ultra-microtome (Leica) and collected onto Formvar-coated nickel grids (Gilder Grids). The grids were floated on PBS-Tween (PBST) buffer (60 mm PBS, 0.1% [v/v] Tween 20, 0.02% [w/v] NaN3, pH 7.2) containing 0.2 m Gly and 1% (w/v) bovine serum albumin to block nonspecific binding, and then in a drop of the primary anti- ADE/LAC (1:50, v/v) for 3 h at 37°C. The grids were then rinsed with PBST three times and floated on 1:50 (v/v) goat anti-rabbit IgG antibody conjugated to 10-nm gold particles (Sigma-Aldrich) for 1 h at 37°C. After washing three times in PBST and then in distilled water, respectively, the grids were air dried and stained with saturated uranyl acetate for 30 min. Control sections were treated similarly except that the primary antibody was substituted with PBS. The samples were observed using a Philip FEI-TECNAI 12 transmission electron microscope (FEI). For the TEM observation of litchi pericarp at different developmental stages, 2- × 2-mm discs of pericarp were fixed with 2% (v/v) glutaraldehyde in pH 7.2 10 mm PBS for 12 h. After being washed four times in 10 mm PBS, the samples were subsequently fixed in 1% (w/v) OsO4 according to the methods by Zhao et al. (2002).
To confirm the subcellular localization detected by immunolgold labeling, the Nb leaves infiltrated with A. tumefaciens strain LBA4404 containing pBI101.1-ADE/LAC-YFP, as described above, were photographed with a confocal laser scanning microscope (LSM 710, Zeiss). Lysotracker red (50 nm; Invitrogen; Barasch et al., 1991) and ER-Tracker Blue-White DPX (100 nm; Invitrogen; Zünkler et al., 2004) were applied on infiltrated leaves for 1 or 0.5 h, and then the epidermis of the infiltrated leaf was layered in a drop of 30% (w/w) Suc on a glass slide for plasmolysis of the cells. The fluorescent markers were visualized with the following excitation and emission wavelengths (excitation/emission): for YFP, 540 nm/590 to 610 nm; for ER-Tracker, 374 nm/430 to 640 nm; for Lysotracker red, 577 nm/590.
Gene Transcript Abundance Analysis
Ten micrograms of total RNA was separated on a 1.2% (w/v) agarose formaldehyde gel and capillary blotted onto a 0.45-μm polyvinylidene difluoride membrane (Biodyne_B). The membrane was UV cross linked at 280 nm. DIG-labeled probes of litchi ADE/LAC were produced using a PCR DIG probe synthesis kit (Roche). The membrane was hybridized with the DIG-labeled probe for 16 h at 45°C in high-SDS buffer (7% [w/v] SDS, 5 × SSC, 50 mm pH 7.0 sodium phosphate, 2% [w/v] blocking reagent [Roche], and 0.1% [w/v] N-lauroylsarcosine, and 50% [v/v] deionized formamide). Blots were washed twice at 37°C in 2 × SSC and 0.1% (w/v) SDS for 10 min, and twice at 62°C in 0.2 × SSC and 0.1% (w/v) SDS for 30 min. The membranes were then subjected to immunological detection following the manufacturer’s instructions (Roche).
qRT-PCR was applied to analyze the gene transcript abundance of the litchi ADE/LAC and PPO after harvest. cDNA was prepared from 1 μg of total RNA with PrimeScript reverse transcriptase (TakaRa) after genomic DNA was removed by RQ1 DNase (Promega). Specific primers for the genes were designed and are described in Supplemental Table S4. Litchi Actin 2 (ACT) was selected as a reference gene. PCRs were performed in a total volume of 20 μL, containing 200 nm each primer and 10 μL of SYBR Green PCR Supermix (Toyobo) on a Bio-Rad CFX96 Real-Time PCR system. The specificity of primes was tested using a melting curve, product sequencing, and PCR efficiency. qRT-PCRs were normalized using the cycle threshold (Ct) value corresponding to the ACT gene. The relative transcript levels of the ADE/LAC or PPO gene to ACT gene were calculated using the equation 2–ΔCt (Livak and Schmittgen, 2001).
