A microRNA regulates laccase transcription to affect the secondary growth of vascular tissues under the control of the transcription factor SQUAMOSA PROMOTER BINDING PROTEIN LIKE7.
Abstract
MicroRNAs (miRNAs) are endogenous small RNAs that repress target gene expression posttranscriptionally, and are critically involved in various developmental processes and responses to environmental stresses in eukaryotes. MiRNA857 is not widely distributed in plants and is encoded by a single gene, AtMIR857, in Arabidopsis (Arabidopsis thaliana). The functions of miR857 and its mechanisms in regulating plant growth and development are still unclear. Here, by means of genetic analysis coupled with cytological studies, we investigated the expression pattern and regulation mechanism of miR857 and its biological functions in Arabidopsis development. We found that miR857 regulates its target gene, Arabidopsis LACCASE7, at the transcriptional level, thereby reducing laccase activity. Using stimulated Raman scattering and x-ray microtomography three-dimensional analyses, we showed that miR857 was involved in the regulation of lignin content and consequently morphogenesis of the secondary xylem. In addition, miR857 was activated by SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7 in response to low copper conditions. Collectively, these findings demonstrate the role of miR857 in the regulation of secondary growth of vascular tissues in Arabidopsis and reveal a unique control mechanism for secondary growth based on the miR857 expression in response to copper deficiency.
MicroRNAs (miRNAs), RNAs of 20 to 22 nucleotides that frequently down-regulate gene expression posttranscriptionally in eukaryotic organisms ranging from animals to plants, are derived from imperfectly paired hairpin precursors (Bartel, 2004; Jones-Rhoades and Bartel, 2004). By binding to transcripts to which they are complementary, miRNAs either trigger mRNA degradation or inhibit their translation. The roles of many miRNAs in plants have been widely confirmed by knockout or overexpression studies and by characterization of their target genes (Guo et al., 2005; Vidal et al., 2010; Yoon et al., 2010). Increasing evidence has revealed that miRNAs play important roles in diverse processes in plant growth and development (Dugas and Bartel, 2004; Chen, 2005; Jover-Gil et al., 2005; Kidner and Martienssen, 2005; Zhang et al., 2006). Due to their biological significance in regulating gene expression, miRNAs have received considerable attention (Krol et al., 2010). In contrast to numerous reports demonstrating their roles in target gene expression, knowledge regarding the regulation of miRNAs is limited.
Secondary growth, one of the most important biological processes in woody plants, results from cell division in the cambia, or lateral meristems. On the xylem side of the cambium, the cells first undergo stages of differentiation that involve cell division, expansion, maturation, lignification, secondary cell wall thickening, and programmed cell death (Chaffey, 1999). However, secondary growth is still poorly defined, mainly because of the inherent problems of tree species as scientific subjects: long generation time, large size, and lack of genetically pure lines (Zhao et al., 2000). Recently, Arabidopsis (Arabidopsis thaliana), the most well-studied herbaceous model species, was used as a model to investigate secondary growth and fiber production. Arabidopsis has been reported to produce a significant quantity of secondary xylem sufficient for various developmental studies (Lev-Yadun, 1994; Beers and Zhao, 2001). Although putative genes and mechanisms regulating cell differentiation were described in this species, less was known about its regulation of miRNA during secondary growth, especially during lignification and secondary cell wall thickening in the past years. Recently, two new findings highlight the roles of miR397a and miR397b in lignin biosynthesis in poplar (Populus trichocarpa) and Arabidopsis, respectively (Lu et al., 2013; Wang et al., 2014). Moreover, recent experiments have shown that miRNA165/6 is required to maintain a stable bisymmetric vascular pattern (Muraro et al., 2014).
Lignin, the second most abundant terrestrial biopolymer after cellulose, is a complex of phenolic polymers that consist principally of guaiacyl (G) and syringyl (S) units, together with trace amounts of p-hydroxyphenyl (H) units. Lignin plays a crucial role in the structural integrity of the cell wall and the stiffness and strength of the stem (Bao et al., 1993; Chabannes et al., 2001). Lignin is formed through dehydrogenative polymerization of monolignols (Christensen et al., 2001), and lignification is the process of lignin deposition in the cell wall. Peroxidase and laccases are known to be involved in the final step of lignin biosynthesis through the oxidative polymerization of monolignols in lignifying cells (Bao et al., 1993; Dittmer et al., 2009). Monolignols are synthesized in the cytosol and transported to the cell wall, where their oxidation produces lignins (Vanholme et al., 2008). In addition, laccases have been speculated to have roles in lignifications in Arabidopsis (Turlapati et al., 2011), and genetic evidence on the involvement of several laccases in lignin biosynthesis has been reported recently (Berthet et al., 2011; Zhao et al., 2013). Arabidopsis contains 17 laccases with some predicted to be the targets of miRNAs. It is noteworthy that Arabidopsis LACCASE7 (AtLAC7) was identified to be the target of miR857, which could just regulate this one laccase among the laccase members (Fahlgren et al., 2007; Abdel-Ghany and Pilon, 2008). MiR857 might be unique for regulating the laccase in lignification.
In this study, we examined the effects of miR857 overexpression and knockdown on secondary growth in Arabidopsis. We found that miR857 is specifically expressed in the vascular tissues of seedlings, and is involved in regulating the morphogenesis of the secondary xylem. Stimulated Raman scattering (SRS) analyses and x-ray microtomography three-dimensional (3D-μCT) imaging showed that miR857 affects the lignin content in the cell wall of the secondary xylem in Arabidopsis. Furthermore, using electrophoretic mobility shift assay (EMSA) analyses, we revealed that SQUAMOSA promoter-binding protein like7 (SPL7) binds to specific motifs in the AtMIR857 promoter. The induction of miR857 in response to low copper (Cu) was blocked by the spl7 mutation. Taken together, these results provide new insight into the essential role of miR857 in regulating vascular tissue morphogenesis in Arabidopsis and lay a foundation for further investigation of the mechanisms of secondary growth in plants.
RESULTS
Expression pattern of MIR857 in Arabidopsis
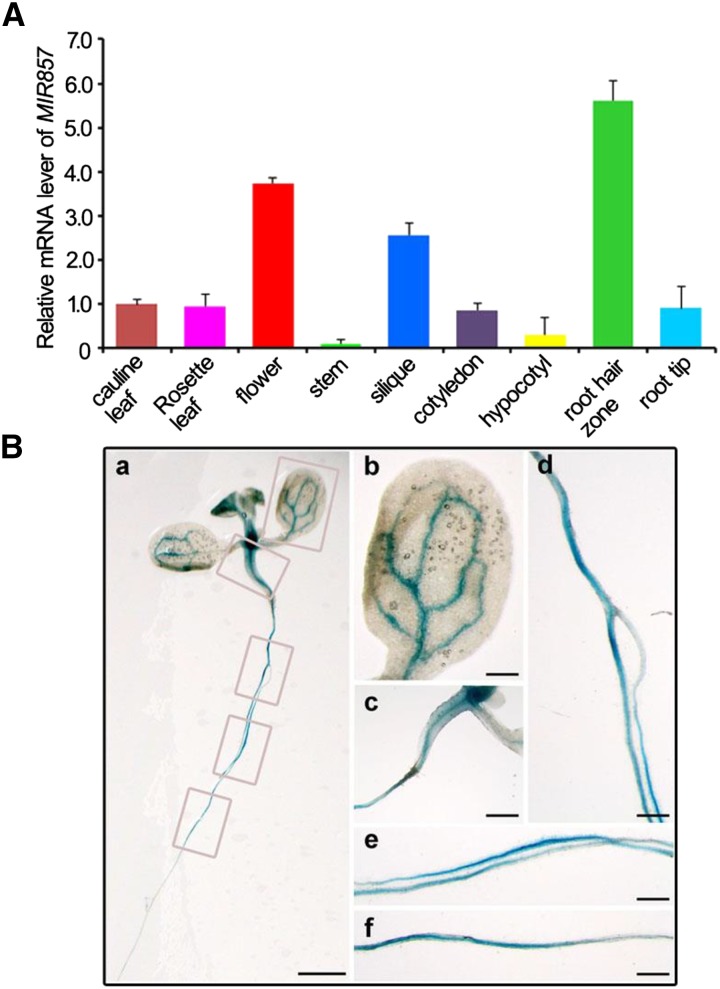
To investigate the evolutionary distribution of miR857 sequences, we performed a homology search in other species. We found miR857 sequences in only three plant species: Arabidopsis, Arabidopsis lyrata, and Citrus sinensis; the sequences share quite high homology (Supplemental Fig. S1). In Arabidopsis, miR857 is encoded by a single gene and has a single genomic location on chromosome 4 (Reinhart et al., 2002; Fahlgren et al., 2007). We examined the expression profile of miR857 using quantitative real-time (qRT)-PCR. MiR857 was readily detectable in both seedlings and 6-week-old plants. Expression was detected in the various organ types, including cotyledon, hypocotyls, root hair zoon, and root tip (7-d-old seedlings), and stem, flower, silique, rosette leaves, and cauline leaves (6-week-old plants) with high expression in flowers and siliques and low expression in stems (Fig. 1A). To confirm the tissue-specific expression of miR857, we generated transgenic plants with a reporter construct containing the GUS gene fused to the native promoter region located 1,259 bp upstream of the miR857 transcription start site (pMIR857::GUS). We found that the AtMIR857 promoter was active mainly in the vascular tissues of the pMIR857::GUS seedlings (Fig. 1B). In summary, these results suggest that miR857 shows limited evolutionary conservation in plants; the expression pattern of MIR857 has space and time specialty in Arabidopsis and is specifically expressed in the vascular tissues of seedlings.
