Cis-regulatory elements and regulatory networks have diverged considerably in duplicated genes.
Abstract
Plant genomes contain large numbers of duplicated genes that contribute to the evolution of new functions. Following duplication, genes can exhibit divergence in their coding sequence and their expression patterns. Changes in the cis-regulatory element landscape can result in changes in gene expression patterns. High-throughput methods developed recently can identify potential cis-regulatory elements on a genome-wide scale. Here, we use a recent comprehensive data set of DNase I sequencing-identified cis-regulatory binding sites (footprints) at single-base-pair resolution to compare binding sites and network connectivity in duplicated gene pairs in Arabidopsis (Arabidopsis thaliana). We found that duplicated gene pairs vary greatly in their cis-regulatory element architecture, resulting in changes in regulatory network connectivity. Whole-genome duplicates (WGDs) have approximately twice as many footprints in their promoters left by potential regulatory proteins than do tandem duplicates (TDs). The WGDs have a greater average number of footprint differences between paralogs than TDs. The footprints, in turn, result in more regulatory network connections between WGDs and other genes, forming denser, more complex regulatory networks than shown by TDs. When comparing regulatory connections between duplicates, WGDs had more pairs in which the two genes are either partially or fully diverged in their network connections, but fewer genes with no network connections than the TDs. There is evidence of younger TDs and WGDs having fewer unique connections compared with older duplicates. This study provides insights into cis-regulatory element evolution and network divergence in duplicated genes.
Gene duplication events during evolutionary history have provided large numbers of new genes that can diverge in function and gain new functions, resulting in new morphological, physiological, and biochemical characteristics of organisms and cellular systems. Whole-genome duplication events provide a major source of duplicated genes (whole-genome duplicates [WGDs]) and are frequent in flowering plants where multiple whole-genome duplications have occurred during the evolutionary history of most lineages (for review, see Van de Peer et al., 2009; Tang et al., 2010; Jiao et al., 2011). Genes may be duplicated by several mechanisms in addition to WGDs, which have been collectively referred to as small-scale duplications (Maere et al., 2005). Tandem duplicate (TD) genes, which are consecutive genes in the genome, are thought to arise from unequal crossing over (Freeling, 2009). There are also other types of duplicates, including proximal duplicates, which are near one another but separated by a few genes; dispersed duplicates, which are neither adjacent to each other in the genome nor within homologous chromosome segments; and retroposed duplicates, formed by RNA-based retroposition (e.g. Wang et al., 2013). A large portion of the Arabidopsis (Arabidopsis thaliana) genome is believed to derive from evolutionarily recent polyploidy events, including the alpha and beta WGDs, and approximately 17% of genes are TDs (Vision et al., 2000; Simillion et al., 2002; Bowers et al., 2003).
Genes can acquire various fates following duplication. The most likely fate is the transformation of one paralog to a pseudogene (Lynch and Conery, 2000; Long and Thornton, 2001; Zhang et al., 2001). Alternatively, one copy can acquire a novel function while the other maintains the ancestral function (neofunctionalization), or one copy acquires a new expression pattern (regulatory neofunctionalization). Multiple functions of the original gene can be divided between the two duplicates (subfunctionalization), or the organ-specific expression pattern can be divided between the two duplicates (regulatory subfunctionalization). Alternatively, duplicated genes may retain the same function and expression pattern, and thus add to the robustness of the regulatory network (Force et al., 1999; Gu et al., 2003). Regulatory neofunctionalization and subfunctionalization are likely caused by mutational changes within the cis-regulatory region of the duplicated genes, which alter the temporal/spatial expression profile as well as responses to various biotic and abiotic stimuli.
Several genome-wide studies have attempted to describe the characteristics of regulatory evolution in model organisms (e.g. Dermitzakis and Clark, 2002; Schmidt et al., 2010). A study of Escherichia coli and Saccharomyces cerevisiae showed that duplicated new genes inherit more than one-third of the regulatory interactions from their ancestral genes (Teichmann and Babu, 2004). In yeast (S. cerevisiae), recently duplicated genes were shown to rapidly gain transcription factor binding sites after duplication (Tsai et al., 2012). In Arabidopsis, Haberer et al. (2004) compared upstream promoter regions between duplicated genes and identified conserved regions that might be cis-regulatory elements. When analyzing a few duplicated pairs that contain experimentally verified cis-regulatory elements, Haberer et al. (2004) demonstrated conservation of these known cis-regulatory elements. Similarly, sequence alignments of the regions surrounding retained WGDs have revealed small (15–255 bp) conserved noncoding DNA sequence (CNS) patterns in close proximity to retained duplicate genes (Freeling et al., 2007; Thomas et al., 2007). The size and similarity of these CNS signatures imply a functional role, and those genes that were most enriched for CNSs were most often associated with transcription factor activity (Freeling et al., 2007). It is likely that many CNSs play cis-regulatory roles shared by WGDs, as reviewed by Freeling and Subramaniam (2009). A significant positive correlation of duplicate pair coexpression with the full-gene CNS count in a number of different tissues was observed, indicating a broad positive effect of CNS signatures on WGD pair coexpression (Spangler et al., 2012).
