Four isoforms of 4-coumaryl CoA ligase have different roles in the biosynthesis of soluble plant metabolites and cell wall polymers.
Abstract
The biosynthesis of lignin, flavonoids, and hydroxycinnamoyl esters share the first three enzymatic steps of the phenylpropanoid pathway. The last shared step is catalyzed by 4-coumarate:CoA ligase (4CL), which generates p-coumaroyl CoA and caffeoyl CoA from their respective acids. Four isoforms of 4CL have been identified in Arabidopsis (Arabidopsis thaliana). Phylogenetic analysis reveals that 4CL1, 4CL2, and 4CL4 are more closely related to each other than to 4CL3, suggesting that the two groups may serve different biological functions. Promoter-GUS analysis shows that 4CL1 and 4CL2 are expressed in lignifying cells. In contrast, 4CL3 is expressed in a broad range of cell types, and 4CL3 has acquired a distinct role in flavonoid metabolism. Sinapoylmalate, the major hydroxycinnamoyl ester found in Arabidopsis, is greatly reduced in the 4cl1 4cl3 mutant, showing that 4CL1 and 4CL3 function redundantly in its biosynthesis. 4CL1 accounts for the majority of the total 4CL activity, and loss of 4CL1 leads to reduction in lignin content but no growth defect. The 4cl1 4cl2 and 4cl1 4cl2 4cl3 mutants are both dwarf but do not have further reduced lignin than the 4cl1 mutant, indicating that either 4CL1 or 4CL2 is required for normal plant growth. Although 4CL4 has a limited expression profile, it does make a modest contribution to lignin biosynthesis. Together, these data show that the four isoforms of 4CL in Arabidopsis have overlapping yet distinct roles in phenylpropanoid metabolism.
Plants are sessile organisms that cannot escape from natural predators or nonideal environments. It is thought that selective pressures associated with defense and abiotic and biotic stresses led to the ability to accumulate the large number of specialized metabolites seen in plants today. Pathways that are required for the biosynthesis of specialized metabolites often intersect with primary metabolism and, in some cases, contribute to plant vigor and fitness. For example, 30% of the total inorganic carbon fixed by plants enters the phenylpropanoid pathway, which produces major specialized metabolites, including lignin, hydroxycinnamoyl esters, and flavonoids (Weisshaar and Jenkins, 1998; Boerjan et al., 2003). The emergence of lignin enabled land plants to stand upright and transport water long distances. In addition, lignin also serves as the first mechanical defense against pathogens (Boerjan et al., 2003; Bonawitz and Chapple, 2010). Although lignin accounts for the vast majority of the carbon flux through this pathway, soluble phenylpropanoids also play vital roles in plant growth, development, and viability. For example, sinapoylmalate, a hydroxycinnamoyl ester primarily found in members of the Brassicaceae, serves as a UV protectant (Chapple et al., 1992; Landry et al., 1995). More than 1,000 flavonoids have been identified to date, some of which play key roles as pigments, in pathogen resistance, and in protection against oxidative stress (Dixon and Paiva, 1995; Landry et al., 1995; Winkel-Shirley, 2001; Broun, 2005; Tanaka et al., 2008). Certain subclasses of flavonoids are better known for their antioxidant properties in the human diet; examples include isoflavonoids in soybean (Glycine max) and stilbenes in red wine (Winkel-Shirley, 2001). Being able to harness and manipulate this pathway would be desirable for many applications; however, we still lack a complete understanding of the genetics and biochemistry underlying the biosynthesis of these compounds, as evidenced by the fact that the biosynthetic pathway continues to be revised (Bonawitz and Chapple, 2010; Weng et al., 2012; Vanholme et al., 2013). Furthering our understanding of the phenylpropanoid pathway not only provides an improved basic understanding of plant specialized metabolism, but also enhances our ability to rationally design plant metabolic pathways for future applications.
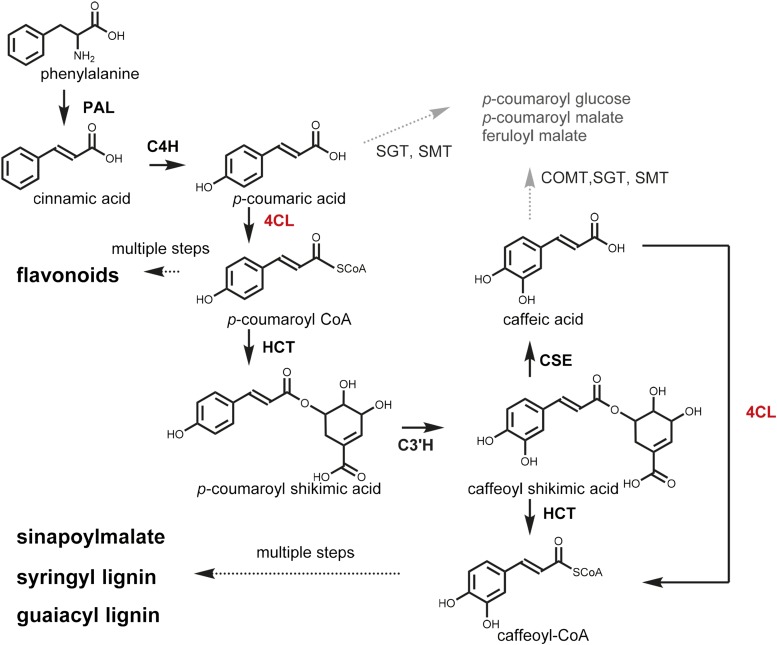
The first three steps, known as the general phenylpropanoid pathway, are conserved in almost all dicots (Bonawitz and Chapple, 2010). Phe is first deaminated by phenyl-Ala ammonia lyase (PAL), a step that is followed by a 4-hydroxylation reaction catalyzed by cinnamate 4-hydroxylase (C4H), the product of which is converted to its CoA ester by 4-coumarate:CoA ligase (4CL; Fig. 1). These reactions have been known since the 1960s and are described in detail in many publications (Russell and Conn, 1967; Russell, 1971; Knobloch and Hahlbrock, 1975; Hahlbrock and Grisebach, 1979). Genetic screens, especially in Arabidopsis (Arabidopsis thaliana), have led to the identification of many genes involved in this pathway. PAL and 4CL are encoded by multiple paralogs in most plant species, consistent with the observation that only two 4CL mutants have been identified from a forward genetic screen (Dobritsa et al., 2011; Saballos et al., 2012). In Arabidopsis, both PAL and 4CL are encoded by four genes (Lee et al., 1995; Rohde et al., 2004; Soltani et al., 2006; Huang et al., 2010). Although studies show that PAL isoforms 1 and 2 are the major PALs and are redundant in function (Rohde et al., 2004; Huang et al., 2010), the functions of 4CL isoforms are less clear. The substrate specificities of the Arabidopsis 4CL and 4CL-like proteins have been reported, and four have been shown to use p-coumaric acid, caffeic acid, and ferulic acid as substrates: 4CL1 (At1g51680), 4CL2 (At3g21240), 4CL3 (At1g65060), and 4CL4 (At3g21230; Ehlting et al., 1999; Hamberger and Hahlbrock, 2004; Costa et al., 2005; Soltani et al., 2006). Interestingly, 4CL4 is the only 4CL capable of activating sinapic acid (Hamberger and Hahlbrock, 2004). The amino acid residues important for recognizing and accommodating substrates at the enzymes’ active sites have been identified and characterized (Ehlting et al., 2001; Schneider et al., 2003).
Figure 1.
The phenylpropanoid pathway. 4CL acts at two steps (indicated in red) of the pathway and is required for the biosynthesis of lignin and soluble metabolites. In mutants containing at least the 4cl1 mutation, p-coumaroyl Glc, p-coumaroyl malate, and feruloyl malate accumulate as atypical phenylpropanoids as indicated in gray. HCT, Hydroxycinnamoyl CoA shikimate:quinate hydroxycinnamoyltransferase; C3′H, p-coumaroyl shikimate 3′-hydroxylase; CSE, caffeoyl shikimate esterase; COMT, caffeic acid/5-hydroxyferulic acid O-methyltransferase; SGT, sinapic acid:UDP-Glc-dependent sinapoyltransferase; SMT, sinapoylglucose malate:sinapoyltransferase.