Figure 1.
Expression pattern of AtMIR857. A, Tissue pattern of MIR857 transcript accumulation. Total RNA was isolated from various tissues of 1- and 6-week-old wild-type plants grown under long-day growth conditions. Real-time reverse transcription (RT)-PCR results were normalized to the expression of tubulin. Error bars represent the se of three independent experiments. B, pMIR857::GUS expression pattern in seedling tissues. Staining was prominent in the vascular tissues of leaves (a and b) and hypocotyls (a and c). Staining was also visible in the root vascular system (a, d–f). Bars = 5 mm (a), 1 mm (b–f).
Overexpression of miR857 Downregulates AtLAC7 Expression
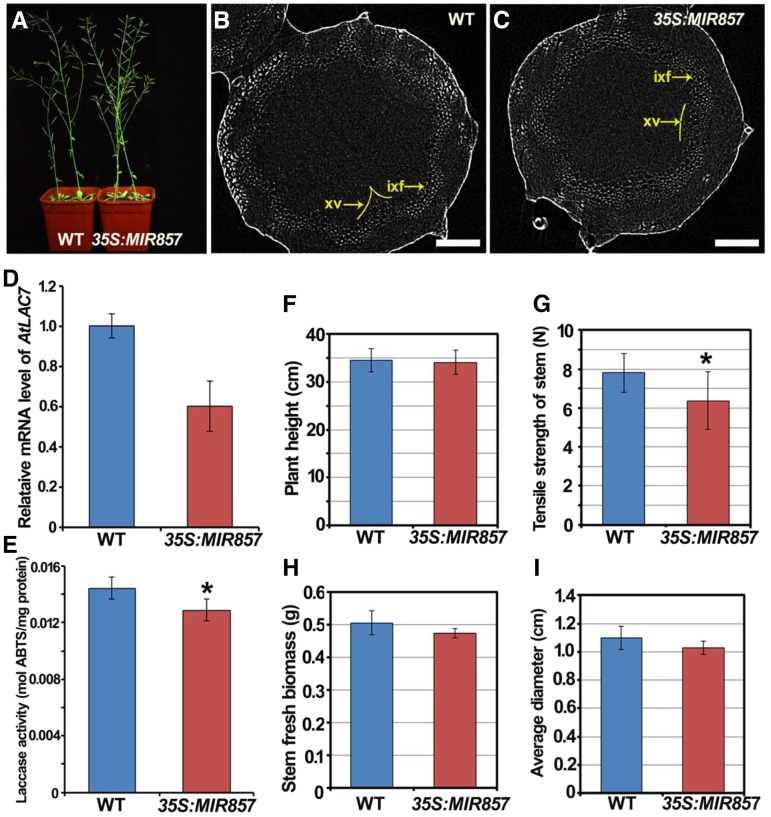
To investigate the function of miR857 in Arabidopsis, we first generated transgenic plants overexpressing premiR857 under the control of the 35S promoter. Real-time PCR analysis showed that the expression of miR857 in two transgenic lines (35S:MIR857-2 and 35S:MIR857-9) was higher than in the wild type. Moreover, the miR857 expression in the 35S:MIR857-9 line was higher than in the 35S:MIR857-2 line (Supplemental Fig. S2). Therefore, we focused on 35S:MIR857-9 as miR857-overexpression (OX) plants to carry out further analyses. Next, using qRT-PCR, we measured the abundance of the target gene AtLAC7 (AT3G09220) to examine whether the AtLAC7 transcript is downregulated in miR857-OX plants. We found that the expression of AtLAC7 was significantly lower in the miR857-OX plants than in the wild type (Fig. 2D). In addition, 5′-RACE analysis was performed to detect cleavage events at predicted sites in AtLAC7 mRNAs. We found that the AtLAC7 could be validated as a target since cleavage products were observed close to the nucleotide that is opposite nucleotide 10 of miR857, counting from the 5′ end (Supplemental Fig. S3). To determine whether a corresponding decrease in laccase activity occurred when the AtLAC7 transcript was downregulated, we evaluated the laccase activity in protein extracts isolated from stems harvested after 6 weeks of growth. The results showed that the laccase activity in the miR857-OX plants was 10.8% lower than that in the wild type (Fig. 2E).
Figure 2.
Phenotypic analysis of 35S:MIR857 plants. A, Six-week-old plants of the wild type (WT) and 35S:MIR857. B and C, 3D-μCT image of stem cross sections of wild-type and 35S:MIR857 plants. ixf, Interfascicular xylem fibers; xv, xylem. Scale bar = 100 μm. D,) Relative AtLAC7 mRNA levels in wild-type and 35S:MIR857 plants as measured by qRT-PCR. E, Quantification of laccase activity in partially purified protein extracts, with 2,2′-azinobis-3-ethylbenzthiazolinesulfonic acid (ABTS) as the substrate. Data represent means ± sd (n = 6). F to I, Quantitative analysis of plant height, tensile strength of the stem, stem fresh biomass, and average stem diameter in 6-week-old wild type and 35S:MIR857. Data represent means ± sd (n = 17). *, 0.01 < P < 0.05.
After 6 weeks of growth under long-day conditions, the size and shape of miR857-OX plants was equivalent to those of the wild type (Fig. 2A). Given the pMIR857::GUS staining pattern in vascular tissues, we measured parameters related to vascular development, including the average plant height, stem tensile strength, fresh weight, and stem diameter. Statistical analyses revealed that in the miR857-OX plants, the tensile strength of the stem was 26.4% lower than that of the wild type, and the average diameter and fresh weight were 6.5% and 18.4%, respectively, lower than those of the wild type (Fig. 2, F–I). Moreover, x-ray 3D-μCT analyses of stem cross sections showed that the cell layers in the secondary xylem were remarkably smaller in the overexpressing plants as compared with the wild type (Fig. 2, B and C). These results indicate that the overaccumulation of miR857 leads to the down-regulation of its target gene AtLAC7 and reduced secondary growth of vascular tissues.
Knocking Down miR857 Results in Increased AtLAC7 Expression
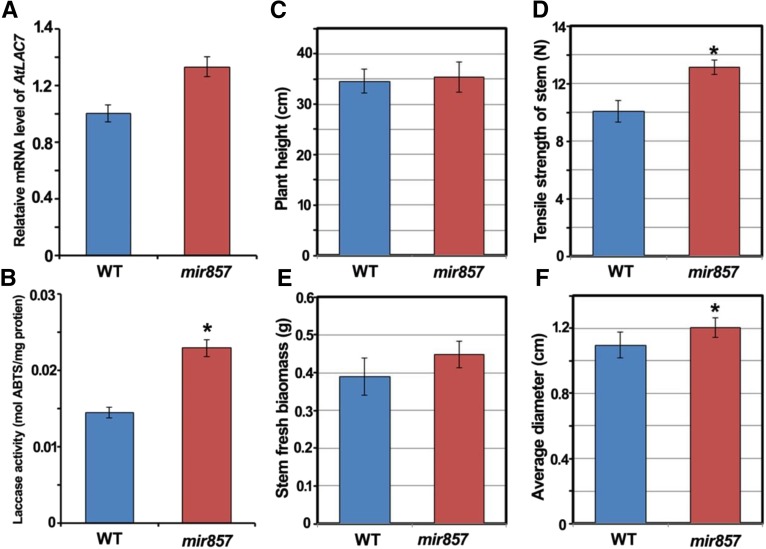
To evaluate the effects of reduced miR857 expression on vascular development, we obtained an miR857 heterozygous transfer DNA (T-DNA) insertion mutant (SALK_072720) from the Arabidopsis Biological Resource Center and screened for a homozygous line (Supplemental Fig. S4). RT-PCR analysis showed that the miR857 transcript level was dramatically lower in the homozygous line than in the wild type (Supplemental Fig. S4). In addition, the level of transcripts of the target gene AtLAC7 was higher in the mir857 insertion mutant than in the wild type (Fig. 3A), and the total laccase activity in the mir857 mutant was markedly higher (33.3%) than in the wild type (Fig. 3B). The qRT-PCR analysis showed that there were no obvious differences in the expression of the other 16 laccase members among the wild type, the overexpression plants, and mir857 mutants (Supplemental Fig. S5).