The interaction of transacting regulatory factors with cis-regulatory sites that lead to chromatin remodeling and changes in gene expression has long been assayed as hypersensitivity to cleavage by the nonspecific endonuclease DNase I (Wu et al., 1979; Wu, 1980). DNase I hypersensitive sites colocalize with a spectrum of regulatory sequences, including enhancers, promoters, silencers, insulators, locus control regions, and domain boundary elements (Stalder et al., 1980; Forrester et al., 1986; Grosveld et al., 1987; Chung et al., 1993). DNase I sequencing, a method that relies on short sequence tags from DNase I-treated genetic material, was adapted and applied to Arabidopsis, revealing a large amount of regulatory information (Sullivan et al., 2014). The study examined the dynamics of the cis-regulatory landscape and found thousands of DNase I-hypersensitive sites. Within those sites were hundreds of thousands of footprints left by regulatory proteins as they bound the underlying DNA and protected it from DNase I cleavage. Although only a handful of the approximately 1,700 predicted Arabidopsis transcription factors (Palaniswamy et al., 2006) have well-described target motifs, Sullivan et al. (2014) used this information to build and describe empirical genome-wide regulatory networks.
In this study, we use DNase I footprints, which are locations of protein binding sites, and regulatory information from Sullivan et al. (2014) to analyze the evolution of cis-regulatory elements in duplicated genes of Arabidopsis. We found that footprint density and regulatory network characteristics vary based on the type of duplicate. WGDs had more footprints and formed denser and more complex regulatory networks than TDs. The regulatory networks of pairs of duplicates appeared more diverged than conserved, regardless of duplication class, and this divergence appeared to be linked to the age of the duplication.
RESULTS
The Regulatory Footprint Landscape Differs between Types of Duplicated Genes
To analyze the evolution of cis-regulatory elements in duplicated genes, the number and distribution of DNase I-protected sites within DNase I-hypersensitive regions, referred to as footprints, between two classes of duplicated genes were compared. The DNase I footprints provide single-base resolution of protein binding sites. (This approach is different from phylogenetic footprinting, where sequences from multiple species are aligned to identify conserved noncoding regions.) Duplicate gene sets analyzed included 3,045 pairs of WGDs from Blanc et al. (2003) and 1,370 pairs of TDs from Liu et al. (2011). A set of 2,301 duplicates of other types, including dispersed, proximal, and retroposed, referred to here as other duplicates, was also included in some analyses for comparison with the WGDs and TDs. DNase I footprint coordinates from Sullivan et al. (2014) were assigned to each duplicate gene. In Arabidopsis, the largest percentage of DNase I-hypersensitive regions falls within 400 bp upstream of a gene’s transcription start site (Sullivan et al., 2014). To confirm that the footprints left behind by regulatory elements in duplicated genes are also mainly found within a few hundred nucleotides upstream of the transcription start site, the distribution of footprints around duplicate genes was calculated. Similar to DNase I-hypersensitive areas, the largest proportion of footprints (31.3%) among duplicate genes could be found within the first 500 bp upstream (Table I). A recent study of single nucleotide polymorphism density profiles also identified this length as the effective promoter length (Korkuc et al., 2014). The regulatory analysis focused on footprints located within the first 500 bp upstream of the transcription start site, and that region is referred to as the promoter region.
Table I. Footprint distribution of WGDs and TDs.
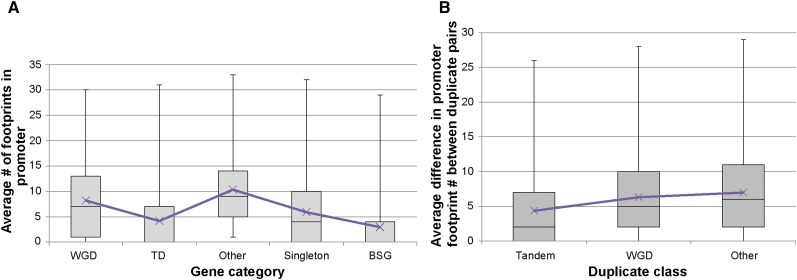
To assess the regulatory landscape of WGD and TD genes, the average number of footprints in the promoter region was calculated. On average, WGD genes had more footprints in their promoters compared with TDs (Fig. 1A), 8.1 versus 4.2 (P = 2.07E−153, Mann-Whitney U test following a Kruskal-Wallis one-way ANOVA). This was the case in 7-d-old seedlings grown in the dark as well as seedlings exposed to heat shock (Supplemental Fig. S1). To compare the WGD and TDs with the other duplicates set, the average number of footprints in the promoters was calculated for the other duplicates and found to be 10.3. This is significantly higher than the WGDs (P = 1.94E−60) and the TDs (P = 7.35E−348). To compare duplicates with singletons, the average number of footprints in singletons was calculated and found to be 5.9 (Fig. 1A). Thus, singletons tend to have fewer footprints than duplicates, except for TDs (P = 3.36E−49 compared with WGDs, 9.56E−38 compared with TDs, and 2.74E−195 compared with other duplicates).
Figure 1.
Regulatory protein footprint numbers differ in classes of duplicates. A, Box plots show the number of footprints within 500 bp of the transcription start site in different classes of duplicates as well as Brassicaceae-specific genes (BSG) and singletons. Purple Xs show the mean (Kruskal-Wallis test and Mann-Whitney U test P values are significant for all comparisons, P < 0.01). B, Box plots show the difference in the number of footprints between two genes in a duplicate pair within each duplicate class. Purple Xs show the mean (Kruskal-Wallis test and Mann-Whitney U test P values are significant for all comparisons, P < 0.01).
In general, the alpha WGDs are believed to represent an older group of genes than TDs (Haberer et al., 2004). However, TD formation is an ongoing process that includes ancient as well as recent events. To assess how the average numbers of footprints in the promoters of duplicate genes compared with recently formed genes, footprints were assigned to 958 genes unique to the Brassicaceae that originated during the evolution of the family (Donoghue et al., 2011). The Brassicaceae-specific genes had a lower average number of footprints in their promoter than any of the three groups of duplicates and the singletons (Fig. 1A; P = 7.19E−202, 6.03E−12, 1.25E−398, and 7.81E−78 compared with WGDs, TDs, other duplicates, and singletons, respectively).