Phylogenetic analyses reveal that 4CL1, 4CL2, and 4CL4 are more closely related to one another than to 4CL3 (Ehlting et al., 1999; Soltani et al., 2006), suggesting that 4CL3 may have evolved a distinct biological function. Coexpression network analysis indicates that 4CL1, 4CL2, and 4CL4 are coexpressed with lignin and sinapoylmalate biosynthetic genes, whereas 4CL3 is associated with flavonoid gene expression (Koopman et al., 2012). Further, 4cl3 mutants were identified in a large-scale genetic screen to find genes involved in pollen exine formation (Dobritsa et al., 2011). In that study, two mutant alleles of 4cl3 were found to have defects in pollen exine and flavonoid production; however, flavonoid biosynthesis in tissues other than pollen was not examined. In addition, promoter-GUS analyses show that 4CL1 and 4CL2 are expressed in lignifying cells, whereas 4CL3 is expressed in a broad range of cell types in Arabidopsis seedlings (Ehlting et al., 1999). It has also been reported that 4CL1, 4CL2, and 4CL4, but not 4CL3, transcripts are induced upon wounding in Arabidopsis leaves (Soltani et al., 2006), consistent with the observation that physical damage can induce lignification (Rittinger et al., 1987; Dixon and Paiva, 1995; Hawkins and Boudet, 1996). Last, antisense suppression of 4CL activity in Arabidopsis alters lignin content and composition (Lee et al., 1997), although it was unclear in this study whether the suppression of 4CL activity was isoform specific. With regard to this issue, a recent study showed that 4cl1, but not 4cl2, mutants contain less total lignin content than wild-type plants, suggesting that 4CL1 is required for wild-type lignin deposition (Van Acker et al., 2013). No 4CL has been implicated in sinapoylmalate production. Based on this evidence, it has been proposed that 4CL1 and 4CL2 are the major 4CLs involved in lignin biosynthesis, whereas 4CL3 has a more important role in flavonoid biosynthesis (Ehlting et al., 1999; Saito et al., 2013). In summary, although some evidence for the roles of 4CLs has been established in Arabidopsis, there has not been a comprehensive genetic study on the impact of these isoforms on overall phenylpropanoid metabolism.
In this article, we provide experimental evidence that describes the biological functions of the Arabidopsis 4CL isoforms. We report that 4CL3 is the predominant 4CL involved in flavonoid biosynthesis, 4CL1 and 4CL3 are both important in sinapoylmalate biosynthesis, 4CL1 contributes the most to lignin biosynthesis, and for normal plant growth, either 4CL1 or 4CL2 is required. Last, 4CL4 contributes modestly to lignin deposition, at least in the absence of 4CL1. These findings together show that, although the four isoforms of 4CL have similar catalytic efficiencies toward their substrate(s), their biological functions in phenylpropanoid metabolism are distinct but overlapping.
RESULTS
Phylogenetic Analysis of 4CL Homologs
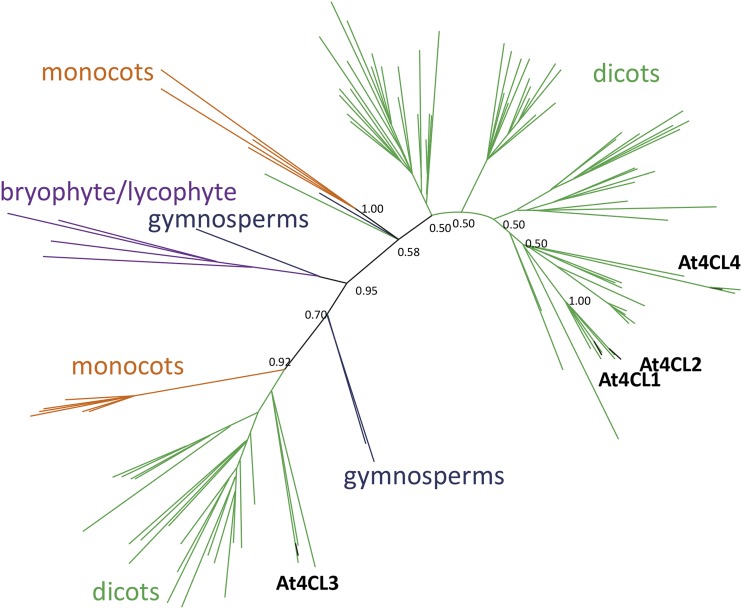
As mentioned previously, there are four 4CL genes annotated in the Arabidopsis genome that encode enzymes with catalytic activity toward hydroxycinnamic acids (Ehlting et al., 1999; Hamberger and Hahlbrock, 2004; Costa et al., 2005; Soltani et al., 2006). Previous phylogenetic analysis shows that the four Arabidopsis 4CLs belong to two distinct classes (Ehlting et al., 1999; Soltani et al., 2006; Chen et al., 2014); however, the scope of these phylogenetic analyses was limited to a few plant species. Using the four Arabidopsis 4CLs as query, we identified 192 4CL protein homologs from across the plant kingdom with 70% or more sequence identity to at least one of the Arabidopsis proteins, and generated a new phylogenetic tree using Bayesian analysis (Fig. 2). Our findings confirm that 4CL3 belongs to a different clade than the other 4CLs, and that 4CL1 and 4CL2 are more closely related to one another than to 4CL4.
Figure 2.
Bayesian phylogenetic analysis of 192 4CL homologs. Selected bootstrap values are shown. The colors represent different plant lineages: green, dicots; orange, monocots; blue, gymnosperms; and purple, bryophyte and lycophyte. 4CL3-like and 4CL1/2/4-like 4CLs are present in both monocots and dicots. At4CLs are indicated in black.
Promoter-GUS Analysis of Four 4CLs in Arabidopsis
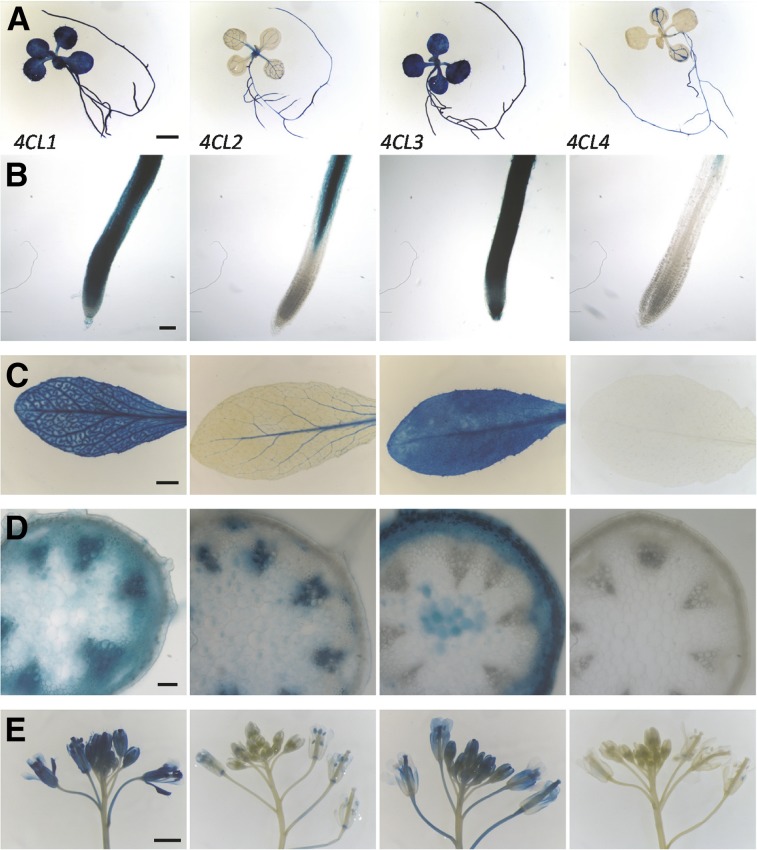
Previous promoter-GUS transcriptional analysis (Soltani et al., 2006) showed that 4CL1 and 4CL2 are expressed predominantly in lignifying tissues, whereas 4CL3 is expressed more broadly. These findings are consistent with the hypothesis that the differential expression of the four 4CLs leads to their different biological functions (Costa et al., 2005; Soltani et al., 2006; Bassard et al., 2012). To examine the spatiotemporal patterns of expression of the four 4CLs, we generated transgenic Arabidopsis plants carrying 4CL promoter-GUS constructs and analyzed plants throughout development. Differences were distinct in both 10-d-old seedlings and 3-week-old leaves: 4CL1 and 4CL3 both have strong overall expression, but 4CL1 is more abundant in the vasculature; 4CL2 is expressed mainly in the midrib; and 4CL4 is limited to roots and the veins of the cotyledons (Fig. 3). All 4CLs are broadly expressed in the root, but 4CL2 and 4CL4 are excluded from the root tip (Fig. 3). In the inflorescence, we observed that both 4CL1 and 4CL2 have strong expression in the xylem, consistent with their proposed role in lignin biosynthesis (Fig. 3). In addition, 4CL1 is expressed in the cortex, phloem, and interfascicular region. Although 4CL4 does not show any detectable stem expression, 4CL3 is expressed in the epidermis, cortex, cambium, phloem, and pith (Fig. 3). We examined the flowers of the same plants and found that 4CL1 is expressed almost everywhere; 4CL2 is present mainly in the stigma and the veins of the sepal; 4CL3 is expressed in the sepal, petal, filament, and stigma; and 4CL4 is essentially undetectable (Fig. 3).
