A transporter orthologous to a yeast vacuole phosphate transporter mediates phosphate efflux from the vacuole into the cytosol.
Abstract
To maintain a stable cytosol phosphate (Pi) concentration, plant cells store Pi in their vacuoles. When the Pi concentration in the cytosol decreases, Pi is exported from the vacuole into the cytosol. This export is mediated by Pi transporters on the tonoplast. In this study, we demonstrate that SYG1, PHO81, and XPR1 (SPX)-Major Facility Superfamily (MFS) proteins have a similar structure with yeast (Saccharomyces cerevisiae) low-affinity Pi transporters Phosphatase87 (PHO87), PHO90, and PHO91. OsSPX-MFS1, OsSPX-MFS2, and OsSPX-MFS3 all localized on the tonoplast of rice (Oryza sativa) protoplasts, even in the absence of the SPX domain. At high external Pi concentration, OsSPX-MFS3 could partially complement the yeast mutant strain EY917 under pH 5.5, which lacks all five Pi transporters present in yeast. In oocytes, OsSPX-MFS3 was shown to facilitate Pi influx or efflux depending on the external pH and Pi concentrations. In contrast to tonoplast localization in plants cells, OsSPX-MFS3 was localized to the plasma membrane when expressed in both yeast and oocytes. Overexpression of OsSPX-MFS3 results in decreased Pi concentration in the vacuole of rice tissues. We conclude that OsSPX-MFS3 is a low-affinity Pi transporter that mediates Pi efflux from the vacuole into cytosol and is coupled to proton movement.
Phosphorus (P) is an important macronutrient for growth and development in plants. As a sessile organism, plants have evolved a sophisticated signaling network to obtain phosphate (Pi) from the environment where Pi concentration varies greatly and is often below that required for optimal growth (Chiou and Lin, 2011; Wu et al., 2013). The difference in Pi concentrations of plant tissues can be up to 10-fold when grown in Pi-sufficient and -deficient conditions. This large variation of Pi concentration in plant tissues is mainly due to differences in the Pi content of the vacuole (Lambers et al., 2011). Under Pi-sufficient conditions, a large amount of Pi is stored in the vacuole. The vacuolar Pi can be transported into the cytosol when external available Pi concentration decreases to a threshold level (Pratt et al., 2009). Although a number of molecular components of Pi uptake, mobilization, and utilization have been identified and functionally characterized, transporters of Pi import and export from the vacuole remain undefined in plants (Chiou and Lin, 2011).
The SYG1, PHO81, and XPR1 (SPX) domain (Pfam PF03105) is named after the Suppressor of Yeast G-protein α-subunit1 (SYG1), the yeast (Saccharomyces cerevisiae) Phosphatase81 (PHO81), and the human Xenotropic and Polytrophic Retrovirus receptor1 (XPR1). Normally, the SPX domain is localized at the N-terminal region of proteins. Depending on the characteristics to the C terminus domain of the SPX proteins, these proteins are divided into four subfamilies in plants (Secco et al., 2012a). Interestingly, members from all subfamilies have been shown to be involved in Pi homeostasis in plants (Hamburger et al., 2002; Wang et al., 2009, 2012, 2014; Kant et al., 2011; Secco et al., 2012a; Puga et al., 2014). Phosphate1 (PHO1), the first members characterized, contains a SPX domain and an EXS domain (named after Early Responsive to Dehydration1 [ERD1], XPR1, and SYG1) and regulates Pi loading into the xylem (Hamburger et al., 2002). A ubiquitin E2 ligase, Ubiquitin Conjugase24 (UBC24), interacts with the SPX domain of PHO1 and mediates the degradation of PHO1 (Liu et al., 2012). The second family of proteins contains only the SPX domain and negatively regulates Pi starvation signaling in plants (Wang et al., 2009; Liu et al., 2010). It has been reported that OsSPX4 inhibits the nuclear localization of transcription factor OsPHR2 by physical interaction with Phosphate Response2 (OsPHR2) in rice (Oryza sativa; Lv et al., 2014). OsSPX1/OsSPX2 was recently shown to directly interact with PHR2 in nucleus through its SPX domain, inhibiting its binding to the PHR1 binding sequence (GNATATNC; Wang et al., 2014). While the third family of SPX proteins, SPX-Major Facility Superfamily (SPX-MFS) subfamily, has been reported to be involved in regulation of Pi homeostasis in rice, the detailed molecular mechanism is unclear (Lin et al., 2010; Wang et al., 2012). The fourth SPX protein family contains a protein called Nitrogen Limitation Adaptation (NLA), first identified by its role in nitrogen starvation resistance (Peng et al., 2007). Later, a combination of physiology and genetic studies demonstrated that NLA regulates nitrate-dependent Pi homeostasis in Arabidopsis (Arabidopsis thaliana; Kant et al., 2011). The NLA protein contains an SPX and ring domain and mediates the degradation of Pi transporters under Pi-sufficient conditions (Lin et al., 2013; Park et al., 2014).
Pi uptake and signaling regulation is well characterized in yeast and involves the participation of several SPX domain proteins (Secco et al., 2012b). There are five Pi:H+ symporters characterized in yeast, including two high-affinity Pi transporters (Pho84 and Pho89) and three low-affinity Pi transporters (Pho87, Pho90, and Pho91). Interestingly, the low-affinity Pi transporters share a similar structure with a SPX domain in the N terminus and a transmembrane (TM) region in the C terminus. PHO87 and PHO90 are plasma membrane proteins and mediate Pi uptake into cells under replete Pi condition. Although the expression of these low-affinity Pi transporters is independent of Pi status, PHO87 and PHO90 are targeted to the vacuole and degraded under Pi starvation conditions (Ghillebert et al., 2011). The PHO91 protein is localized on the tonoplast and is responsible for transport of Pi from the vacuole lumen to the cytosol (Hürlimann et al., 2007). Deletion of all five Pi transporters causes lethality in yeast but viability can be rescued by overexpression of any one of the five Pi:H+ transporters (Wykoff and O’Shea, 2001).
Plants appear to have more complex Pi transport systems than yeast. A number of Pi:H+ symporters have been identified and characterized in plants. The majority of these transporters are Phosphate Transporter1 (PHT1)-type Pi transporters that are localized on the plasma membrane (Javot et al., 2007; Nussaume et al., 2011), and although most are implicated in uptake Pi into cells, some have also shown Pi efflux activity and regulate Pi remobilization (Preuss et al., 2010). Two other families of Pi:H+ symporters are the plastidic PHT2-type Pi transporters and the mitochondrial PHT3-type Pi transporters (Versaw and Harrison, 2002; Hamel et al., 2004). In addition to Pi:H+ symporters, mitochondria and plastid contain Pi antiporters or translocators to exchange Pi with various phosphorylated intermediates (Javot et al., 2007; Palmieri et al., 2008). The PHT4-type Pi transporters share similarity with the human Solute Carrier17 (SLC17) and exhibit Na+-dependent Pi transport (Guo et al., 2008). Five of the PHT4-type Pi transporters reside in the plastidic envelope, while the PHT4;6 is localized on the Golgi membrane (Guo et al., 2008). In addition to the Pi transporters mentioned above, it is reported that PHO1 and PHO1-like proteins influence cellular Pi homeostasis and export inorganic Pi from cells (Stefanovic et al., 2011). Although these proteins were localized in the membrane of Golgi and endoplasmic reticulum, the exact biochemical properties of these proteins are lacking.
The vacuole represents a dynamic Pi storage organelle, which may contain up to 90% of inorganic Pi of a plant cell under Pi-sufficient conditions. Under Pi starvation, Pi is exported from the vacuole into the cytosol of specific cells that store Pi (mostly epidermal cells in eudicots and mesophyll in Poales; Conn and Gilliham, 2010), and this export maintains Pi equilibrium and metabolism in the cytosol (Pratt et al., 2009). Although Pi:H+ symporters localized on tonoplast were proposed to mediate Pi import and export from the vacuole for some time (Martinoia et al., 2007), no distinct vacuolar Pi transporter has been identified at the molecular level.