We next examined whether duplicate pairs of different origin show a difference in the number of footprints when comparing genes within a pair with each other. We calculated the difference in footprint number between genes in each duplicate pair. WGDs had a larger average difference in footprint number compared with TDs: 6.32 and 4.37, respectively (Fig. 1B; P = 1.67E−278). The other duplicates set had the largest average change in footprint number between duplicate pairs, 6.98 (Fig. 1B; Supplemental Table S1), which was significantly different from the TDs (P = 2.37E−202) and the WGDs (P = 1.92E−4). Thus, one duplicate in a pair often has a higher or lower number of footprints than the other. This suggests that, in addition to having a higher average number of promoter footprints in their promoters under various conditions, WGD pairs are also more different from each other than their TD counterparts.
Network Connectivity and Divergence Is Greater in WGD Pairs
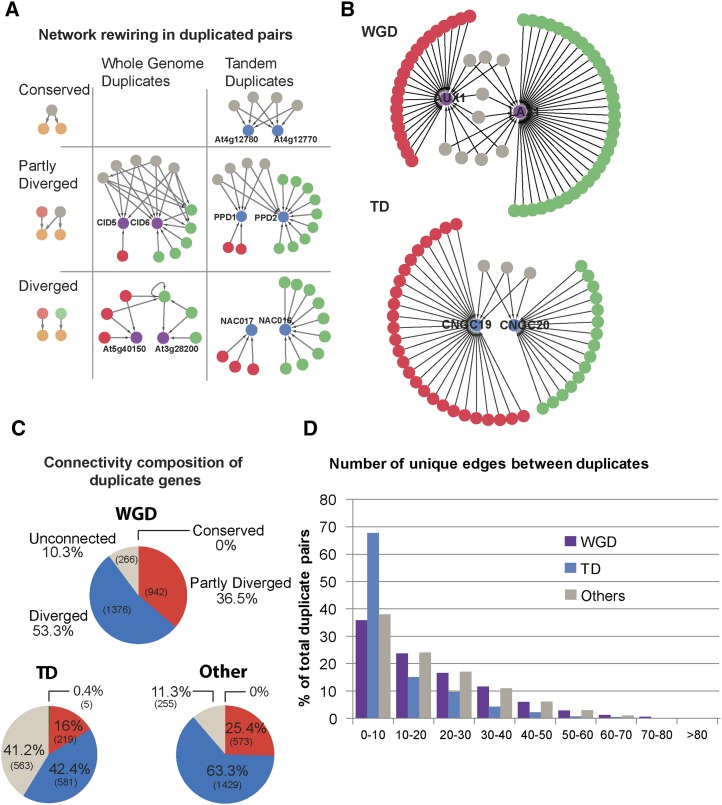
Duplicated genes can acquire new expression patterns by regulatory neofunctionalization and subfunctionalization. Changes in gene regulation are likely preceded by changes in the cis-regulatory landscape. To analyze regulatory network rewiring in duplicated pairs, transcription factor/binding site interactions in the tandem and WGD pair sets were compared using network regulatory connections from Sullivan et al. (2014). These connections allowed for the visualization of regulatory connections between a WGD or TD gene and any gene in the Arabidopsis genome. When considering regulatory network rewiring following a duplication event, there are three possible outcomes. The two duplicates may continue to have identical network connections; this is the conserved scenario. At4g12780 and At4g12770, which are a gene pair in the chaperone DnaJ domain superfamily, are an example of a TD pair in which all four connections are conserved. There were no conserved examples among the WGDs (Fig. 2A). Alternatively, the duplicates may share some common connections and may have some unique connections, which we refer to as partly diverged. Conversely, they may not have any shared connections, which we refer to as diverged (Fig. 2A). Examples are given in Figure 2A. The WGD pair of CTC-IINTERACTING DOMAIN5 (CID5) and CID6 is partly diverged, sharing five connections along with six unique connections. NAC DOMAIN CONTAINING PROTEIN17 (NAC017) and NAC016 are a tandem pair that had completely divergent regulatory networks. NAC017 has three unique connections, whereas NAC016 had nine (Fig. 2A).
Figure 2.
WGD and TD pairs have differing wiring outcomes following duplication. A, Network rewiring between pairs may not change after duplication (conserved), pairs may have some common and some unique connections (partly diverged), or they may have no connections in common (diverged). Examples from each scenario from the two classes of duplicates are presented; there are no conserved examples from the WGDs. Within the specific examples, purple and blue circles represent WGD and TD pairs, respectively. Red and green circles are genes and interactions unique to each gene within the pair, and gray circles are shared genes and interactions. Lines with arrows indicate regulatory interactions, with the arrow pointing toward the target gene. PPD1, PEAPOD1; PPD2, PEAPOD2. B, Examples of a partially diverged WGD and TD pair. Purple circles are WGD pair, and blue are TD pair. Red and green circles are genes and interactions unique to each gene within the pair, and gray circles are shared genes and interactions. Lines with arrows indicate regulatory interactions, with the arrow pointing toward the target gene. C, Connectivity composition in the WGD, tandem, and other duplicates sets. The number of gene pairs is in brackets. D, The number of unique edges between members of a duplicate pair.