Extraction and Purification of the Endogenous Substrate for LAC from Litchi Pericarp
Frozen litchi pericarp (200 g) was blanched with 2 L of 0.1 m HCl in water. All extracts were combined and filtered through two layers of microcloth. The filtrate was then subjected to Amberlite XAD-7 resin (Sigma-Aldrich) column chromatography according to Zhang et al. (2004). Fractions with the highest A510 were pooled and concentrated by removing the methanol at 40°C in a rotary evaporator (Heidolph). The concentrated substrates were then loaded onto a Sephadex LH-20 column (1.0 × 60 cm; Sigma-Aldrich), eluted with aqueous 1% (v/v) formic acid with a methanol linear gradient (0%–100% [v/v]) at 24 mL h−1, and collected using a fraction collector (2 mL per tube). The fractions of eight peaks, based on the absorbance values at 510 and 280 nm, were pooled and concentrated by a rotary evaporator at 40°C. Fractions were then diluted in 0.1 m sodium acetate buffer (pH 4.0) to an A280 of around 1.0, then used as substrates for the activity assay by incubation with the purified ADE/LAC enzyme. The reactions that turned brown were considered to contain endogenous substrates for the ADE/LAC.
Identification of Endogenous Substrates by HPLC and LC-MS Analysis after the Laccase Reactions
The activity assay of the ADE/LAC enzyme on the potential substrates obtained above was carried out by incubating 2.95 mL of the substrates with 50 μL of the purified enzyme, with the boiled enzyme serving as the control. A standard, 10 mm epicatechin in the same buffer, was also tested as substrate for the reactions. The reactions were terminated by addition of 3 mL of chloroform. After centrifugation at 12,396g for 5 min, the upper aqueous phase was collected and analyzed using HPLC analysis (Martinez et al., 2012) on an Agilent 1200 Series HPLC system (Agilent Technologies) equipped with an autosampler, diode array detector, and ChemStation software. The reaction mixture was loaded onto a C18 (2) column (Luna, 5 μm, 250 × 4.6 mm; Phenomenex) and eluted with a linear gradient of acetonitrile (0%–100% [v/v]) over 35 min. The flow rate was 1 mL min−1 and the injection volume was 15 μL. The monitoring wavelength was 280 or 510 nm. The products of ADE/LAC enzyme on highly purified anthocyanins with and without epicatechin were analyzed by HPLC as described above.
Identification of potential substrates was achieved by comparing the retention times of the standards and the elution profiles between the reactions with native and denatured enzyme preparations. Compounds that decreased in concentration significantly due to the enzymatic reaction were collected and injected to the HPLC-MS system (Thermo Finnigan LCQ DECA) for MS analysis with acetonitrile with 0.5% (v/v) formic acid as mobile phase at a flow rate of 1 mL min−1. The MS was recorded with a heat capillary voltage of 5 kV, a heat capillary temperature of 275°C, sheath gas flow rate of 80 units, and auxiliary gas flow rate of 20 units. The scan range of molecular mass was 50 to 1,000 D.
Epicatechin Content Measurement
Two grams of pericarp tissue from 30 fruits was ground in liquid N2 and extracted with 40 mL of 60% (v/v) ethanol for 30 min in a 30°C ultrasonic bath. The extract was then filtered through a 0.45-μm polyvinylidene difluoride membrane (ANPEL Scientific Instruments) and injected into the HPLC system. Elution was done with a linear gradient of 0.1% (v/v) formic acid and methanol (0%–100% [v/v]) over 35 min. The epicatechin peak was identified by comparison with a standard. The content of epicatechin in the extraction was calculated based on the integral of the identified peak and the standard curve.
Sequence data from this article can be found in the GenBank data libraries (http://www.ncbi.nlm.nih.gov) under the following accession numbers: L. chinensis, GU367910.1 (ADE/LAC); DQ990337.1 (ACT); JF926153.1 (PPO). Liriodendron tulipifera, U73104.1 (LAC2-2), U73106.1 (LAC2-4), U73105.1 (LAC2-3), U73103.1 (LAC2-1); Populus trichocarpa, Y13773.1 (Lac110), Y13769.1 (Lac1), Y13771.1 (Lac3), Y13772.1 (Lac90); Zea mays, AM086217.1 (Lac5); A. pseudoplatanus, U12757.1; Castanea mollissima, FJ231469.1; Arabidopsis, At5g48100 (Laccase 15), NM_128574.3 (Laccase 3); Picea abies, JX500690.1 (Lac4); Cryphonectria parasitica, S38903.1; and Neurospora crassa, M18333.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Determination of the extinction coefficient of epicatechin oxidation product by ADE/LAC.