Figure 3.
Promoter GUS reporter gene analysis of 4CL gene expression. A, Ten-day-old seeding. B, Root tip of 10-d-old seeding. C, Three-week-old rosette. D, Cross section of 5-week-old inflorescence stem. E, Flower on a 5-week-old plant. Scale bars = 2 mm (A, D, and E); and 100 µm (B and D).
4CL1 Accounts for the Majority of Total 4CL Activity
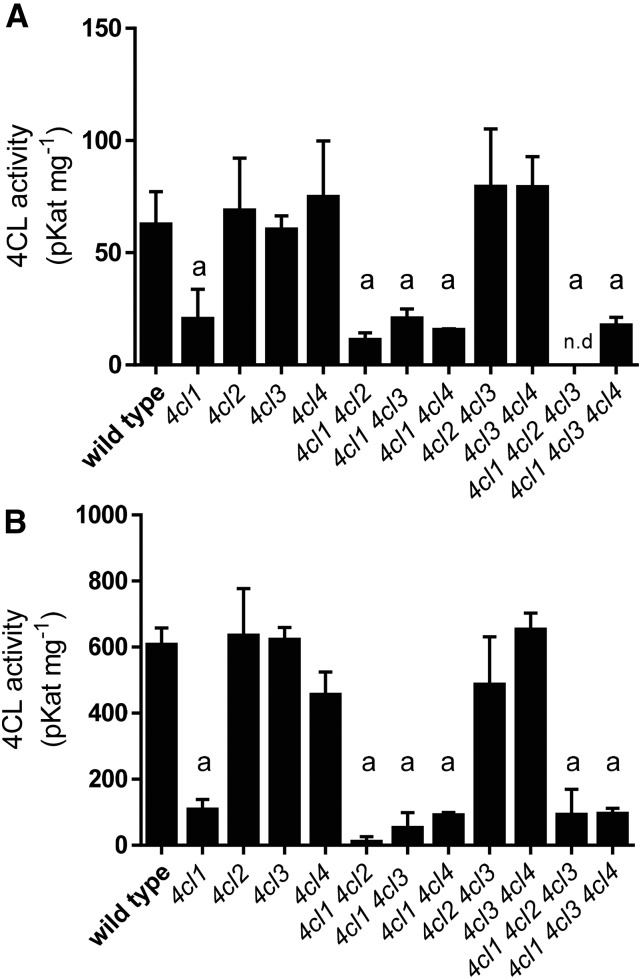
To further dissect the biological functions of the 4CL isoforms, we identified single and higher-order mutants of the four 4CLs. We were unable to identify mutants with both 4cl2 and 4cl4 mutations due to the tight genetic linkage between the two adjacent genes. We then measured the total 4CL activity in the set of mutants generated and found that the 4cl1 single mutant shows a 90% reduction in both 2-week-old rosettes and 5-week-old inflorescences as compared with the wild type, whereas 4CL activity in extracts of the other single mutants is unaffected (Fig. 4). Of the higher-order mutants, 4cl2 4cl3 and 4cl3 4cl4 both have wild-type levels of 4CL activity, but all of the double and triple mutants containing the 4cl1 mutation show reductions similar to 4cl1. These data indicate that 4CL1 alone accounts for the majority of the total 4CL activity in the organs examined, and the other three 4CLs contribute little overall, at least as measured in vitro.
Figure 4.
4CL enzyme activity measuring product formation. A, Two-week-old rosette. B, Five-week-old inflorescence. n.d., Not detected. Error bars represent the sd of the means of three biological replicates. Using GraphPad Prism, the means were compared by one-way ANOVA (fixed model), and statistically significant differences (P < 0.05) were identified by Dunnett’s test and are indicated by the letter a.
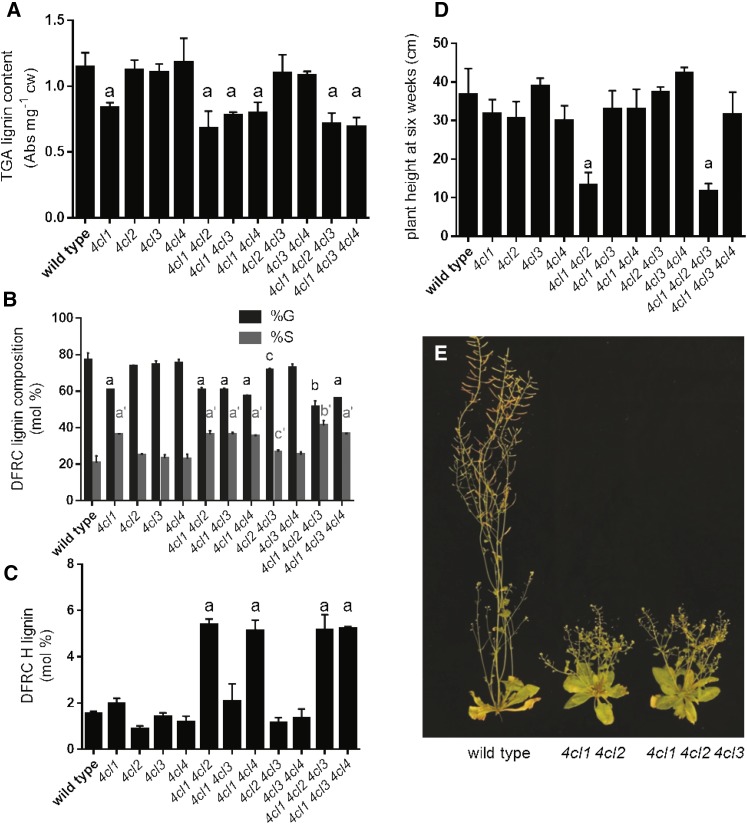
Lignin Modification and Dwarfing in 4cl Mutants Are Not Directly Correlated
To determine how reductions in 4CL activity impact the various branches of phenylpropanoid metabolism, we analyzed lignin content in the various single and multiple mutants. 4cl1 plants show an approximate 30% reduction in total lignin content compared with the wild type; however, addition of other 4cl mutations does not further exacerbate this phenotype (Fig. 5), consistent with the impact of the 4cl1 mutation on total 4CL activity. Despite the fact 4CL1 is required for wild-type levels of lignin biosynthesis, we did not observe any growth defect in the 4cl1 mutant. In contrast, although 4cl2 mutants grow normally, 4cl1 4cl2 and 4cl1 4cl2 4cl3 plants exhibit a dwarf and bushy phenotype at the same developmental stage even though they deposit similar levels of lignin as the 4cl1 mutant (Fig. 5). These data suggest that the reduction of lignin content is not the cause of dwarfism in these mutants, consistent with other findings that deficiency in lignin production often, but not always, leads to a dwarf phenotype (Bonawitz and Chapple, 2013). Although still fertile, both mutants produce fewer seeds than the wild type.
Figure 5.
Plant growth and lignin analysis. A, Thioglycolic acid (TGA) quantification of lignin content. B, Derivatization followed by reductive cleavage (DFRC) lignin composition. Total guaiacyl (G)+syringyl (S) is ≥94% in all samples. C, DFRC p-hydroxyphenyl (H) lignin. D, Plant heights at 6 weeks of age. E, Plant images of 4cl1 4cl2 and 4cl1 4cl2 4cl3 mutants compared with the wild type at 8 weeks of age. Error bars represent the sd of the means of three biological replicates. Using GraphPad Prism, the means were compared by one-way ANOVA (fixed model), and statistically significant differences (P < 0.05) were identified by Dunnett’s test and are indicated by a to c or a′ to c′.
Two previous studies have shown that down-regulation of 4CL can impact lignin monomer composition (Lee et al., 1997; Van Acker et al., 2013), but multiple 4CL knockouts have not been examined. We measured lignin composition by the DFRC method and found that the G monomer content was decreased from 77 mol% in the wild type to 60 mol% in a 4cl1 single mutant (Fig. 5). The 4cl2 4cl3 and 4cl3 4cl4 mutants have wild-type lignin deposition, but higher-order mutants, including the 4cl1 mutation, all show a similar trend as 4cl1 plants (Fig. 5). The mol% G lignin in a 4cl1 4cl2 4cl3 triple mutant was further decreased to 50. In addition, the H lignin monomer content increased from 2 mol% in the wild type to 6 mol% in mutants containing either the 4cl1 and 4cl2 mutations or the 4cl1 and 4cl4 mutations (Fig. 5).