There are examples in which a divergence in connectivity between duplicated genes revealed in our analysis is accompanied by divergence in expression as well as potential functional divergence. For example, the TD pair CYCLIC NUCLEOTIDE GATED CHANNEL19 (CNGC19; AT3G17690) and CNGC20 (At3g17700) is part of the family of cyclic nucleotide-gated channels. Members of this family have been shown to be involved in control of growth processes and response to stress (Kaplan et al., 2007). We found that CNGC19 has 27 unique connections, CNGC20 has 15 unique connections, and only three are shared (Fig. 2B). Reporter assays used to study the expression of CNGC19 and CNGC20 showed the two genes were differentially expressed in roots and shoots (Kugler et al., 2009). The CNGC19 gene was predominantly active in roots already at early growth stages. CNGC20 expression increased during development and was maximal in mature and senescent leaves. With respect to function, both genes appeared to respond to changes in salt concentration in the shoot, but in different cell types and at different times after treatment (Kugler et al., 2009). Therefore, the divergence in connectivity in our analysis seems to agree with observed divergence in expression in the seedling during early development as well as some potential divergence in function between these two duplicated genes. In another example, AUXIN RESISTANT1 (AUX1; At2g38120) and LIKE AUXIN RESISTANT1 (LAX1; At5g01240) are a whole genome duplicate pair that appears to have distinct functions during Arabidopsis development and show a divergence in connectivity. AUX1 and LAX1 have nine shared and 21 and 34 unique connections in the seedling, respectively (Fig. 2B). These genes belong to a family of auxin influx transporters, and both demonstrate auxin uptake function. AUX1 is localized within the lateral root primordia during all stages of development (Marchant et al., 2002). In contrast, LAX1 expression is first detected in stage I primordia and then mainly persists at the primordium base throughout lateral root formation (Péret et al., 2012). LAX1 expression also appears stronger in the presence of auxin and is detected much closer to the root apex compared with untreated controls. Conversely, AUX1 expression does not appear altered in the presence of auxin (Péret et al., 2012). These results indicate that the regulation of AUX/LAX gene expression has diverged, suggesting that they have acquired distinct roles in different developmental/physiological processes. Despite experiments showing both AUX1 (Yang et al., 2006) and LAX1 (Péret et al., 2012) have auxin uptake activity, a mutation in aux1 affected root gravitropic responses and sensitivity to a synthetic auxin, but this was not the case with the lax1 mutant (Péret et al., 2012). This suggests that, along with divergent regulation of these two duplicates, there is also evidence for a divergence in function.
When comparing the WGD and TDs, a number of striking differences appear. There appear to be a larger percentage of partly diverged and diverged pairs in the WGD set compared with tandem pairs: 36.5% and 53.3% versus 16% and 42.4%, respectively. Conserved pairs are completely absent from the WGD group, whereas five pairs are conserved among the tandems. Unconnected duplicates (those with no regulatory connections) make up 41.2% of the tandem group while only comprising 10.3% of the WGDs in the seedling data set. To compare the WGDs and tandems with other types of duplicates, the other duplicates set was analyzed. The other duplicates set had more fully diverged connections than WGDs and tandems, fewer partly diverged than WGDs, and a comparable number of genes with no connections to WGDs (Fig. 2C; Supplemental Table S2). Thus, the WGDs and other duplicates are more similar to each other than either is to the TDs.
Pseudogenization is an outcome following gene duplication and may play a role in the observed lack of TD connectivity. The Arabidopsis Information Resource (TAIR)-annotated pseudogenes were filtered out in our analysis, and thus characterized pseudogenes should not influence our results. However, there could be other pseudogenes among the TDs that have not been annotated as such by TAIR. For example, Yang et al. (2011) looked at the expression of Arabidopsis genes and found 1,939 pseudogene candidates that lacked expression in EST, full-length complementary DNA, and 17 libraries of massively parallel signature sequencing data (Yang et al., 2011). Although some of those genes may be expressed in other tissue types or environmental conditions not analyzed in that study, others are likely to be pseudogenes. In any case, all of the pseudogene candidates have very limited expression patterns if they are expressed at all. In our analysis, the TDs were found to contain 283 of the candidate pseudogenes, whereas the WGDs set only contained 41. Of the 283 candidate pseudogenes in the TDs set, 209 (73.8%) were unconnected.
When a footprint harboring a known binding site of a transcription factor (TF) is found in the promoter region of one duplicate but not in the other, we considered this a unique connection. When comparing unique connections between duplicate pairs, TD pairs have fewer unique connections between pairs, with almost 70% of pairs having between 0 and 10 unique connections. Conversely, less than 40% of WGDs and other duplicates fall within this group, with more pairs having higher numbers of unique connections (Fig. 2D). Together, these data suggest that WGD pairs, as well as the other duplicates, are more different from each other and have more connections than TD pairs. TD pairs seem less diverged but have a larger number of unconnected pairs than either WGDs or the other duplicates.
