Single and combined Fe and S starvation of plants induces a complex partially overlapping regulatory system to coordinate the starvation response.
Abstract
Plant response mechanisms to deficiency of a single nutrient, such as sulfur (S) or iron (Fe), have been described at agronomic, physiological, biochemical, metabolomics, and transcriptomic levels. However, agroecosystems are often characterized by different scenarios, in which combined nutrient deficiencies are likely to occur. Soils are becoming depleted for S, whereas Fe, although highly abundant in the soil, is poorly available for uptake because of its insolubility in the soil matrix. To this end, earlier reports showed that a limited S availability reduces Fe uptake and that Fe deficiency results in the modulation of sulfate uptake and assimilation. However, the mechanistic basis of this interaction remains largely unknown. Metabolite profiling of tomato (Solanum lycopersicum) shoots and roots from plants exposed to Fe, S, and combined Fe and S deficiency was performed to improve the understanding of the S-Fe interaction through the identification of the main players in the considered pathways. Distinct changes were revealed under the different nutritional conditions. Furthermore, we investigated the development of the Fe deficiency response through the analysis of expression of ferric chelate reductase, iron-regulated transporter, and putative transcription factor genes and plant sulfate uptake and mobilization capacity by analyzing the expression of genes encoding sulfate transporters (STs) of groups 1, 2, and 4 (SlST1.1, SlST1.2, SlST2.1, SlST2.2, and SlST4.1). We identified a high degree of common and even synergistic response patterns as well as nutrient-specific responses. The results are discussed in the context of current models of nutrient deficiency responses in crop plants.
Several biotic and abiotic stresses provoke unfavorable environmental fluctuations, which require plants to reprogram and adjust metabolism, growth, and development to adapt and survive. In particular, crop species are continuously challenged by inadequate supply of nutrients. In recent years, the decrease of atmospheric SO2 emissions and lower sulfur (S) supply through mineral fertilization (McGrath et al., 2002) have led to the occurrence of S deficiency, rendering S an important issue for plant nutrition. As a result, the effects of S deficiency have become a major subject area, particularly for studies focused on regulatory aspects of S assimilation. Responses of plants under S deficiency have been described at the levels of agronomy, physiology, biochemistry, metabolomics, and transcriptomics (for review, see Lewandowska and Sirko, 2008; Watanabe et al., 2010). However, little is known concerning the physiological responses during S deficiency conditions in relation to how they are modified by the interactions that S has with other nutrients in the rhizosphere. The consequences of nutrient interactions are largely unknown, although such scenarios commonly occur in agroecosystems. In particular, excess or deficiency of one nutrient could modify the internal demand and consequently, the uptake and assimilation rate of other nutrients.
Recently, it was shown that the iron (Fe) use efficiency in maize (Zea mays; Astolfi et al., 2003; Bouranis et al., 2003), barley (Hordeum vulgare; Kuwajima and Kawai, 1997; Astolfi et al., 2006a), tomato (Solanum lycopersicum; Zuchi et al., 2009), and durum wheat (Triticum durum; Zuchi et al., 2012; Ciaffi et al., 2013) increased under adequate S supply. In these studies, it has been suggested that, in grasses (strategy II plants), this effect could be ascribed to a decrease in the production and release of phytosiderophores induced by S deficiency, whereas in tomato (a strategy I plant), the effect was rather caused by an impaired ethylene and nicotianamine (NA) production.
Strategy I plants cope with low Fe availability in the soil by mobilizing and taking up Fe3+ ions from soil particles by acidification of the rhizosphere, likely driven by an increased plasma membrane proton-pumping ATPase activity (Dell’Orto et al., 2000), by enhancing the activity of a plasma membrane-bound Fe(III)-chelate reductase, and by overexpression of an Fe2+ transporter of root epidermal cells (Eide et al., 1996; Robinson et al., 1999). Romera et al. (1999) showed that pea (Pisum sativum), cucumber (Cucumis sativus), and tomato roots produced more ethylene under Fe deficiency, with ethylene production being correlated with Fe(III) reducing capacity. Furthermore, after inside root cells, NA plays an important role for Fe distribution within the plant (Rudolph et al., 1985; Douchkov et al., 2002; Takahashi et al., 2003).
The link between S and Fe mainly arises from the fact that ethylene and NA share a common precursor, namely S-adenosyl-methionine (SAM), and Met is required for the synthesis of SAM (Hesse and Hoefgen, 2003). Furthermore, it has been shown in plants that Fe is normally linked with S when it is bound in Fe-S proteins, suggesting that Fe-S clusters are the biggest sink for Fe within the plant (Balk and Pilon, 2011).
As described above for S deficiency, most studies aimed at understanding the physiological basis of the plant response to Fe deficiency have largely been focused on manipulating this single nutrient in isolation and on model plants, such as Arabidopsis (Arabidopsis thaliana). Fewer studies have concentrated on crop plants, such as barley, rice (Oryza sativa), tomato, and other economically important species, although there are indications that adaptive mechanisms of crops might differ from those of model plants (Møller and Tester, 2007). Furthermore, even fewer studies dealt with responses of plants to combinations of nutrient deficiencies (Pii et al., 2015), although it is reasonable to suggest that simultaneous imposition of two stresses will trigger response mechanisms different than the response to each stress applied separately.
Previously, Zuchi et al. (2009) showed that, in tomato plants exposed to both S and Fe starvation, no induction of Fe(III)-chelate reductase activity and ethylene production occurred. Thus, S deficiency seems to prevent the development of the typical responses to Fe deficiency in tomato. However, it was also recently shown that Fe deficiency significantly up-regulated most of the sulfate transporter (ST) genes belonging to groups 1, 2, and 4 in tomato roots (Paolacci et al., 2014). This interaction between S and Fe triggers the activation of complex response mechanisms to assumingly maintain plant primary metabolism function and maintain adequate levels of Cys, Met, and their derivatives as well as Fe-S clusters as essential building blocks of various enzymes. Especially, the latter asks for a coordinated control of Fe and S metabolism. However, the metabolic mechanisms involved in coordination are far from being understood and will likely require considerable research effort to be ultimately unraveled. As a first step toward this goal, the changes in the metabolome of tomato plants subjected to simultaneous S and Fe deficiency were evaluated and compared with the results for the metabolic response to S or Fe starvation alone. The development of the Fe deficiency stress response was followed through the analysis of the expression of the ferric chelate reductase (SlFRO1), iron-regulated transporter (SlIRT1), and putative transcription factor (SlFER) genes, and plant sulfate uptake and mobilization capacity were investigated by analyzing the expression of genes encoding STs of the groups 1, 2, and 4 (SlST1.1, SlST1.2, SlST2.1, SlST2.2, and SlST4.1).
RESULTS
Plant Growth and S and Fe Accumulation
As previously observed (Zuchi et al., 2009), the overall growth of tomato seedlings was severely reduced by either S or Fe deficiency (Table I), which is perhaps unsurprising given that both are essential elements. Total protein concentration was additionally significantly lower in plants exposed to nutrient deficiency than in plants grown in complete nutrient solution (Table I). In particular, after the imposition of sole Fe starvation, protein concentrations were approximately 15% and 25% lower than the control in shoots and roots, respectively. S deficiency affected this parameter even more dramatically, provoking approximately 65% and 35% decreases with respect to the control in shoots and roots, respectively (Table I). Under the combined deficiency condition, it reached its lowest value, being approximately 70% and 50% lower than the control in shoots and roots, respectively (Table I).
Table I. Shoot and root dry weight (grams plant−1) and protein, S, and Fe contents (milligrams plant−1) of tomato plants subjected to varying nutritional stress.
Values given in parentheses are percentages of control. Data are means of six independent replicates run in triplicate. C, Control; D, dual deficiency; F, Fe deficiency; S, S deficiency.
| Tissue and Treatment | Dry Weight | Protein Content | S Content | Fe content |
|---|---|---|---|---|
| g plant−1 | mg plant−1 | |||
| Shoot | ||||
| C | 0.185 (100)a | 12.729 (100)a | 0.236 (100)a | 0.098 (100)a |
| F | 0.112 (60)b | 10.570 (83)b | 0.115 (58)b | 0.026 (25)b |
| S | 0.066 (35)c | 4.605 (36)c | 0.023 (4)c | 0.016 (15)c |
| D | 0.059 (32)c | 3.484 (27)d | 0.014 (3)c | 0.008 (8)d |
| Root | ||||
| C | 0.024 (100)a | 1.494 (100)a | 2.199 (100)a | 0.750 (100)a |
| F | 0.016 (66)b | 1.126 (75)b | 1.292 (49)b | 0.186 (27)b |
| S | 0.009 (40)c | 0.961 (64)c | 0.099 (10)c | 0.111 (16)c |
| D | 0.008 (32)c | 0.764 (51)d | 0.075 (6)c | 0.061 (8)d |
Significant differences between samples (P < 0.05).
Significant differences between samples (P < 0.05).
Significant differences between samples (P < 0.05).
Significant differences between samples (P < 0.05).
In accordance with the plant phenotype, S deficiency drastically reduced the accumulation of total S in both shoot and root tissues (−96% and −90%, respectively). However, Fe-deficient plants also showed markedly reduced levels of total S accumulation in both shoots (−40%) and roots (−50%; Table I).
Furthermore, as would be anticipated, Fe deficiency led to large decreases in Fe content (−75% in both shoots and roots); however, surprisingly, greater reductions were found under S deficiency (−85% in both shoots and roots) and dual deficiency (−92% in both shoots and roots; Table I).
Changes in Inorganic Anion Concentrations
In addition to total S and Fe contents, the concentrations of sulfate, phosphate, and nitrate were measured to estimate whether the different treatments differentially altered the accumulation of other essential macroelements within the tomato seedlings (Fig. 1).
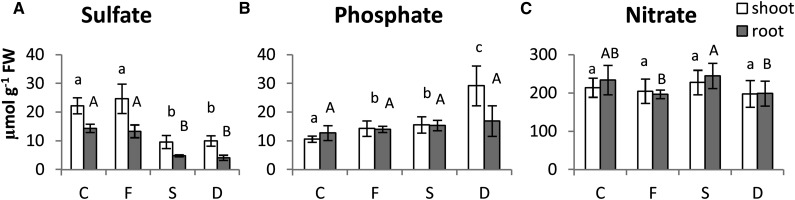
Figure 1.
Sulfate (A), phosphate (B), and nitrate (C) concentrations (micromoles gram−1 fresh weight [FW]) in shoots (white bars) and roots (gray bars) of tomato plants subjected to varying nutritional stress. Data are means ± sd of six independent replicates run in triplicate. Significant differences between samples are indicated by different letters: different uppercase letters indicate significant differences in roots (P < 0.05), and different lowercase letters indicate significant differences in shoots (P < 0.05). C, Control; D, dual deficiency; F, Fe deficiency; S, S deficiency.
As expected, lower concentrations of sulfate were measured in both shoots and roots of S-deficient tomato seedlings. Fe deficiency did not affect the sulfate content in shoots and roots, and a combined deficiency of Fe and S did not further reduce the sulfate content than under sulfate-deplete conditions (Fig. 1A). This finding reveals a distinctively different behavior of sulfate accumulation in the tissue compared with total S amounts, which are, indeed, negatively affected by Fe depletion.
By contrast, shoot concentrations of phosphate seemed sensitive to both Fe and S deficiency. For instance, an increase in phosphate concentration was observed in shoots of −Fe (+34%) and −S (+46%) plants, and the increase became more evident with the imposition of the dual deficiency, reaching an almost 3-fold increase compared with control plants (Fig. 1B). However, at the root level, neither Fe nor S starvation significantly affected phosphate concentrations (Fig. 1B). Shoot nitrate concentrations were not significantly different between nutrient-deprived and control plants (Fig. 1C), whereas −Fe and −Fe-−S plants showed slightly reduced nitrate concentrations in roots (approximately −15%), and −S plants displayed control or even slightly increased nitrate levels (Fig. 1C).