Figure 3.
Phenotypic analysis of mir857 plants. A, Relative AtLAC7 mRNA levels in the wild type (WT) and the mir857 mutant as measured by qRT-PCR. B, Quantification of laccase activity in partially purified protein extracts, with ABTS as the substrate. Data represent means ± sd (n = 6). C to F, Quantitative analysis of plant height, tensile strength of the stem, stem fresh biomass, and average diameter in the 6-week-old wild-type and mir857 plants. Data represent means ± sd (n = 16). *, 0.01 < P < 0.05.
Since T-DNA insertion in SALK_072720 may also influence MIR397expression, we carried out experiments to detect the expression level of MIR397b and its target gene LAC4 through qRT-PCR analysis in the mir857 mutant. The data showed that the expression level of MIR397b was reduced, and accordingly, LAC4 was slightly up-regulated, but not obviously, as shown in Supplemental Figure S6. However, there is no significant difference between the down-regulation of LAC4 induced by miR397 and the down-regulation of LAC7 induced by miR857. Additionally, we also tested the expression of mature miR857 in the wild type, miR857-OX plants, and mir857 T-DNA insertion mutants using qRT-PCR (Supplemental Fig. S7). The results showed that the expression of mature miR857 increased in miR857-OX plants compared with the wild type, whereas it was reduced in mir857 mutants.
Notably, the mir857 mutant showed high levels of the measured parameters related to vascular development. Further statistical analysis revealed that in the mir857 mutant, the tensile strength of the stem was 26.4% higher than that in the wild type, and the average diameter and fresh weight were 10% and 14.7% higher, respectively (Fig. 3, C–F). These data suggest that the loss of function of miR857 leads to the up-regulation of its target gene AtLAC7 and improves the development of vascular tissues.
MiR857 Is Involved in the Regulation of Secondary Xylem Morphogenesis
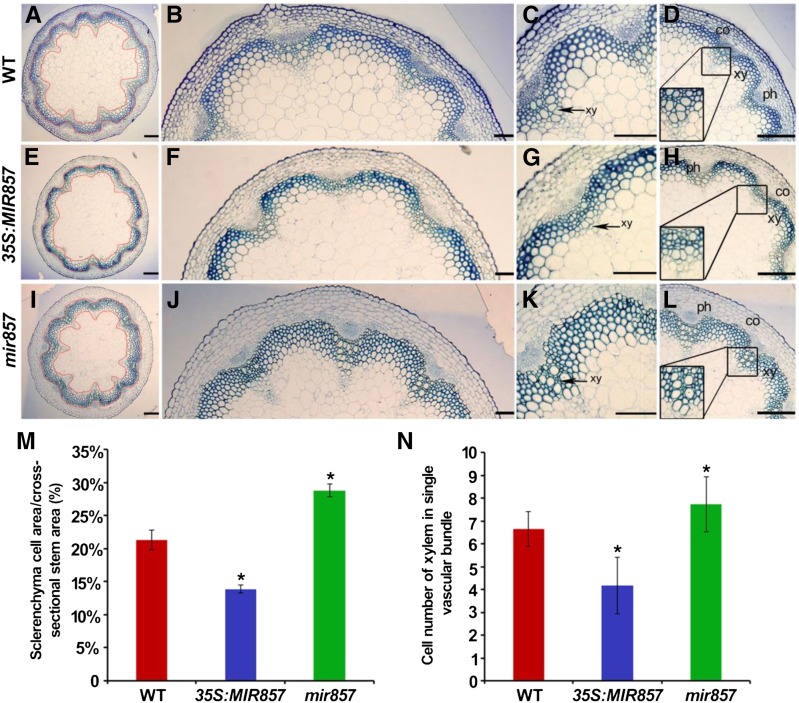
To further examine the roles of miR857 in Arabidopsis vascular development, we prepared semithin cross sections to investigate the anatomical features of inflorescence stems. As shown in Figure 4, the region containing sclerenchyma cells, including interfascicular fibers and vascular bundles, was thinner in stems of the miR857-OX plants than in wild-type stems (Fig. 4, A–D and E–H, respectively). In contrast, the mir857 mutant plants exhibited a thicker region of sclerenchyma cells than the wild type (Fig. 4, I–L). To compare these differences quantitatively, the area containing sclerenchyma cells was divided by the entire area of inflorescence stem in the cross section. The ratios were 21.3% in the wild type, 13.9% in miR857-OX plants, and 28.9% in the mir857 mutant (Fig. 4M). Next, we counted the numbers of secondary xylem cells in the inflorescence stem. In the miR857-OX plants, the cells in the secondary xylem were arranged more loosely, and 37.2% fewer cells were observed than in the wild type (Fig. 4N). However, in mir857 mutant plants, the cells in the secondary xylem were arranged tightly, with 16.7% more cells than in the wild type (Fig. 4N). We also examined the anatomical structure and cell numbers in roots of these lines. As shown in Supplemental Figure S8, the miR857-OX plants exhibited 16.7% fewer cells in the secondary xylem than in the wild type (Supplemental Fig. S8), whereas the mir857 mutant plants showed 5.0% more cells (Supplemental Fig. S8). In addition, the average cell diameter of the secondary xylem in overexpressing plants was only 87.3% of that in the wild type, whereas the corresponding diameter in the mir857 mutant was 1.5 times larger than in the wild type (Supplemental Fig. S8).
Figure 4.
Anatomical structures of stems of the wild type (WT), 35S:MIR857, and the mir857 mutant. A to L, Cross sections of 6-week-old stems of wild-type, 35S:MIR857, and mir857 mutant plants. co, Cortex; ph, phloem; xy, xylem. Scale bar = 100 μm (A, E, and I), and 50 μm (B–D, F–H, and J–L). The area within the red-yellow circles corresponds to the region containing sclerenchyma cells in A, E, and I. M, Sclerenchyma development was impaired in 35S:MIR857 and mir857 plants. The definition of the ratio was used to evaluate the development of sclerenchyma. The ratios of the different genotypes are plotted. Data represent means ± sd (n = 10). *, 0.01 < P < 0.05. N, Xylem cell numbers in single vascular bundles of inflorescence stems. Data represent means ± sd (n = 15).
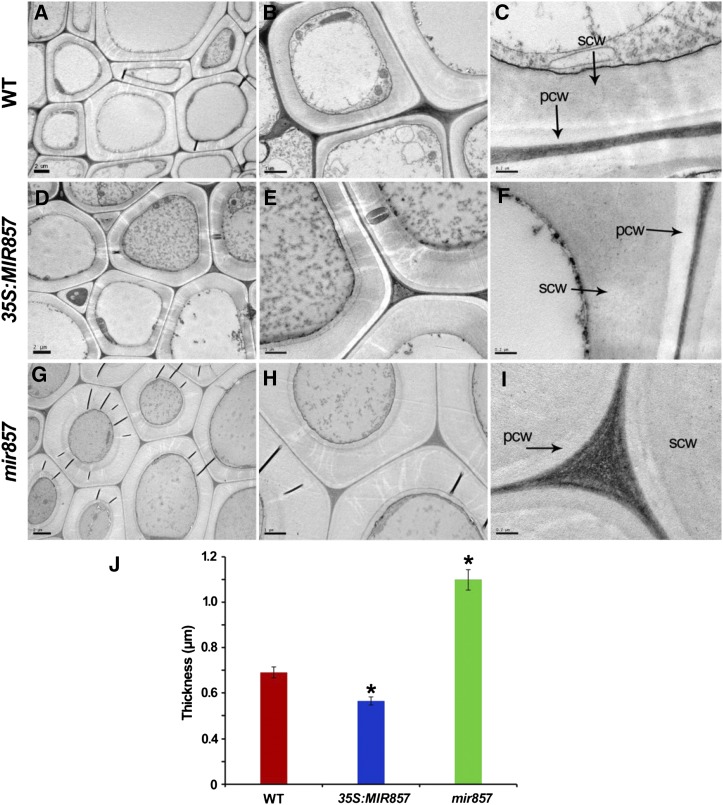
Furthermore, we examined whether miR857 affects the synthesis of secondary cell walls, which are the dominant feature of xylem cells. An obvious difference was detected between the wild type (Fig. 5, A–C) and the mir857 mutant (Fig. 5, G–I) when the secondary cell wall was visualized directly by transmission electron microscopy (TEM). To quantify this difference, we measured the cell wall thickness of secondary xylem cells by using Image-Pro Plus software (Media Cybernetics). The secondary cell wall in the stems of mir857 mutants was found to be 1.8 times thicker than in the wild type (Fig. 5J), whereas the thickness of secondary cell wall was decreased in overexpressing plants compared with the wild type (Fig. 5, D–F and J, respectively). In addition, MIR857 expression in the mir857 mutants rescued their phenotypes, regarding the expression level of MIR857 mRNA and the structure of secondary xylem (Supplemental Fig. S9). These results indicate that miR857 participates in the regulation of morphogenesis of the secondary xylem in Arabidopsis.