WGDs and TDs Form Distinctly Different Regulatory Networks
To analyze the role that WGDs and TDs play in the larger regulatory network of Arabidopsis, 1,000 WGD pairs and 1,000 pairs of TDs were randomly selected. All of the genes to which each duplicate was directly connected were identified, and then the networks formed were analyzed. These networks inevitably involved genes that did not belong to either WGD or TD groups, and therefore, the number of total nodes is greater than 1,000. Duplicates that did not have footprints in their promoters are considered unconnected and were included in the networks. Three independent networks of connected genes as well as isolated genes were considered for each group. All genes (duplicate and singleton) within the network comprised the nodes of the network. If the binding motif of a transcription factor within this network was found within 500 bp upstream of the start codon of another gene also within this network, an edge was created. Because the source of the edge was known (the TF) as well as the target (the target gene), the derived networks were directed. The networks were analyzed with TFs excluded as well as included from the sample. This was because the WGDs have enrichment in TFs compared with the TDs (127 TFs with known binding sites are found among the WGDs, whereas only five are found in the TDs), and TFs can disproportionally alter the structure of a network. With TFs excluded, the 1,000 WGD subset formed a more densely connected network when compared with the 1,000 TD subset (Table II). The 1,000 WGD subsets created a network with an average of 1,135.3 nodes and 9,218.7 edges compared with 1,247.0 nodes and 55,184.3 edges created by TDs (Table II). Network density is defined as the number of observed edges divided by the number of possible edges [n*(n − 1)/2]. When the TFs are excluded, the WGDs had an average network density twice that of the TDs: 0.014 and 0.007, respectively. The network diameter is the largest number of edges connecting any two nodes within the network. This value is 1 for both WGD and TD networks when TFs are excluded (Table II). When TFs are included in the analysis, the results are very different. The number of nodes and edges increases. WGD networks had an average 9,397.0 nodes and 25,219.7 edges, whereas the TD networks had 2,082.3 nodes and 6,200.0 edges. TD networks had a higher network density (0.003) compared with WGDs (0.0006). However, the diameter of WGD networks increased drastically to 9.3 compared with 3 for TD networks. Collectively, these results suggest that WGD-derived networks can form larger and denser networks than their TD counterparts when TFs are excluded, but when included in network analyses, TFs can drastically alter the structure and attributes of a regulatory network.
Table II. Comparison of networks formed using 1,000 randomly selected WGDs and TDs.
Numbers indicate parameters from an average of three subsets of 1,000 pairs of each type.
Relationships between Divergence in Connectivity and Duplication Age
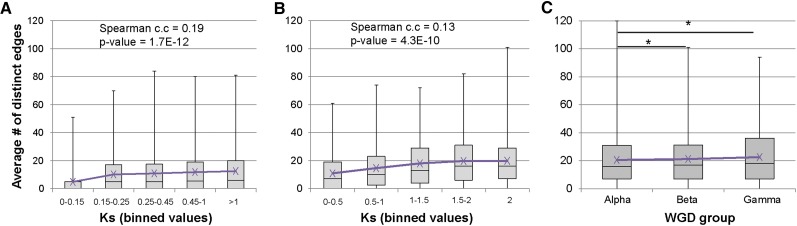
The above analyses indicate that TDs tend to show different footprint and regulatory network properties from WGDs and other duplicates. Unlike WGDs, TDs have occurred continuously during the evolutionary history of the Arabidopsis lineage. To examine the effect of time on pair connectivity in networks, Ks values were calculated for all TDs and compared with the number of shared and unique edges (regulatory connections) between duplicate pairs. Synonymous substitution rates (Ks) are used as a rough proxy for duplication age, as commonly done in molecular evolution studies. When considering edges, multiple instances of the same TF motif within the promoter of a duplicate gene were ignored and designated as a single edge. Among the TDs, the average number of unique edges increased from 4.8 in duplicates with Ks values between 0 and 0.15 to 11.8 in duplicates with Ks values above 1 (Fig. 3A). The correlation between Ks value and the number of unique edges between duplicates was significant (Spearman correlation coefficient = 0.19; P = 1.7E−12 for TDs). These results suggest that unique edges between duplicates increase soon after formation, as the pair likely diverges in their respective regulatory landscape. However, the number of unique edges then is relatively stable, with only small and statistically insignificant increases observed (Fig. 3A).
Figure 3.
Relationships between divergence in connectivity and duplication age. Box plots of the average number of unique edges between duplicate pairs within binned Ks ranges. A, TDs. B, Other duplicates. C, WGDs (asterisks indicate Mann-Whitney U test P values < 0.05).
To determine if other types of duplicates show similar patterns, we performed a similar analysis with the other duplicates set. The other duplicates set contains duplicates that tend to be older than the TDs. Thus, the Ks bins were adjusted to increase the sample size per bin. The average number of distinct edges showed a steady increase up to the Ks values of 1.5 to 2 (Fig. 3B). The correlation between Ks value and the number of unique edges between duplicates was again significant (Spearman correlation coefficient = 0.13; P = 4.3E−10). Thus, the younger duplicates have fewer distinct edges. As with the TDs, the number of shared edges stayed constant at approximately 1.0 average shared connections (data not shown).
In addition, we analyzed gene pairs derived from three successively older WGD events: the alpha, beta, and gamma WGDs. The average number of distinct edges showed a statistically significant increase when comparing the alpha WGDs (youngest) to the beta and gamma WGDs (P = 0.03 and 0.02, respectively; Mann-Whitney U test; Fig. 3C). There was an increase in distinct edges when comparing the beta and gamma (oldest) WGDs, but it was not statistically significant (Fig. 3C). Like the TDs and other duplicates, the number of shared edges remained constant at approximately 1.0 average connections (data not shown).
DISCUSSION
Evolution of Duplicated Genes by Divergence in Regulatory Elements and Networks
In this work, we show that duplicate gene pairs in Arabidopsis, as a whole, show extensive divergence in DNase 1 regulatory footprints. WGDs and TDs vary greatly in the number and distribution of regulatory footprints (protein binding sites) within 500 bp upstream of their respective transcription start sites. WGDs have approximately twice as many footprints as TDs across different conditions (Fig. 1; Supplemental Fig. S1). They appear to have more footprints than either singleton genes or Brassicaceae-specific genes and slightly more than the set of other duplicates. Further, we show that this increase in footprints translates to an increase in connectivity within larger regulatory networks. Because only a small subset of Arabidopsis transcription factors have known and characterized binding sites, only a fraction of footprints can be paired with a potential transcription factor that would bind the underlying DNA. Nonetheless, WGDs have fewer unconnected genes compared with TDs, 10.3% compared with 41.2% (Fig. 2). Furthermore, sample networks show more incoming and outgoing regulatory interactions (edges) compared with TDs (Table II). This increased connectivity leads to the formation of larger, and in certain cases more densely connected, networks (Table II).