Transcriptional Analysis of ST Genes
To explain the reduction in the accumulation of total S in Fe-deprived plants, the expression patterns of five genes coding for STs belonging to three different groups (1, 2, and 4) were evaluated (Figs. 2 and 3).
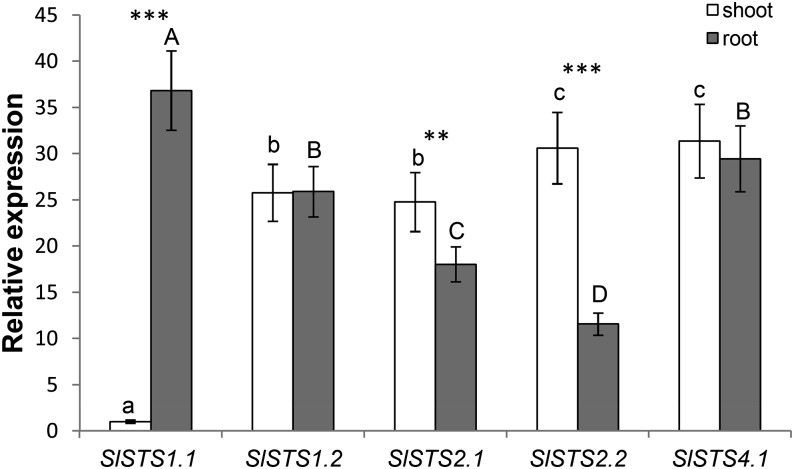
Figure 2.
Relative expression levels by qRT-PCR of the five ST genes in shoots (white bars) and roots (gray bars) of control tomato plants. The six cDNA pools from shoots and roots of three control plants (three biological replicates) were tested in triplicate and normalized using the geometric average of the relative expression of the two reference genes SlTIP4l and SlCAC. Relative expression levels of the five genes were referred to those of a calibrator set to the value of 1, which was represented by the gene in the two tissues with the lowest expression (SlSTS1.1 in shoots), and are given as averages ± sd. Significant differences between samples are indicated by different letters: different uppercase letters indicate significant differences in roots (P < 0.05), and different lowercase letters indicate significant differences in shoots (P < 0.05). Asterisks indicate significant differences between different tissues at the same nutritional condition (**, P < 0.05; and ***, P < 0.01).
Figure 3.
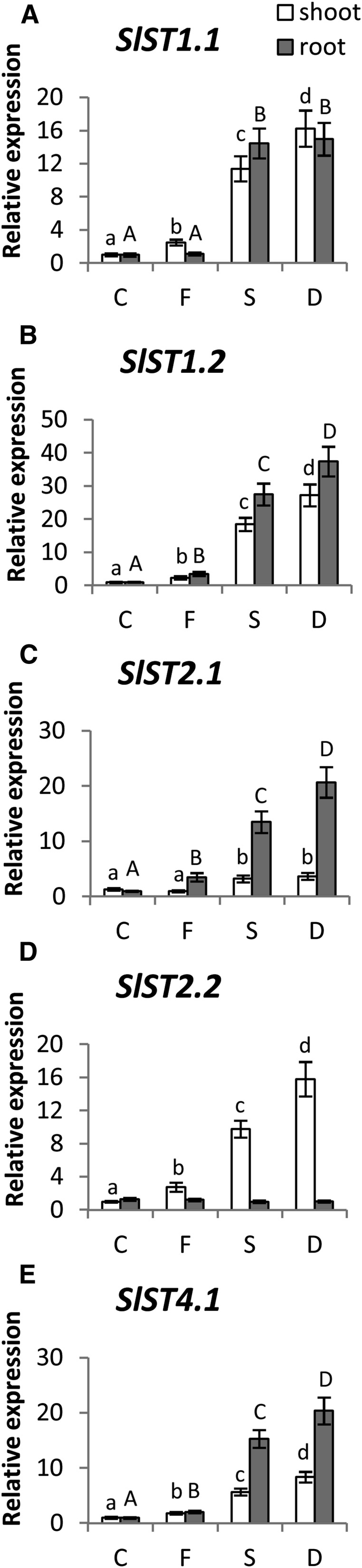
Relative expression levels by real-time qRT-PCR of SlST1.1 (A), SlST1.2 (B), SlST2.1 (C), SlST2.2 (D), and SlST4.1 (E) genes in shoots (white bars) and roots (gray bars) of tomato plants subjected to varying nutritional stress. Data are means ± sd of three independent replicates run in triplicate. Significant differences between samples are indicated by different letters: different uppercase letters indicate significant differences in roots (P < 0.05), and different lowercase letters indicate significant differences among in shoots (P < 0.05). C, Control; D, dual deficiency; F, Fe deficiency; S, S deficiency.
All five ST genes were found to be expressed in both roots and shoots of control plants, although their relative expression was different among the two tissues (Fig. 2). SlST1.1 was preferentially expressed in roots, with a transcriptional level about 37 times higher than in shoots, whereas the abundance of transcripts of the other ST of the group 1, SlST1.2, was approximately the same in shoots and roots of control plants and about 1.5 times lower than that of SlST1.1 in roots (Fig. 2). In contrast, the expression levels of the two tomato group 2 STs, SlST2.1 and SlST2.2, were higher in shoots than in roots of control plants, with the highest difference observed for SlST2.2 (approximately 3-fold higher; Fig. 2). Finally, the transcript levels of SlST4.1 were equally high in both shoots and roots of control plants and similar or even higher compared with the respective highest expression levels of the other four ST genes (Fig. 2). In agreement with previous results, the five ST genes belonging to groups 1, 2, and 4 were significantly up-regulated by S deficiency in both shoots and roots of tomato seedlings, with the exception of SlST2.2, which was unaffected in root tissues by exposure to S deprivation (Fig. 3). For instance, the transcript levels of SlST1.1 were approximately 11 and 15 times higher in shoots and roots, respectively, compared with control plants, whereas the relative amounts of transcripts of the other high-affinity ST gene SlST1.2 increased approximately 18 and 27 times in shoots and roots, respectively (Fig. 3, A and B). Similarly, the expression of SlST4.1 was significantly up-regulated in response to S deprivation, with transcript abundance approximately 6 and 15 times higher in shoots and roots, respectively, than in control plants (Fig. 3E). Moreover, although SlST2.1 was induced in shoots by about 3-fold, the transcript levels of this gene were strongly up-regulated in root tissues upon sulfate starvation (approximately 13 times higher than in control plants), whereas S deprivation induced significant changes in the expression of the other low-affinity ST gene SlST2.2 only in shoots, with a transcriptional level approximately 10 times higher than in control plants (Fig. 3, C and D).
Interestingly, with the exceptions of SlST2.1 in shoots (Fig. 3C) and both SlST1.1 and SlST2.2 in roots (Fig. 3, A and D), all of the tomato ST genes analyzed were already significantly up-regulated after the imposition of Fe limitation under S-sufficient conditions, and their expression was increased to an even greater extent when S-deficient plants were additionally deprived of Fe (Fig. 3). At the root level, the high-affinity ST gene SlST1.2 was up-regulated by Fe deficiency (transcription levels approximately 3 times higher than in control plants; Fig. 3B), whereas the expression of the two low-affinity STs SlST2.1 and SlST4.1 was approximately 3 and 2 times higher than in control plants, respectively (Fig. 3, C and E). However, the expression of four of five analyzed ST genes was up-regulated in shoots upon Fe deprivation, with transcriptional levels about 2- to 3-fold higher than in control plants (Fig. 3).
It is worth noting that, despite these increases in expression of ST genes, which indicate that the plants sense sulfate deprivation, total S levels are reduced under S starvation, dual starvation, and even exclusive Fe starvation (Table I).
Transcriptional Analysis of Fe Stress-Related Genes
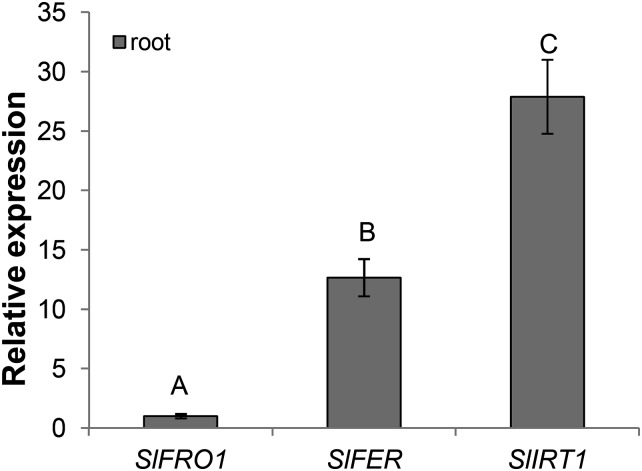
The three genes SlFRO1, SlIRT1, and SlFER were used as representative Fe deficiency-responsive genes. Their expressions in response to the Fe availability and function have been previously described (Eckhardt et al., 2001; Ling et al., 2002; Bereczky et al., 2003; Li et al., 2004; Brumbarova and Bauer, 2005; Schikora et al., 2006). SlFRO1 encodes ferric chelate reductase, which reduces Fe3+ at the root-soil interface, and the SlIRT1 gene encodes a high-affinity Fe2+ transporter. Together, the products of these two genes mobilize Fe2+ across the root epidermal plasma membrane into root cells. The expression of both SlFRO1 and SlIRT1 genes is controlled by the basic helix-loop-helix (bHLH) transcription factor SlFER. Their expression was detected only in roots under all nutritional conditions analyzed, and as previously described (Paolacci et al., 2014), the relative amounts of their transcripts in roots of control plants were markedly different (Fig. 4). In particular, the transcript level of SlIRT1 was approximately 2 times that of SlFER in control plants, whereas SlFRO1 was expressed at a very low level (transcription levels approximately 28 and 13 times lower than those of SlIRT1 and SlFER, respectively; Fig. 4).
Figure 4.
Relative expression levels by qRT-PCR of the three iron stress-related genes (SlFRO1, SlIRT1, and SlFER) in roots of control tomato plants. The three cDNA pools from roots of three control plants (three biological replicates) were tested in triplicate and normalized using the geometric average of the relative expression of the two reference genes SlTIP4l and SlCAC. Relative expression levels of the three genes were referred to those of a calibrator set to the value of one, which was represented by the gene with the lowest expression (SlFRO1). Statistics are the same as in Figure 2.
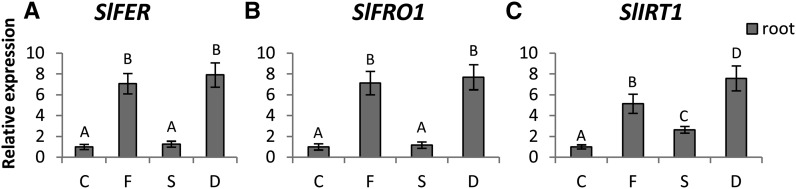
In agreement with previous results, the expression of SlFRO1, SlIRT1, and SlFER was strongly up-regulated by Fe deprivation in roots of tomato seedlings (Fig. 5). In particular, SlFER and SlFRO1 displayed very similar expression patterns, with significant increases under Fe deficiency alone (approximately 7-fold) or combined with S deficiency (approximately 8-fold), whereas their expression was unaffected by S starvation alone (Fig. 5, A and B).
Figure 5.
Relative expression levels by real-time qRT-PCR of SlFER (A), SlFRO1 (B), and SlIRT1 (C) genes in roots of tomato plants subjected to varying nutritional stress. Statistics are the same as in Figure 3. C, Control; D, dual deficiency; F, Fe deficiency; S, S deficiency.