Supplemental Figure S2. The original gel images of ADE/LAC protein and activity assays in different litchi tissues.
Supplemental Figure S3. TEM images of litchi pericarp at different stages.
Supplemental Figure S4. MS and MS/MS profiles of the active compound of the substrate I.
Supplemental Table S1. Purification of the ADE from litchi fruit pericarp.
Supplemental Table S2. Comparison of inhibitor responses for ADE activity from litchi fruit pericarp.
Supplemental Table S3. Primers used for full length cDNA fragment cloning of ADE/LAC gene.
Supplemental Table S4. Primers used for construction of the vector for transient expression ADE/LAC and ADE/LAC-YFP protein in Nb leaves.
Supplemental Table S5. Primers used for qRT-PCR analysis.
Supplementary Material
Acknowledgments
The authors thank Maor Bar-Peled (University of Georgia) for helping to set up the activity assay for ADE/LAC on substrate and anthocyanins by HPLC and LC-MS, Xiao-yi Wei (South China Botanical Garden, Chinese Academy of Sciences) for detailed suggestions for the identification of the endogenous substrate for the litchi ADE/LAC, and Yao-guang Liu and Le-tian Chen (South China Agricultural University) for helping with the potential 3D structure prediction.
Glossary
- POD
peroxidase
- MS/MS
tandem mass spectrometry
- cDNA
complementary DNA
- ABTS
2,2′-azinobis-3-ethylbenzthiazolinesulfonic acid
- NCBI
National Center for Biotechnology Information
- 3D
three-dimensional
- DIG
digoxigenin
- DAN
1,8-diaminonaphthalene
- CTAB
cetyl-trimethyl-ammonium bromide
- TEM
transmission electron microscopy
- Phs
phenolic substance
- ICS
intercellular space
- LC
liquid chromatography
- MS
mass spectrometry
- PBS
phosphate-buffered saline
- Nb
Nicotiana benthamiana
- PBST
to come
- Phs
phenolic grains
- A510
absorbance at 510 nm
- PBST
phosphate-buffered saline-Tween
Footnotes
This work was supported by the National Key Basic Research Program of China (grant no. 2013CB127105), the National Natural Science Foundation of China (grant no. 31171988), and the China Litchi and Logan Research System (grant no. CARS–33–14).
Articles can be viewed without a subscription.
References
- Bar-Akiva A, Ovadia R, Rogachev I, Bar-Or C, Bar E, Freiman Z, Nissim-Levi A, Gollop N, Lewinsohn E, Aharoni A, et al. (2010) Metabolic networking in Brunfelsia calycina petals after flower opening. J Exp Bot 61: 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barasch J, Kiss B, Prince A, Saiman L, Gruenert D, al-Awqati Q (1991) Defective acidification of intracellular organelles in cystic fibrosis. Nature 352: 70–73 [DOI] [PubMed] [Google Scholar]
- Barbagallo RN, Palmeri R, Fabiano S, Rapisarda P, Spagna G (2007) Characteristic of beta-glucosidase from sicilian blood oranges in relation to anthocyanin degradation. Enzyme Microb Technol 41: 570–575 [Google Scholar]
- Beckman CH, Mueller WC (1970) Distribution of phenols in specialized cells of Banana roots. Phytopathology 70: 79–82 [Google Scholar]
- Burke RM, Cairney JW (2002) Laccases and other polyphenol oxidases in ecto- and ericoid mycorrhizal fungi. Mycorrhiza 12: 105–116 [DOI] [PubMed] [Google Scholar]
- Cai X, Davis EJ, Ballif J, Liang M, Bushman E, Haroldsen V, Torabinejad J, Wu Y (2006) Mutant identification and characterization of the laccase gene family in Arabidopsis. J Exp Bot 57: 2563–2569 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116 [Google Scholar]
- Cheynier V, Souquet JM, Kontek A, Moutounet M (1994) Anthocyanin degradation in oxidising grape musts. J Sci Food Agric 66: 283–288 [Google Scholar]
- Dela G, Or E, Ovadia R, Nissim-Levi A, Weiss D, Oren-Shamir M (2003) Changes in anthocyanin concentration and composition in ‘Jaguar’ rose flowers due to transient high-temperature conditions. Plant Sci 164: 333–340 [Google Scholar]