In this study, we show that rice SPX-MFS proteins are localized on the vacuolar membrane. OsSPX-MFS3 complements the yeast mutant EY917 under high external Pi conditions. Moreover, the transport properties of OsSPX-MFS3 when expressed in Xenopus laevis oocytes and transgenic plants overexpressing the OsSPX-MFS3 are consistent with OsSPX-MFS3 functioning to export Pi from the vacuole into the cytosol.
RESULTS
OsSPX-MFS Family Proteins Localized on Tonoplast
In yeast, three SPX domain proteins, PHO87, PHO90, and PHO91, contain 12 membrane-spanning domains in their C terminus (Secco et al., 2012b). OsSPX-MFS1, OsSPX-MFS2, and OsSPX-MFS3 share 86% to 93% similarity with each other and share 25% to 26% similarity with PHO87, PHO90, and PHO91. Protein sequence alignments predict that OsSPX-MFS1, OsSPX-MFS2, and OsSPX-MFS3 contain 12 putative TM domains, similar to PHO87, PHO90, and PHO91 (Supplemental Fig. S1). Hydrophobicity plots (http://www.cbs.dtu.dk/services/TMHMM/) predict 12 TM domains in OsSPX-MFS1, OsSPX-MFS2, and OsSPX-MFS3 consisting of two partially duplicated subdomains of six TM segments similar to other MFS family Pi transporters (Supplemental Fig. S2). Thus, it was hypothesized that OsSPX-MFS1, OsSPX-MFS2, and OsSPX-MFS3 may be localized on the plasma membrane as PHO87 and PHO90 or on the tonoplast as PHO91 (Secco et al., 2012a, 2012b).
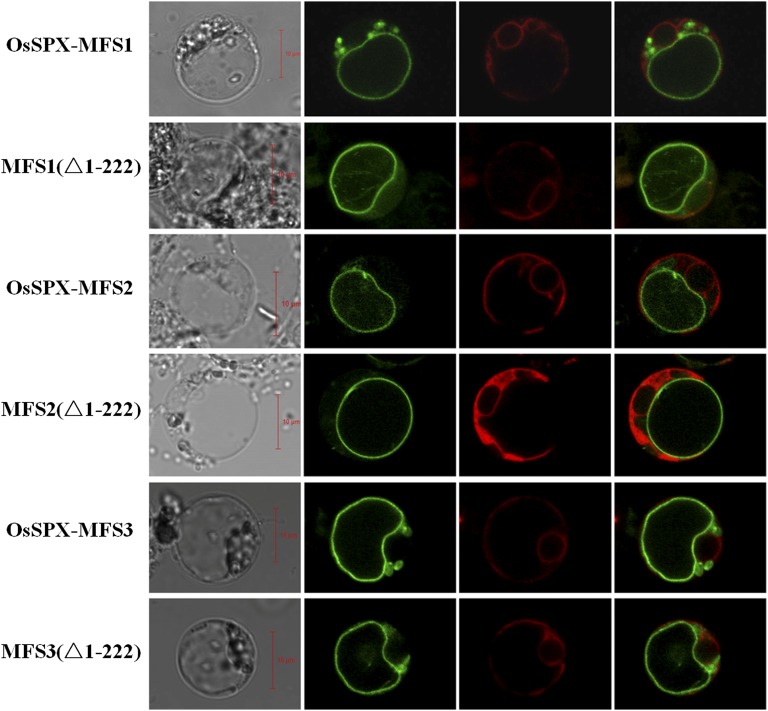
To determine the membrane location, full-length complementary DNAs (cDNAs) of OsSPX-MFS1, OsSPX-MFS2, and OsSPX-MFS3 were fused to GFP and transiently expressed in rice protoplasts. OsSPX-MFS1-GFP, OsSPX-MFS2-GFP, and OsSPX-MFS3-GFP all localized on the tonoplast (Fig. 1). As a control, the Arabidopsis Plasma Membrane Intrinsic Protein 2A::mCherry was localized on the plasma membrane and endoplasmic reticulum (Hachez et al., 2013). Moreover, the truncation versions of these three proteins, where the SPX domains were deleted, designated △MFS1 (1-222), △MFS2 (1-222), and △MFS3 (1-222), showed the same localization as the full-length proteins, suggesting the TM domains or C termini were sufficient for the correct localizations of these proteins (Fig. 1).
Figure 1.
Subcellular localization of full-length OSPSX-MFS proteins and truncated MFS domains in rice protoplasts. N-terminal GFP fusion constructs were transformed with isolated rice protoplasts. The green signals indicate GFP, and the red signals indicate plasma membrane marker AtPIP2A::mCherry. The ∆MFS1 (1-222), ∆MFS2 (1-222), and ∆MFS3 (1-222) lack the N-terminal 222 amino acid corresponding to the SPX domain. Bars = 10 µm.
A digital expression analysis showed that the transcript abundance of OsSPX-MFS3 is significantly higher than that of OsSPX-MFS1 and OsSPX-MFS2 in all tested tissues (Supplemental Fig. S3). Thus, OsSPX-MFS3 is likely the major vacuole Pi transporter among the SPX-MFS family. Thus the functional analyses will focus on OsSPX-MFS3.
OsSPX-MFS3 Partially Complements the Yeast Pi Transporter Mutants at High Pi Concentration
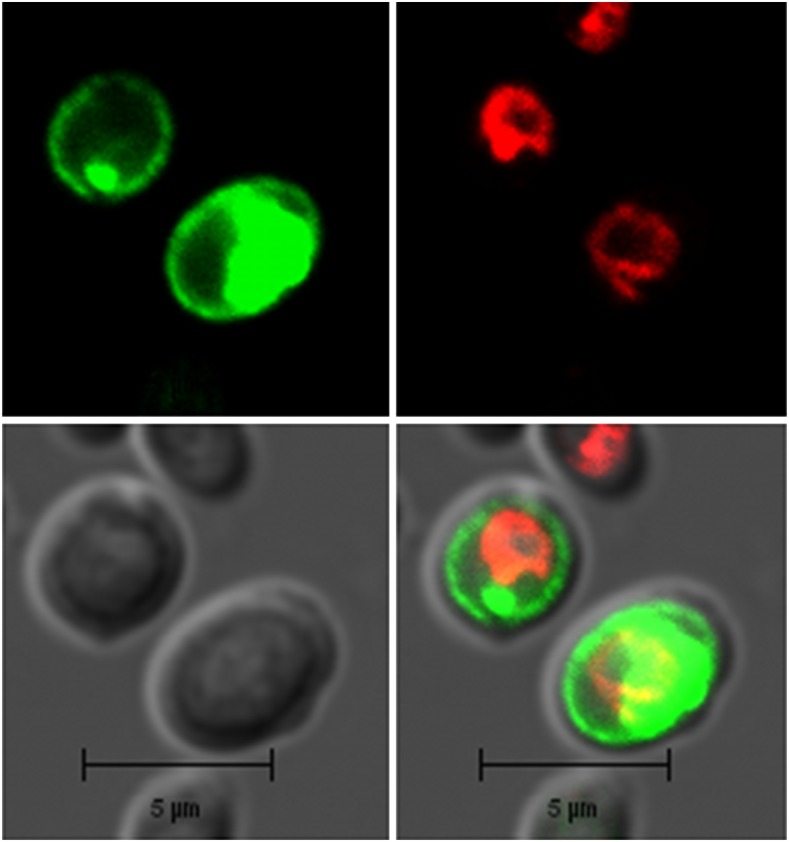
Previously, we have shown OsSPX-MFS1 could weakly support the growth of a yeast PAM2 mutant at pH 4.5, which lacks the high-affinity Pi transporters PHO84 and PHO89 (Wang et al., 2012). To study the function of OsSPX-MFS3 in yeast, the N-terminal GFP fusion of OsSPX-MFS was transformed into yeast to determine the subcellular localization of OsSPX-MFS3. The results showed that the OsSPX-MFS3 protein localized to the plasma membrane and cytosol of yeast but not to the tonoplast, which was stained by the tonoplast marker FM4-64 (Fig. 2).
Figure 2.
Subcellular localization of OSPSX-MFS3 proteins in yeast cells. Localization of N-terminal GFP of OsSPX-MFS proteins in yeast by confocal laser scanning microscopy. The green signals indicate GFP, and the red signals indicate vacuolar membranes that were specifically stained with the dye FM4-64. Bars = 5 µm.