However, the expression of the SlIRT1 gene in roots of tomato seedlings under Fe deficiency treatment also showed a sharp increase with a transcriptional level approximately 5 times higher than in control plants (Fig. 5C). Interestingly, SlIRT1 was also significantly up-regulated in roots harvested from S-deficient plants, even if the expression level of SlIRT1 induced by S starvation (approximately 2-fold higher than in control plants) was significantly lower than that found under Fe deficiency (Fig. 5C). Moreover, we found that the transcriptional level of SlIRT1 in roots exposed to both Fe and S deficiency was approximately 7 times higher than in control plants, which corresponded to the sum of the transcript increases observed in the −Fe and −S plants (Fig. 5C).
Determination of Metabolite Levels
More than 40 metabolites were extracted and identified from both shoots and roots of tomato seedlings (Table II). We describe changes in metabolites involved in the tricarboxylic acid cycle (Table II), changes in primary metabolites and ions involved in the S assimilation pathway (Fig. 6; Table II), and changes in the content of the amino acid NA (Fig. 7), which plays an important role in Fe transport within the plant (von Wiren et al., 1999). Although fully quantitative data are displayed in Figures 6 and 7 and Table II, allowing the comparisons between both tissues and conditions, all data were summarized as fold changes normalized to the control level in Figure 8, which facilitates the comparison of relative changes and tendencies in relation to the full nutrient state. A principal component analysis of this data set (Supplemental Fig. S1A) placed root and shoot samples from fully nutrient-supplied seedlings at a similar position, whereas all other nutrient conditions were clearly separated. Principal component accounts for 71.6%, separating the data sets on the basis of sulfate availability. The respective PCA loadings (Supplemental Fig. S1B) display that the main metabolites driving this separation are the known sulfate starvation response metabolite O-acetyl-serine (OAS); the N-rich amino acids Lys, Arg, Asn, Orn, and Gln, and the aromatic amino acids Tyr, Trp, and Phe, all of which accumulate. Furthermore, down-regulated metabolites contributing strongly to PC1 are glutathione, reduced (GSH), γ-glutamyl-cysteine (GEC), sulphide, total S, Cys, and sulfate (i.e. all S-containing metabolites) as well as pyruvate (Hirai et al., 2004; Nikiforova et al., 2005; Sieh et al., 2013; Bielecka et al., 2014).
Table II. Relative amounts of metabolic compounds in shoots and roots of tomato plants subjected to varying nutritional stress.
Peak height in GC-TOF/MS analysis was normalized to sample fresh weight. Statistics are the same as in Table I. C, Control; D, dual deficiency; F, Fe deficiency; na, not analyzed; S, S deficiency.
| Compound | Shoot |
Root |
||||||
|---|---|---|---|---|---|---|---|---|
| C | F | S | D | C | F | S | D | |
| Ala | 3.73a | 3.52a | 9.79b,c | 16.44c,d | 1.20a | 1.40a | 3.07b,c | 2.06a,b |
| Ala, β- | 0.02a | 0.02a | 0.05b,c | 0.08c,d | 0.02a | 0.02a | 0.05b,c | 0.03a,b |
| Arg | 0.11a | 0.08a,b | 1.12a,d | 3.43c,e | 0.03a | 0.01a | 0.19b,c | 0.41c,d |
| Asn | 0.13a | 0.11a | 0.95a | 3.89b,c | 0.10a | 0.06a | 0.67a | 2.39b,c |
| Asp | 2.95a | 2.55b,f | 2.55b,f | 2.25b,f | 2.49a | 1.96a | 3.71b,c | 2.56a |
| Citric acid | 5.47a | 5.99b,c | 4.26b,f | 4.23b,f | 1.45a | 4.71b,c | 1.80a | 2.24a |
| Fru | 3.24a | 4.33a | 4.73a | 4.20a | 0.18a | 0.22a | 0.92b,c | 0.71b,c |
| Fumaric acid | 0.53a | 0.66b,c | 0.36b,f | 0.19e,f | 0.16a | 0.14a | 0.15a | 0.09b,f |
| γ-Aminobutyrate | 3.01a | 2.56b,f | 2.84b,d,f | 3.26a,d | 1.18a | 1.28a | 1.78b,c | 1.71b,c |
| Glc | 3.91a | 5.24a | 6.10a | 5.05a | 0.16a | 0.23a | 0.70b,c | 0.43c,d |
| Glc-6-P | 0.02a | 0.02b | 0.02b | 0.01b,f | 0.04a | 0.04a | 0.05b,c | 0.03b,f |
| Glu | 5.23a | 5.34b,c | 6.02b,c,d | 4.64b,e,f | 3.51a | 3.44a | 6.57b,c | 5.78b,c |
| Gln | 1.78a | 1.71a | 21.16b,c | 28.46c,d | 1.33a | 1.07a | 7.82b,c | 14.07c,d |
| Glutaric acid, 2-oxo- | 0.07a | 0.09a | 0.03b,f | 0.02d,f | 0.32a | 0.44a | 0.30a | 0.31a |
| Inositol, myo- | 2.93a | 3.45a,b | 4.82a,d | 5.19c,d | 0.54a | 0.73a | 1.16a | 2.70b,c |
| Ile | 0.30a | 0.33a | 1.40a | 4.53b,c | 0.48a | 0.26a | 0.83b,c | 1.63c,d |
| Lys | 0.07a | 0.09a | 0.94a | 3.66b,c | 0.11a,b | 0.07a | 0.25b,c | 0.65c,d |
| Malic acid | 6.71a | 7.94b,c | 4.76d,f | 3.93d,f | 5.19a | 4.70a | 4.13a,b | 3.50b,f |
| Met | 0.05a | 0.04b,f | 0.02d,f | 0.04b,f | 0.09a | 0.06a | 0.09a | 0.11a |
| Orn | 0.10a | 0.07a | 1.03b,c | 3.62b,c | 0.07a | 0.07a | 0.27b,c | 0.53c,d |
| Phe | 0.15a | 0.15a | 0.93b,c | 2.08c,d | 0.25a | 0.13a | 0.41b,c | 0.45b,c |
| Phosphoric acid | 1.34a | 1.76a | 2.22a | 4.76b,c | 8.16a | 8.36a | 9.27a | 10.63a |
| Pro | 0.63a | 0.48b,f | 0.73a | 0.81a,b | 0.45a | 0.36a | 0.55a,b | 0.76b,c |
| Pro, 4-hydroxy | 0.01a | 0.01a | 0.04a | 0.11b,c | n.a.a | n.a.a | 0.03b,c | 0.12c,d |
| Putrescine | 0.30a | 0.30a | 0.47b,c | 0.60b,c | 0.32a | 0.23b,f | 0.28b,f | 0.39a |
| Pyruvic acid | 0.04a | 0.04a | 0.02b,f | 0.01b,f | 0.03a | 0.03a | 0.02b,f | 0.02b,f |
| Quinic acid | 0.59a | 0.70a,b | 0.87a | 1.14a,d | 0.32a | 0.29a | 0.44b,c | 0.54b,c |
| Saccharic acid | 0.08a | 0.09a | 0.09a | 0.09a | 1.68a | 0.80b,f | 0.77b,f | 0.38d,f |
| Ser | 1.03a | 1.34a | 5.28b,c | 8.93c,d | 0.01a | 0.01a | 0.02b,c | 0.02b,c |
| Ser, O-acetyl- | 0.02a | 0.02a | 0.47a | 2.03b,c | 0.01a | 0.01a | 0.20b,c | 0.17b,c |
| Succinic acid | 0.13a | 0.22a | 0.08b,f | 0.04b,f | 0.19a | 0.17a | 0.13b,f | 0.08d,f |
| Suc | 4.93a | 5.67a | 5.88a | 5.70a | 0.35a | 0.33a | 0.52b,c | 0.46c,d |
| Thr | 0.51a | 0.62a | 1.78a | 3.85b,c | 0.24a | 0.21a | 0.46b,c | 0.82c,d |
| Trp | 0.12a | 0.08a | 0.56a | 1.71b,c | 0.02a | 0.01a | 0.02a | 0.05b,c |
| Tyramine | 1.41a | 1.32a | 1.04a | 1.51a | 0.44a | 0.30a,b | 0.54a | 0.28b,f |
| Tyr | 0.08a | 0.07a | 0.43a | 2.03b,c | 0.23a | 0.12a | 0.47b,c | 0.58b,c |
| Val | 0.60a | 0.62a | 2.64a | 7.75b,c | 0.60a | 0.48a | 1.35b,c | 2.19c,d |
Significant differences between samples (P < 0.05).
Significant differences between samples (P < 0.05).
Increased levels.
Significant differences between samples (P < 0.05).
Significant differences between samples (P < 0.05).
Decreased levels.
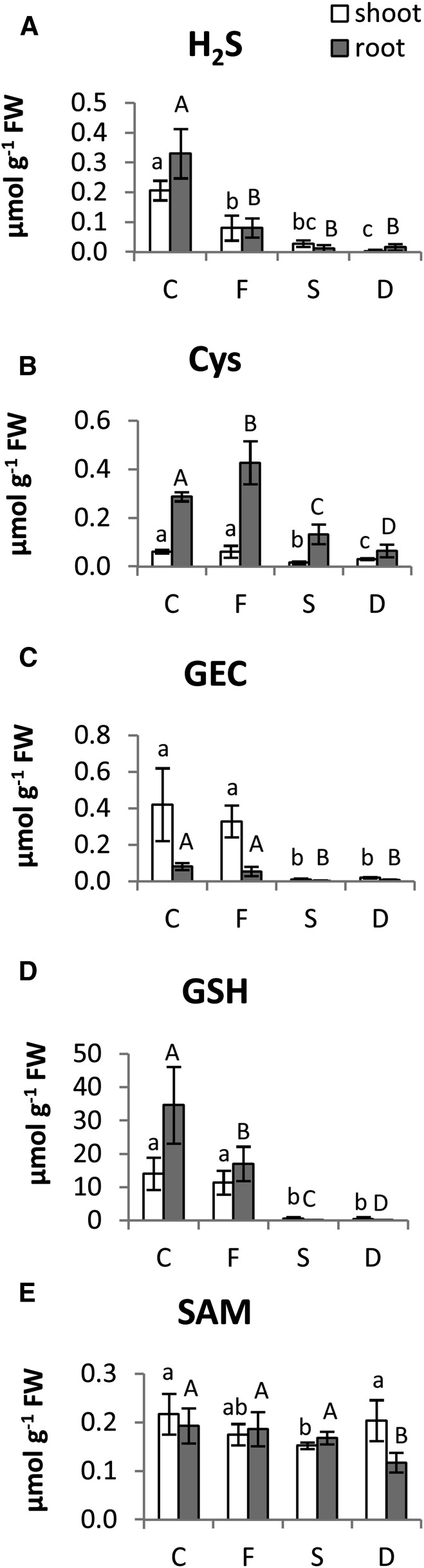
Figure 6.
Sulfide (A), Cys (B), GEC (C), GSH (D), and SAM (E) concentrations (micromoles gram−1 fresh weight [FW]) in shoots (white bars) and roots (gray bars) of tomato plants subjected to varying nutritional stress. Data are means ± sd of six independent replications run in triplicate. Statistics are the same as in Figure 1. C, Control; D, dual deficiency; F, Fe deficiency; S, S deficiency.
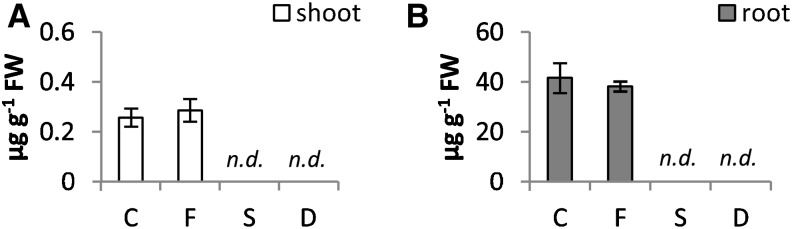
Figure 7.