Together, these data indicate that 4CL1 is required for normal lignin deposition and composition, and that 4CL2 is important for some aspect of metabolism that is required for normal plant growth. 4CL2, 4CL3, and 4CL4 contribute to phenylpropanoid flux in a way that determines H, G, and S lignin deposition, but their impact is only revealed in the absence of 4CL1.
4CL1 and 4CL3 Are the Major 4CLs Required for Sinapoylmalate Biosynthesis
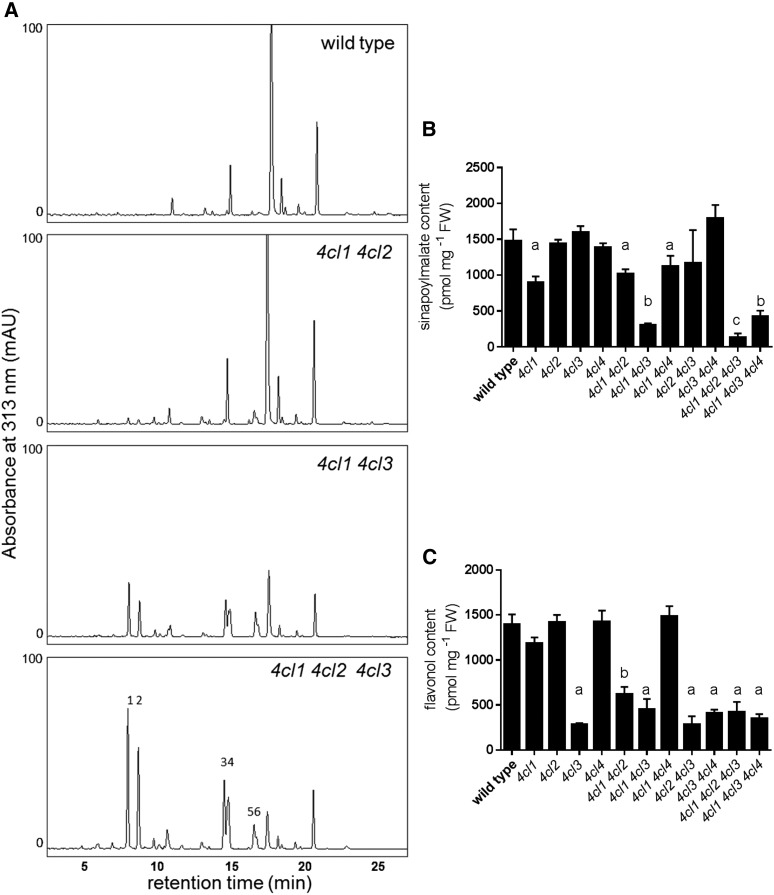
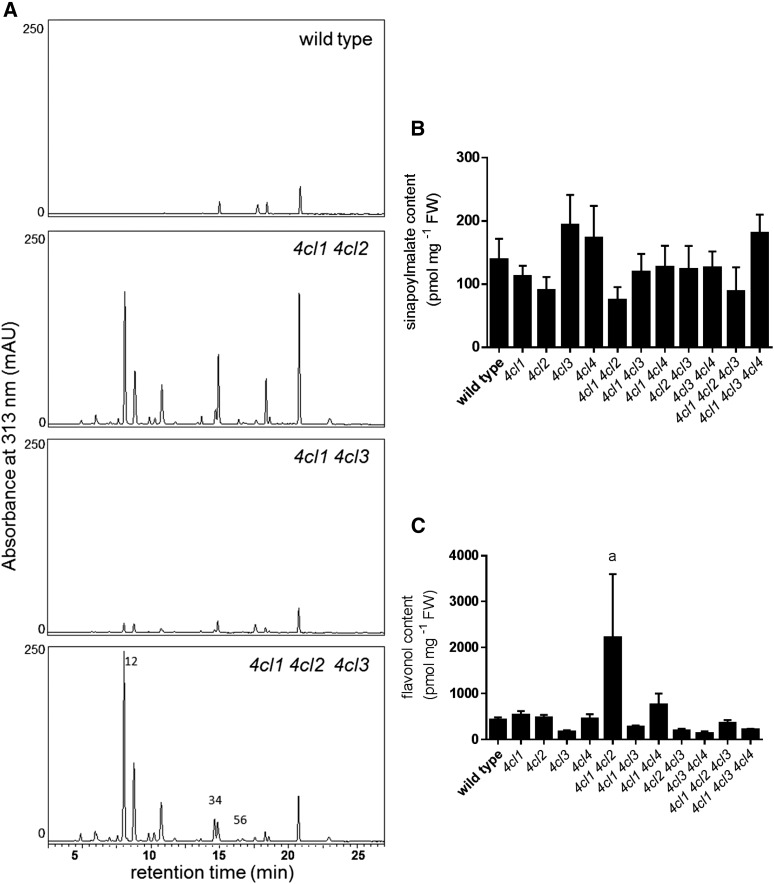
Sinapoylmalate accumulates in the epidermis of Arabidopsis as a UV protectant, and its biosynthesis is thought to require 4CL activity. To test whether one or more isoforms of 4CL have a major role in its biosynthesis, we measured sinapoylmalate content in rosette leaves and found that the 4cl1 single mutant accumulates 40% less than the wild type (Fig. 6). This reduction is further exacerbated in the 4cl1 4cl3 (80%) and 4cl1 4cl2 4cl3 (93%) mutants (Fig. 6), indicating that these three isoforms contribute to sinapoylmalate biosynthesis. It is notable that, in the mutants that contain less sinapoylmalate than the 4cl1 single mutant, none shows any less 4CL activity than the 4cl1 single mutant, at least as measured in extracts in vitro (Fig. 4), suggesting that the epidermal cells in which sinapoylmalate is made may only contribute a small portion of the total 4CL activity measured in leaf homogenates. Sinapoylmalate content was also quantified in the inflorescence, and no significant change was observed in any of the genotypes (Fig. 7).
Figure 6.
Measurements of soluble metabolites in 2-week-old rosettes. A, HPLC chromatograms of rosette methanol extracts with UV absorbance measured at 313 nm. 1,2: p-coumaroyl Glc; 3,4: p-coumaroylmalate; 5,6: feruloylmalate. B, Sinapoylmalate content quantification by HPLC. C, Flavonol content quantification by HPLC. Error bars represent the sd of the means of at least three biological replicates. Using GraphPad Prism, the means were compared by one-way ANOVA (fixed model), and statistically significant differences (P < 0.05) were identified by Dunnett’s test and are indicated by a to c.
Figure 7.
Measurements of soluble metabolites in 5-week-old inflorescences. A, HPLC chromatograms of rosette methanol extracts with UV absorbance measured at 313 nm. 1,2: p-coumaroyl Glc; 3,4: p-coumaroylmalate; 5,6: feruloylmalate. B, Sinapoylmalate content quantification by HPLC. C, Flavonol content quantification by HPLC. Error bars represent the sd of the means of at least three biological replicates. Using GraphPad Prism, the means were compared by one-way ANOVA (fixed model), and statistically significant differences (P < 0.05) were identified by Dunnett’s test and are indicated by a.
Atypical Soluble Phenylpropanoid Metabolites Accumulate in 4cl Mutants
The Arabidopsis 4CLs can use p-coumarate, caffeate, ferulate, and, in the case of 4CL4, sinapate as substrates (Ehlting et al., 1999; Hamberger and Hahlbrock, 2004; Costa et al., 2005). The canonical view of the phenylpropanoid pathway (Fig. 1) is that 4CL activates p-coumarate; however, caffeoyl CoA is now also thought to be generated by 4CL using caffeic acid produced by CSE (Vanholme et al., 2013). To further evaluate this model, we analyzed soluble metabolites of 2-week-old rosettes and 5-week-old inflorescences by liquid chromatography-mass spectrometry. We found that substances with masses that match those of p-coumaroyl Glc, p-coumaroylmalate, and feruloylmalate accumulate in 4cl mutants, particularly in 4cl1 4cl2, 4cl1 4cl3, and 4cl1 4cl2 4cl3 plants (Fig. 6; Supplemental Fig. S1). In their inflorescences, 4cl1 4cl2 and 4cl1 4cl2 4cl3 plants accumulate higher levels of these atypical compounds than does the 4cl1 4cl3 mutant. Further, the levels of these compounds exceed those of the sinapoylmalate found in the wild type (Fig. 7; Supplemental Fig. S1). In contrast, in the rosette, knocking out 4CL1 and 4CL3 has the largest impact on the accumulation of p-coumaroyl Glc, p-coumaroylmalate, and feruloylmalate (Fig. 6; Supplemental Fig. S1). These data are consistent with the involvement of 4CLs in both the activation of p-coumarate to form p-coumaroyl CoA and in the activation of CSE-generated caffeate to form caffeoyl CoA.