The increase in footprints may reflect a more complex and tissue-specific level of transcription of WGDs compared with TDs. An increase in tissue-specific expression and perhaps a higher overall level of expression means more transcription factors binding to the promoter, which in turn manifests as an increase in detectable footprints by DNase I sequence. This may be explained by the difference in ages of the two sets of genes. WGDs have been shown to be, on average, older than TDs in Arabidopsis (Haberer et al., 2004). In Arabidopsis, as well as in animal and yeast model systems, older genes have been shown to be transcribed at higher levels (Liao and Zhang, 2006; Wolf, 2006; Donoghue et al., 2011). The functional classes of genes retained among tandem and WGDs may also explain the reason for increased footprint number. TDs have been shown to be more involved in responses to environmental factors and stresses (Parniske et al., 1997; Michelmore and Meyers, 1998; Lucht et al., 2002; Kovalchuk et al., 2003; Rizzon et al., 2006). Genes retained through WGDs tend to be TFs, genes involved in signal transduction, and those involved in development (Blanc and Wolfe, 2004; Maere et al., 2005). WGDs may, on average, have a broader expression pattern and, in turn, may be more likely to be expressed in the 7-d-old seedling, resulting in an increase in footprints that were detected in the promoter in this study. Conversely, TDs may be less likely to be expressed at a single developmental stage, thus resulting in fewer footprints on average in the seedling data set. When comparing the number of footprints in tandems and WGDs under heat stress and in the dark, TDs did not appear to have a significantly more pronounced response to heat stress and dark in terms of footprint numbers compared with WGDs (Supplemental Fig. S1). Both WGD and TD footprint numbers remained unchanged under heat shock, and both decreased significantly in the dark. This reduction in the dark is expected, because in the absence of light, expression is generally reduced across the genome, and promoter accessibility is low (Sullivan et al., 2014).
In cases where the binding motif of a TF is known, that motif can subsequently be mapped to a footprint, and a regulatory connection can be made. When these regulatory connections are made genome wide, a dense network begins to emerge. However, in Arabidopsis, the number of TFs whose binding site is known is incomplete, and this is a limiting factor in the depth of the analysis. In the case of WGDs, the larger number of footprints appears to translate to an increase in the overall network. Whether comparing sample networks that include or exclude TFs, WGDs form larger networks with more nodes and edges than TDs (Table II). TFs have many targets and therefore quickly increase the number of edges in a network and, thus, its density and connectivity. The distribution of TF within any randomly selected set may significantly affect the distribution of edges/nodes. When TFs are excluded, the WGD networks had twice the density of TD networks. However, when included in the analysis, TD networks had a higher density (Table II). The WGD set had an enrichment of TFs compared with TDs, and therefore always had more TFs within the 1,000 random duplicates set. This resulted in a lower density because each TF would introduce a large number of nodes to the network but relatively few edges. For example, if one of the 1,000 randomly selected WGDs was a TF with 500 target genes, then this would result in an additional 500 nodes and 500 edges added to the network. Unless these targets are also WGDs, they do not provide any additional edges to the network because only genes with direct interactions to WGDs are considered. Introducing so many nodes to the network drastically increases the number of possible edges while only slightly increasing the number of observed edges. The high numbers of unconnected genes also likely contribute to the lower node and edge numbers of the tandem networks. Of tandem genes, 41.2% have no connections in the seedling DNase 1 data set, whereas in the WGDs, this number is only 10.3%. This may again be due to the nature of the types of genes found among the TDs. As mentioned previously, TDs tend to be genes involved in stress responses, and thus may have a restricted expression pattern manifested here as fewer footprints and thus decreased network involvement.
Regulatory Divergence and Functional Differentiation
Regulatory divergence in the form of expression pattern changes appears frequently when comparing pairs of duplicates (for review, see DeSmet and Van de Peer, 2012). In yeast, only a small fraction of duplicated gene pairs showed no or little expression variance, whereas most duplicated genes quickly diverged in their expression patterns (Gu et al., 2002). Expression of duplicated genes seems to be initially coupled and subsequently diverges rapidly, suggesting rapid neo- and/or subfunctionalization (Gu et al., 2002). In Arabidopsis, a majority of duplicated genes show a divergence in their expression patterns (e.g. Blanc and Wolfe 2004; Haberer et al., 2004; Casneuf et al., 2006; Liu et al., 2011). If regulatory divergence precedes expression divergence, then it is not surprising that we find even more extreme differentiation in regulatory elements between duplicates in our study. Only five (0.1%) pairs of our analyzed duplicates share the same regulatory connections, whereas the vast majority (82.4%) are either partly or fully diverged (Fig. 2). In this study, we were not able to directly link the changes in regulatory footprints and networks found between duplicated genes in the seedlings with divergence in expression patterns. However, many of them are likely to be involved in the extensive divergence in expression patterns that has been found in many duplicated genes in Arabidopsis. Divergence in the cis-regulatory element landscape could potentially generate differing expression patterns and eventually lead to neofunctionalization and subfunctionalization on a transcriptional level.
This study has shown extensive divergence in regulatory elements and networks between duplicated genes in Arabidopsis, using new high-throughput data that allow for single-base resolution of the cis-regulatory elements. It would be interesting to extend our analyses of regulatory elements found in seedlings to other organ types and developmental stages once such data sets are available.