NA concentration (micrograms gram−1 fresh weight [FW]) in shoots (A) and roots (B) of tomato plants subjected to varying nutritional stress. Data are means ± sd of six independent replications run in triplicate. Statistics are the same as in Figure 1. C, Control; D, dual deficiency; F, Fe deficiency; n.d., not detectable; S, S deficiency.
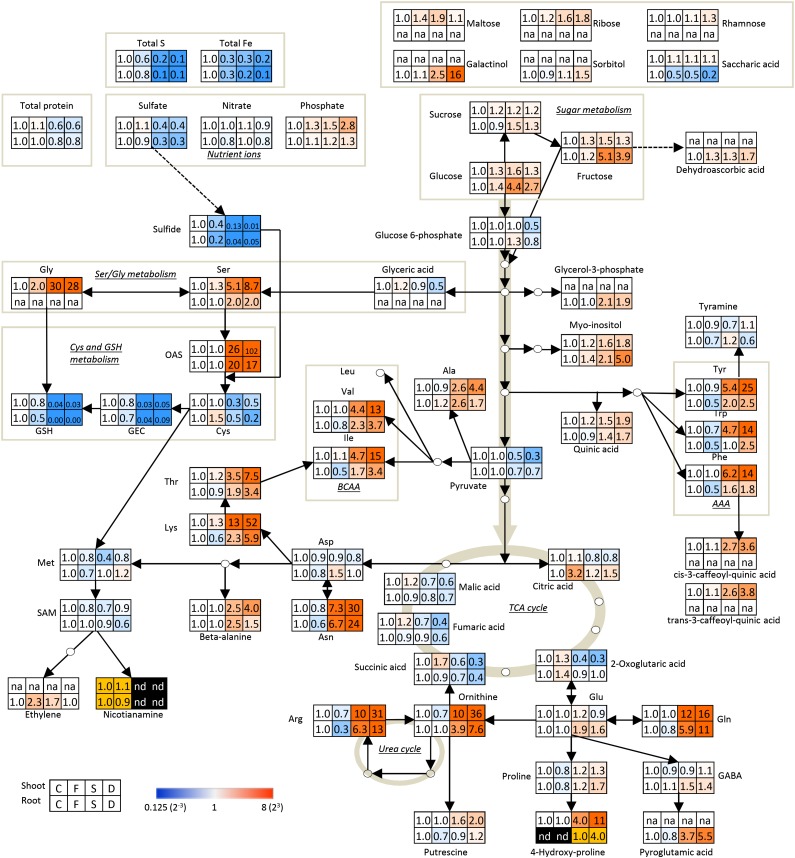
Figure 8.
Changes in the metabolite levels in shoots and roots of tomato plants subjected to varying nutritional stress. Differences in metabolite abundance on a fresh weight basis were calculated by dividing the metabolite levels in the plants subjected to nutritional stress (Fe deficiency [F], S deficiency [S], and dual deficiency [D]) by the levels in the control plants (C). Blue and red represent decreases and increases, respectively, compared with C plants. Fold changes of NA and 4-hydroxy-Pro in roots between two conditions were calculated and colored orange, because the metabolites were not at detectable levels in other two conditions. Data of ethylene production in roots were published (Zuchi et al., 2009). AAA, Aromatic amino acid; BCAA, branched chain amino acid; GABA, γ-aminobutyric acid; na, not analyzed; nd, not detected (below detection limit); TCA, tricarboxylic acid.
In tomato shoots, organic acid metabolism (the tricarboxylic acid cycle) was differentially affected by Fe and S availability. In particular, fumarate, citrate, and malate increased significantly with sole Fe deficiency, whereas sole S deficiency led to large and significant decreases of their relative amounts (Table II). When tomato seedlings were exposed to dual deficiency, the relative amounts of malate and citrate did not change significantly compared with S-deprived plants, whereas fumarate was further reduced (Table II). When −Fe plants were also exposed to S deficiency, the greatest change was found in tricarboxylic acid cycle intermediates, all of which significantly declined (Table II).
In tomato roots, Fe deficiency resulted in a 3-fold increase of citrate, but the same compound did not change significantly with either S deficiency alone or dual deficiency (Table II), especially compared with shoots where citrate is slightly reduced compared with control. However, the relative amounts of fumarate and malate did not change when plants were exposed to a single deficiency but decreased significantly on exposure to dual deficiency (Table II).
S-containing compounds can be assumed to react in a sensitive manner to depletion of the availability of the element. Figure 6 shows the effect of different nutrient availabilities on the concentrations of sulfide and selected S-containing metabolites (the thiols Cys, GEC, and GSH and the downstream product of the Cys Met pathway SAM), whereas changes in Met content are reported in Table II; all data are summarized in Figure 8. As already observed for total S content (Table I) and sulfate concentration (Fig. 1), the sulfide concentrations were strongly reduced by S starvation in both shoots and roots of tomato plants (Fig. 6). Moreover, changes in sulfide concentration closely followed the pattern of total S content when tomato seedlings were grown under Fe deprivation, showing significant decreases (−60% and −80% in shoots and roots, respectively) with respect to the control plants (Figs. 6 and 8). As expected, the withdrawal of S from the nutrient solution resulted in a significant decrease of pools of selected S-containing compounds, with the exception of SAM and at least in root tissues, Met, which seemed to be less affected by S deficiency (Fig. 6). Met levels were differently affected by S starvation in shoots and roots (Table II). In particular, in root tissues, Met levels were almost unaffected but significantly decreased in shoots of plants exposed to single or combined S deficiency compared with control plants (Fig. 8; Table II). This reduced pool size was coupled to increases of the Cys precursors Ser, Gly, and OAS (Fig. 8; Table II).
As noted previously for total S and sulfide contents, exposure to Fe deficiency alone also resulted in a pronounced decrease of S-containing metabolites, with the exception of Cys, which was unaffected in shoots and even increased (+50% compared with the control) in roots under Fe stress (Fig. 6). Surprisingly, the increase in Cys accumulation was not associated with a corresponding reduction of either Ser or OAS (Table II). However, the reduction of sulfide and the major thiols, GEC and GSH (Fig. 6A), indicate an existing sulfate starvation stress condition under which Cys contents would be anticipated to be reduced (Fig. 6D).
The majority of amino acids increased under either S or dual deficiency (Table II). Among amino acid compounds, NA plays a key role in Fe transport within the plant and is produced from the precursor SAM, which is synthesized directly from the S-containing amino acid Met. NA levels were found to be very low in shoots compared with roots, and also, NA accumulation was not affected by Fe deficiency alone (Fig. 7). Interestingly, under S deficiency, whether alone or in combination, NA accumulation in both shoots and roots was not detectable (Fig. 7). Despite this effect, the levels of the NA precursor SAM remained available (Fig. 6E).
DISCUSSION
Is There a Differential Metabolic Response to S Deficiency when Plants Are Also Fe Deficient?
The effects of S deficiency on Arabidopsis seedlings have been described by Nikiforova et al. (2003, 2004, 2005), Maruyama-Nakashita et al. (2004), and Hirai et al. (2005) using transcriptome and metabolome-wide approaches. Growth of Arabidopsis plants on S-free nutrient medium has been shown to lead to large decreases in chlorophyll, proteins, and RNA, suggesting severe alterations of plant metabolism. Accordingly, in tomato plants, it was also previously shown that S deficiency resulted in a significant decrease of plant growth and a diminution of chlorophyll content (Zuchi et al., 2009). In this study, those effects observed at the macroscopic level when tomato was exposed to S starvation were correlated with a significant reduction of protein abundance (Table I). Data shown in Table I confirm previous findings (Zuchi et al., 2009) that sulfate nutrition affects not only S accumulation in plant tissues but also Fe accumulation, with S deficiency resulting in a decrease of the total S content, which would be expected, and interestingly, a massive decrease in Fe content.
Sulfate uptake and assimilation rates are modulated in response to both plant S demand for growth and external S supply (Hawkesford and De Kok, 2006). Plant exposure to limited S supply results in an up-regulation of the sulfate uptake capacity by an increased expression of STs (for review, see Lewandowska and Sirko, 2008; Takahashi et al., 2011). Accordingly, most of the ST genes belonging to groups 1, 2, and 4 showed a significant increase in their expression upon S starvation (Fig. 3). In particular, a major increment of transcript abundance in response to S deprivation was detected for SlST1.1 and SlST1.2 in both roots and shoots (Fig. 3, A and B). Previously, such high-affinity group 1 STs were characterized as being responsible for the primary uptake of sulfate by the root (Buchner et al., 2010). However, several studies also showed the presence of their transcripts in shoots of S-deficient plants, suggesting a further function for S distribution into different cell types in different tissues (Koralewska et al., 2007; Ciaffi et al., 2013). In contrast to what has been suggested before (Parmar et al., 2007; Koralewska et al., 2008), SlST1.1 and SlST1.2 showed the same pattern of regulation in both shoots and roots of tomato plants (Fig. 3, A and B), suggesting that these transporters could be involved in the widespread and tissue-independent response to S deficiency. Otherwise, S deprivation promoted the expression of other transporters in a tissue-dependent manner. In particular, the low-affinity group 2 ST, SlST2.2, seems to play a specific role in S deficiency response at the shoot level (Fig. 3D), whereas both SlST2.1 and SlST4.1 seem to be predominantly root specific (Fig. 3, C and E). There were also some interesting additional trends in the expression profiles of STs. First, there was a significant increase in the abundance of transcripts after imposition of sole Fe deficiency. Second, S and Fe deficiency in most cases interacts synergistically, resulting in an even more increased expression of STs in shoots and roots of plants exposed to dual deficiency than in those deprived for a single nutrient (Fig. 3).
As indicated in Figure 3, apart from SlST2.1, all of the tomato ST genes analyzed were significantly up-regulated after the imposition of Fe limitation alone under S-sufficient conditions, clearly suggesting an increased uptake and translocation capacity of sulfate within the plant induced by Fe deprivation. However, upon Fe deprivation, some transporters showed expression changes, which were common to the shoots and the roots (SlST1.2 and SlST4.1), whereas others were specific to shoots (SlST1.1 and SlST2.2) or roots (SlST2.1). The up-regulation of the high-affinity ST gene SlST1.2 by Fe deficiency in roots suggests an increased sulfate uptake capacity, whereas the elevated expression of the two low-affinity STs SlST2.1 and SlST4.1 is probably an indication of an increased sulfate translocation capacity from root to shoot induced by Fe deprivation. The finding that SlST4.1 transcripts were less abundant in shoots than in roots under S and dual deficiency conditions, while being at least as abundant in shoots as in roots under Fe starvation suggests an important function of this transporter in the mobilization of sulfate from the vacuoles to the cytosol in roots to regulate the flux of sulfate toward the xylem, which was previously shown in Arabidopsis by Kataoka et al. (2004). It should, however, be noted that, despite the increased uptake capacity and sulfate availability in the medium, total S levels were reduced under Fe starvation and not just under S and dual starvation (Table I).