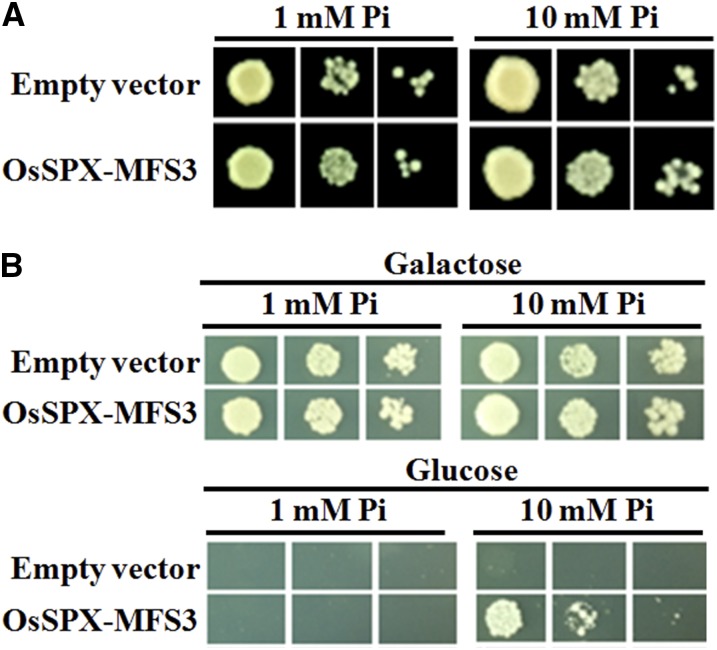
Two Pi concentrations, 1 and 10 mm, were used to determine the influence of external Pi on the complementation effect of OsSPX-MFS3 on growth of the PAM2 yeast at pH 5.5. OsSPX-MFS3 could slightly improve the growth of PAM2 cells at 10 mm Pi (Fig. 3A). At 1 mm Pi concentration at pH 5.5, OsSPX-MFS3 did not support the growth of PAM2. Also, when the medium pH was increased to 6.5 or 7.5, OsSPX-MFS3 inhibited the growth of the PAM2 mutant (Supplemental Fig. S4).
Figure 3.
Complementation of yeast Pi transporter mutant. A, Complementation of yeast mutant PAM2 (∆pho84∆pho89) by the OsSPX-MFS3 gene. B, Complementation of yeast mutant EY917 (∆pho84∆pho87∆pho89 ∆pho90∆pho91) by the OsSPX-MFS3 gene. Yeast cells harboring either an empty expression vector (control) or OsSPX-MFS3 cDNA construct were grown in YPD medium with Gal to an OD600 of 1. Equal volumes of 10-fold serial dilutions were applied to YPD (pH 5.5) medium with 1 or 10 mm Pi and then incubated at 30°C for 4 d.
To exclude the interference of OsSPX-MFS3 with endogenous Pi transporters in PAM2 mutant, the yeast EY917 strain was used, which lacks all of the five functional Pi transporters, PHO84, PHO87, PHO89, PHO90, and PHO91 (Wykoff and O’Shea, 2001). This strain contains the plasmid harboring a Pho84 gene under the GAL1 promoter, which allows the normal growth of this strain on media using Gal as the only carbon resource. When the yeast grows on media with Gal as the sole carbon resource, the growth of EY917 transformed with the SPX-MFS3 gene was similar to the empty vector control under both 1 and 10 mm Pi at pH 5.5 (Fig. 3B). When Glc was used as the carbon resource, the EY917 strain transformed with the OsSPX-MFS3 gene did not grow on the media containing 1 mm Pi (Fig. 3B). However, OsSPX-MFS3 transformed cells, but not the empty vector control, and restored the growth of EY917 when the media Pi concentration was increased to 10 mm. By contrast, both the OsSPX-MFS3-transformed EY917 cells and empty vector control could not grow at pH 7.5 on Glc medium, irrespective of the Pi concentration (data not shown).
Pi Transporter Activity of OsSPX-MFS3 in X. laevis Oocytes
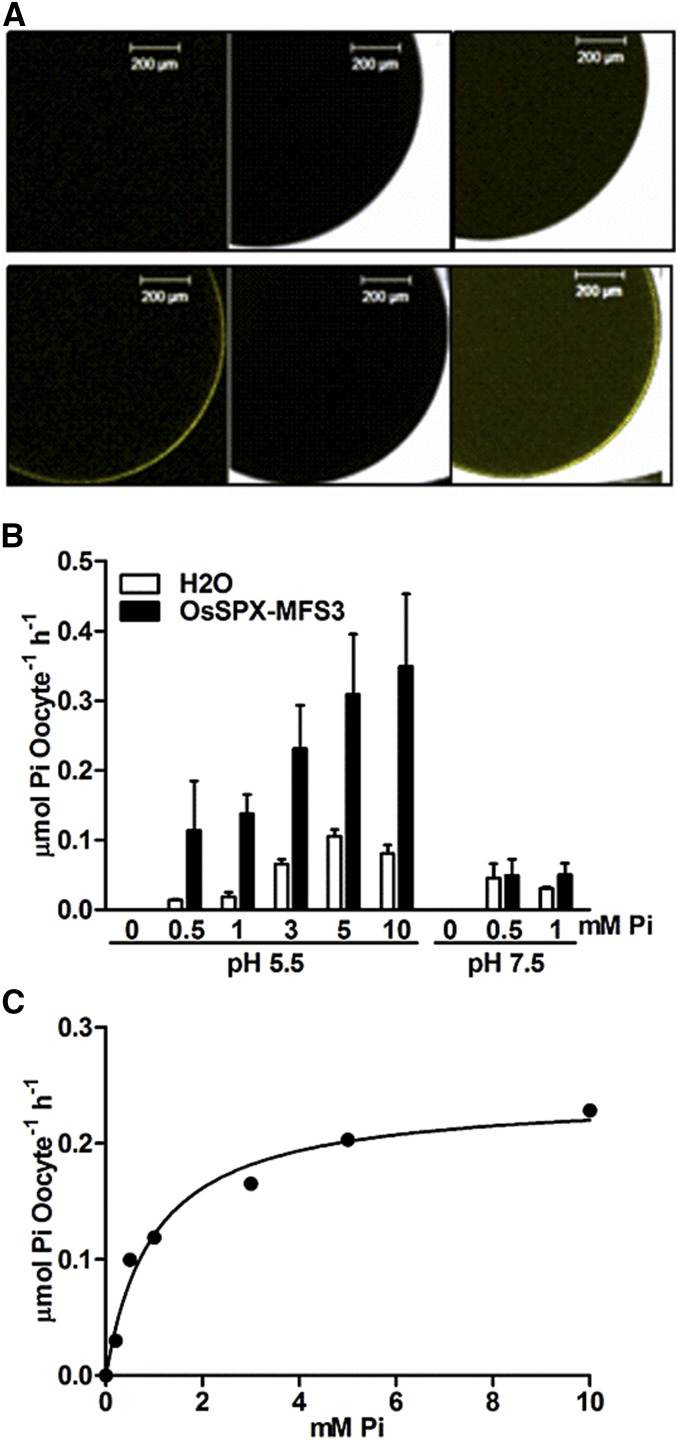
To investigate the Pi transport characteristics of OsSPX-MFS3, it was expressed in X. laevis oocytes. Because oocytes do not have a vacuole or related structures, it is proposed that tonoplast-localized proteins should target to the plasma membrane of oocytes, as observed with Tonoplast Intrinsic Protein aquaporins (Daniels et al., 1994). An N-terminal Yellow Fluorescence Protein (YFP) fused to OsSPX-MFS3 targeted to the plasma membrane in oocytes as predicted (Fig. 4A). Chemical flux was examined using 33P to determine Pi uptake activity of OsSPX-MFS3. The OsSPX-MFS3-expressing oocytes showed a significant 33P influx activity at pH 5.5 relative to water-injected controls when the Pi concentration was 0.5 mm in the solution (Fig. 4B). The 33P influx activities increased with higher Pi concentration ranging from 0.5 to 5 mm and achieved a maximum influx at 5 mm Pi (Fig. 4B). However, when the external pH was increased to 7.5, no significant 33P influx activity was observed at Pi concentrations of 0.5 and 1 mm (Fig. 4B). Because Ca2+ in the bath solution (BS) will precipitate with Pi when the Pi concentration is higher than 1.5 mm at pH 7.5, no further increased Pi concentration could be measured under this condition.