Figure 5.
Micrographs showing the secondary cell wall of stems in wild-type (WT), 35S:MIR857, and mir857 mutant plants. A to I, Stem cross sections of 6-week-old wild-type, 35S:MIR857, and mir857 mutant plants. pcw, Primary cell wall; scw, secondary cell wall. Scale bar = 2 μm (A, D, and G); 1 μm (B, E, and H); and 0.1 μm (C, F, and H). J, Thickness of secondary cell wall of secondary xylem in stems of wild-type, 35S:MIR857, and mir857 plants. Data represent means ± sd (n = 46 cells). *, 0.01 < P < 0.05.
MiR857 Is Involved in the Regulation of Lignin Content
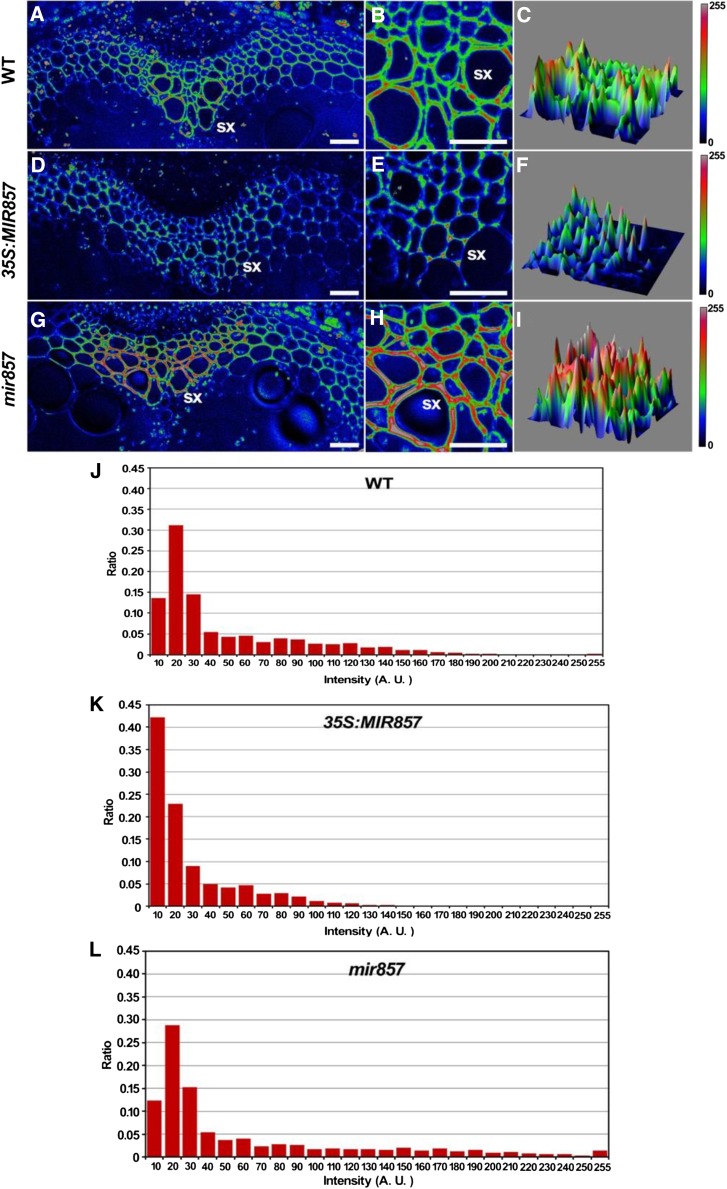
Given that miR857 is involved in the regulation of morphogenesis of the Arabidopsis secondary xylem, we next examined the content and distribution of lignin to gain further insight into the role of miR857 in secondary growth. Using Fourier transform infrared (FTIR) spectroscopy, we found that the lignin content in secondary xylem cells was higher in the mir857 mutant and lower in overexpressing plants than in the wild type (Supplemental Fig. S10). In addition, we used SRS analysis, a label-free microscopic imaging system, to analyze the lignin distribution in stems (Fig. 6). The fluorescence intensity of the cell wall in the secondary xylem in overexpressing plants (Fig. 6, D–F) was clearly lower than in the wild type (Fig. 6, A–C). Overexpressing plants showed an intensity profile between 10 and 120 (Fig. 6K), whereas the intensity profile of the wild type was between 0 and 180 (Fig. 6J), demonstrating decreased lignin deposition in overexpressing plants. In contrast, the fluorescence intensity in the mir857 mutant was remarkably enhanced (Fig. 6, G–I), with an intensity profile between 10 and 255 (Fig. 6L), indicating increased lignin deposition. We also examined the lignin distribution in roots (Supplemental Fig. S11). Consistent with the results in stems, the fluorescence intensity of cell walls in the secondary xylem in roots of overexpressing plants was lower than in the wild type, whereas that in mir857 mutants was enhanced, but not significantly.
Figure 6.
SRS chemical images of lignin in stems. A to I, SRS images of stem cross sections based on the deconvoluted lignin band at 1,600 cm–1 in 6-week-old wild-type (WT), 35S:MIR857, and mir857 mutant plants. sx, Secondary xylem. Scale bar = 50 μm. J to L, Distribution of fluorescence intensity of the SRS images in A to I in stems of the wild type, 35S:MIR857, and mir857.
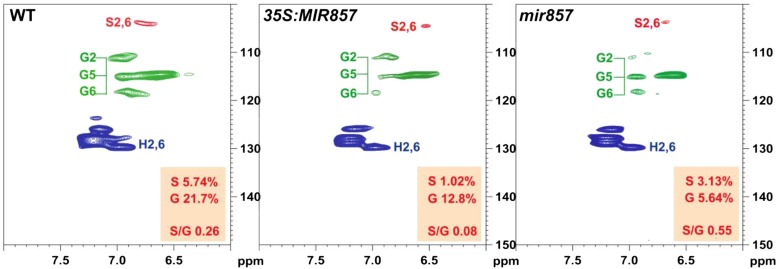
To further compare lignin composition in the wild type, overexpression plants, and mir857 mutants, lignin was extracted and subjected to NMR spectroscopy. Analysis of 13C–1H heteronuclear singular quantum correlation (HSQC) spectra revealed similar carbon-proton correlation signal patterns in both the aromatic and aliphatic regions of the lignin from the wild type, overexpression plants, and mir857 mutants, but was indicative of significant changes in lignin contents (Fig. 7). The aromatic carbons in S and G units were readily observed with the presence of their diagnostic correlation signals around 103.8/6.67 (S2/6), 110.5/6.94 (G2), 114.8/6.88 (G5), and 118.6/6.81 (G6) ppm, respectively. The semiquantitative analysis of subunit contours in HSQC spectra showed that the lignin in mir857 mutants appeared to have a higher relative abundance of S units and S/G, whereas overexpression plants appeared to have a lower abundance of S units and S/G (Fig. 7).
Figure 7.
Two-dimensional NMR spectra revealing lignin unit compositions. Partial short-range 13C-1H (HSQC) correlation spectra (aromatic regions only) of cell wall gels in 4:1 (v/v) dimethyl sulfoxide (DMSO)-d6/pyridine-d5 from 6-week-old plants of the wild type (WT), 35S:MIR857, and mir857 mutants.
Furthermore, to examine the trend in accumulation of lignin content and secondary wall deposition, in various tissues of mutant lines and the wild type, we measured the lignin content in stem, cauline leaves, rosette leaves, and root at stages 4, 5, and 7 weeks (Supplemental Fig. S12). These results indicated that the accumulation trends of lignin content in stem is consistent within other tissues, such as caulinar leaves, rosette leaves, and roots.
We then evaluated whether ectopic expression of miR857 affected the expression of other enzymes in the lignin synthesis pathway. A qRT-PCR analysis showed that overexpression of miR857 did not affect expression of the cellulose synthase genes CesA7 and CesA8 or the lignin biosynthetic genes 4CL1 (for hydroxycinnamate CoA ligase), CCoAOMT (for caffeoyl CoA O-methyltransferase) CAD (cinnamyl alcohol dehydrogenase), HCT (hydroxycinnamoyl CoA:shikimate/quinate hydroxycinnamoyl transferase), 4CL1 (4-coumarate CoA ligase), CCR1 (Cinnamoyl-CoA reductase) and F5H2 (ferulic acid 5-hydroxylase; Supplemental Fig. S13).