The differential expression patterns of genes encoding the two high-affinity STs (SlST1.1 and SlST1.2) observed in roots after Fe starvation likely indicate that the mechanisms of sulfate uptake regulation under Fe and S deficiency are distinct in tomato. This cross influence cannot be explained currently. Although sulfide levels get reduced under Fe starvation, Cys levels even increase without a corresponding decrease of the Cys precursor molecules Gly, Ser, and OAS (Figs. 3 and 8; Table II), thus displaying a combination of S starvation and S sufficiency metabolic signatures. One possible explanation is that, despite the transcriptional induction of the uptake system, the STs do not get activated posttranslationally by phosphorylation of the Sulfate Transporter and Anti-Sigma factor antagonist domain, which has been shown to be essential (Rouached et al., 2005). This would require distinct regulatory processes targeting STs upon Fe or S starvation. Under Fe starvation, the flux toward SAM and NA or ethylene might be favored to secure Fe uptake and in planta transport rather than GSH biosynthesis. It is also noteworthy that Fe depletion additionally results in a reduction of shikimate pathway intermediates (Fig. 8). Thus, it seems that, under Fe depletion, Reactive Oxygen Species scavenging is compromised in favor of using resources for safeguarding Fe uptake.
It has been suggested that the pathway intermediate OAS is a regulator of S metabolism (Hubberten et al., 2012). However, OAS levels were not affected by sole Fe starvation (Fig. 8; Table II), thus excluding the OAS cluster-controlled response module from Fe starvation control. This is in concordance with the fact that the control of expression of STs seems to be independent from OAS accumulation (Hopkins et al., 2005). With respect to the regulation of sulfate uptake, it has been proposed that the induction of STs is most likely triggered by changes in sulfate concentration (Koralewska et al., 2008), other than those in thiol compounds (Maruyama-Nakashita et al., 2004; Davidian and Kopriva, 2010). However, the molecular details underlying this mechanism remain unknown. This prompted us to measure the concentration of these compounds in the root and shoot of tomato plants upon exposure to the different nutritional stresses. Clearly, the up-regulation of ST genes after imposition of S deficiency could be ascribed to a significant decrease of sulfate concentration (Fig. 1A) as well as reductions in S metabolites, particularly sulfide, Cys, GEC, and GSH (Fig. 6), in both shoots and roots. The up-regulation of the ST genes after exposure of tomato seedlings to Fe deficiency, however, did not depend on changes in sulfate levels (which were not affected by this treatment) but may result from associated alterations in sulfide, GEC, and GSH concentrations (Fig. 6). In particular, these compounds decreased in Fe-deficient plants compared with control plants, and the largest reduction was observed in levels in roots.
Plants suffering from S limitation exhibit a reduction of photosynthetic rate as well as decreased synthesis of SAM and otherwise increased photorespiratory pathway as evidenced by transcript, metabolite, or protein profiling studies in Arabidopsis (Nikiforova et al., 2005). Here, we report that, when tomato plants cope with S deficiency at the shoot level, pools of the S-containing compounds (Cys, GSH, Met, and SAM) were decreased, whereas Ser and OAS were increased, corresponding to the typical responses to S deficiency alone in other plants. However, when S-deficient plants were additionally exposed to Fe deficiency, SAM levels were not reduced, potentially retaining flux to SAM as discussed above.
The importance of Met and SAM as precursors of ethylene has been suggested (Zuchi et al., 2009). Interestingly, in this current study, Met and SAM levels did not change in S-deficient roots, most likely mirroring the increase in ethylene production in the same roots and adding further support to what was initially hypothesized. However, the significant reduction of root ethylene production because of the imposition of additional Fe deficiency to −S plants did not seem to be associated to a reduction in root Met levels (Fig. 8; Table II), although a decrease of SAM and associated metabolites, as Cys and GSH, was observed (Fig. 6). This stability in the amount of Met is in concordance with previous findings in Arabidopsis that was exposed to continued or exposed sulfate starvation (Nikiforova et al., 2005).
The majority of amino acids increased under either S or dual deficiency. This finding might be explained by either an increased biosynthetic capacity or an increased protein degradation rate. Previously, it was shown that plants compensate for amino acid imbalances, here caused by unavailability of S, by enhancing amino acid biosynthesis (Höfgen et al., 1995). Furthermore, protein degradation may contribute to amino acid increases (Table I), and the excess pools of nitrogen need to be channeled into N-rich amino acids (Fig. 8; Table II), such as Orn, Arg, Gln, Asn, and Lys (Nikiforova et al., 2006).
A previous report showed that the polyamine precursor putrescine accumulates under sulfate starvation conditions (Nikiforova et al., 2005). The lack of SAM providing the aminopropyl moiety to generate spermidine and spermine likely accounts for this accumulation (Bielecka et al., 2014). However, under the starvation conditions imposed in this experiment, SAM levels remained unchanged, suggesting the operation of a different, as yet unknown mechanism. At this point, it might be hypothesized that Fe and S starvation induces distinct response mechanisms that exhibit partial overlap, which has indeed been shown by comparing the transcriptome of either Fe- or S-starved Arabidopsis plants (Watanabe et al., 2010, 2012). This might provide an explanation of why we find both synergistic and contrasting changes at the transcript and metabolite levels. Physiologically, Fe and S metabolism needs to be correlated to a certain extent, because for example, maintenance of Fe-S cluster formation under conditions of inadequate nutrient supply needs coordinated responses, whereas S metabolism is additionally involved in multiple processes of primary and secondary metabolism. Thus, we assume a control independent from Fe metabolism of these especially sulfate-related processes. In part, this is mirrored by the intensive overlap but also, by distinct responses of the S and Fe starvation transcriptome (Watanabe et al., 2010, 2012). Furthermore, we need to state that the tomato system shows differences to the standard Arabidopsis system.
Is There a Differential Metabolic Response to Fe Deficiency when Plants Are Also S Deficient?
The characterization of the response to limited Fe nutrition in strategy I plants showed that the mobilization of Fe3+ ions from soil particles is favored by an increased proton extrusion, likely driven by an increase in plasma membrane H+ATPase activity, the induction of a ferric chelate reductase activity, which allows higher reduction rates of Fe3+ to Fe2+, and increased citrate and ethylene production in roots (Schmidt et al., 1999; López-Millán et al., 2001; Waters et al., 2002; Curie and Briat, 2003; Kabir et al., 2012). The Arabidopsis FRO2 and IRT1 genes, encoding the ferric reductase and the Fe transporter involved in Fe2+ uptake from the soil, respectively, are up-regulated in the root epidermis in response to low Fe availability, with the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR (FIT) transcription factor (bHLH29) coordinating their responses (Colangelo and Guerinot, 2004; Walker and Connolly, 2008).
In this study, we found that exposure of tomato seedlings to Fe deprivation inhibited protein synthesis (Table I) and furthermore, that the reduction of Fe accumulation in both shoot and root tissues (Table I) was closely related to the up-regulation of SlFER, SlFRO1, and SlIRT1 (Fig. 5), similar to what is reported for their orthologous genes FIT, FRO2, and IRT1 from Arabidopsis (Colangelo and Guerinot, 2004; Walker and Connolly, 2008). More importantly, we found that Fe deficiency additionally strongly reduced total S content in both shoots and roots of tomato plants (−40% and −50%, respectively), which led to an increased transcription of STs, albeit that they seem to be inactive.
Some have proposed that increased root ethylene production is required in strategy I plants under Fe deficiency to positively regulate the expression of FIT-like genes (Lucena et al., 2006; García et al., 2010). It has been previously shown that, in tomato, the amount of ethylene released by Fe-deficient roots was higher than in the control plants (Zuchi et al., 2009), and accordingly, in this study, an enhanced expression of SlFER (Fig. 5) was found, which could be assumed to be essential for the high-level induction of both SlFRO1 and SlIRT1 (Fig. 5). Surprisingly, S deficiency treatment alone has also been shown to increase the root ethylene production (Zuchi et al., 2009), which has been suggested to be because of a marked decrease of protein synthesis (Table I) and consequently, the accumulation of ammonia in plant tissue (Nikiforova et al., 2005). However, the increased ethylene release observed under S deprivation may not be accompanied by a complete activation of the Fe deficiency response mechanisms, because among the three Fe-responsive genes analyzed in this study, only the expression of SlIRT1 was significantly affected in roots of −S plants (Fig. 5). This finding indicates that the two components of the Fe deficiency response (reduction and transport) are differentially sensitive to or regulated by ethylene levels, confirming and extending previous findings in tomato (Zuchi et al., 2009). Most interestingly, we found that the transcriptional level of SlIRT1 in roots exposed to both Fe and S deficiency was approximately 7 times higher than in control plants, which corresponded to the sum of the transcript increases observed in the −Fe (5 times higher than in control plants) and −S (approximately 2-fold higher than in control plants) plants. This finding suggests that IRT1 gene expression could be regulated by complex mechanisms that might differ from Fe supply. This hypothesis is further supported by the finding that, in the roots of the tomato Fe-inefficient mutant T328fer, SlFRO1 transcripts were not detected, whereas SlIRT1 expression may still be observed under both Fe-deficient and -sufficient conditions (Li et al., 2004), suggesting that the SlIRT1 expression is under the control of different transcriptional regulators. Moreover, as recently reviewed by Brumbarova et al. (2015), several studies suggested that the expression of IRT1 in Arabidopsis requires more than just the core strategy I transcriptional regulators, which may explain the strong connection between Fe uptake and other mineral stress responses, including S deprivation (Cailliatte et al., 2010; Abel, 2011; Bernal et al., 2012; Forieri et al., 2013), and its integration into the plant developmental program (Giehl et al., 2012; Blum et al., 2014).
Synthesis of ethylene has been shown to require Met as a precursor of SAM, which is also the precursor of molecules, such as NA and polyamines (Hesse and Hoefgen, 2003). In strategy I plants, it has been shown that NA plays a key role in symplastic and phloem Fe transport (Rudolph et al., 1985; Douchkov et al., 2002). In higher plants, Met is produced by S assimilation, and the Asp pathway provides the carbon backbone (Ravanel et al., 1998; Hesse and Hoefgen, 2003). A constant level of free Met is guaranteed by the Yang cycle, which recycles the methylthio group for regeneration of Met when SAM is used for synthesis of polyamines or ethylene (Yang and Hoffman, 1984). Thus, it seems likely that a limited S supply in tomato might impair both ethylene and NA synthesis, thus inhibiting both uptake and translocation of Fe to the shoot (Table I). Our previous finding supports this hypothesis, because under S-deprived conditions in tomato, the expression of the NA synthase gene is completely blocked independently from the availability of Fe (Zuchi et al., 2009). However, in this study, we showed that the levels of the two precursors of ethylene and NA, Met and SAM, remain constant in tomato seedlings under the observed starvation conditions (Fig. 6; Table II). Thus, on the basis of these results, it is difficult to interpret the significance of this response, even if it is undoubtedly attributable to regulatory mechanisms in which tomato plants cope with changes in S availability in their environment by tightly regulating Fe homeostasis (Table I). However, considering the fact that the major sinks of Fe are Fe-S clusters and heme, impaired Fe-S clusters assembly occurring in the absence of S might induce plants to limit excessive Fe accumulation to avoid resulting toxicity effects. Moreover, recently, it was proposed that the demand of Fe and S for the biosynthesis of Fe-S clusters in the organelles may constitute a feedback signal that coordinates the uptake and reduction of both nutrients (Balk and Pilon, 2011). In support of this hypothesis, retrograde signals have been suggested that regulate the uptake and metabolism of Fe during Fe deficiency (Vigani et al., 2013).