Figure 4.
A, Subcellular localization of OsSPX-MFS3 in X. laevis oocytes. cRNAs of nYFP-fused OsSPX-MFS3 were injected into oocytes and incubated for 48 h in BS before imaging with confocal laser scanning microscopy. B, Pi influx activity of OsSPX-MFS3. Oocytes were incubated for 48 h in BS (without Pi) and then transferred into BS solution containing different Pi concentrations containing 32P (2 mCi mL–1) for 1 h. The radioactivity response to increasing concentrations of externally applied Pi was recorded at pH 5.5 and 7.5. Mean ± se of the mean (n = 10 oocytes) is shown. C, Nonlinear regression of Pi uptake of OsSPX-MFS3 versus external Pi concentration at pH 5.5 was used to estimates the Km value. Oocytes were incubated for 48 h in BS (without Pi) and then transferred into BS solution containing different Pi concentrations containing 33P (5 mCi mL–1) for 1 h. The radioactivity response to increasing concentrations of externally applied Pi was recorded at pH 5.5. Estimated Km and Vmax were 1.101 ± 0.03 mm and 0.24 ± 0.01 μmol Pi oocyte–1 h–1, respectively. Bars = 200 µm.
Time course experiments at pH 5.5 showed that the rate of net transport was linear with the time for the first 1.5 h (Supplemental Fig. S5). To estimate Km and Vmax values, the component of net transport contributed by the oocyte endogenous transport activity was subtracted before Michaelis-Menten analysis. The apparent Km value and Vmax of OsSPX-MFS3 was 1.101 ± 0.03 mm and 0.24 ± 0.01 μmol Pi oocyte–1 h–1 according to Pi uptake velocities at different Pi concentrations at pH 5.5 (Fig. 4C).
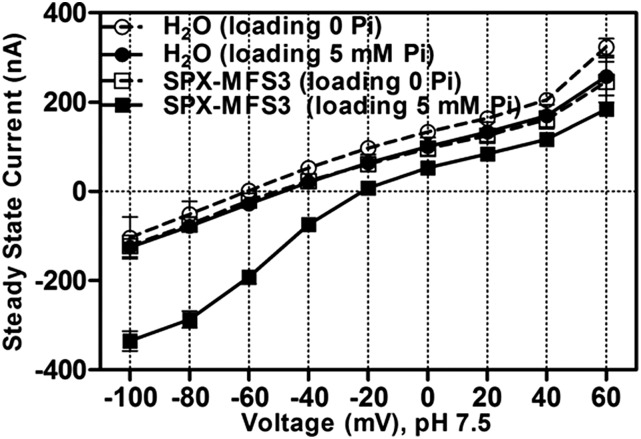
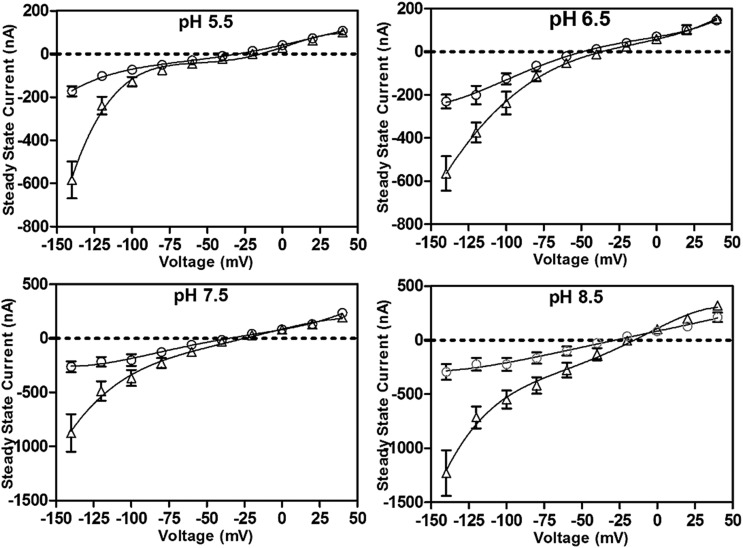
Two-electrode voltage clamp was used to investigate if Pi-dependent currents could be observed when OsSPX-MFS3 was expressed in oocytes. Although a significant 33P uptake activity was observed for OsSPX-MFS3 oocytes at pH 5.5, no Pi-dependent currents could be detected in OsSPX-MFS3 oocytes (Supplemental Fig. S6). To test whether Pi-dependent currents via OsSPX-MFS3 occurred when internal Pi was increased, OsSPX-MFS3-injected oocytes were preloaded with Pi by incubating the oocytes with 5 mm Pi for 2 d. Interestingly, OsSPX-MFS3-injected oocytes showed larger inward currents and more positive current reversal voltage than water-injected controls at pH 7.5 (Fig. 5). The result indicated that OsSPX-MFS3 may mediate Pi efflux from the oocytes at pH 7.5 (Fig. 5). To explore whether this Pi efflux was dependent on pH, OsSPX-MFS3 oocytes were further measured under different pH conditions. The results showed that inward currents decreased as the pH of the solutions decreased, from pH 8.5 to 5.5 (Fig. 6).
Figure 5.
OsSPX-MFS3 mediates Pi-dependent inward current in X. laevis oocytes. Steady-state current/voltage plots of water-injected control oocytes (filled and unfilled circles) and OsSPX-MFS3-expressing oocytes (filled and unfilled squares). The oocytes were preincubated in BS with 0 (dash line) or 5 mm (full line) Pi for 2 d. The currents were recorded with Pi-free ND-10 solution (pH 7.5). Mean currents ± se of the mean (n = 4) are present.
Figure 6.
OsSPX-MFS3 induced Pi-dependent inward currents is pH dependent. Steady-state current/voltage plots of OsSPX-MFS3-expressing oocytes (triangle) and water-injected control (circle) under different pH conditions. The oocytes were preincubated in BS with 5 mm Pi for 2 d. The currents were recorded with Pi-free ND-10 solution with pH 8.5, 7.5, 6.5, and 5.5. Mean currents ± se of the mean (n = 4) are present.