4CL3 Has an Important Role in Flavonoid Biosynthesis
Flavonoids are accumulated as flavonol glycosides in the aerial part of the Arabidopsis plant, and 4CL3 has been shown to be important in floral flavonol biosynthesis (Dobritsa et al., 2011). To more broadly test the role of 4CL3, we analyzed the flavonol glycoside content of Arabidopsis 2-week-old rosettes. We found that, among the single mutants, only in 4cl3 is flavonol glycoside content reduced as compared with wild-type plants (Fig. 6). Higher-order mutants containing the 4cl3 mutation exhibit similar phenotypes as 4cl3 plants (Fig. 6). The 4cl1 4cl2 mutant accumulates fewer flavonols than the wild type but more than the 4cl3 mutant alone or mutants containing the 4cl3 mutation (Fig. 6), suggesting that 4CL1 and 4CL2 also contribute to flavonol biosynthesis. Flavonol content was also quantified in the inflorescence where there was no significant difference among the genotypes except 4cl1 4cl2, in which total flavonol content is increased (Fig. 7). It seems likely that, when both 4CL1 and 4CL2 are blocked, phenylpropanoid flux is redirected to flavonol biosynthesis, resulting in a 4CL3-dependent hyperaccumulation of flavonols.
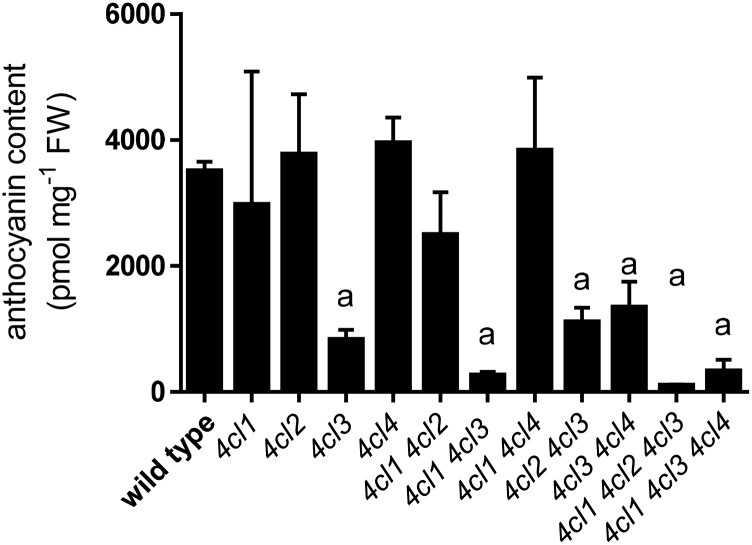
Anthocyanins are flavonoid pigments transiently accumulated under high light and/or Suc induction conditions in Arabidopsis (Kubasek et al., 1992; Shirley et al., 1995). To further test the role of 4CL3 in flavonoid biosynthesis, we grew seedlings under anthocyanin-inducing conditions (2% [w/v] Suc). The 4cl3-containing mutants fail to accumulate anthocyanins compared with wild-type seedlings when quantified by HPLC (Fig. 8). Together, these data show that 4CL3 is the predominant 4CL in flavonoid biosynthesis under a variety of conditions.
Figure 8.
Anthocyanin induction of Arabidopsis seedlings. Arabidopsis seedlings were grown under 2% (w/v) Suc stress and 24-h continuous light. Anthocyanin content was quantified by HPLC at day 4. Error bars represent the sd of the means of three biological replicates. Using GraphPad Prism, the means were compared by one-way ANOVA (fixed model), and statistically significant differences (P < 0.05) were identified by Dunnett’s test and are indicated by a.
DISCUSSION
The Arabidopsis genome contains a large number of genes encoding enzymes that can activate carboxylic acids, including UDP-glucosyltransferases (Lim et al., 2001), long-chain fatty acyl-CoA synthetases (LACSs), acetate CoA ligases, and 4CL and 4CL-like proteins (Cukovic et al., 2001; Kienow et al., 2008), and for each class, there are multiple paralogs. The generation of these paralogous genes is thought to be largely due to gene duplication events. Although the majority are silenced post gene duplication, the small portion of genes that are retained have either neofunctionalized or subfunctionalized (Lynch and Conery, 2000; Rensing, 2014). Many of these enzymes are promiscuous and have evolved diverse functions: many of the LACSs are involved in fatty acid biosynthesis; however, one LACS member has neofunctionalized to participate in jasmonate biosynthesis (Kienow et al., 2008). It has also been reported that one 4CL-like protein encoded by At4g19010 is important for ubiquinone biosynthesis (Block et al., 2014). Among the 4CL genes that are thought to be involved in phenylpropanoid metabolism in Arabidopsis, four encode proteins that can catalyze the formation of hydroxycinnamoyl CoA esters (Ehlting et al., 1999; Hamberger and Hahlbrock, 2004). These 4CL isoforms appear to have subfunctionalized in that their expression patterns have diverged even though their enzyme activities remain similar, with the exception that 4CL4 has gained the ability to activate sinapate (Cukovic et al., 2001; Ehlting et al., 2001; Stuible and Kombrink, 2001; Hamberger and Hahlbrock, 2004).
Isoforms of 4CL Have Diverged in Function
The presence of multiple 4CL isoforms is common in plant lineages, and the divergence of their expression patterns and functions has also been reported. For example, four 4CLs identified in sorghum exhibit similar catalytic efficiency toward p-coumaric acid, caffeic acid, and ferulic acid, but their expression patterns have diverged in leaves, stems, and roots (Saballos et al., 2012). From kudzu, a member of the Fabaceae, two 4CLs have been isolated. Although both can use p-coumaric acid as a substrate, only one 4CL can activate caffeic acid. In addition, their expression patterns are different in that one is expressed highly in the root, whereas the other is in the stem (Li et al., 2014). Functional analysis showed that Pinta4CL1 and Pinta4CL3 from loblolly pine have similar catalytic efficiencies toward p-coumaric acid and caffeic acid; however, Pinta4CL1 has the highest expression in xylem, where it presumably has a role in lignin biosynthesis, and Pinta4CL3 functions mainly in the biosynthesis of soluble metabolites (Chen et al., 2014). Similarly, a recent study showed that 4CL1 in Populus spp. is involved in lignin biosynthesis in the stem, and 4CL2 is involved in flavonoid biosynthesis in the roots (Zhou et al., 2015). Together, these examples show that the enzymatic functions of 4CL isoforms often remain similar, and that the divergence of expression patterns is widespread and may be a major factor contributing to their different roles in phenylpropanoid metabolism among plant lineages. In Arabidopsis, our findings are consistent with this model in that, although the four 4CL isoforms have similar enzymatic functions, 4CL1,4CL2, and 4CL4 contribute to lignin synthesis in the xylem, whereas 4CL3 is expressed more broadly and is a key isoform in flavonoid and sinapoylmalate biosynthesis.
Loss of 4CL Isoforms Leads to the Elevation of Certain Classes of Metabolites
When we examined the soluble metabolite content of 4cl1 4cl2 inflorescences, we found that they contain elevated levels of flavonols (Fig. 7). We also noted in other 4CL multiple mutants the accumulation of atypical metabolites,including p-coumaroyl Glc, p-coumaroylmalate, and feruloylmalate (Figs. 6 and 7). It has been observed previously that, when earlier steps of the phenylpropanoid pathway are blocked, flux that normally enters lignin and sinapoylmalate biosynthesis is redirected to the biosynthesis of other metabolites. For example, C4H-deficient mutants accumulate cinnamoyl esters (Schilmiller et al., 2009), and when p-coumaroyl shikimate 3′-hydroxylase is blocked, the plants accumulate high levels of flavonols, anthocyanins, and atypical soluble phenylpropanoids, including p-coumaroyl Glc, p-coumaroylmalate, and additional metabolites derived from p-coumaric acid (Schoch et al., 2001; Franke et al., 2002). These findings again demonstrate the plasticity of plant secondary metabolism. Perhaps most importantly, the presence of feruloylmalate is consistent with the model showing that CSE generates caffeic acid, which is used by 4CL to make caffeoyl CoA (Vanholme et al., 2013; Fig. 1), and suggests that when 4CL activity is reduced, caffeic acid generated by CSE can be methylated by caffeic acid O-methyl transferase to generate ferulate, which is subsequently used by sinapic acid:UDP-Glc-dependent sinapoyltransferase and sinapoylglucose malate:sinapoyltransferase (Fig. 1), the two subsequent and promiscuous enzymes of sinapoylmalate biosynthesis, to make feruloylmalate (Do et al., 2007).