It is well known that exposure to Fe deficiency induces several metabolic changes occurring at the whole-plant level (Zocchi, 2006). An increase in organic acid metabolism (tricarboxylic acid cycle) with Fe deficiency was observed at the shoot level, most likely to sustain Fe acquisition by providing organic acids (hence, the increase of citrate but no other tricarboxylic acid cycle intermediates in roots under Fe starvation [Fig. 8; Table II] and protons for rhizosphere acidification). When −Fe plants were also exposed to S deficiency, the greatest change was found in the tricarboxylic acid cycle, which did not increase, and especially, no significant citrate accumulation occurred in root tissues. Furthermore, we found an accumulation of both Ser and OAS upon Fe starvation alone as well as a strong reduction of sulfide, suggesting that Cys biosynthesis was prevented, even if, surprisingly, the Cys concentration was increased. Most likely, an increased rate of protein degradation could explain the increased Cys accumulation. The most likely explanation for the elevated Cys content would be an increase in the protein degradation rate (Table I). Another possibility could be a specific regulation that favors the Cys-Met-SAM branch for eventual NA biosynthesis under Fe-depleted conditions. Furthermore, the reduction of available sulfide, although sulfate is available, could be speculated to be the result of regulatory systems keeping Fe and S levels in balance when Fe is not available (Fig. 8).
In addition to the increased accumulation of organic acids, particularly citric acid, in roots facilitating Fe mobilization (Rellán-Álvarez et al., 2011; Valentinuzzi et al., 2015), metabolite profiles reported in Figure 8 and Table II showed that Fe deficiency induced an accumulation of Glc and Fru. A drop of pyruvic acid in plants exposed to sulfate deficiency indicates a slowdown of the glycolytic pathway, especially prevalent under combined Fe and S starvation, where additionally Glc-6-P levels were reduced (Fig. 8). These observations are in contrast to previous studies in sugar beet (Beta vulgaris), which have generally shown that metabolites related to glycolysis increased in Fe-deficient roots (López-Millán et al., 2000a, 2000b). Increase of both tricarboxylic acid cycle and glycolytic activities is commonly coupled with increased phosphoenolpyruvate carboxylase activity, which is involved in C fixation (De Nisi and Zocchi, 2000), most likely to provide extra C skeletons for leaves with reduced photosynthetic activity (López-Millán et al., 2000a, 2000b). We additionally assume that Glc and Fru accumulation is related to stress responses and the onset of senescence, which is in concordance with the observed protein degradation, amino acid accumulation, and chlorophyll breakdown (Watanabe et al., 2013).
Other important Fe deficiency-induced changes were the increase in Cys concentration and ethylene production, whereas GSH decreased, and Met and SAM were not changed. An increase in root Cys content under Fe stress conditions was also found previously in barley, a strategy II plant (Astolfi et al., 2006b). Such findings would indicate a role of this amino acid in both strategies of Fe deficiency response. However, the imposition of Fe deficiency to −S plants caused a strong reduction of Cys and a moderate reduction of SAM (Fig. 8), which resulted in a lower ethylene production compared with sole Fe depletion, thus likely hindering plant capability to develop the response mechanisms to cope with Fe deficiency.
NA levels remained relatively stable in roots of Fe-deficient plants (Fig. 7), regardless of a significant decrease of Fe content in the same tissue (Table I), whereas its level drastically dropped when S was removed from the nutrient solution (Figs. 7 and 8). The same held true for NA concentrations in shoots, although levels were clearly much lower than those observed in roots (Figs. 7 and 8). It is interesting to note that the reduction of Fe content caused by S deficiency was greater than that induced by Fe deficiency and also, that changes in NA level show a correlation with previous observation that S deficiency virtually abolished expression of the Nicotianamine Synthase (NAS) gene, independent of the Fe growth conditions (Zuchi et al., 2009). NA is involved in both xylem and phloem Fe transport (von Wiren et al., 1999; Briat et al., 2007), and it has been reported that an elevated internal concentration of Fe can significantly promote NA synthesis (Pich et al., 2001). This response would be complementary to the overall alleviation of Fe-induced oxidative damage.
As hypothesized above, we have to assume a complex regulatory system, wherein Fe and S starvation induces partially overlapping and partially distinct response mechanisms that coordinate plant response to joint starvations. The detailed wiring of these response modules still needs to be resolved. It is further noteworthy that distinct differences occur between the Arabidopsis model system and tomato.
MATERIALS AND METHODS
Plant Material
Tomato (Solanum lycopersicum ‘Gimar’) seedlings were grown hydroponically in plastic pots (six seedlings per pot) containing 2.2 L of nutrient solution (Zhang et al., 1991) for 7 d and exposed to 1.2 mm sulfate and 40 µm FeIII-EDTA. One-half of the plants were then transferred for another week to an S-free nutrient solution. Thereafter, one-half of the plants derived from the two S growth conditions (+S and −S) were transferred to an Fe-free nutrient solution.
In S-free nutrient solution, sulfate salts (K+, Mn2+, Zn2+, and Cu2+) were replaced by appropriate amounts of the corresponding chloride salts (K+, Mn2+, Zn2+, and Cu2+). Nutrient solution was continuously aerated and changed every 2 d. Plants were grown in a growth chamber under 200 μmol photons m−2 s−1 photosynthetic photon flux and a 14-h/10-h day/night regime (27°C/20°C day/night temperature cycling and 80% relative humidity). Plants were harvested 17 d after sowing.
Extraction of Total RNA, Complementary DNA Preparation, and Expression Analyses
Total RNA was extracted from shoots and roots of control and treated (dual deficiency, Fe deficiency, and S deficiency) tomato plants using the method described by Verwoerd et al. (1989). The resulting RNA was treated with RNase-Free DNase I (Promega) according to the manufacturer’s protocol. After digestion, nucleotides were removed from RNA using a G50 Sepharose Buffer Exchange Column (Amersham). RNA concentration and integrity were checked using a NanoDrop ND-1000 Spectrophotometer (Labtech). The quality of RNA samples was also assessed by electrophoresis on 1% (w/w) formaldehyde agarose gels. First-strand complementary DNA (cDNA) was synthesized from 1 μg of RNA by the M-MLV (H-) Reverse Transcriptase (Invitrogen), and the resulting cDNA was diluted 1:5 for real-time quantitative reverse transcription (qRT)-PCR analyses.
The expression of genes involved in Fe homeostasis (SlFRO1, SlIRT1, and SlFER) and coding for STs of groups 1, 2, and 4 (SlST1.1, SlST1.2, SlST2.1, SlST2.2, and SlST4.1) were analyzed by qRT-PCR as described in Paolacci et al., 2014. In particular, qRT-PCR analyses were performed using an Mx3000PTM Real-Time PCR System with Brilliant SYBR Green QPCR Master Mix (STRATAGENE) according to manufacturer’s protocols in 25-μL reaction volumes containing 1 μL of each 5-fold diluted cDNA and 150 nm forward and reverse primers (reported for each analyzed gene in Paolacci et al., [2014]). Standard curves based on five points, corresponding to a 5-fold dilution series (1:1–1:625) from pooled cDNA, were used to compute the PCR efficiency of each primer pair. PCR efficiency (E) is given by the equation E = (10 [−1/m] − 1) × 100 (Radonić et al., 2004), where m is the slope of linear regression model fitted over log-transformed data of the input cDNA concentration versus Cycle threshold (Ct) values according to the linear equation y = m × log(x) + b. Three biological replicates, resulting from three different RNA extractions and reverse transcription PCR and qRT-PCR reactions from three separate plants, were used in quantification analysis; three technical replicates were analyzed for each biological replicate.
Raw Ct values were transformed to relative quantities using the ΔCt formula Q = EΔCt, where E is the efficiency of the primer pair used in the amplification of a particular gene, and ΔCt is the difference between the sample with the lowest Ct (highest expression) from the data set and the Ct value of the sample in question. The expression data of the genes of interest were normalized using the geometric average of the two reference genes Tonoplast intrinsic protein4-like family protein (SlTIP4I) and Clatrin adaptor complexes medium subunit (SlCAC; primer pairs reported in Paolacci et al., 2014), and their normalized relative values were given as mean value ± sd; sds on normalized expression levels were computed according to the geNorm User Manual (geNorm Manual; updated July 8, 2008). Relative expression levels of the genes of interest were referred to those of a calibrator set to the value 1, which was represented by the treatment with the lowest expression.
Metabolite Extraction and Preparation for Gas Chromatography-Time of Flight/Mass Spectrometry Metabolite Profiling
Samples of shoots and roots grown in the conditions described above were pulverized in liquid nitrogen, and aliquots of 100 to 150 mg of fresh weight were prepared and used for metabolite extraction, derivatization, and analysis exactly as described in Lisec et al., 2006.
Briefly, plant tissues were homogenized in a ball mill for 2 min at −60°C. Thereafter, 1,400 μL of 100% methanol to stop enzymatic activity and 60 μL of ribitol (0.2 mg mL−1) as internal quantitative standard were added. Samples were shaken for 15 min at 70°C in a thermomixer and then centrifuged for 10 min at 11,000g. The supernatant of each sample was transferred to a glass vial; then, 750 μL of chloroform and 1,500 μL of deionized water were added, and samples were centrifuged for 15 min at 2,200g. The polar upper phase (150 μL) was collected from each sample, transferred into fresh Eppendorf tubes, and dried in a vacuum concentrator without heating.
Resulting extracts were derivatized before gas chromatography-time of flight/mass spectrometry (GC-TOF/MS) metabolite profiling as follow: 40 μL of methoxyamination reagent constituted of 20 mg mL−1 methoxyaminhydrochloride in pure pyridine freshly prepared was added to each sample, which was then shaken for 2 h at 37°C. Thereafter, 70 μL of a mixture containing N-methyl-N-(trimethylsilyl)trifluoroacetamide used as derivatization reagent and an excess of the retention time index standard (0.8 mg mL−1 fatty acid methylesters in chloroform) was added, and samples were shaken for 30 min at 37°C. Finally, samples were transferred to glass vials that are suitable for GC-TOF/MS analysis.
GC-TOF/MS Metabolite Profiling
The metabolite profiling was conducted on the polar phase of samples using GC-TOF/MS analysis. In this experiment, an MDN-35 Column was used with fatty acid methylesters as retention time standards and N-methyl-N-(trimethylsilyl)trifluoroacetamide as derivatization reagent. Injection, chromatography, and mass spectrometer parameters used are described in Lisec et al., 2006. The TagFinder Software described in Luedemann et al., 2008 was used for the quantitative analysis of the metabolite profiling experiment.
NA Quantification
Extraction, derivatization, and analysis of NA levels were carried out using a standard protocol for GC-TOF/MS (Lisec et al., 2006). NA was identified by coelution with an authentic standard (provided by R. Hell). The ion with a mass-to-charge ratio value of 186 was used for quantification. Recovery experiments, in which the amount of authentic NA added at the start of the experiment was doubled, yielded a result of 90.2 ± 3.5%, indicating a high stability of the metabolite and IR derivative throughout extraction, derivatization, and analytical processes.
Thiol Extraction and Determination
Thiols extract was prepared from shoot and root tissues of plants grown as described above; 50 mg of frozen homogenized plant material was homogenized in 100 μL of a solution containing 0.1 m HCl and 1% (w/v) polyvinylpolypyrrolidone and then put on ice. To each sample, 150 μL of 0.25 m N-cyclohexyl-2-aminoethanesulfonic acid buffer, pH 9.4 and 20 μL of 25 mm monobromobimane were added. Derivatization was carried out for 15 min in the dark at 4°C. Then, the reaction was stopped by adding 100 μL of 100 mm methanosulfonic acid, and after a centrifugation step of 15 min at 14,000g and 4°C, samples were diluted 1:3 with 92% (w/w) running buffer constituted of 0.25% (w/w) acetic acid, pH 4.0 and 8% (w/w) methanol; finally, they were loaded to HPLC. HPLC analysis was conducted with an increasing methanol gradient at pH 4.0 as described in Hubberten et al., 2012. Column eluent was monitored by fluorescence detection (λex = 380 nm/λem = 480 nm). Mixed standards treated exactly as the sample supernatants were used as a reference for the quantification of free sulfide, Cys, GEC, and GSH.