Overexpression of OsSPX-MFS3 Resulted in a Lower Vacuole Pi Concentration in Rice
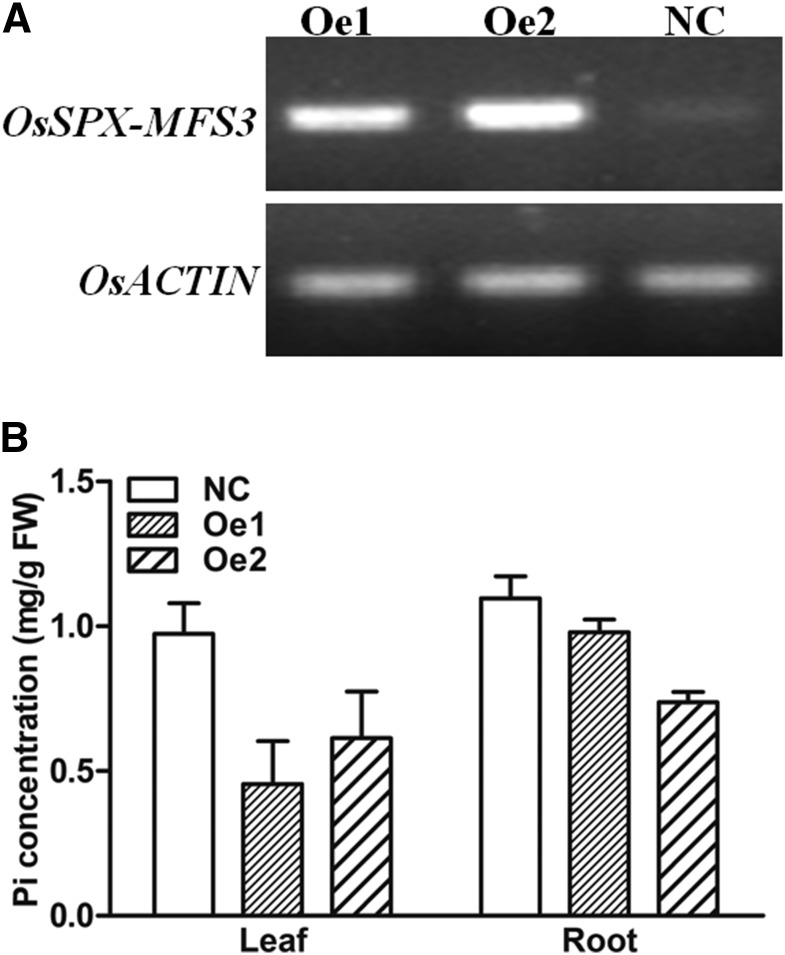
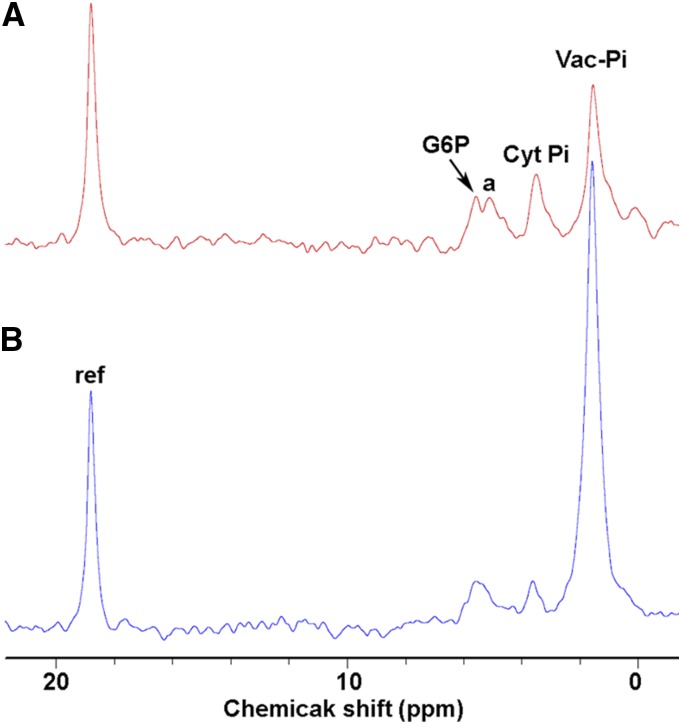
To test whether OsSPX-MFS3 functions as a Pi transporter on the tonoplast in rice, transgenic rice plants overexpressing OsSPX-MFS3 were produced via Agrobacterium tumefaciens-mediated transformation (Fig. 7A). Compared with that of the nontransformed counterparts, Pi concentrations in both the leaves and roots of OsSPX-MFS3 overexpression plants were significantly decreased (Fig. 7B). Vacuolar Pi concentrations were compared between the OsSPX-MFS3 overexpression and wild-type plants using NMR spectroscopy. The vacuolar Pi concentration of OsSPX-MFS3 overexpression plants was significantly lower than that of wild-type plants, while the cytosolic Pi concentration was slightly higher in the OsSPX-MFS3 overexpression plants compared with the wild-type plants (Fig. 8).
Figure 7.
RT-PCR (A) and Pi concentration (B) of OsSPX-MFS3 overexpression plants. Homozygous seeds of the T2 generation were geminated for 3 d and hydroponically cultured for 3 weeks. Oe1 and Oe2 represent two independent OsSPX-MFS3 transgenic lines. The leaves of the plants were sampled for RT-PCR. Leaves and roots of positive and negative segregants were sampled for Pi concentration detection. Mean ± se of the mean (n = 3) is present. NC, Negative control; FW, fresh weight.
Figure 8.
In vivo proton-decoupled 31P-NMR spectra of OsSPX-MFS3 overexpression (A) and negative segregants (B). Peak assignment are as follows: G6P, Glc-6-Pi; peak A, position of Fru-6-Pi, ribose-5-Pi, other sugar Pi and phosphomonoesters; Cyt Pi, cytoplasmic Pi; Vac Pi, vacuolar Pi; and ref, reference (methylenediphosphonate) used to measure chemical shifts and for quantifications.
DISCUSSION
Under Pi-sufficient conditions, the excessive cellular Pi is stored in vacuoles. When the external Pi concentration decreases, the stored vacuole Pi can be exported to the cytosol. These processes are mediated by Pi transporters on the tonoplast (Pratt et al., 2009), but plant tonoplast Pi transporters are yet to be identified. As a number of SPX domain proteins are involved in Pi signaling, it is proposed that SPX-MFS proteins may be involved in vacuole Pi transport (Lin et al., 2010; Secco et al., 2012a). In this study, we have shown that OsSPX-MFS1, OsSPX-MFS2, and OsSPX-MFS3 are localized on the tonoplast by GFP fusion proteins in rice tonoplast. The transcript abundance of OsSPX-MFS3 is higher than those of OsSPX-MFS1 and OsSPX-MFS2 in all tissues under sufficient Pi condition (Supplemental Fig. S3). Short-term Pi deficiency (1 d) suppressed the expression of OsSPX-MFS1 and induced OsSPX-MFS2 (Wang et al., 2012). By contrast, the expression of OsSPX-MFS3 remained stable for 5 d of Pi starvation and only slightly decreased with longer Pi starvation treatment (Wang et al., 2012). Based on the expression patterns of the three OsSPX-MFS genes, it seems that OsSPX-MFS3 has a general function in rice plants, while OsSPX-MFS1 and OsSPX-MFS2 may function under specific conditions or in cell- or tissue-specific tissues. Consistent with this proposal, an organelle proteome detected only AtSPX-MFS3, the homolog of OsSPX-MFS3, in Arabidopsis under sufficient Pi condition (Dunkley et al., 2006).
Previously, we have observed a moderate effect on promotion of growth of the PAM2 mutant in liquid solution at pH 4.5 by transformation with OsSPX-MFS1 (Wang et al., 2012). In this study, OsSPX-MFS3 also slightly improved the growth of PAM2 cells at pH 5.5 with a high Pi concentration (Fig. 3A). The weak complementation effect of OsSPX-MFS3 on the yeast mutants may be attributed to incorrect localization of OsSPX-MFS3 protein in yeast or weak Pi flux activity, as it is a low-affinity Pi transporter. OsSPX-MFS3 is localized on the plasma membrane of yeast but not the tonoplast as in plant (Fig. 2). Because the PAM2 mutant still contains three low-affinity Pi transporters, PHO87, PHO90, and PHO91, these proteins may mask the function of OsSPX-MFS3 in PAM2 as well. Therefore, the yeast mutant EY917, which lacks all five inorganic Pi transporters and could not grow on Glc medium, was used to test the function of OsSPX-MFS3. Growth of the mutant can be restored by any Pi transporters in yeast, including the yeast vacuole Pi transporter PHO91 and glycerphosphoinositol permease1 (GIT1). It is proposed that both of them could mediate inorganic Pi uptake from the growth medium into yeast under the alcohol dehydrogenase1 (ADH1) promoter (Wykoff and O’Shea, 2001). Transformed EY917 with OsSPX-MFS3 restored the growth of the yeast mutant at a high Pi concentration (10 mm) but not at lower Pi concentrations (1 mm), indicating SPX-MFS may play a similar function as ScPHO91, i.e., a low-affinity inorganic Pi transport activity in yeast. The estimated Km value proved this hypothesis that OsSPX-MFS3 is a low-affinity Pi transporter.
To define the biochemical activity of this Pi transporter, OsSPX-MFS3 was expressed in X. laevis oocytes. In oocytes, a chemical influx of Pi was observed at pH 5.5 (Fig. 4B). However, no electrical currents were detected. It is possible that at low pH, Pi influx is coupled with H+ to produce an electroneutral influx that would not be detected electrically. Similarly, at a low-pH and high-Pi environment, OsSPX-MFS3 showed influx activity in both PAM2 and EY917 yeast strains (Fig. 3, A and B). When the pH was increased to 7.5, the uptake of 32Pi in OsSPX-MFS3 oocytes was similar to controls. Interestingly, OsSPX-MFS3 oocytes generated an inward current in pH 7.5 and shifted the reverse potential to more positive values than control cells when the oocytes were preloaded with Pi (Fig. 5). A detailed analysis showed that the inward current increased at pH 8.5 and decreased at pH 5.5 compared with pH 7.5 (Fig. 6). This suggests that under a high-pH condition, OsSPX-MFS3 works as a Pi efflux in oocytes, either uncoupled from H+ or with a stoichiometry that produces an electrical current. Similarly, the suppression of the growth of OsSPX-MFS3 complemented the PAM2 strain at low Pi concentration and high-pH (pH 6.5 and 7.5) conditions may also result from its Pi efflux activity of OsSPX-MFS3 (Supplemental Fig. S4). The shift between a Pi influx and efflux transporter of OsSPX-MFS3 in oocytes and yeast depends on the electrochemical gradient by external pH conditions, which may change the orientation of OsSPX-MFS3.