Growth Defects and Lignin Deposition in 4cl Mutants
Targeted engineering of the phenylpropanoid genes can result in changes in both lignin content and composition (Lee et al., 1997; Franke et al., 2002; Sibout et al., 2005; Coleman et al., 2008; Li et al., 2010; Van Acker et al., 2013). 4CL activity in plants considerably exceeds what is needed to support normal lignin biosynthesis. Consistent with this hypothesis, RNA interference poplar (Populus spp.) with 50% wild-type 4CL activity shows no reduction in lignin content (Voelker et al., 2010). In tobacco (Nicotiana tabacum), even when total 4CL activity is reduced by 99%, there is only a 30% reduction in lignin content (Kajita et al., 1996). There have been similar reports on the role of 4CL in lignin biosynthesis in pine (Pinus spp.), poplar, and switchgrass (Panicum virgatum). In these studies, when the total 4CL activity is reduced by 70% to 100%, lignin content is reduced by 20% to 40% and the plants are dwarf (Li et al., 2003; Wagner et al., 2009; Xu et al., 2011; Zhou et al., 2015). Similarly, all of our 4cl1-deficient mutants have 90% reductions in 4CL activity but only a 30% reduction in lignin content; however, only two of these six multiple mutant lines are dwarf.
Disruptions in the phenylpropanoid pathway often lead to plant growth defects. It was thought that the dwarf phenotype associated with lignin-deficient mutants is due to the inability of these plants to transport water (Bonawitz and Chapple, 2010; Bonawitz et al., 2014). In general, plants defective in the earlier steps of the pathway have more severe consequences: the pal1 pal2 pal3 pal4 quadruple mutant is dwarf and sterile and contains only one-third of the amount of lignin and 10% of the PAL activity found in the wild type (Huang et al., 2010). Similarly, an allelic series of C4H mutants have increasingly severe reductions in phenylpropanoid metabolites and cell wall components. The strongest alleles, which have an 80% reduction in lignin content, also have a dwarf phenotype (Schilmiller et al., 2009). Dwarf phenotypes were also observed in 4cl1 4cl2 and 4cl1 4cl2 4cl3 plants even though they contain 70% of the lignin found in the wild type (Fig. 5). Further, 4cl1 plants contain the same level of lignin as these dwarf lines but grow normally, indicating that the degree of growth perturbation observed is not proportional to the extent of lignin reduction. Recent studies show that dwarfism in some lignin-deficient plants may involve sensing (directly or indirectly) of metabolic (lack of or hyperaccumulation of an intermediate) or cell wall changes via the mediator complex, a multisubunit transcriptional coregulator that mediates the recruitment of RNA polymerase II during transcription (Bonawitz et al., 2012, 2014). If dwarfism in 4CL-deficient plants occurs through this mechanism, the molecules that are sensed must be 4CL1 and 4CL2 dependent or accumulate in the absence of their activities.
In addition to the lignin deficiency phenotype, 4cl mutants exhibit changes in lignin composition. In wild-type plants, the ratio of G:S subunits is approximately 3:1, but in a 4cl1 single mutant, G lignin decreases to 60 mol% and is further reduced to 50 mol% in a 4cl1 4cl2 4cl3 mutant (Fig. 5). Similar phenotypes have been reported in 4CL knockdowns of Arabidopsis (Lee et al., 1997; Van Acker et al., 2013) and other species (Xu et al., 2011; Xiang et al., 2015), although this is not always the case (Li et al., 2003). Because G lignin is preferentially deposited in the xylem and S lignin is primarily deposited in the interfascicular tissues (Chapple et al., 1992), it has been proposed that 4CL knockouts or antisense knockdowns may affect one tissue more than another, resulting in a changed G:S ratio. For example, 4CL1 may have a major role in the deposition of G lignin in the xylem, and reducing its activity may preferentially down-regulate xylem lignification but leave S-rich fiber lignification unaffected. Alternatively, the changes in lignin monomer composition may be related to reduced flux through the pathway and the kinetic properties of the enzymes involved. If reduced flux leads to decreased pools of coniferaldehyde and coniferyl alcohol, ferulate 5-hydroxylase, with a KM of 3 to 5 µm (Humphreys et al., 1999), may outcompete cinnamyl alcohol dehydrogenase (KM of approximately 20–50 µm; Kim et al., 2004) and the yet-to-be-identified monolignol transporters, and divert a higher proportion of phenylpropanoid intermediates into S lignin. This model is supported by the observation that the lignin of Arabidopsis C4H mutants is also decreased in G monomer content (Schilmiller et al., 2009). C4H is encoded by a single gene in Arabidopsis, thus this phenotype cannot be explained by isoform tissue-specific expression and is more readily explained in terms of reduced flux and altered intermediate partitioning, although we cannot rule out the formal possibility that there is a 4CL-independent pathway leading to S subunits.
Finally, we have identified a role for 4CL4 in Arabidopsis. In wild-type plants, H lignin accounts for only 2 mol%; however, it is increased to 6 mol% when 4CL1 and 4CL4 are blocked, indicating that 4CL4 contributes to lignin deposition. Further, the elevated H lignin phenotype of 4cl1 4cl2 and 4cl1 4cl4 plants is a partial phenocopy of the cse mutant (Vanholme et al., 2013), providing additional evidence for the role of 4CL in the activation of caffeic acid generated by CSE in lignin biosynthesis.
In summary, the four isoforms of Arabidopsis 4CLs have overlapping yet distinctive roles in phenylpropanoid metabolism, probably due to their divergent expression patterns. The subfunctionalization of Arabidopsis 4CL isoforms adds further biochemical understanding that could be broadly applicable in other plant species. This study revises previous predictions and extends our understanding of the roles of the 4CL isoforms in soluble metabolism and lignin biosynthesis, and may assist in further rational engineering of the phenylpropanoid pathway.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants were grown in a chamber using a 16-h-light and 8-h-dark cycle under a light intensity of 120 µE sec−1 m−2 at 22°C (Redi-Earth Plug and Seedling Mixture; Sun Gro Horticulture). The soil was supplemented with Scotts Osmocote Plus controlled-release fertilizer (Hummert International). For plate-grown Arabidopsis plants, seeds were surface sterilized at room temperature while shaking for 10 min in a mixture of 2 parts 0.1% (v/v) Triton X-100 and 1 part household bleach. Seeds were washed five times with sterile water and plated onto regular or ammonia-free Murashige and Skoog (MS) medium. The plants were grown at 22°C in a growth chamber with continuous light or a 16-h-light/8-h-dark photoperiod. For anthocyanin induction experiments, Arabidopsis seedlings were grown on ammonia-free MS medium supplemented with 0.5% (w/v) or 2% (w/v) Suc under continuous light for 3 d.
Phylogenetic Analysis
The phylogenetic tree of plant 4CL homologs was constructed using MrBayes version 3.1.1 (Huelsenbeck and Ronquist, 2001), and protein sequences with at least 70% sequence identity to at least one of the four Arabidopsis 4CLs were aligned using MEGA version 5 (Tamura et al., 2011). The protein sequences were obtained from the National Center for Biotechnology Information using BLAST. The sequences are listed in Supplemental Table S1. The parameters used for generating the tree were set to lset = 6, rates = invgamma, and a sampling frequency of every 100th generation for 1 million generations.
GUS Analysis
Promoter regions of 4CL1 (2,548 bp), 4CL2 (952 bp), 4CL3 (1,589 bp), and 4CL4 (2,416 bp) were amplified by PCR and cloned into the destination vector pHGWFS7 (Karimi et al., 2002) using Gateway technology (Invitrogen). Arabidopsis plants were transformed with these promoter-GUS constructs via Agrobacterium tumefaciens-mediated transformation using the floral dip method (Clough and Bent, 1998). Transformed seedlings were selected on MS medium containing 50 µg mL−1 hygromycin. GUS activity in T3 plants was determined by staining at 37°C for 12 to18 h in 50 mm sodium phosphate buffer at pH 7 containing 0.5 mg mL−1 X-Gluc (5-bromo-4-chloro-3-indolyl β-d-glucuronide), 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, and 0.3% (v/v) Triton X-100. The samples were washed with 70% (v/v) ethanol until clear (Jefferson et al., 1987) and were visualized using either a dissecting microscope (Leica DFC450 C), using LAS version 4.5.0 or a light microscope (Leica MZ12 5), using SPOT 5.0.