Inorganic Ions Extraction and Determination
Frozen and ground shoot and root tissues (approximately 50 mg) from plants grown as described above were homogenized in 500 μL of 0.1 mm HCl. Samples were centrifuged twice for 5 min at 14,000g at 4°C, and finally, the supernatant was transferred to Amicon Ultra-3K Millipore Centrifugal Filters and centrifuged again at 5,000g at 4°C until complete filtration. The flow through diluted 1:20 with deionized water was loaded to the Dionex ICS-2000 System. Elution was conducted with a linear gradient of KOH as described in Hubberten et al. (2012). The concentrations of sulfate, nitrate, and phosphate were determined using standard reference.
SAM Extraction and Determination
Frozen and ground shoot and root tissues (approximately 50 mg) from plants grown as described above were homogenized in 400 μL of 0.4 m perchloric acid. Samples were shaken for 15 min at 4°C and then centrifuged for 10 min at 14,000g at 4°C. Thereafter, the supernatant was transferred to Amicon Ultra-3K Millipore Centrifugal Filters and centrifuged at 5,000g at 4°C until complete filtration. The flow through was subjected to reversed-phase HPLC using an ODS Column (Hyperclone C18; 250- × 4.6-mm i.d.; 5 μm; Phenomenex) connected to an HPLC system (Dionex) with a UV detector. Chromatography was performed using an increasing methanol gradient comprising buffer A (50 mm sodium dihydrogen phosphate and 8 mm octanesulfonic acid) and buffer B (100% methanol) as follows: 0 to 1 min, 100% A and 0% B; 2 to 10 min, 80% A and 20% (w/w) B; 10.5 to 20.5 min, 60% (w/w) A and 40% (w/w) B; 23 to 25 min, 80% (w/w) A and 20% (w/w) B; and 26 to 29 min, 100% A and 0% B with a flow rate of 1.0 mL min−1. SAM was detected by UV absorption at 254 nm.
Amino Acid Extraction and Determination
Soluble amino acids were determined following a protocol modified from Scheible et al., 1997. Frozen and ground shoot and root tissues (approximately 100 mg) from plants grown as described above were extracted three times for 20 min at 80°C: one time with 400 μL of 80% (v/v) aqueous ethanol (buffered with 2.5 mm HEPES-KOH, pH 7.5) and 10 μL of 20 μm nor-Val (as an internal standard), one time with 400 μL of 50% (v/v) aqueous ethanol (buffered as before), and one time with 200 μL of 80% (v/v) aqueous ethanol. Between the extraction steps, samples were centrifuged for 10 min at 13,000g, and the supernatants were collected. The combined ethanol-water extracts were stored at −20°C or directly subjected to reversed-phase HPLC using an ODS Column (Hyperclone C18; 150- × 4.6-mm i.d.; 3 μm; Phenomenex) connected to an HPLC system (Dionex). Amino acids were measured by precolumn derivatization with ortho-phthaldehyde in combination with fluorescence detection (Lindroth and Mopper, 1979) as described by Kreft et al. (2003). Peak areas were integrated using Chromeleon 6.30 Software (Dionex) and subjected to quantification by means of calibration curves made from amino acid standard mixtures.
Other Measurements
To determine total S content, 1 g of each shoot or root sample was dried at 105°C and ashed in a muffle furnace at 600°C. The ashes were dissolved in 10 mL of 3 n HCl and filtered through Whatman Number 42 Paper. After mixing 2.5 mL of sieved mixture with 2.5 mL of 2% (w/v) BaCl2, the amount of BaSO4 precipitate was determined turbidimetrically (Bardsley and Lancaster, 1962).
Fe content in shoots and roots was determined after dry ashing (500°C) of plant tissue and 1:30 HCl extraction by atomic absorption spectrometry.
Protein concentration was determined according to the work by Bradford (1976) using bovine serum albumin as the standard.
Statistics
Each reported value represents the mean ± sd of measurements carried out in triplicate and was obtained from six independent experiments.
For expression analysis, each reported value represents the mean ± sd of measurements carried out in triplicate and was obtained from three independent experiments.
Statistical analyses of data were carried out by ANOVA with the GraphPad InStat Program (version 3.06). Significant differences were established by post hoc comparisons (Tukey’s honestly significant difference mean-separation test) at P < 0.05.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Principal component analysis of metabolite data.
Supplementary Material
Glossary
- cDNA
complementary DNA
- GC-TOF/MS
gas chromatography-time of flight/mass spectrometry
- GEC
γ-glutamyl-cysteine
- GSH
glutathione, reduced
- NA
nicotianamine
- OAS
O-acetyl-serine
- qRT
quantitative reverse transcription
- SAM
S-adenosyl-methionine
Footnotes
This work was supported by the Organisation for Economic Co-operation and Development (fellowship to S.Z.), the Max Planck Society, the Spanish Ministerio de Ciencia e Innovación (S.O.), the Ministerio de Economía y Competitividad (S.O.), and the University of Málaga (Grant Ramón y Cajal Program to S.O.).
Articles can be viewed without a subscription.
References
- Abel S. (2011) Phosphate sensing in root development. Curr Opin Plant Biol 14: 303–309 [DOI] [PubMed] [Google Scholar]
- Astolfi S, Cesco S, Zuchi S, Neumann G, Roemheld V (2006a) Sulphur starvation reduces phytosiderophores release by Fe-deficient barley plants. Soil Sci Plant Nutr 52: 80–85 [Google Scholar]
- Astolfi S, Zuchi S, Cesco S, Sanità di, Toppi L, Pirazzi D, Badiani M, Varanini Z, Pinton R (2006b) Fe deficiency induces sulphate uptake and modulates redistribution of reduced sulphur pool in barley plants. Funct Plant Biol 33: 1055–1061 [DOI] [PubMed] [Google Scholar]
- Astolfi S, Zuchi S, Passera C, Cesco S (2003) Does the sulphur assimilation pathway play a role in the response to Fe deficiency in maize (Zea mays L.) plants? J Plant Nutr 26: 2111–2121 [Google Scholar]
- Balk J, Pilon M (2011) Ancient and essential: the assembly of iron-sulfur clusters in plants. Trends Plant Sci 16: 218–226 [DOI] [PubMed] [Google Scholar]
- Bardsley CE, Lancaster JD (1962) Determination of reserve sulphur and soluble sulphate in soils. Soil Sci Soc Am Proc 24: 265–268 [Google Scholar]
- Bereczky Z, Wang HY, Schubert V, Ganal M, Bauer P (2003) Differential regulation of nramp and irt metal transporter genes in wild type and iron uptake mutants of tomato. J Biol Chem 278: 24697–24704 [DOI] [PubMed] [Google Scholar]
- Bernal M, Casero D, Singh V, Wilson GT, Grande A, Yang H, Dodani SC, Pellegrini M, Huijser P, Connolly EL, et al. (2012) Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. Plant Cell 24: 738–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielecka M, Watanabe M, Morcuende R, Scheible WR, Hawkesford MJ, Hesse H, Hoefgen R (2014) Transcriptome and metabolome analysis of plant sulfate starvation and resupply provides novel information on transcriptional regulation of metabolism associated with sulfur, nitrogen and phosphorus nutritional responses in Arabidopsis. Front Plant Sci 5: 805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum A, Brumbarova T, Bauer P, Ivanov R (2014) Hormone influence on the spatial regulation of IRT1 expression in iron-deficient Arabidopsis thaliana roots. Plant Signal Behav 9: e28787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouranis DL, Chorianopoulou SN, Siyiannis VF, Protonotarios VE, Hawkesford MJ (2003) Aerenchyma formation in roots of maize during sulphate starvation. Planta 217: 382–391 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Briat JF, Curie C, Gaymard F (2007) Iron utilization and metabolism in plants. Curr Opin Plant Biol 10: 276–282 [DOI] [PubMed] [Google Scholar]
- Brumbarova T, Bauer P (2005) Iron-mediated control of the basic helix-loop-helix protein FER, a regulator of iron uptake in tomato. Plant Physiol 137: 1018–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbarova T, Bauer P, Ivanov R (2015) Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci 20: 124–133 [DOI] [PubMed] [Google Scholar]
- Buchner P, Parmar S, Kriegel A, Carpentier M, Hawkesford MJ (2010) The sulfate transporter family in wheat: tissue-specific gene expression in relation to nutrition. Mol Plant 3: 374–389 [DOI] [PubMed] [Google Scholar]
- Cailliatte R, Schikora A, Briat JF, Mari S, Curie C (2010) High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell 22: 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciaffi M, Paolacci AR, Celletti S, Catarcione G, Kopriva S, Astolfi S (2013) Transcriptional and physiological changes in the S assimilation pathway due to single or combined S and Fe deprivation in durum wheat (Triticum durum L.) seedlings. J Exp Bot 64: 1663–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Briat JF (2003) Iron transport and signaling in plants. Annu Rev Plant Biol 54: 183–206 [DOI] [PubMed] [Google Scholar]
- Davidian JC, Kopriva S (2010) Regulation of sulfate uptake and assimilation: the same or not the same? Mol Plant 3: 314–325 [DOI] [PubMed] [Google Scholar]
- Dell’Orto M, Santi S, De Nisi P, Cesco S, Varanini Z, Zocchi G, Pinton R (2000) Development of Fe-deficiency responses in cucumber (Cucumis sativus L.) roots: involvement of plasma membrane H(+)-ATPase activity. J Exp Bot 51: 695–701 [PubMed] [Google Scholar]
- De Nisi PD, Zocchi G (2000) Phosphoenolpyruvate carboxylase in cucumber (Cucumis sativus L.) roots under iron deficiency: activity and kinetic characterization. J Exp Bot 51: 1903–1909 [DOI] [PubMed] [Google Scholar]
- Douchkov D, Herbik A, Koch G, Mock HP, Melzer M, Stephan UW, Bäumlein H (2002) Nicotianamine synthase: gene isolation, gene transfer and application for the manipulation of plant iron assimilation. Plant Soil 241: 115–119 [Google Scholar]
- Eckhardt U, Mas Marques A, Buckhout TJ (2001) Two iron-regulated cation transporters from tomato complement metal uptake-deficient yeast mutants. Plant Mol Biol 45: 437–448 [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forieri I, Wirtz M, Hell R (2013) Toward new perspectives on the interaction of iron and sulfur metabolism in plants. Front Plant Sci 4: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García MJ, Lucena C, Romera FJ, Alcántara E, Pérez-Vicente R (2010) Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis. J Exp Bot 61: 3885–3899 [DOI] [PubMed] [Google Scholar]
- Giehl RFH, Lima JE, von Wirén N (2012) Localized iron supply triggers lateral root elongation in Arabidopsis by altering the AUX1-mediated auxin distribution. Plant Cell 24: 33–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkesford MJ, De Kok LJ (2006) Managing sulphur metabolism in plants. Plant Cell Environ 29: 382–395 [DOI] [PubMed] [Google Scholar]
- Hesse H, Hoefgen R (2003) Molecular aspects of methionine biosynthesis. Trends Plant Sci 8: 259–262 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Klein M, Fujikawa Y, Yano M, Goodenowe DB, Yamazaki Y, Kanaya S, Nakamura Y, Kitayama M, Suzuki H, et al. (2005) Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J Biol Chem 280: 25590–25595 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K (2004) Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfgen R, Laber B, Schüttke I, Klonus AK, Streber W, Pohlenz HD (1995) Repression of Acetolactate Synthase Activity through antisense inhibition. Plant Physiol 107: 469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins L, Parmar S, Błaszczyk A, Hesse H, Hoefgen R, Hawkesford MJ (2005) O-acetylserine and the regulation of expression of genes encoding components for sulfate uptake and assimilation in potato. Plant Physiol 138: 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubberten HM, Klie S, Caldana C, Degenkolbe T, Willmitzer L, Hoefgen R (2012) Additional role of O-acetylserine as a sulfur status-independent regulator during plant growth. Plant J 70: 666–677 [DOI] [PubMed] [Google Scholar]