MATERIALS AND METHODS
Duplicate Genes and DNase I Footprint Data
DNase I footprint data from 7-d-old seedlings of Arabidopsis (Arabidopsis thaliana) for three conditions, normal (DS21094), heat shock (DS20423), and dark (DS22138), were obtained from Sullivan et al. (2014). WGDs were from Blanc et al. (2003). TD pairs were from Liu et al. (2011). The set of other duplicates was identified by pairs with reciprocal best BLAST hits that did not overlap with the WGDs and TDs, and as such, contain proximal as well as dispersed duplicates. Brassicaceae-specific genes in Arabidopsis that arose at some point during the evolutionary history of the family were from Donoghue et al. (2011). Singleton genes were identified as those genes that had zero nonself hits with a BLAST e-value lower than 1e-3. WGDs derived from the gamma, beta, and alpha whole genome duplications, used in the analyses shown in Figure 3, were from Wang et al. (2013).
Analysis of Footprint Data to Assign and Count Cis-Regulatory Elements
The software BEDTools (Quinlan and Hall, 2010) was used in combination with Python scripts developed in house to assign and count the footprints that overlap with the following genomic regions of our genes of interest: 500 and 1,000 bp upstream of the transcription start site, 5′ UTR, exons, introns, 3′ UTR, and 500 and 1,000 bp downstream of the transcribed region. Sequences for the genomic features from TAIR9 were downloaded from the file transfer protocol site (ftp://ftp.arabidopsis.org/Genes/TAIR9_genome_release/; July 2013). BEDTools’ intersection tool was used along with the footprint data to produce footprints 500 bp upstream of the transcription start site for each condition.
Calculating Ks Values
Pairwise synonymous substitution rates (Ks) were estimated using the software KaKs Calculator (Zhang et al., 2006), with the MYN (Modified version of the Yang-Nielsen method) approximation method. Protein alignments were performed using Clustal (Sievers et al., 2011) and were translated into codon alignments using PAL2NAL (Suyama et al., 2006).
Regulatory Network Analyses of Duplicate Pairs
An entire (not TF filtered) regulatory TF and target gene network from light-grown 7-d-old seedlings was obtained from Sullivan et al. (2014). Duplicate gene pairs were assigned as being conserved, partly diverged, diverged, and unconnected based on the conservation of their regulatory connections, using a Python script.
Three random samples of 1,000 WGDs and 1,000 TDs were extracted from the entire network. Subnetworks composed only of the genes in our WGD and TD lists and their first neighbors (genes having a direct regulatory interaction) were derived. These networks inevitably involved genes that did not belong to either WGD or TD groups, and therefore, the number of nodes is greater than 1,000. Duplicates that did not have footprints in their promoters are considered unconnected and were included in the networks. The resulting networks of connected genes as well as isolated genes were considered. All genes (duplicated and singleton) within the network comprised the nodes of the network. If the binding motif of a transcription factor within this network was found within 500 bp upstream of the start codon of another gene also within this network, an edge was created. Because the source of the edge was known (the TF) as well as the target (the target gene), the derived networks were directed. The resulting subnetworks were further analyzed using the Cytoscape v. 3.0.1 software (Shannon et al., 2003). Network diameter or maximum eccentricity is the maximum noninfinite length of a shortest path between any two nodes in the network. If a node is unconnected, the value is zero. Network density was calculated as (number of observed edges) / number of possible edges [n*(n − 1)/2]. A Python script was used to identify distinct and shared incoming connections for duplicates. Shared and unique connections were calculated such that the number of binding sites is not considered (i.e. if gene 1 has two inputs from gene X, and gene 2 has one input from gene X, they have 1 shared input).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. WGD and TD promoter footprints in various conditions.
Supplemental Table S1. Differences in promoter footprints.
Supplemental Table S2. Connectivity composition of duplicated genes.
Acknowledgments
We thank Sishuo Wang and Allen Zhang for assistance with analyses.
Glossary
- WGD
whole-genome duplicate
- TD
tandem duplicate
- CNS
conserved noncoding DNA sequence
- UTR
untranslated region
- TF
transcription factor
Footnotes
This work was supported by the Natural Science and Engineering Research Council of Canada (a grant to K.L.A. and an undergraduate student research award to J.P.).
Articles can be viewed without a subscription.