- Driouich A, Lainé A, Vian B, Faye L (1992) Characterization and localization of laccase forms in stem and cell cultures of sycamore. Plant J 2: 13–24 [Google Scholar]
- Fang Z, Zhang M, Du W, Sun J (2007) Effect of fining and filtration on the haze formation in bayberry (Myrica rubra Sieb. et Zucc.) juice. J Agric Food Chem 55: 113–119 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenetics: an approach using the bootstrap. Evolution 4: 783–791 [DOI] [PubMed] [Google Scholar]
- Fuchs Y, Zauberman G, Ronen R, Rot I, Weksler A, Akerman M (1993) The physilological basis of lichi fruit pericarp color retention. Acta Hortic 343: 29–33 [Google Scholar]
- Galuszka P, Frébortová J, Luhová L, Bilyeu KD, English JT, Frébort I (2005) Tissue localization of cytokinin dehydrogenase in maize: possible involvement of quinone species generated from plant phenolics by other enzymatic systems in the catalytic reaction. Plant Cell Physiol 46: 716–728 [DOI] [PubMed] [Google Scholar]
- Gavnholt B, Larsen K, Rasmussen SK (2002) Isolation and characterisation of laccase cDNAs from meristematic and stem tissues of ryegrass (Lolium perenne). Plant Sci 162: 873–885 [Google Scholar]
- Giardina P, Faraco V, Pezzella C, Piscitelli A, Vanhulle S, Sannia G (2010) Laccases: a never-ending story. Cell Mol Life Sci 67: 369–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks CB, Reid PE, Clement PB, Owen DA (1988) Simple procedure for assessing relative quantities of neutral and acidic sugars in mucin glycoproteins: its use in assessing cyclical changes in cervical mucins. J Clin Pathol 41: 1021–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbach RJ. (2005) Biochemistry and genetics of flower color. Plant Breed Rev 25: 89–114 [Google Scholar]
- Holcroft DM, Mitcham EJ (1996) Postharvest physiology and handling of litchi (Litchi chinensis Sonn.). Postharvest Biol Technol 9: 265–281 [Google Scholar]
- Hoopes JT, Dean JF (2004) Ferroxidase activity in a laccase-like multicopper oxidase from Liriodendron tulipifera. Plant Physiol Biochem 42: 27–33 [DOI] [PubMed] [Google Scholar]
- Huang HT. (1956) The Kinetics of the Decolorization of Anthocyanins by Fungal “Anthocyanase”. J Am Chem Soc 78: 2390–2393 [Google Scholar]
- Jiang Y. (2000) Role of anthocyanins, polyphenol oxidase and phenols in lychee pericarp browning. J Sci Food Agric 80: 305–310 [Google Scholar]
- Jiang Y, Fu J (1998) Inhibition of polyphenol oxidase and the browning control of litchi fruit by glutathione and citric acid. Food Chem 62: 49–52 [Google Scholar]
- Jiang YM, Wang Y, Song L, Liu H, Lichter A, Kerdchoechuen O, Joyce DC, Shi J (2006) Postharvest characteristics and handling of litchi fruit - an overview. Aust J Exp Agric 46: 1541–1556 [Google Scholar]
- Kader F, Haluk J, Nicolas J, Metche M (1998) Degradation of cyanidin 3-glucoside by blueberry polyphenol oxidase: Kinetic studies and mechanisms. J Agric Food Chem 46: 3060–3065 [Google Scholar]
- Kennedy JA, Jones GP (2001) Analysis of proanthocyanidin cleavage products following acid-catalysis in the presence of excess phloroglucinol. J Agric Food Chem 49: 1740–1746 [DOI] [PubMed] [Google Scholar]
- Kitamura S, Shikazono N, Tanaka A (2004) TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J 37: 104–114 [DOI] [PubMed] [Google Scholar]