MiR857 Is Activated by SPL7 in Response to Low Cu Conditions
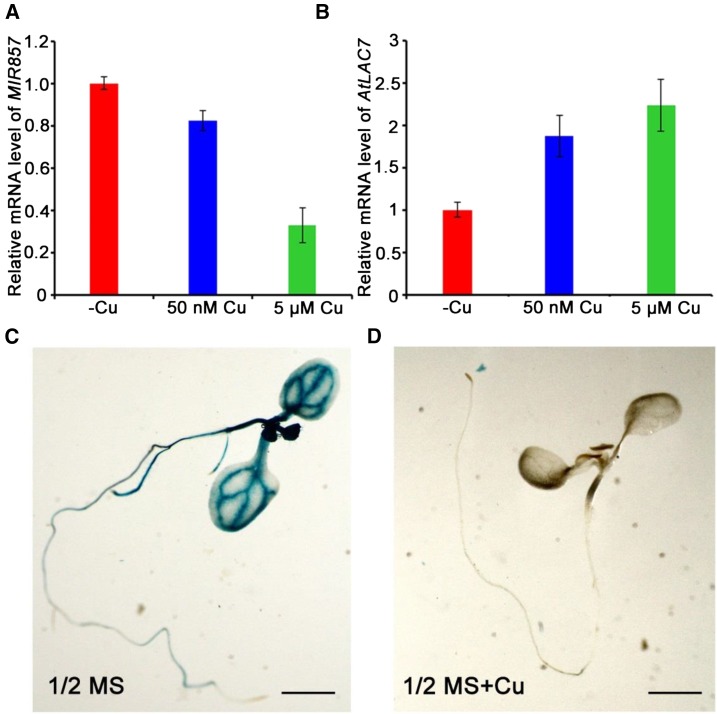
We next studied the regulation of miR857 at the seedling stage to gain further insights into the mechanisms of its roles. Sequence analysis revealed an array of seven GTAC motifs, the core sequence of Cu-response elements, in the proximal promoter regions of AtMIR857. To examine whether miR857 expression is induced by varying Cu levels in Arabidopsis seedlings, we performed real-time PCR analyses on plants treated with increasing Cu concentrations. The results showed that the expression of miR857 was repressed by Cu, whereas the expression of the target gene AtLAC7 increased (Fig. 8, A and B). We also analyzed the pMIR857::GUS transgenic plants to test whether miR857 accumulation is controlled at the transcriptional level. Strong GUS activity throughout the seedlings was observed under lower Cu supplementation (plants grown in one-half-strength MS medium), whereas treatment with higher Cu (plants grown in one-half-strength MS with added Cu) effectively prevented the accumulating miR857 in seedlings (Fig. 8, C and D). These results indicate that the AtMIR857 promoter is induced under low-Cu conditions, and transcriptional control is important for proper AtMIR857 expression in response to varying Cu levels.
Figure 8.
AtMIR857 responds to Cu2+ at the transcriptional level. qRT-PCR expression levels of the miR857 precursor (A) and its target LAC7 (B) in roots grown under deficient (–Cu), limited (50 nm), or sufficient (5 μm) Cu conditions. C and D, GUS staining of transgenic seedlings expressing the pMIR857::GUS reporter gene in the wild-type background (pMIR408::GUS/wild type). Seedlings were grown in one-half-strength Murashige and Skoog (MS) medium (C) or one-half-strength MS medium containing supplemental Cu (D) as indicated. Scale bar = 5 mm.
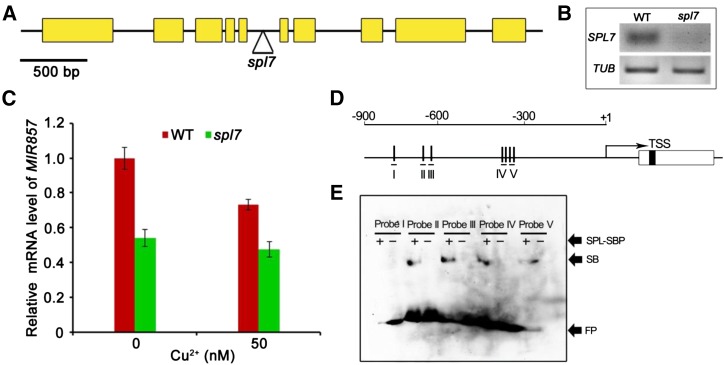
The Arabidopsis gene SPL7 is homologous to copper-response regulator (CRR1) of Chlamydomonas spp., which is specifically activated under Cu deficiency and may bind GTAC motifs (Kropat et al., 2005). To genetically test whether SPL7 is required for AtMIR857 expression, we used an spl7 null mutant containing a T-DNA inserted into the SPL7 locus (Fig. 9, A and B; Yamasaki et al., 2009). Using real-time PCR analyses, we examined the transcription levels of miR857 in spl7 and wild-type plants grown on medium containing 0.1 or 5 μm CuSO4. Little difference was observed between spl7 seedlings grown under low (0.1 μm) and high (5 μm) Cu concentrations, indicating that the induction of miR857 in response to low Cu was blocked by the spl7 mutation (Fig. 9C).
Figure 9.
SPL7 directly regulates miR857 expression. A, SPL7 gene structure. Yellow boxes and bars represent exons and introns, respectively. The triangle indicates the T-DNA insertion site. B, Semiquantitative RT-PCR analysis of SPL7 expression in the wild type (WT) and spl7. β-TUBULIN (TUB) was used as a control. C, Relative levels of miR857 mRNA in wild-type and spl7 plants grown in medium containing 0 or 50 nm Cu2+. Data represent means ± sd. *, 0.01 < P < 0.05. D, Schematic presentation of the MIR857 proximal promoter region in Arabidopsis. The annotated premiRNA is depicted as an open horizontal bar in which the position of the mature miRNA is shaded. The transcription start site (TSS; +1) was determined by mapping the full-length complementary DNA (cDNA) sequence to the genome. The vertical lines indicate the positions of the GTAC motifs. The positions of the probes used in the subsequent EMSA are depicted as short horizontal lines. E, EMSA of the recombinant SBP domain of SPL7. The ability of SPL7-SBP to retard the migration of five GTAC-containing regions of the MIR857 promoter (probes I–V) was tested. +, Labeled probe incubated with SBP; –, excess unlabeled probe added; FP, free probe; SB, shifted band.
Additionally, we examined whether SPL7 directly activates the expression of miR857. First, we performed an EMSA to detect any interaction between SPL7 and the AtMIR857 promoter. Based on the seven GTAC motifs in the upstream region (–900 to –1 bp) of the AtMIR857 promoter region, we designed five probes (Fig. 9D): three probes containing one GTAC (probes I, II, and III) and two probes containing two GTAC motifs (probes IV and V). The N-terminal portion of SPL7, which contains a conserved DNA binding domain (SQUAMOSA promoter binding protein [SBP]), was fused to a sequence encoding a His tag. The resulting recombinant SPL7-SBP protein purified from Escherichia coli was used in EMSA. The results showed that electrophoresis of the biotin-labeled probes II, III, IV, and V, but not probe I, was retarded by the binding of the SPL7-SBP recombinant protein (Fig. 9E). Notably, probes IV and V both affected the migration even though each contains only one GTAC motif. These results demonstrate that SPL7 directly interacts with AtMIR857, mainly through binding to the GTAC motifs in the proximal promoter region (–553 to –317 bp). Taken together, these results demonstrate that SPL7 binds to the GTAC motifs in the AtMIR857 promoter in response to Cu deficiency.
DISCUSSION
Precise temporal and spatial gene expression is essential during plant development. Previous studies have shown that miRNAs play indispensable roles in different gene regulatory networks (Shalgi et al., 2007; Yu et al., 2008; Re et al., 2009). Some miRNAs only exist in one or two species, suggesting that these pathways represent unique mechanisms for controlling gene expression. MiRNA857 is not widely distributed in plants and is encoded by a single gene in Arabidopsis. In the current study, we showed that miR857 is readily detectable in both 7-d-old seedlings and 6-week-stage plants. A ubiquitous pattern of expression appeared in various organ types: seedling, stem, flower, silique, rosette leaves, and cauline leaves. Furthermore, GUS staining experiments revealed that the GUS signal was mainly present in vascular tissues of the young transgenic pMIR857::GUS seedlings. These results suggest that miR857 shows limited evolutionary conservation in plants; it is expressed throughout the life cycle of Arabidopsis and is specifically expressed in the vascular tissues of seedlings.