References
- Blanc G, Hokamp K, Wolfe KH (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Casneuf T, De Bodt S, Raes J, Maere S, Van de Peer Y (2006) Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol 7: R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G (1993) A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74: 505–514 [DOI] [PubMed] [Google Scholar]
- Dermitzakis ET, Clark AG (2002) Evolution of transcription factor binding sites in Mammalian gene regulatory regions: conservation and turnover. Mol Biol Evol 19: 1114–1121 [DOI] [PubMed] [Google Scholar]
- De Smet R, Van de Peer Y (2012) Redundancy and rewiring of genetic networks following genome-wide duplication events. Curr Opin Plant Biol 15: 168–176 [DOI] [PubMed] [Google Scholar]
- Donoghue MT, Keshavaiah C, Swamidatta SH, Spillane C (2011) Evolutionary origins of Brassicaceae specific genes in Arabidopsis thaliana. BMC Evol Biol 11: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester WC, Thompson C, Elder JT, Groudine M (1986) A developmentally stable chromatin structure in the human beta-globin gene cluster. Proc Natl Acad Sci USA 83: 1359–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. (2009) Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 60: 433–453 [DOI] [PubMed] [Google Scholar]
- Freeling M, Rapaka L, Lyons E, Pedersen B, Thomas BC (2007) G-boxes, Bigfoot genes, and environmental response: characterization of intragenomic conserved noncoding sequences in Arabidopsis. Plant Cell 19: 1441–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Subramaniam S (2009) Conserved noncoding sequences (CNSs) in higher plants. Curr Opin Plant Biol 12: 126–132 [DOI] [PubMed] [Google Scholar]
- Grosveld F, van Assendelft GB, Greaves DR, Kollias G (1987) Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell 51: 975–985 [DOI] [PubMed] [Google Scholar]
- Gu Z, Nicolae D, Lu HHS, Li WH (2002) Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet 18: 609–613 [DOI] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH (2003) Role of duplicate genes in genetic robustness against null mutations. Nature 421: 63–66 [DOI] [PubMed] [Google Scholar]
- Haberer G, Hindemitt T, Meyers BC, Mayer KFX (2004) Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiol 136: 3009–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Kaplan B, Sherman T, Fromm H (2007) Cyclic nucleotide-gated channels in plants. FEBS Lett 581: 2237–2246 [DOI] [PubMed] [Google Scholar]
- Korkuc P, Schippers JHM, Walther D (2014) Characterization and identification of cis-regulatory elements in Arabidopsis based on single-nucleotide polymorphism information. Plant Physiol 164: 181–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk I, Kovalchuk O, Kalck V, Boyko V, Filkowski J, Heinlein M, Hohn B (2003) Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature 423: 760–762 [DOI] [PubMed] [Google Scholar]
- Kugler A, Köhler B, Palme K, Wolff P, Dietrich P (2009) Salt-dependent regulation of a CNG channel subfamily in Arabidopsis. BMC Plant Biol 9: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BY, Zhang J (2006) Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during mammalian evolution. Mol Biol Evol 23: 1119–1128 [DOI] [PubMed] [Google Scholar]
- Liu SL, Baute GJ, Adams KL (2011) Organ and cell type-specific complementary expression patterns and regulatory neofunctionalization between duplicated genes in Arabidopsis thaliana. Genome Biol Evol 3: 1419–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Thornton K (2001) Gene duplication and evolution. Science 293: 1551. [DOI] [PubMed] [Google Scholar]
- Lucht JM, Mauch-Mani B, Steiner HY, Metraux JP, Ryals J, Hohn B (2002) Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat Genet 30: 311–314 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y (2005) Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA 102: 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Bhalerao R, Casimiro I, Eklöf J, Casero PJ, Bennett M, Sandberg G (2002) AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8: 1113–1130 [DOI] [PubMed] [Google Scholar]
- Palaniswamy SK, James S, Sun H, Lamb RS, Davuluri RV, Grotewold E (2006) AGRIS and AtRegNet. a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol 140: 818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BB, Jones JD (1997) Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91: 821–832 [DOI] [PubMed] [Google Scholar]
- Péret B, Swarup K, Ferguson A, Seth M, Yang Y, Dhondt S, James N, Casimiro I, Perry P, Syed A, et al. (2012) AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 24: 2874–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon C, Ponger L, Gaut BS (2006) Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput Biol 2: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. (2010) Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J. , et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 13627–13632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler JB, Ficklin SP, Luo F, Freeling M, Feltus FA (2012) Conserved non-coding regulatory signatures in Arabidopsis co-expressed gene modules. PLoS One 7: e45041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder J, Larsen A, Engel JD, Dolan M, Groudine M, Weintraub H (1980) Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell 20: 451–460 [DOI] [PubMed] [Google Scholar]
- Sullivan AM, Arsovski AA, Lempe J, Bubb KL, Weirauch MT, Sabo PJ, Sandstrom R, Thurman RE, Neph S, Reynolds AP, et al. (2014) Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Reports 8: 2015–2030 [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34: W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Paterson AH (2010) Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc Natl Acad Sci USA 107: 472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann SA, Babu MM (2004) Gene regulatory network growth by duplication. Nat Genet 36: 492–496 [DOI] [PubMed] [Google Scholar]
- Thomas BC, Rapaka L, Lyons E, Pedersen B, Freeling M (2007) Arabidopsis intragenomic conserved noncoding sequence. Proc Natl Acad Sci USA 104: 3348–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai ZTY, Tsai HK, Cheng JH, Lin CH, Tsai YF, Wang D (2012) Evolution of cis-regulatory elements in yeast de novo and duplicated new genes. BMC Genomics 13: 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A (2009) The evolutionary significance of ancient genome duplications. Nat Rev Genet 10: 725–732 [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD (2000) The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117 [DOI] [PubMed] [Google Scholar]
- Wang Y, Tan X, Paterson AH (2013) Different patterns of gene structure divergence following gene duplication in Arabidopsis. BMC Genomics 14: 652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI. (2006) Coping with the quantitative genomics ‘elephant’: the correlation between the gene dispensability and evolution rate. Trends Genet 22: 354–357 [DOI] [PubMed] [Google Scholar]
- Wu C. (1980) The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature 286: 854–860 [DOI] [PubMed] [Google Scholar]
- Wu C, Wong YC, Elgin SCR (1979) The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell 16: 807–814 [DOI] [PubMed] [Google Scholar]
- Yang L, Takuno S, Waters ER, Gaut BS (2011) Lowly expressed genes in Arabidopsis thaliana bear the signature of possible pseudogenization by promoter degradation. Mol Biol Evol 28: 1193–1203 [DOI] [PubMed] [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E (2006) High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol 16: 1123–1127 [DOI] [PubMed] [Google Scholar]
- Zhang L, Gaut BS, Vision TJ (2001) Gene duplication and evolution. Science 293: 1551. [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao XQ, Wang J, Wong GKS, Yu J (2006) KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4: 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]