Genotyping
The transfer DNA insertional lines were obtained from the Arabidopsis Biological Resource Center. Homozygous transfer DNA insertional mutants were isolated by genotyping PCR. WiscDsLox473B01 is designated to be the 4cl1 mutant. Left primer (LP), right primer (RP), and border primers used were cc 3991 (5′-ATC ACC CTC AGC AAA ATC ATG-3′), cc 3992 (5′-CTG GAT CAG CTC CAA TAG CAG-3′), and cc 4208 (5′-GTG TGA GTA GTT CCC AGA TAA G-3′). Salk_110197 is named the 4cl2 mutant. LP, RP, and border primers used were cc 4123 (5′-AAC GGG TTC GCG GAG GTG GTG-3′), cc 4124 (5′-GCA GAG GAA ACT CTT CTC AGA ATC-3′), and cc 4125 (5′-GGA ACA ACA CTC AAC CCT ATC TCG-3′). Sail_636_b07 is used as the 4cl3 mutant. LP, RP, and border primers used were cc 4036 (5′-GCA TAA GTT TGA GAT CGG TGC-3′), cc 4364 (5′-TCG AGA CCC GCA TCT ACT TT-3′), and cc 1973 (5′-CAA GCC TCG CTA GTC AAA AGT GT-3′). Salk_063824 is termed the 4cl4 mutant. LP, RP, and border primers used were cc 4365 (5′-GGA CAG CTT CTC CAC TTT GTC TT-3′), cc 4366 (5′-GAG CTG ATG ACC TCG GAC GCA-3′), and cc 4125.
Soluble Metabolite Analysis
Flavonol glycosides and sinapoylmalate were extracted from rosettes or inflorescences with 50% (v/v) methanol at a concentration of 100 mg fresh weight mL−1 for 30 min at 65°C. The extracts were separated by HPLC using a Shim-pack XR-ODS column (Shimadzu; column dimensions: 3.0-mm i.d. × 75-mm length; bead size, 2.2 µm). The gradient for HPLC separation was 2.0% to 25% acetonitrile during 29.5 min in 0.1% formic acid at a flow rate of 0.9 mL min−1. Anthocyanins in seedlings were extracted with a solution containing 50% (v/v) methanol and 4% acetic acid at a concentration of 100 mg fresh weight mL−1 and separated by HPLC using an acetonitrile gradient from 10% to 20% over 10.5 min in 10% formic acid at a flow rate of 0.7 mL min−1. Kaempferol, sinapic acid, and cyanidin were used as standards for quantification of flavonol glycosides, sinapoylmalate, and anthocyanins, respectively, using UV-diode array detection at their respective absorption maxima. For the identification of atypical soluble metabolites, the same methanol extracts were first separated using an Agilent 1100 HPLC system with the same Shimadzu column at a gradient from 2.0% to 25% (v/v) of 0.1% (v/v) formic acid in acetonitrile during 45 min in 0.1% (v/v) formic acid at a flow rate of 0.6 mL min−1. Coupled with liquid chromatography, an Agilent MSD time-of-flight mass spectrometer was then used under negative electrospray ionization mode at 3.2 kV with a 125-V fragmentation voltage. Mass data were analyzed using Agilent MassHunter B.02 software.
Lignin Analysis
For plant cell wall preparation, eight-week-old Arabidopsis inflorescence tissue was ground to powder in liquid nitrogen as described previously (Meyer et al., 1998). DFRC lignin content was determined as described previously (Peng et al., 1998). For the thioglycolic acid lignin analysis, approximately 20 mg of dried plant cell wall was incubated at 80°C in 750 µL of water, 250 µL of concentrated HCl, and 100 µL thioglycolic acid for 3 h. The mixture was centrifuged and the pellet was washed with 1 mL of water and resuspended in 1 mL of 1 m NaOH on a rocking plate at room temperature overnight. The mixture was centrifuged again, and the supernatant was collected and mixed with 200 µL of concentrated HCl. After vortexing and incubating at 4°C for 4 h, the mixture was centrifuged, and the pellet was dissolved in 1 mL of 1 m NaOH. The absorbance of a 50-fold dilution of the supernatant in 1 m NaOH was measured at 280 nm.
4CL Enzyme Assay
4CL activity assays were conducted according to conditions previously described, with slight modifications (Klempien et al., 2012): 25 µL of final volume contained 100 mm Tris-HCl (pH 8.0), 5 mm MgCl2, 5 mm ATP, 25 µm CoA, and 4 mm p-coumaric acid. The reaction was started by adding 5 µL of desalted protein extract from plants and continued for 40 min at room temperature. The reaction was stopped by adding 4 µL of 40% (w/v) TCA. Samples were then centrifuged, and the supernatant was analyzed by HPLC using an Agilent SB-C18 column (4.6 × 250 mm) and a methanol gradient from 10% to 40% (w/v) over 20 min using 50 mm potassium phosphate buffer at pH 7.0 as mobile phase. Protein concentration was determined by the bicinchoninic acid assay method (www.piercenet.com) using bovine serum albumin (Sigma-Aldrich) as a standard. p-coumaryl CoA was used as a standard for product identification and quantification.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers 4CL1 (At1g51680), 4CL2 (At3g21240), 4CL3 (At1g65060), and 4CL4 (At3g21230).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Measurements of atypical soluble metabolites.
Supplemental Table S1. Sequences used to generate the phylogenetic tree.
Supplementary Material
Acknowledgments
We thank Joanne Cusumano for critical review of this article; Drs. Natalia Dudareva, Mary Alice Webb, and Joseph Ogas for kindly sharing instruments; and Drs. Jane Glazebrook and Gerit Bethke for providing the initial seed stocks.
Glossary
- DFRC
derivatization followed by reductive cleavage
- G
guaiacyl
- H
p-hydroxyphenyl
- LACS
long-chain fatty acyl-CoA synthetase
- MS
Murashige and Skoog
Footnotes
This work was supported by the National Science Foundation (grant no. MCB–1121925 to C.C.).
Articles can be viewed without a subscription.
References
- Bassard JE, Richert L, Geerinck J, Renault H, Duval F, Ullmann P, Schmitt M, Meyer E, Mutterer J, Boerjan W, et al. (2012) Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell 24: 4465–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A, Widhalm JR, Fatihi A, Cahoon RE, Wamboldt Y, Elowsky C, Mackenzie SA, Cahoon EB, Chapple C, Dudareva N, et al. (2014) The origin and biosynthesis of the benzenoid moiety of ubiquinone (Coenzyme Q) in Arabidopsis. Plant Cell 26: 1938–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C (2010) The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet 44: 337–363 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C (2013) Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr Opin Biotechnol 24: 336–343 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Kim JI, Tobimatsu Y, Ciesielski PN, Anderson NA, Ximenes E, Maeda J, Ralph J, Donohoe BS, Ladisch M, et al. (2014) Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509: 376–380 [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Soltau WL, Blatchley MR, Powers BL, Hurlock AK, Seals LA, Weng J-K, Stout J, Chapple C (2012) REF4 and RFR1, subunits of the transcriptional coregulatory complex mediator, are required for phenylpropanoid homeostasis in Arabidopsis. J Biol Chem 287: 5434–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P. (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin Plant Biol 8: 272–279 [DOI] [PubMed] [Google Scholar]
- Chapple CC, Vogt T, Ellis BE, Somerville CR (1992) An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4: 1413–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-Y, Babst BA, Nyamdari B, Hu H, Sykes R, Davis MF, Harding SA, Tsai CJ (2014) Ectopic expression of a loblolly pine class II 4-coumarate:CoA ligase alters soluble phenylpropanoid metabolism but not lignin biosynthesis in Populus. Plant Cell Physiol 55: 1669–1678 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coleman HD, Park J-Y, Nair R, Chapple C, Mansfield SD (2008) RNAi-mediated suppression of p-coumaroyl-CoA 3′-hydroxylase in hybrid poplar impacts lignin deposition and soluble secondary metabolism. Proc Natl Acad Sci USA 105: 4501–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MA, Bedgar DL, Moinuddin SGA, Kim K-W, Cardenas CL, Cochrane FC, Shockey JM, Helms GL, Amakura Y, Takahashi H, et al. (2005) Characterization in vitro and in vivo of the putative multigene 4-coumarate:CoA ligase network in Arabidopsis: syringyl lignin and sinapate/sinapyl alcohol derivative formation. Phytochemistry 66: 2072–2091 [DOI] [PubMed] [Google Scholar]
- Cukovic D, Ehlting J, VanZiffle JA, Douglas CJ (2001) Structure and evolution of 4-coumarate:coenzyme A ligase (4CL) gene families. Biol Chem 382: 645–654 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7: 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do CT, Pollet B, Thévenin J, Sibout R, Denoue D, Barrière Y, Lapierre C, Jouanin L (2007) Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226: 1117–1129 [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Geanconteri A, Shrestha J, Carlson A, Kooyers N, Coerper D, Urbanczyk-Wochniak E, Bench BJ, Sumner LW, Swanson R, et al. (2011) A large-scale genetic screen in Arabidopsis to identify genes involved in pollen exine production. Plant Physiol 157: 947–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J, Büttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E (1999) Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J 19: 9–20 [DOI] [PubMed] [Google Scholar]
- Ehlting J, Shin JJ, Douglas CJ (2001) Identification of 4-coumarate:coenzyme A ligase (4CL) substrate recognition domains. Plant J 27: 455–465 [DOI] [PubMed] [Google Scholar]