The Arabidopsis AtMIR408, AtMIR857, and AtMIR397 gene families have been shown to regulate a subset of AtLAC members (Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Fahlgren et al., 2007; Abdel-Ghany and Pilon, 2008). A recent study reported that the overaccumulation of miR408 leads to the down-regulation of its target genes LAC13 and ARPN (PLANTACYANIN) and the promotion of vegetative growth (Zhang and Li, 2013). In this study, we isolated miR857 from Arabidopsis and generated miR857-overexpressing plants. Real-time PCR experiments showed that the expression of AtLAC7 was significantly repressed in the overexpressing plants as compared with the wild type, whereas AtLAC7 transcripts accumulated to much higher levels in an mir857 mutant (SALK_072720). Since the T-DNA insertion in SALK_072720 is also in the exon of miR397, we further evaluated the effects of miR397 on the phenotype of this mutant. The qRT-PCR analysis showed that the expression level of miR397b was indeed reduced, and accordingly, LAC4 was up-regulated (Supplemental Fig. S11). However, lac4 mutants had growth and development characteristics similar to those of the wild-type line grown under long-day conditions (Berthet et al., 2011). Therefore, we considered that there might be a gene dosage effect related to the mir857 mutant phenotype. Although the possibility that miR397b may have some effects on the mutant phenotype that could not be excluded, miR857 might have an indispensible effect on the phenotype. In addition, the laccase activity in the overexpressing plants was lower than in the wild type, whereas the mir857 mutant showed obviously higher laccase activity. These results indicate that miR857 represses the expression of its target gene AtLAC7 posttranscriptionally, negatively regulating laccase activity.
Peroxidases and laccases are potentially involved in the oxidation of lignin precursors in plant cell walls (Tognolli et al., 2002). Analyses of the physiological functions of laccases involved in lignification in higher plants are scarce. Only a few studies have reported alterations to lignification in mutants or transgenic plants with impaired laccase gene expression. The seed coat of the AtLAC15-deficient transparent testa10 Arabidopsis mutant has been found to have lignin contents 30% lower than the wild type, as determined by a thioglycolic acid assay (Pourcel et al., 2005). Using the same method, Wang et al. (2008) reported 19.6% higher ultraviolet absorbance in the thioglycolic acid extract recovered from a transgenic poplar line overexpressing a cotton laccase gene as compared with the control (Wang et al., 2008). Both AtLAC4 and AtLAC17 contribute to the constitutive lignification of Arabidopsis stems (Berthet et al., 2011). In addition, the regulation of laccases at the transcriptional level has only rarely been reported.
Our findings are in agreement with those of previous reports. Our results showed that the average stem diameter and mechanical strength of mir857-overexpressing plants were significantly lower than in the wild type, whereas the average stem diameter and mechanical strength were higher in the mir857 mutant. x-ray 3D-μCT analyses showed that the cell layers in the secondary xylem were remarkably thinner in the stem cross sections of overexpressing plants. Thin cross-sectional specimens were prepared to quantify the anatomical variations in stems and roots. The area of sclerenchyma cells was found to be significantly lower in overexpressing plants than in the wild type, whereas that in the mir857 mutant plants was significantly higher. Moreover, in overexpressing plants, the cells in the secondary xylem were more loosely arranged, and fewer cells were observed than in the wild type. However, in mir857 mutant plants, the cells in the secondary xylem were arranged tightly and were higher in number. These results suggest that miR857 is involved in regulating morphogenesis of the secondary xylem in Arabidopsis by regulating the expression of AtLAC7.
Lignin polymerization and deposition are important factors in secondary growth in plants (Plomion et al., 2001). The activity of laccase is closely connected with the lignin content. Disruption of laccase genes in Arabidopsis results in tissue-specific alterations to lignification (Berthet et al., 2011). We found that the lignin content of secondary xylem cells was higher in the mir857 mutant but lower in overexpressing plants as compared with the wild type. In addition, SRS analysis showed that the fluorescence intensity of the cell wall in secondary xylem in overexpressing plants was clearly lower than in the wild type, indicating decreased lignin deposition. However, the fluorescence intensity in the mir857 mutant was remarkably enhanced due to increased lignin deposition, suggesting that mir857 affects the lignin content in the cell wall of the secondary xylem in Arabidopsis. In some sense, there is a potential dosage efficiency between the lignin content and the thickness of secondary cell wall (Goicoechea et al., 2005; Day et al., 2009). TEM showed that the secondary cell wall in stems of the mir857 mutant was 1.8 times thicker than in the wild type, whereas the secondary cell wall thickness in overexpressing plants was smaller than the wild type.
Previous investigations of Arabidopsis have revealed that Cu availability is a factor in miR857 regulation (Yamasaki et al., 2007; Abdel-Ghany and Pilon, 2008). Cu is one of the most important cofactors due to its transference of a valence state between Cu2+ and reduced Cu+ in plants (Burkhead et al., 2009; Palmer and Guerinot, 2009). Plastocyanin, which transfers electrons from the cytochrome b complex to the photosystem, is the most abundant Cu-containing protein in plants. The experiment described here showed that increasing Cu concentrations repressed the expression of miR857 but increased the transcript levels of the target gene AtLAC7. We also analyzed pMIR857::GUS transgenic plants to test whether miR857 accumulation is controlled at the transcriptional level. We found that strong GUS activity occurred throughout the seedling grown in medium containing lower Cu concentrations, indicating that the AtMIR857 promoter can be induced under low-Cu conditions.
One of the best-studied models of Cu homeostasis in plants is CRR1, a transcription factor that contains an SBP domain (Sommer et al., 2010). CRR1 is thought to be a Cu sensor and is activated under Cu-deficient conditions (Kropat et al., 2005). Active CRR1 binds to the GTAC motif in the promoter region of the target gene to further regulate its expression (Quinn and Merchant, 1995; Quinn et al., 1999; Kropat et al., 2005; Nagae et al., 2008; Yamasaki et al., 2009). As with CRR1, recent studies have revealed that the binding activity of SPL7 to the GTAC motif is inhibited by Cu2+ (Sommer et al., 2010). SPL7 is thus probably a Cu sensor in plants, in addition to CRR1. Given the involvement of miR857 in Cu homeostasis, that the expression of miR857 requires SPL7 is not surprising. Using EMSA, we found that SPL7 transcriptionally regulates miR857 expression by binding to the GTAC motifs in its promoter. Notably, the induction of miR857 in response to low Cu was blocked by the spl7 mutation. These results demonstrate that SPL7 binds to the GTAC motifs in the AtMIR857 promoter in response to Cu deficiency. AtMIR857 is activated by SPL7 in response to low-Cu conditions and is also involved in the maintenance of Cu homeostasis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used as the wild type, including all transformations. The lac7 (S_003265c) and mir857 (S_072720) mutants were obtained from the Salk Institute T-DNA insertion collection. Homozygous plants were identified using genomic PCR. A list of primers is provided in Supplemental Table S1. A homozygous mutant defective in the SPL7 gene was obtained from the Toshiharu Shikanai laboratory (Yamasaki et al., 2009). For soil-grown plants, seeds were cold stratified at 4°C for 2 d in darkness and subsequently grown in a chamber at 120 μmol photons m−2 s−1 light intensity under long-day conditions (16 h of light/8 h of dark) at 22°C. For young tissues (germinating seeds to 14-d-old seedlings), seeds were germinated on solid one-half-strength MS medium. For RT-PCR and microarray analyses, plants were arranged at random in the same growth chamber and grown under long-day conditions.
Plasmid Construction and Plant Transformation
AtMIR857 on chromosome 4 was PCR amplified from genomic DNA isolated from wild-type Arabidopsis ecotype Columbia. The amplified DNA was ligated downstream of the Cauliflower mosaic virus 35S promoter in the binary vector pBI121 (Clontech) and confirmed by sequencing. The Arabidopsis Information Resource database was used to retrieve promoter region sequence information for the AtMIR857 gene. The AtMIR857 promoter region was PCR amplified using the primers AtMIR857 pro-F and AtMIR857 pro-R for AtMIR857pro::GUS, inserted into pBI101 (Clontech), and confirmed by sequencing. The constructs were introduced into wild-type Arabidopsis plants (ecotype Columbia) by Agrobacterium tumefaciens-mediated transformation (Bechtold and Pelletier, 1998). Transgenic plants were selected using kanamycin, and third-generation plants were used for gene expression analysis and phenotypic characterization.
qRT-PCR
Total RNA was extracted from seedlings and plants using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. RT was performed using the First-Strand cDNA Synthesis Kit (Thermo Scientific) and an oligo(dT) primer. Specific primers were designed for AtMIR857 and AtLAC7 (see Supplemental Table S1). PCR was performed using the SYBR Green Mix (TaKaRa) in an optical 36-well plate with the Rotor-Gene 3000 system (Qiagen). In each reaction, 0.3 μm primer and 10 ng of cDNA were used. For each of three biological replicates, PCR was performed in triplicate. The initial denaturing time was 30 s, followed by 40 cycles at 95°C for 15 s, 58°C for 15 s, and 72°C for 15 s, with a final extension at 72°C for 10 min. A melting curve was run after the PCR cycles.
Measurement of the Breaking Force
The basal portion of inflorescence stems of 6-week-old plants of the wild type, miR857-overexpressing plants, and the mir857 mutant was used for measurements. The ends of each stem segment were clamped at the same distance between two clamps and torn apart at the same speed. The force required to break the samples was recorded by a microtester (Instron 55R1122; http://www.instron.us/wa/home/ default_en.aspx). Seventeen plants of each genotype were examined, and all samples were treated under identical conditions.