- Konarska A. (2014) Differences in the structure of fruit buds in two apple cultivars with particular emphasis on features responsible for fruit storability and quality. Acta Sci Pol Hortorum Cultus 13: 91–105 [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Martinez V, Ingwers M, Smith J, Glushka J, Yang T, Bar-Peled M (2012) Biosynthesis of UDP-4-keto-6-deoxyglucose and UDP-rhamnose in pathogenic fungi Magnaporthe grisea and Botryotinia fuckeliana. J Biol Chem 287: 879–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer AM. (2006) Polyphenol oxidases in plants and fungi: going places? A review. Phytochemistry 67: 2318–2331 [DOI] [PubMed] [Google Scholar]
- Messerschmidt A. (1997) Multi-copper oxidases. World Scientific, Singapore [Google Scholar]
- Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3: 212–217 [Google Scholar]
- Mori K, Goto-Yamamoto N, Kitayama M, Hashizume K (2007) Loss of anthocyanins in red-wine grape under high temperature. J Exp Bot 58: 1935–1945 [DOI] [PubMed] [Google Scholar]
- Nothwehr SF, Gordon JI (1990) Targeting of proteins into the eukaryotic secretory pathway: signal peptide structure/function relationships. BioEssays 12: 479–484 [DOI] [PubMed] [Google Scholar]
- Oren-Shamir M. (2009) Does anthocyanin degradation play a significant role in determining pigment concentration in plants? Plant Sci 177: 310–316 [Google Scholar]
- Otto B, Schlosser D (2014) First laccase in green algae: purification and characterization of an extracellular phenol oxidase from Tetracystis aeria. Planta 240: 1225–1236 [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I (2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17: 2966–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranocha P, McDougall G, Hawkins S, Sterjiades R, Borderies G, Stewart D, Cabanes-Macheteau M, Boudet AM, Goffner D (1999) Biochemical characterization, molecular cloning and expression of laccases - a divergent gene family - in poplar. Eur J Biochem 259: 485–495 [DOI] [PubMed] [Google Scholar]
- Raynal J, Moutounet M, Souquet JM (1989) Intervention of phenolic compounds in plum technology. 1. Changes during drying. J Agric Food Chem 37: 1046–1050 [Google Scholar]
- Rowan DD, Cao M, Lin-Wang K, Cooney JM, Jensen DJ, Austin PT, Hunt MB, Norling C, Hellens RP, Schaffer RJ, et al. (2009) Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol 182: 102–115 [DOI] [PubMed] [Google Scholar]
- Sainsbury F, Thuenemann EC, Lomonossoff GP (2009) pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J 7: 682–693 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniais T (1989) Molecular Cloning: A Laboratory Manual (2nd ed.). Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- Sarni P, Fulcrand H, Souillol V, Souquet J, Cheynier V (1995) Mechanisms of anthocyanin degradation in grape must-like model solutions. J Sci Food Agric 69: 385–391 [Google Scholar]
- Schuetz M, Benske A, Smith RA, Watanabe Y, Tobimatsu Y, Ralph J, Demura T, Ellis B, Samuels AL (2014) Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol 166: 798–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterjiades R, Dean J, Gamble G, Himmelsbach D, Eriksson K (1993) Extracellular laccases and peroxidases from sycamore maple (Acer pseudoplatanus) cell-suspension cultures. Planta 190: 75–87 [Google Scholar]
- Sterjiades R, Dean JF, Eriksson KE (1992) Laccase from Sycamore Maple (Acer pseudoplatanus) polymerizes monolignols. Plant Physiol 99: 1162–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn WJ, Holcroft DM, Wand SJE, Jacobs G (2004) Anthocyanin degradation in detached pome fruit with reference to preharvest red color loss and pigmentation patterns of blushed and fully red pears. J Am Soc Hortic Sci 129: 13–19 [Google Scholar]
- Steyn WJ, Wand SJE, Holcroft DM, Jacobs G (2002) Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol 155: 349–361 [DOI] [PubMed] [Google Scholar]