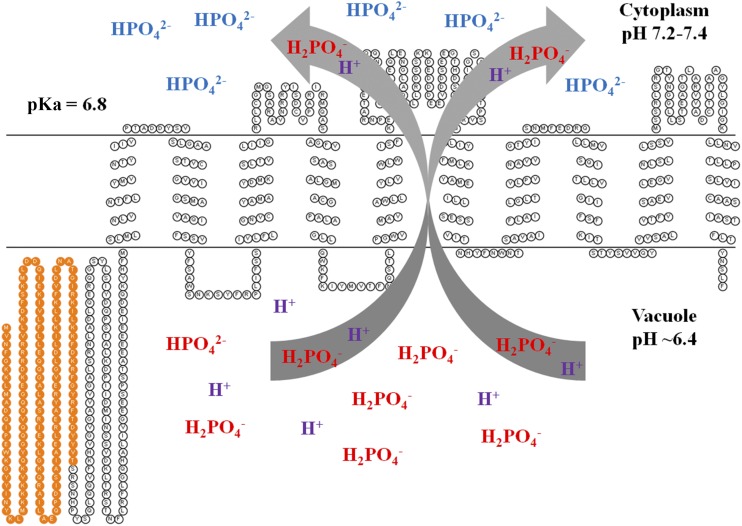
Unlike the plasma membrane localization in yeast or oocytes, OsSPX-MFS3 is localized on tonoplast in rice cells. It was reported that the pH of cytosol and vacuole in Arabidopsis is 7.3 and 5.2, respectively (Shen et al., 2013). It is also known that the Pi concentration in tonoplast is much higher than that in cytosols (Fig. 8). Thus, localized in the tonoplast, OsSPX-MFS3 should transport Pi only in one direction, that is, from tonoplast to cytosol. The hypothesis is supported by the fact that overexpression of OsSPX-MFS3 significantly reduced the vacuole Pi (Fig. 8). The working model for the Pi transporter activity of OsSPX-MFS3 is illustrated in Figure 9. OsSPX-MFS3 mediates Pi transport from the vacuole into the cytoplasm by coupling with protons. As the pKa value of H2PO4– and HPO42– is 6.8, the form of Pi in vacuole would be predominately H2PO4–, while in cytosol, there would be a mix of HPO42– and H2PO4– in a ratio of approximately 3.2:1. If the transported ratio between H2PO4– and H+ was 1:1, no inward current would be detected, with OsSPX-MFS3 using a voltage clamp at pH 5.5 as observed in X. laevis oocytes (if it is assumed that the external medium for X. laevis oocytes corresponds to the vacuole in planta). However, preloading of X. laevis oocytes with 5 mm Pi gave an inward ion current at more alkaline pH. Here, OsSPX-MFS3 may act as an anion (HPO42–) efflux in oocytes. If HPO42– efflux is coupled with H+ in a ratio of 1:1 as discussed above, it will still display a negative current. Because the OsSPX-MFS3 localized on the plasma membrane of oocyte and yeast, efflux of Pi out of cells would correspond to Pi loading into the vacuole in plants. However, normally, this phenomenon will not happen in the plants, because the vacuole usually has a higher Pi concentration and lower pH than the cytosol.
Figure 9.
A working model of OsSPX-MFS3 in plant. Predicted topology profile of OsSPX-MFS3 was made and displayed using the TMHMM and Topo2 software (http://www.sacs.ucsf.edu/TOPO2/). OsSPX-MFS3 contains an N-terminal SPX domain (orange color) and C-terminal TM domain. The pH of the cytoplasm and vacuole usually are nearly pH 7.2 to 7.4 and 6.4, respectively. As the acid dissociation constant (pKa) value of H2PO4– and HPO42– was 6.8, the major form of Pi in cytoplasm and vacuole would be H2PO4– and HPO42–, respectively. OsSPX-MFS3 is proposed to transport H2PO4– from vacuole into cytoplasm coupled to a proton in a ratio of 1:1.
Previous results have shown that the SPX domain of ScPHO87 and ScPHO90 is involved in interaction with suppressor of phosphoinositide-specific phospholipase C2 that regulates both the Pi influx and efflux activity of these transporters (Hürlimann et al., 2009). Deletion of the SPX domain of OsSPX-MFS1, OsSPX-MFS2, and OSPX-MFS3 proteins did not change the subcellular localization of these proteins. Thus, the SPX domain may regulate SPX-MFS Pi transporters by interaction with other unidentified proteins or ligands.
MATERIALS AND METHODS
Construction of Vectors
Primers were designed according to the full-length cDNA of OsSPX-MFS1, OsSPX-MFS2, and OsSPX-MFS3 genes. The open reading frame of OsSPX-MFS1, OsSPX-MFS2, and OsSPX-MFS3 genes were amplified from cDNA of rice (Oryza sativa) leaves and cloned into pENTR/D-TOPO vector (Invitrogen). The clones were selected by PCR and sequenced to confirmation. The corrected vectors were recombinant with destination vectors pH7WGF2 and pAG426GPD-ccdB using the Gateway system, respectively. GFP was fused at the N terminus with target genes and driven by the 35S promoter in the pH7WGF2 vector. Vector pAG426GPD ccdB was used for the yeast (Saccharomyces cerevisiae) complementation with a constitutively expressed promoter. OsSPX-MFS3 was recombinant into destination vectors pGEMHE and nYFP-pGEMHE and driven by the T7 promoter. The resultant vector was used by complementary RNA (cRNA) synthesis.
For the overexpression vector of OsSPX-MFS3 genes, a BamHI site was introduced into the upper and lower primers. The full length of the OsSPX-MFS3 gene was amplified and purified. The product fragment was digested with BamHI and introduced into pTF101.1-ubi as previously described. The resultant vector was confirmed by sequence and used for Agrobacterium tumefaciens-mediated rice transformation as described previously. The primers used for all of the amplification are listed in Supplemental Table S1.
Subcellular Localization of SPX-MFS Proteins in Rice Protoplast
One hundred rice seeds were germinated on one-half-strength Murashige and Skoog media under light for 3 d and grew seedlings at 26°C in the dark for about 12 to 14 d. Using a razor blade, the stems and leaves of the seedlings were cut into approximately 0.5-mm strips. The strips were put into a petri dish with 10 mL of enzyme solution (1.5% [w/v] cellulose and 0.3% [w/v] macerozyme in K3 medium). We applied a vacuum for 1 h for infiltration of the enzyme solution and incubated it for about 4 h in the dark with gentle shaking (approximately 40 rpm) at room temperature. The enzyme solution was removed and added 10 mL of W5 medium (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, and 2 mm MES, pH 5.7) with gentle shaking (approximately 80 rpm) for 1 h to release the protoplast. The protoplasts were filtered through a 35-µm nylon mesh and centrifuged at 1,500 rpm for 4 min to collect the protoplasts.
Protein fusion constructs were transformed rice protoplasts by the polyethylene glycol-mediated method. In brief, 10 μg of plasmid DNA of each construct was transformed into 0.2 mL of protoplast suspension. Two hundred twenty microliters of 40% (v/v) polyethylene glycol was added and mixed immediately by gently shaking. The mixture was incubated 20 min at room temperature, and 1 mL of K3 medium was added to the tube. After incubation at 30°C dark for 13 to 15 h, fluorescence signals in rice protoplasts were detected using a LSM710nlo confocal laser scanning microscope (Zeiss). Excitation/emission wavelengths were 488/506 to 538 nm for GFP and 561/575 to 630 nm for red fluorescent protein. The plasmid CD3-1007 containing the plasma membrane marker (AtPIP2A::mCherry fusion) was used as a control.