Characterization of Growth Phenotype
For adult plant phenotypes, the wild type, miR857-overexpressing plants, and the mir857 mutant were grown side by side in a growth chamber. Mature (6-week-old) plants were examined for the growth phenotype and photographed. To investigate the cellular architecture of the xylem, shoots and roots were fixed with 2.5% (w/v) glutaraldehyde in 0.1 m cacodylate buffer (pH 7.2) for 48 h at 4°C. The samples were then rinsed in 0.1 m cacodylate buffer, dehydrated in a graded ethanol series, infiltrated, and embedded in LR White resin (Electron Microscopy Sciences). The resin blocks were polymerized for 24 h at 60°C. Thick sections (1 mm) were cut using an ultramicrotome (Leica). The sections were stained with 1% (w/v) toluidine blue in 1% (w/v) borax, and images were taken using a Zeiss Axioplan II compound microscope with a Maximal Ratio Combiner digital camera and AxioVision software (Carl Zeiss).
EMSA
The sequence encoding the SBP domain of SPL7 was amplified by RT-PCR using the high-fidelity Pfusion DNA polymerase (New England Biolabs; http://www.neb.com) with the primers listed in Supplemental Table S1. The PCR products were inserted into the vector pET-28a (+) (Novagen, now EMD Millipore; http://www.emdmillipore.com) between the NcoI and XhoI restriction sites and sequenced. The resulting plasmid was introduced into the Escherichia coli strain BL-21 (Stratagene, now Agilent Technologies; http://www.genomics.agilent.com). Production of the SPL7-SBP protein fused to a His tag was induced by the addition of 1 mm isopropyl-β-d-thiogalactopyranoside to the medium followed by growth at 37°C. The recombinant proteins were purified with the Ni-NTA Agarose system (Qiagen).
Fragments of the AtMIR857 promoter containing the GTAC motifs were synthesized as pairs of complementary oligonucleotides and labeled with biotin using the second-generation Biotin Gel Shift Kit (Biyuntian; http://www.beyotime.com). In brief, the SBP domain protein was incubated together with biotin-labeled probes and various concentrations of competitor DNA in 20-μL reaction mixtures containing 20 mm HEPES (pH 7.6), 1 mm EDTA, 10 mm (NH4)2SO4, 1 mm dithiothreitol, 0.2% (w/v) Tween 20, 30 mm KCl, 0.05 µg μL−1 poly [d (I–C)], and 0.005 μg μL−1 poly-l-Lys for 15 min at room temperature (22°C–25°C). The samples were loaded onto 6% (w/v) polyacrylamide gels and run at 4°C at 0.8 V cm2 in one-half-strength Tris-borate/EDTA electrophoresis buffer (44.5 mm Tris, 44.5 mm boric acid, 1 mm EDTA). The electrophoresed DNA was blotted onto a Hybond+ nylon membrane (GE Healthcare Life Sciences; http://www.gelifesciences.com) and detected with a biotin-specific antibody. The probes used are listed in Supplemental Table S2.
Image-Pro Plus Analysis
Image-Pro Plus 6.0 (Media Cybernetics) software was used to quantify cell wall area for TEM image of 6-week-old plants. The cell wall area was subtracted from the total cell area. The software was performed according to the procedure as described previously (Liang et al., 2007).
FTIR Analysis
Six-week-old wild-type and 35S:MIR857 plants were collected and washed with deionized water three times. The samples were dried at room temperature on a barium fluoride window (13-mm diameter × 2 mm). Infrared spectra were obtained from the stem region using aMAGNA750 FTIR spectrometer (Nicolet) equipped with a mercury-cadmium-telluride detector. The spectra were recorded at a resolution of 8 cm−1 with 128 coadded interferograms, and were normalized to obtain the relative absorbance.
NMR Analysis
The whole-cell wall samples (ball-milled sample) for NMR spectroscopy analysis were prepared according to the procedure described previously (Mansfield, et al., 2012). NMR spectra were acquired using a Bruker Avance-III 400 MHz spectrometer operating at a frequency of 100.59 MHz for 13C nucleus. Deuterated DMSO was used as solvent for plant cell wall samples (15 mg/0.5 mL). The 13C-1H HSQC correlation spectra were recorded using a Bruker standard pulse sequence (‘hsqcetgpsi2’) with the following acquisition parameters: The number of collected complex points was 1,024 for the 1H dimension with a recycle delay (d1) of 1.5 s, number of transients for the HSQC spectra was 64, and 256 time increments were always recorded in the 13C dimension. The NMR coupling constant was 145 Hz, and the central solvent peak (δC 39.5 ppm; δH 2.49 ppm) was used for chemical shift calibration. NMR data were processed using the TopSpin 2.1 (Bruker BioSpin) software.
SRS Microscopy
The SRS imaging microscope using a mode-locked Nd: YVO4 laser (High Q Laser) was used to generate a 7-ps, 76-MHz pulse train of both 1,064-nm (1 W average power) and 532-nm (5 W average power) laser beams. The 1,064-nm output was used as the Stokes light. The 532-nm beam was 50/50 split to pump two optical parametric oscillators (Levante Emerald, A∙P∙E Angewandte Physik und Elektronik GmbH). The output wavelengths of the optical parametric oscillators were selected at 812 and 909 nm to use as pump beams to induce the stimulated Raman signal for the 2,900 cm−1 carbohydrate C-H vibration and the 1,600 cm−1 lignin aromatic ring vibration, respectively. All pump and Stokes beams were directed into an Olympus laser scanning microscope scanning unit (BX62WI/FV300; Olympus) and focused by a high numerical aperture water-immersion objective (UPLSApo 60X 1.20 NA W; Olympus). The light transmitted through the sample was collected by an oil-immersion condenser (1.45 NA O; Nikon). The stimulated Raman loss signals were detected by silicon PIN photodiodes (FDS1010; Thorlabs) and a lock-in amplifier (SR844; Stanford Research Systems) as previously described (Ding et al., 2012).
In Vitro Laccase Activity Assays
Mature stems from 6-week-old plants were harvested for lignin analysis using previously published procedures (Berthet, et al., 2011). About 10 g of Arabidopsis stems was homogenized in 7 mL of extraction buffer (25 mm BisTris [pH 7], 200 mm CaCl2, 10% [v/v] glycerol, 4 mm sodium cacodylate, and 1/200 [v/v] protease inhibitor cocktail [P-9599; Sigma-Aldrich]) for 5 min in a blender. The homogenate was centrifuged twice at 88°C, for 5 min each, at 3,000g and once at 48°C for 5 min at 13,000g. The supernatant was centrifuged at 88°C for 45 min at 15,000g. Proteins were purified by affinity chromatography on a 0.5- × 3-cm column filled with 1 mL of Concanavalin-A Sepharose (Sigma-Aldrich) and washed with 3 mL of 20 mm Tris-HCl and 0.5 m NaCl buffer, pH 7.4. The soluble protein extract was loaded and the column was washed with 10 mL of buffer. The proteins were eluted with 0.2 m methyl-a-glucopyranoside in the same buffer. The eluates were collected (1 mL per fraction), and 3- or 5-mL samples from each fraction were tested for laccase activity. Pooled fractions showing laccase activity were equilibrated in 25 mm Tris-HCl buffer, pH 7.4, supplemented with 5% (v/v) glycerol and 0.015% (v/v) Triton X-100. Glycerol was added to the buffer to prevent partial inactivation of the enzymes. An 11-mg mL−1 solution of the substrate, ABTS, was prepared in DMSO and stored in aliquots at −20°C. Laccase activity was determined at 30°C by the oxidation of ABTS to generate a stable cationic radical assayed by spectrometry at 420 nm. The reaction mixture contained 100 mm acetate buffer (pH 5), 1 mm ABTS, and 20 mL of protein extract in a total volume of 200 mL.
GUS Assays
GUS staining was performed according to Vielle-Calzada et al. (2000). Seedlings were incubated in GUS staining solution (1 mg mL−1 X-Gluc [Biosynth], 2 mm K4Fe(CN)6, 2 mm K3Fe(CN)6, 10 mm EDTA, 0.1% (v/v) Triton X-100, and 100 mg mL−1 chloramphenicol in 50 mm sodium phosphate buffer, pH 7.0) for 2 to 3 d at 37°C. The stained sample was fixed with 70% (v/v) ethanol. Stained tissues were observed on a Zeiss Axioplan microscope with Nomarski and dark-field optics.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AtMIR857 (GU125421), AtLAC7 (NM_111756), and AtSPL7 (NM_001203404).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic analyses of miR857 in plants.
Supplemental Figure S2. Overexpression of AtMIR857 in 35S:MIR857 plants.