- Kabir AH, Paltridge NG, Able AJ, Paull JG, Stangoulis JCR (2012) Natural variation for Fe-efficiency is associated with upregulation of Strategy I mechanisms and enhanced citrate and ethylene synthesis in Pisum sativum L. Planta 235: 1409–1419 [DOI] [PubMed] [Google Scholar]
- Kataoka T, Watanabe-Takahashi A, Hayashi N, Ohnishi M, Mimura T, Buchner P, Hawkesford MJ, Yamaya T, Takahashi H (2004) Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koralewska A, Posthumus FS, Stuiver CEE, Buchner P, Hawkesford MJ, De Kok LJ (2007) The characteristic high sulfate content in Brassica oleracea is controlled by the expression and activity of sulfate transporters. Plant Biol (Stuttg) 9: 654–661 [DOI] [PubMed] [Google Scholar]
- Koralewska A, Stuiver CEE, Posthumus FS, Kopriva S, Hawkesford MJ, De Kok LJ (2008) Regulation of sulfate uptake, expression of the sulfate transporters Sultr1;1 and Sultr1;2, and APS reductase in Chinese cabbage (Brassica pekinensis) as affected by atmospheric H2S nutrition and sulfate deprivation. Funct Plant Biol 35: 318–327 [DOI] [PubMed] [Google Scholar]
- Kreft O, Hoefgen R, Hesse H (2003) Functional analysis of cystathionine γ-synthase in genetically engineered potato plants. Plant Physiol 131: 1843–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwajima K, Kawai S (1997) Relationship between sulfur metabolism and biosynthesis of phytosiderophores in barley roots. In Ando T, Fujita K, Mae T, Matsumoto H, Mori S, Sekiya J, eds, Plant Nutrition for Sustainable Food Production and Environment. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 285–286 [Google Scholar]
- Lewandowska M, Sirko A (2008) Recent advances in understanding plant response to sulfur-deficiency stress. Acta Biochim Pol 55: 457–471 [PubMed] [Google Scholar]
- Li L, Cheng X, Ling HQ (2004) Isolation and characterization of Fe(III)-chelate reductase gene LeFRO1 in tomato. Plant Mol Biol 54: 125–136 [DOI] [PubMed] [Google Scholar]
- Lindroth P, Mopper K (1979) High performance liquid chromatographic determination of subpicomole amounts of amino acids by precolumn fluorescence derivatization with o-phthaldialdehyde. Anal Chem 51: 1667–1674 [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M (2002) The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA 99: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- López-Millán AF, Morales F, Abadía A, Abadía J (2000a) Effects of iron deficiency on the composition of the leaf apoplastic fluid and xylem sap in sugar beet. Implications for iron and carbon transport. Plant Physiol 124: 873–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Millán AF, Morales F, Abadía A, Abadía J (2001) Iron deficiency-associated changes in the composition of the leaf apoplastic fluid from field-grown pear (Pyrus communis L.) trees. J Exp Bot 52: 1489–1498 [DOI] [PubMed] [Google Scholar]
- López-Millán AF, Morales F, Andaluz S, Gogorcena Y, Abadía A, De Las Rivas J, Abadía J (2000b) Responses of sugar beet roots to iron deficiency. Changes in carbon assimilation and oxygen use. Plant Physiol 124: 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena C, Waters BM, Romera FJ, García MJ, Morales M, Alcántara E, Pérez-Vicente R (2006) Ethylene could influence ferric reductase, iron transporter, and H+-ATPase gene expression by affecting FER (or FER-like) gene activity. J Exp Bot 57: 4145–4154 [DOI] [PubMed] [Google Scholar]
- Luedemann A, Strassburg K, Erban A, Kopka J (2008) TagFinder for the quantitative analysis of gas chromatography–mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 24: 732–737 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Yamaya T, Takahashi H (2004) A novel regulatory pathway of sulfate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J 38: 779–789 [DOI] [PubMed] [Google Scholar]
- McGrath SP, Zhao FJ, Blake-Kalff M (2002) History and outlook for sulphur fertilisers in Europe. In Proceedings Number 497 of the International Fertilizer Society. International Fertilizer Society, York, United Kingdom [Google Scholar]
- Møller IS, Tester M (2007) Salinity tolerance of Arabidopsis: a good model for cereals? Trends Plant Sci 12: 534–540 [DOI] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R (2003) Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J 33: 633–650 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Bielecka M, Gakière B, Krueger S, Rinder J, Kempa S, Morcuende R, Scheible WR, Hesse H, Hoefgen R (2006) Effect of sulfur availability on the integrity of amino acid biosynthesis in plants. Amino Acids 30: 173–183 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Gakière B, Kempa S, Adamik M, Willmitzer L, Hesse H, Hoefgen R (2004) Towards dissecting nutrient metabolism in plants: a systems biology case study on sulphur metabolism. J Exp Bot 55: 1861–1870 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R (2005) Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol 138: 304–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolacci AR, Celletti S, Catarcione G, Hawkesford MJ, Astolfi S, Ciaffi M (2014) Iron deprivation results in a rapid but not sustained increase of the expression of genes involved in iron metabolism and sulfate uptake in tomato (Solanum lycopersicum L.) seedlings. J Integr Plant Biol 56: 88–100 [DOI] [PubMed] [Google Scholar]
- Parmar S, Buchner P, Hawkesford MJ (2007) Leaf developmental stage affects sulfate depletion and specific sulfate transporter expression during sulfur deprivation in Brassica napus L. Plant Biol (Stuttg) 9: 647–653 [DOI] [PubMed] [Google Scholar]
- Pich A, Manteuffel R, Hillmer S, Scholz G, Schmidt W (2001) Fe homeostasis in plant cells: does nicotianamine play multiple roles in the regulation of cytoplasmic Fe concentration? Planta 213: 967–976 [DOI] [PubMed] [Google Scholar]
- Pii Y, Cesco S, Mimmo T (2015) Shoot ionome to predict the synergism and antagonism between nutrients as affected by substrate and physiological status. Plant Physiol Biochem 94: 48–56 [DOI] [PubMed] [Google Scholar]
- Radonić A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 313: 856–862 [DOI] [PubMed] [Google Scholar]
- Ravanel S, Gakière B, Job D, Douce R (1998) Cystathionine gamma-synthase from Arabidopsis thaliana: purification and biochemical characterization of the recombinant enzyme overexpressed in Escherichia coli. Biochem J 331: 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rellán-Álvarez R, El-Jendoubi H, Wohlgemuth G, Abadía A, Fiehn O, Abadía J, Alvarez-Fernández A (2011) Metabolite profile changes in xylem sap and leaf extracts of strategy I plants in response to iron deficiency and resupply. Front Plant Sci 2: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Romera FJ, Alcantara E, de la Guardia MD (1999) Ethylene production by Fe‐deficient roots and its involvement in the regulation of Fe-deficiency stress responses by Strategy I plants. Ann Bot 83: 51–55 [Google Scholar]
- Rouached H, Berthomieu P, El Kassis E, Cathala N, Catherinot V, Labesse G, Davidian JC, Fourcroy P (2005) Structural and functional analysis of the C-terminal STAS (sulfate transporter and anti-sigma antagonist) domain of the Arabidopsis thaliana sulfate transporter SULTR1.2. J Biol Chem 280: 15976–15983 [DOI] [PubMed] [Google Scholar]
- Rudolph A, Becker R, Scholz G, Prochazka Z, Toman J, Macek T, Herout V (1985) The occurrence of the amino acid nicotianamine in plants and microorganisms: a re-investigation. Biochem Physiol Pflanzen 180: 557–563 [Google Scholar]
- Scheible WR, González-Fontes A, Morcuende R, Lauerer M, Geiger M, Glaab J, Gojon A, Schulze ED, Stitt M (1997) Tobacco mutants with a decreased number of functional nia genes compensate by modifying the diurnal regulation of transcription, post-translational modification and turnover of nitrate reductase. Planta 203: 304–319 [DOI] [PubMed] [Google Scholar]
- Schikora A, Thimm O, Linke B, Buckhout TJ, Müller M, Schmidt W (2006) Expression, localization, and regulation of the iron transporter LeIRT1 in tomato roots. Plant Soil 2984: 101–108 [Google Scholar]
- Schmidt JS, Harper JE, Hoffman TK, Bent AF (1999) Regulation of soybean nodulation independent of ethylene signaling. Plant Physiol 119: 951–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieh D, Watanabe M, Devers EA, Brueckner F, Hoefgen R, Krajinski F (2013) The arbuscular mycorrhizal symbiosis influences sulfur starvation responses of Medicago truncatula. New Phytol 197: 606–616 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R (2011) Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol 62: 157–184 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Terada Y, Nakai I, Nakanishi H, Yoshimura E, Mori S, Nishizawa NK (2003) Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell 15: 1263–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentinuzzi F, Pii Y, Vigani G, Lehmann M, Cesco S, Mimmo T (2015) Phosphorus and iron deficiencies induce a metabolic reprogramming and affect the exudation traits of the woody plant Fragaria × ananassa. J Exp Bot 66: 6483–6495 [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigani G, Zocchi G, Bashir K, Philippar K, Briat JF (2013) Signals from chloroplasts and mitochondria for iron homeostasis regulation. Trends Plant Sci 18: 305–311 [DOI] [PubMed] [Google Scholar]
- von Wiren N, Klair S, Bansal S, Briat JF, Khodr H, Shioiri T, Leigh RA, Hider RC (1999) Nicotianamine chelates both FeIII and FeII: implications for metal transport in plants. Plant Physiol 119: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EL, Connolly EL (2008) Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Curr Opin Plant Biol 11: 530–535 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Balazadeh S, Tohge T, Erban A, Giavalisco P, Kopka J, Mueller-Roeber B, Fernie AR, Hoefgen R (2013) Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiol 162: 1290–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Hubberten HM, Hoefgen R (2012) Plant response to mineral ion availability: transcriptome responses to sulfate, selenium and iron. In De Kok LJ, Tabe L, Tausz M, Hawkesford MJ, Hoefgen R, McManus MT, Norton RM, Rennenberg H, Saito K, Schnug E, eds, Sulfur Metabolism in Plants, Proceedings of the International Sulfur Workshop 1. Springer, Dordrecht, The Netherlands, pp. 123–134 [Google Scholar]
- Watanabe M, Hubberten HM, Saito K, Hoefgen R (2010) General regulatory patterns of plant mineral nutrient depletion as revealed by serat quadruple mutants disturbed in cysteine synthesis. Mol Plant 3: 438–466 [DOI] [PubMed] [Google Scholar]
- Waters BM, Blevins DG, Eide DJ (2002) Characterization of FRO1, a pea ferric-chelate reductase involved in root iron acquisition. Plant Physiol 129: 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35: 155–189 [Google Scholar]
- Zhang FS, Römheld V, Marschner H (1991) Role of the root apoplasm for iron acquisition by wheat plants. Plant Physiol 97: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zocchi G. (2006) Metabolic changes in iron-stressed dicotyledonous plants. In Barton LL, Abadía J, eds, Iron Nutrition in Plants and Rhizospheric Microorganisms. Springer, Dordrecht, The Netherlands, pp 359–370 [Google Scholar]
- Zuchi S, Cesco S, Astolfi S (2012) High S supply improves Fe accumulation in durum wheat plants grown under Fe limitation. Env Exp Bot 77: 25–32 [Google Scholar]
- Zuchi S, Cesco S, Varanini Z, Pinton R, Astolfi S (2009) Sulphur deprivation limits Fe-deficiency responses in tomato plants. Planta 230: 85–94 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.