Yeast Mutant Complement Growth Assay and Confocal Microscopy
Full-length OsSPX-MFS1, OsSPX-MFS2, and OsSPX-MFS3 genes were cloned into vector pAG426GPD-ccdB as described above. These constructs were transformed into the growth defect of the yeast Pi transport-deficient strain PAM2 (Martinez and Persson, 1998) or EY917 (Wykoff and O’Shea, 2001). Transformed yeast was grown in synthetic dropout(-Ura) medium containing 0.22 mm Pi and 25 mm Na-citrate buffer, pH 4.5, to an optical density at 600 nm (OD600) of 1. Cells were harvested by centrifuge, washed with water three times, and suspended to an OD600 of 1. Five-microliter aliquots of serial dilutions were spotted to agar plate with different pH or Pi concentration. Plates were incubated at 30°C.
Yeast vacuoles were stained with FM4-64 as previously described (Hürlimann et al., 2007). In short, yeast cells were harvested as described above, concentrated in 100 µL of yeast peptone dextrose (YPD), and stained at 30°C for 15 to 20 min with 60 µm FM4-64. Cells were pelleted and suspended in 5 mL of YPD, and shaken at 30°C for 60 min. Cells were collected, washed three times with 1 mL of Pi-buffered saline (pH 7.3), and then applied on microscope slide. Fluorescence signals were detected using an LSM710nlo confocal laser scanning microscope (Zeiss). Excitation/emission wavelengths were 488 and 568 nm.
cRNA Synthesis and Xenopus laevis Preparation and Injection
Full-length OsSPX-MFS1, OsSPX-MFS2, and OsSPX-MFS3 genes were cloned into vector pGEMHE-DEST as described above. cRNA was synthesized using the T7 RNA polymerase kit (Ambion) as the instructions described. The quality and size of the synthesized RNA was checked on RNA-free agarose gels. The oocytes were isolated from X. laevis frogs (NASCO Biology) and maintained at 18°C as described previously (Preuss et al., 2010). Healthy stage IV and V oocytes were selected for injection with 23 ng of cRNA (46 nL of 500 µg µL–1 cRNA). The injected oocytes were incubated at 18°C in Barth's solution with or without 5 mm NaH2PO4 for 48 to 94 h prior to ion flux and electrophysiological measurement. The Barth's solution contained 96 mm NaCl, 2 mm KCl, 1 mm MgCl2, 0.6 mm CaCl2, 10 mm HEPES, adjusted to pH 7.6 with Tris-base, 2.5 mL of horse serum was added in 50 mL of solution, and 0.5 mL per 50 mL of penicillin and tetracycline (5 mg mL–1 stock, used 0.5/50 mL).
Electrophysiology and Pi Fluxes
Individual healthy oocytes were selected for voltage clamp experiments. ND-10 solutions (10 mm NaCl, 2 mm KCl, 1 mm MgCl2, 1.8 mm CaCl2, and 10 mm HEPES or MES) containing different Pi concentrations or pH gradients were used. All solutions were adjusted with mannitol to a final osmolarity of 205 mosmol kg–1. Two-electrode voltage clamp experiments were performed with a GeneClamp500 amplifier under the control of the Clampex8 program. Impaling electrodes were filled with 0.22-µm-filtered 3 m KCl. The voltage clamp protocol for the analysis was –40 mV for 0.23 s, then a different voltage ranging from 40 to –140 mV for 1.4 s in a –20-mV increment, and then –40 mV for 0.43 s.
For the Pi influx experiment, either 5 µL of H333PO4 or H333PO4 (2 or 5 mCi mL–1, corresponding to figures) was added into 5 mL of ND-10 BS containing 0, 0.5, 1, 3, 5, and 10 mm Pi. Ten healthy oocytes were added into each treatment and incubated for 1 h. Then, the oocytes were washed with ice-cold ND-10 BS three successive times and loaded in scintillation counting tubes. Scintillation counting was conducted (S6500, multifunction scintillation counter, Beckman and Coulter) on 50 µL of final wash solution containing a single oocyte with 4 mL of IRGA-Safe Plus scintillation fluid (Perkin Elmer).
Plant Materials and Growth Conditions
The japonica variety of cv Nipponbare was used for all physiological experiments and rice transformations. Hydroponic experiments were conducted using a modified rice culture solution containing 1.425 mm NH4NO3, 0.323 mm NaH2PO4, 0.513 mm K2SO4, 0.998 mm CaCl2, 1.643 mm MgSO4, 0.25 mm NaSiO3, 0.009 mm MnCl2, 0.075 μm (NH4)6Mo7O24, 0.019 μm H3BO3, 0.155 μm CuSO4, and 0.152 μm ZnSO4 with 0.125 mm EDTA-Fe (II; Wang et al., 2012). pH of the solution was adjusted to 5.5. Rice plants were grown in growth chambers with a 12-h photoperiod (200 µmol photons m–2 s–1) and a day/night temperature of 30°C/22°C after germination. Humidity was controlled at approximately 60%. For phenotype and Pi concentration measurements, seeds were germinated and grown for 3 weeks.
Measurement of Pi Concentration in Plants
Leaves and roots of the wild-type and transgenic seedlings from either Pi-sufficient or Pi-deficient treatments were sampled separately. The Pi concentration was measured using the procedure described previously (Wang et al., 2009). Briefly, 50 µg of fresh tissue samples was homogenized with 50 µL of 5 m H2SO4 and 3 mL of water. The homogenate was transferred to 1.5-mL tubes and centrifuged at 10,000g for 10 min at 4°C. The supernatant was collected and diluted to an appropriate concentration. The diluted supernatant was mixed with a malachite green reagent in a 3:1 ratio and analyzed 20 min afterward. The absorption values for the solution at 650 nm were determined using a Spectroquant NOVA60 spectrophotometer (Merck). Pi concentration was calculated from a standard curve generated with varying concentrations of KH2PO4.
RNA Isolation and RT-PCR
Total RNA was extracted from plant samples using RNeasy Mini Kits (Qiagen) according to the manufacturer’s recommendations. First-strand cDNAs were synthesized from total RNA using SuperScript II reverse transcriptase (Invitrogen). The ACTIN cDNA is an endogenous control that is used to normalize the samples. The primers that were used for the reverse transcription (RT)-PCR are given in Supplemental Table S1.
NMR Spectroscopy
Nearly 0.07 g (fresh weight) of roots of 3-week-old plants were packed into a 5-mm-diameter NMR tube equipped with a perfusion system connected to a peristaltic pump. In vivo 31P-NMR spectra of the roots were recorded on a Bruker Ascend 600 spectrometer with MestReNova software version 6.1.1-6384. The 31P-NMR spectra were recorded at a 242.9-MHz lock with deuteroxide in the capillary and water. The 31P-NMR acquisition conditions were as follows: 30° pulse angle, 1,500 scans, and a spectral window of 16 kHz. Chemical shifts were measured relative to the signal from a glass capillary containing 10 mm methylenediphosphonic acid as a reference, which is at 18.9 ppm relative to the signal from 85% H3PO4.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers OsSPX-MFS1, Loc_Os04g48390; OsSPX-MFS2, Loc_Os02g45520; and OsSPX-MFS3, Loc_Os06g03860.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Multiple sequence alignment between OsSPX-MFS genes and ScPHO87, ScPHO90, and ScPHO91.
Supplemental Figure S2. Prediction of transmembrane helices of OsSPX-MFS, ScPHO87, ScPHO90, and ScPHO91 proteins by TMHMM.
Supplemental Figure S3. Digital expression analysis of OsSPX-MFS1, OsSPX-MFS2, and OsSPX-MFS3 in different tissues in rice.
Supplemental Figure S4. Complementation of yeast high-affinity Pi transporter mutant PAM2 (Δpho84Δpho89) by OsSPX-MFS3 genes at different pH conditions.
Supplemental Figure S5. Time course of phosphate influx activity of OsSPX-MFS3.
Supplemental Figure S6. Voltage clamp of OsSPX-MFS3 injected oocyte under 0 and 5 mM Pi solution.
Supplemental Table S1. Sequences of the primer sets used in the study.