- Sun J, Jiang Y, Wei X, Shi J, You Y, Liu H, Kakuda Y, Zhao M (2006) Identification of (-)-epicatechin as the direct substrate for polyphenol oxidase isolated from litchi pericarp. Food Res Int 39: 864–870 [Google Scholar]
- Sun J, Jiang Y, Wei X, Zhao M, Shi J, You Y, Yi C (2007) Identification of procyanidin A2 as polyphenol oxidase substrate in pericarp tissues of litchi fruit. J Food Biochem 31: 300–313 [Google Scholar]
- Sun J, Shi J, Zhao M, Xue SJ, Ren J, Jiang Y (2008) A comparative analysis of property of lychee polyphenoloxidase using endogenous and exogenous substrates. Food Chem 108: 818–823 [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2002) PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods), Version 4.0b10. Sunderland, MA: Sinauer. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaknin H, Bar-Akiva A, Ovadia R, Nissim-Levi A, Forer I, Weiss D, Oren-Shamir M (2005) Active anthocyanin degradation in Brunfelsia calycina (yesterday--today--tomorrow) flowers. Planta 222: 19–26 [DOI] [PubMed] [Google Scholar]
- Wang GD, Li QJ, Luo B, Chen XY (2004) Ex planta phytoremediation of trichlorophenol and phenolic allelochemicals via an engineered secretory laccase. Nat Biotechnol 22: 893–897 [DOI] [PubMed] [Google Scholar]
- Wang J, Liu B, Xiao Q, Li H, Sun J (2014) Cloning and expression analysis of litchi (Litchi Chinensis Sonn.) polyphenol oxidase gene and relationship with postharvest pericarp browning. PLoS One 9: e93982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5: 218–223 [DOI] [PubMed] [Google Scholar]
- Wrolstad RE, Culbertson JD, Cornwell CJ, Mattick LR (1982) Detection of adulteration in blackberry juice concentrates and wines. J Assoc Off Anal Chem 65: 1417–1423 [PubMed] [Google Scholar]
- Wu GL, Liu QL, Teixeira Da Silva JA (2009) Ultrastructure of pericarp and seed capsule cells in the developing walnut (Juglans regia L.) fruit. S Afr J Bot 75: 128–136 [Google Scholar]
- Xiang L, Etxeberria E, Van den Ende W (2013) Vacuolar protein sorting mechanisms in plants. FEBS J 280: 979–993 [DOI] [PubMed] [Google Scholar]
- Yoshida H. (1883) Chemistry of lacquer (urushi). Part I. J Chem Soc 43: 472–486 [Google Scholar]
- Zenner K, Bopp M (1987) Anthocyanin Turnover in Sinapis alba L. J Plant Physiol 126: 475–482 [Google Scholar]
- Zhang D, Quantick PC, Grigor JM (2000) Changes in phenolic compounds in Litchi (Litchi chinensis Sonn.) fruit during postharvest storage. Postharvest Biol Technol 19: 165–172 [Google Scholar]
- Zhang Z, Pang X, Ji Z, Jiang Y (2001) Role of anthocyanin degradation in litchi pericarp browning. Food Chem 75: 217–221 [Google Scholar]
- Zhang Z, Xuequn P, Yang C, Ji Z, Jiang Y (2004) Purification and structural analysis of anthocyanins from litchi pericarp. Food Chem 84: 601–604 [Google Scholar]
- Zhang ZQ, Pang XQ, Duan XW, Ji ZL, Jiang YM (2005) Role of peroxidase in anthocyanin degradation in litchi fruit pericarp. Food Chem 90: 47–52 [Google Scholar]
- Zhao J, Yu FL, Liang SP, Zhou C, Yang HY (2002) Changes of calcium distribution in egg cells, zygotes and two-celled proembryos of rice (Oryza sativa L.). Sex Plant Reprod 14: 331–337 [Google Scholar]
- Zipor G, Duarte P, Carqueijeiro I, Shahar L, Ovadia R, Teper-Bamnolker P, Eshel D, Levin Y, Doron-Faigenboim A, Sottomayor M, Oren-Shamir M et al. (2015) In planta anthocyanin degradation by a vacuolar class III peroxidase in Brunfelsia calycina flowers. New Phytol 205: 653–665 [DOI] [PubMed] [Google Scholar]
- Zünkler BJ, Wos-Maganga M, Panten U (2004) Fluorescence microscopy studies with a fluorescent glibenclamide derivative, a high-affinity blocker of pancreatic beta-cell ATP-sensitive K+ currents. Biochem Pharmacol 67: 1437–1444 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.