- Franke R, Hemm MR, Denault JW, Ruegger MO, Humphreys JM, Chapple C (2002) Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J 30: 47–59 [DOI] [PubMed] [Google Scholar]
- Hahlbrock K, Grisebach H (1979) Enzymic controls in the biosynthesis of lignin and flavonoids. Annu Rev Plant Physiol 30: 105–130 [Google Scholar]
- Hamberger B, Hahlbrock K (2004) The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc Natl Acad Sci USA 101: 2209–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins S, Boudet A (1996) Wound-induced lignin and suberin deposition in a woody angiosperm (Eucalyptus gunnii Hook.): Histochemistry of early changes in young plants. Protoplasma 191: 96–104 [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, Chen Z (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153: 1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Humphreys JM, Hemm MR, Chapple C (1999) New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci USA 96: 10045–10050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita S, Katayama Y, Omori S (1996) Alterations in the biosynthesis of lignin in transgenic plants with chimeric genes for 4-coumarate: coenzyme A ligase. Plant Cell Physiol 37: 957–965 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kienow L, Schneider K, Bartsch M, Stuible H-P, Weng H, Miersch O, Wasternack C, Kombrink E (2008) Jasmonates meet fatty acids: functional analysis of a new acyl-coenzyme A synthetase family from Arabidopsis thaliana. J Exp Bot 59: 403–419 [DOI] [PubMed] [Google Scholar]
- Kim SJ, Kim MR, Bedgar DL, Moinuddin SGA, Cardenas CL, Davin LB, Kang C, Lewis NG (2004) Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc Natl Acad Sci USA 101: 1455–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempien A, Kaminaga Y, Qualley A, Nagegowda DA, Widhalm JR, Orlova I, Shasany AK, Taguchi G, Kish CM, Cooper BR, et al. (2012) Contribution of CoA ligases to benzenoid biosynthesis in petunia flowers. Plant Cell 24: 2015–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobloch KH, Hahlbrock K (1975) Isoenzyme der p-Cumarat: CoA Ligase aus Zellsuspensionskulturen von Glycine Max (in German). Planta Med 28(Suppl): 102–111 [DOI] [PubMed] [Google Scholar]
- Koopman F, Beekwilder J, Crimi B, van Houwelingen A, Hall RD, Bosch D, van Maris AJA, Pronk JT, Daran JM (2012) De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Microb Cell Fact 11: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubasek WL, Shirley BW, McKillop A, Goodman HM, Briggs W, Ausubel FM (1992) Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell 4: 1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry LG, Chapple CCS, Last RL (1995) Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol 109: 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Ellard M, Wanner LA, Davis KR, Douglas CJ (1995) The Arabidopsis thaliana 4-coumarate:CoA ligase (4CL) gene: stress and developmentally regulated expression and nucleotide sequence of its cDNA. Plant Mol Biol 28: 871–884 [DOI] [PubMed] [Google Scholar]
- Lee D, Meyer K, Chapple C, Douglas CJ (1997) Antisense suppression of 4-coumarate:coenzyme A ligase activity in Arabidopsis leads to altered lignin subunit composition. Plant Cell 9: 1985–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhou Y, Cheng X, Sun J, Marita JM, Ralph J, Chiang VL (2003) Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc Natl Acad Sci USA 100: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Bonawitz ND, Weng JK, Chapple C (2010) The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22: 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZB, Li CF, Li J, Zhang YS (2014) Molecular cloning and functional characterization of two divergent 4-coumarate : coenzyme A ligases from Kudzu (Pueraria lobata). Biol Pharm Bull 37: 113–122 [DOI] [PubMed] [Google Scholar]
- Lim EK, Li Y, Parr A, Jackson R, Ashford DA, Bowles DJ (2001) Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J Biol Chem 276: 4344–4349 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Meyer K, Shirley AM, Cusumano JC, Bell-Lelong DA, Chapple C (1998) Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc Natl Acad Sci USA 95: 6619–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Lu F, Ralph J (1998) The DFRC method for lignin analysis. 4. Lignin dimers isolated from DFRC-degraded loblolly pine wood. J Agric Food Chem 46: 553–560 [DOI] [PubMed] [Google Scholar]
- Rensing SA. (2014) Gene duplication as a driver of plant morphogenetic evolution. Curr Opin Plant Biol 17: 43–48 [DOI] [PubMed] [Google Scholar]
- Rittinger PA, Biggs AR, Peirson DR (1987) Histochemistry of lignin and suberin deposition in boundary layers formed after wounding in various plant species and organs. Can J Bot 65: 1886–1892 [Google Scholar]
- Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M, et al. (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW. (1971) The metabolism of aromatic compounds in higer plants. X. Properties of the cinnamic acid 4-hydroxylase of pea seedlings and some aspects of its metabolic and developmental control. J Biol Chem 246: 3870–3878 [PubMed] [Google Scholar]
- Russell DW, Conn EE (1967) The cinnamic acid 4-hydroxylase of pea seedlings. Arch Biochem Biophys 122: 256–258 [DOI] [PubMed] [Google Scholar]
- Saballos A, Sattler SE, Sanchez E, Foster TP, Xin Z, Kang C, Pedersen JF, Vermerris W (2012) Brown midrib2 (Bmr2) encodes the major 4-coumarate:coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor (L.) Moench). Plant J 70: 818–830 [DOI] [PubMed] [Google Scholar]
- Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR (2013) The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol Biochem 72: 21–34 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Stout J, Weng JK, Humphreys J, Ruegger MO, Chapple C (2009) Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J 60: 771–782 [DOI] [PubMed] [Google Scholar]
- Schneider K, Hövel K, Witzel K, Hamberger B, Schomburg D, Kombrink E, Stuible HP (2003) The substrate specificity-determining amino acid code of 4-coumarate:CoA ligase. Proc Natl Acad Sci USA 100: 8601–8606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch G, Goepfert S, Morant M, Hehn A, Meyer D, Ullmann P, Werck-Reichhart D (2001) CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J Biol Chem 276: 36566–36574 [DOI] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Séguin A (2005) CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltani BM, Ehlting J, Hamberger B, Douglas CJ (2006) Multiple cis-regulatory elements regulate distinct and complex patterns of developmental and wound-induced expression of Arabidopsis thaliana 4CL gene family members. Planta 224: 1226–1238 [DOI] [PubMed] [Google Scholar]
- Stuible HP, Kombrink E (2001) Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase allows the rational design of mutant enzymes with new catalytic properties. J Biol Chem 276: 26893–26897 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A (2008) Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J 54: 733–749 [DOI] [PubMed] [Google Scholar]
- Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W (2013) Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, et al. (2013) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Voelker SL, Lachenbruch B, Meinzer FC, Jourdes M, Ki C, Patten AM, Davin LB, Lewis NG, Tuskan GA, Gunter L, et al. (2010) Antisense down-regulation of 4CL expression alters lignification, tree growth, and saccharification potential of field-grown poplar. Plant Physiol 154: 874–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Donaldson L, Kim H, Phillips L, Flint H, Steward D, Torr K, Koch G, Schmitt U, Ralph J (2009) Suppression of 4-coumarate-CoA ligase in the coniferous gymnosperm Pinus radiata. Plant Physiol 149: 370–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshaar B, Jenkins GI (1998) Phenylpropanoid biosynthesis and its regulation. Curr Opin Plant Biol 1: 251–257 [DOI] [PubMed] [Google Scholar]
- Weng JK, Li Y, Mo H, Chapple C (2012) Assembly of an evolutionarily new pathway for α-pyrone biosynthesis in Arabidopsis. Science 337: 960–964 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001) Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Sen SK, Roy A, Min D, Savithri D, Jameel H, Chiang V, Chang HM (2015) Wood characteristics and enzymatic saccharification efficiency of field-grown transgenic black cottonwood with altered lignin content and structure. Cellulose 22: 683–693 [Google Scholar]
- Xu B, Escamilla-Treviño LL, Sathitsuksanoh N, Shen Z, Shen H, Zhang YHP, Dixon RA, Zhao B (2011) Silencing of 4-coumarate:coenzyme A ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol 192: 611–625 [DOI] [PubMed] [Google Scholar]
- Zhou X, Jacobs TB, Xue LJ, Harding SA, Tsai CJ (2015) Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-coumarate:CoA ligase specificity and redundancy. New Phytol 208: 298–301 10.1111/nph.13470 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.