Supplementary Material
Acknowledgments
We thank Wendy Sullivan for X. laevis oocyte harvesting, Dr. Yves Poirier for providing the EY917 yeast strain, and Dr. Maria Hrmova for help with radioactivity detection and counting.
Glossary
- Pi
phosphate
- TM
transmembrane
- cDNA
complementary DNA
- YPD
yeast peptone dextrose
- RT
reverse transcription
- BS
bath solution
- OD600
optical density at 600 nm
Footnotes
This work was supported by the Key Basic Research Special Foundation of China (grant no. 2011CB100303), the National Science Foundation of China (grant nos. 31201675, 31471929, and 31572189), the Ministry of Science and Technology of China (2014ZX08003005), the 111 project (grant no. B14027), the Fundamental Research Funds for the Central Universities (grant no. 2014FZA6003), and Australian Research Council Centre of Excellence (grant nos. CE140100008 and CEO561495 to J.W. and S.D.T.).
C.W., S.D.T., and H.S. conceived the original research plans; C.W., W.Y., Y.H., S.W., and Y.L. performed the experiments; H.S. and S.D.T. supervised the experiments; H.S. and S.D.T. conceived the project and wrote the article with contributions of all the authors; D.S. and J.W. helped with the discussion and complemented the writing.
Articles can be viewed without a subscription.
References
- Chiou TJ, Lin SI (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Conn S, Gilliham M (2010) Comparative physiology of elemental distributions in plants. Ann Bot (Lond) 105: 1081–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MJ, Mirkov TE, Chrispeels MJ (1994) The plasma membrane of Arabidopsis thaliana contains a mercury-insensitive aquaporin that is a homolog of the tonoplast water channel protein TIP. Plant Physiol 106: 1325–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley TP, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al. (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103: 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghillebert R, Swinnen E, De Snijder P, Smets B, Winderickx J (2011) Differential roles for the low-affinity phosphate transporters Pho87 and Pho90 in Saccharomyces cerevisiae. Biochem J 434: 243–251 [DOI] [PubMed] [Google Scholar]
- Guo B, Jin Y, Wussler C, Blancaflor EB, Motes CM, Versaw WK (2008) Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol 177: 889–898 [DOI] [PubMed] [Google Scholar]
- Hachez C, Besserer A, Chevalier AS, Chaumont F (2013) Insights into plant plasma membrane aquaporin trafficking. Trends Plant Sci 18: 344–352 [DOI] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel P, Saint-Georges Y, de Pinto B, Lachacinski N, Altamura N, Dujardin G (2004) Redundancy in the function of mitochondrial phosphate transport in Saccharomyces cerevisiae and Arabidopsis thaliana. Mol Microbiol 51: 307–317 [DOI] [PubMed] [Google Scholar]
- Hürlimann HC, Pinson B, Stadler-Waibel M, Zeeman SC, Freimoser FM (2009) The SPX domain of the yeast low-affinity phosphate transporter Pho90 regulates transport activity. EMBO Rep 10: 1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hürlimann HC, Stadler-Waibel M, Werner TP, Freimoser FM (2007) Pho91 Is a vacuolar phosphate transporter that regulates phosphate and polyphosphate metabolism in Saccharomyces cerevisiae. Mol Biol Cell 18: 4438–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H, Pumplin N, Harrison MJ (2007) Phosphate in the arbuscular mycorrhizal symbiosis: transport properties and regulatory roles. Plant Cell Environ 30: 310–322 [DOI] [PubMed] [Google Scholar]
- Kant S, Peng M, Rothstein SJ (2011) Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H, Finnegan PM, Laliberté E, Pearse SJ, Ryan MH, Shane MW, Veneklaas EJ (2011) Update on phosphorus nutrition in Proteaceae. Phosphorus nutrition of proteaceae in severely phosphorus-impoverished soils: Are there lessons to be learned for future crops? Plant Physiol 156: 1058–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SI, Santi C, Jobet E, Lacut E, El Kholti N, Karlowski WM, Verdeil JL, Breitler JC, Périn C, Ko SS, et al. (2010) Complex regulation of two target genes encoding SPX-MFS proteins by rice miR827 in response to phosphate starvation. Plant Cell Physiol 51: 2119–2131 [DOI] [PubMed] [Google Scholar]
- Lin WY, Huang TK, Chiou TJ (2013) Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 25: 4061–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wang Z, Ren H, Shen C, Li Y, Ling HQ, Wu C, Lian X, Wu P (2010) OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J 62: 508–517 [DOI] [PubMed] [Google Scholar]
- Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ (2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q, Zhong Y, Wang Y, Wang Z, Zhang L, Shi J, Wu Z, Liu Y, Mao C, Yi K, et al. (2014) SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26: 1586–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P, Persson BL (1998) Identification, cloning and characterization of a derepressible Na+-coupled phosphate transporter in Saccharomyces cerevisiae. Mol Gen Genet 258: 628–638 [DOI] [PubMed] [Google Scholar]
- Martinoia E, Maeshima M, Neuhaus HE (2007) Vacuolar transporters and their essential role in plant metabolism. J Exp Bot 58: 83–102 [DOI] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC (2011) Phosphate import in plants: focus on the PHT1 transporters. Front Plant Sci 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri L, Picault N, Arrigoni R, Besin E, Palmieri F, Hodges M (2008) Molecular identification of three Arabidopsis thaliana mitochondrial dicarboxylate carrier isoforms: organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. Biochem J 410: 621–629 [DOI] [PubMed] [Google Scholar]
- Park BS, Seo JS, Chua NH (2014) NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26: 454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Hannam C, Gu H, Bi YM, Rothstein SJ (2007) A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J 50: 320–337 [DOI] [PubMed] [Google Scholar]
- Pratt J, Boisson AM, Gout E, Bligny R, Douce R, Aubert S (2009) Phosphate (Pi) starvation effect on the cytosolic Pi concentration and Pi exchanges across the tonoplast in plant cells: an in vivo 31P-nuclear magnetic resonance study using methylphosphonate as a Pi analog. Plant Physiol 151: 1646–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss CP, Huang CY, Gilliham M, Tyerman SD (2010) Channel-like characteristics of the low-affinity barley phosphate transporter PHT1;6 when expressed in Xenopus oocytes. Plant Physiol 152: 1431–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga MI, Mateos I, Charukesi R, Wang Z, Franco-Zorrilla JM, de Lorenzo L, Irigoyen ML, Masiero S, Bustos R, Rodríguez J, et al. (2014) SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc Natl Acad Sci USA 111: 14947–14952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Wang C, Arpat BA, Wang Z, Poirier Y, Tyerman SD, Wu P, Shou H, Whelan J (2012a) The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol 193: 842–851 [DOI] [PubMed] [Google Scholar]
- Secco D, Wang C, Shou H, Whelan J (2012b) Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett 586: 289–295 [DOI] [PubMed] [Google Scholar]
- Shen J, Zeng Y, Zhuang X, Sun L, Yao X, Pimpl P, Jiang L (2013) Organelle pH in the Arabidopsis endomembrane system. Mol Plant 6: 1419–1437 [DOI] [PubMed] [Google Scholar]
- Stefanovic A, Arpat AB, Bligny R, Gout E, Vidoudez C, Bensimon M, Poirier Y (2011) Over-expression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J 66: 689–699 [DOI] [PubMed] [Google Scholar]
- Versaw WK, Harrison MJ (2002) A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14: 1751–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Huang W, Ying Y, Li S, Secco D, Tyerman S, Whelan J, Shou H (2012) Functional characterization of the rice SPX-MFS family reveals a key role of OsSPX-MFS1 in controlling phosphate homeostasis in leaves. New Phytol 196: 139–148 [DOI] [PubMed] [Google Scholar]
- Wang C, Ying S, Huang H, Li K, Wu P, Shou H (2009) Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J 57: 895–904 [DOI] [PubMed] [Google Scholar]
- Wang Z, Ruan W, Shi J, Zhang L, Xiang D, Yang C, Li C, Wu Z, Liu Y, Yu Y, et al. (2014) Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc Natl Acad Sci USA 111: 14953–14958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Shou H, Xu G, Lian X (2013) Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr Opin Plant Biol 16: 205–212 [DOI] [PubMed] [Google Scholar]
- Wykoff DD, O’Shea EK (2001) Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159: 1491–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.