Supplemental Figure S3. Experimental validation of the predicted mRNA target of miR857 in Arabidopsis.
Supplemental Figure S4. RT-PCR analysis of AtMIR857 in mir857 T-DNA insertion mutants.
Supplemental Figure S5. Relative mRNA levels of AtLAC1-AtLAC6 and AtLAC8-AtLAC17 in wild-type, 35S:MIR857, and mir857 plants.
Supplemental Figure S6. Relative mRNA levels of MIR397 and AtLAC4 in wild-type and mir857 plants.
Supplemental Figure S7. Relative mRNA levels of mature miR857 in wild-type, 35S:MIR857, and mir857 mutant plants.
Supplemental Figure S8. Root anatomical structures of wild-type, 35S:MIR857, and mir857 mutant plants.
Supplemental Figure S9. Phenotypic and molecular analysis of miR857/mir857 transgenic plants.
Supplemental Figure S10. FTIR analyses.
Supplemental Figure S11. SRS chemical images in roots.
Supplemental Figure S12. Lignin content in stem, root, caulin leaf, and rosette leaf in wild-type and mir857 plants at 4, 5, and 7 weeks.
Supplemental Figure S13. Relative mRNA levels of AtCesA7, AtCesA8, AtCAD, AtHCT, AtCCoAOMT, At4CL1, AtCCR1, and AtF5H2 in wild-type and 35S:MIR857 plants.
Supplemental Table S1. List of primers.
Supplemental Table S2. List of probes in EMSA analysis.
Supplementary Material
Acknowledgments
We thank Hiroaki Yamasaki (Graduate School of Agriculture, Kyushu University) for providing the seeds of the spl7 mutant.
Glossary
- SRS
stimulated Raman scattering
- qRT
quantitative real-time
- RT
reverse transcription
- ABTS
2,2′-azinobis-3-ethylbenzthiazolinesulfonic acid
- T-DNA
transfer DNA
- TEM
transmission electron microscopy
- FTIR
Fourier transform infrared
- DMSO
dimethyl sulfoxide
- MS
Murashige and Skoog
- cDNA
complementary DNA
Footnotes
This work was supported by the Program of Introducing Talents of Discipline to Universities (111 project, B13007), the Program for Changjiang Scholars and Innovative Research Team in University of Ministry of Education of China (grant no. IRT13047), Major Science Foundation of Ministry of Education of China (grant no. 313008), the National Natural Science Foundation of China (grant nos. 31270224 and 31370212), and the Fundamental Research Funds for the Central Universities (grant no. BLX2012039).
References
- Abdel-Ghany SE, Pilon M (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J Biol Chem 283: 15932–15945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, O’malley DM, Whetten R, Sederoff RR (1993) A laccase associated with lignification in loblolly pine xylem. Science 260: 672–674 [DOI] [PubMed] [Google Scholar]
- Bartel DP. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Beers E, Zhao C (2001) Arabidopsis as a model for investigating gene activity and function in vascular tissues. Prog Biotechnol 18: 43–52 [Google Scholar]
- Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S. , et al. (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhead JL, Reynolds KA, Abdel-Ghany SE, Cohu CM, Pilon M (2009) Copper homeostasis. New Phytol 182: 799–816 [DOI] [PubMed] [Google Scholar]
- Chabannes M, Ruel K, Yoshinaga A, Chabbert B, Jauneau A, Joseleau JP, Boudet AM (2001) In situ analysis of lignins in transgenic tobacco reveals a differential impact of individual transformations on the spatial patterns of lignin deposition at the cellular and subcellular levels. Plant J 28: 271–282 [DOI] [PubMed] [Google Scholar]
- Chaffey N. (1999) Cambium: old challenges–new opportunities. Trees (Berl) 13: 138–151 [Google Scholar]
- Chen X. (2005) MicroRNA biogenesis and function in plants. FEBS Lett 579: 5923–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JH, Overney S, Rohde A, Diaz WA, Bauw G, Simon P, Van Montagu M, Boerjan W (2001) The syringaldazine-oxidizing peroxidase PXP 3-4 from poplar xylem: cDNA isolation, characterization and expression. Plant Mol Biol 47: 581–593 [DOI] [PubMed] [Google Scholar]
- Day A, Neutelings G, Nolin F, Grec S, Habrant A, Crônier D, Maher B, Rolando C, David H, Chabbert B. , et al. (2009) Caffeoyl coenzyme A O-methyltransferase down-regulation is associated with modifications in lignin and cell-wall architecture in flax secondary xylem. Plant Physiol Biochem 47: 9–19 [DOI] [PubMed] [Google Scholar]
- Ding SY, Liu YS, Zeng Y, Himmel ME, Baker JO, Bayer EA (2012) How does plant cell wall nanoscale architecture correlate with enzymatic digestibility? Science 338: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Dittmer NT, Gorman MJ, Kanost MR (2009) Characterization of endogenous and recombinant forms of laccase-2, a multicopper oxidase from the tobacco hornworm, Manduca sexta. Insect Biochem Mol Biol 39: 596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas DV, Bartel B (2004) MicroRNA regulation of gene expression in plants. Curr Opin Plant Biol 7: 512–520 [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL. , et al. (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goicoechea M, Lacombe E, Legay S, Mihaljevic S, Rech P, Jauneau A, Lapierre C, Pollet B, Verhaegen D, Chaubet-Gigot N. , et al. (2005) EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J 43: 553–567 [DOI] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF, Chua NH (2005) MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17: 1376–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Jover-Gil S, Candela H, Ponce MR (2005) Plant microRNAs and development. Int J Dev Biol 49: 733–744 [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA (2005) The developmental role of microRNA in plants. Curr Opin Plant Biol 8: 38–44 [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597–610 [DOI] [PubMed] [Google Scholar]
- Kropat J, Tottey S, Birkenbihl RP, Depège N, Huijser P, Merchant S (2005) A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci USA 102: 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev-Yadun S. (1994) Induction of sclereid differentation in the pith of Arabidopsis thaliana(L.) Heynh. J Exp Bot 45: 1845–1849 [Google Scholar]
- Liang CC, Park AY, Guan JL (2007) In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc 2: 329–333 [DOI] [PubMed] [Google Scholar]
- Lu S, Li Q, Wei H, Chang MJ, Tunlaya-Anukit S, Kim H, Liu J, Song J, Sun YH, Yuan L, et al. (2013) Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc Natl Acad Sci USA 110: 10848–10853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SD, Kim H, Lu F, Ralph J (2012) Whole plant cell wall characterization using solution-state 2D NMR. Nat Protoc 7: 1579–1589 [DOI] [PubMed] [Google Scholar]
- Muraro D, Mellor N, Pound MP, Help H, Lucas M, Chopard J, Byrne HM, Godin C, Hodgman TC, King JR, et al. (2014) Integration of hormonal signaling networks and mobile microRNAs is required for vascular patterning in Arabidopsis roots. Proc Natl Acad Sci USA 111: 857–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae M, Nakata M, Takahashi Y (2008) Identification of negative cis-acting elements in response to copper in the chloroplastic iron superoxide dismutase gene of the moss Barbula unguiculata. Plant Physiol 146: 1687–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CM, Guerinot ML (2009) Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat Chem Biol 5: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomion C, Leprovost G, Stokes A (2001) Wood formation in trees. Plant Physiol 127: 1513–1523 [PMC free article] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I (2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17: 2966–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JM, Merchant S (1995) Two copper-responsive elements associated with the Chlamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell 7: 623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JM, Nakamoto SS, Merchant S (1999) Induction of coproporphyrinogen oxidase in Chlamydomonas chloroplasts occurs via transcriptional regulation of Cpx1 mediated by copper response elements and increased translation from a copper deficiency-specific form of the transcript. J Biol Chem 274: 14444–14454 [DOI] [PubMed] [Google Scholar]
- Re A, Corá D, Taverna D, Caselle M (2009) Genome-wide survey of microRNA-transcription factor feed-forward regulatory circuits in human. Mol Biosyst 5: 854–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Lieber D, Oren M, Pilpel Y (2007) Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput Biol 3: e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Kropat J, Malasarn D, Grossoehme NE, Chen X, Giedroc DP, Merchant SS (2010) The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell 22: 4098–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognolli M, Penel C, Greppin H, Simon P (2002) Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288: 129–138 [DOI] [PubMed] [Google Scholar]
- Turlapati PV, Kim KW, Davin LB, Lewis NG (2011) The laccase multigene family in Arabidopsis thaliana: towards addressing the mystery of their gene function(s). Planta 233: 439–470 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Morreel K, Ralph J, Boerjan W (2008) Lignin engineering. Curr Opin Plant Biol 11: 278–285 [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Baskar R, Grossniklaus U (2000) Delayed activation of the paternal genome during seed development. Nature 404: 91–94 [DOI] [PubMed] [Google Scholar]
- Wang CY, Zhang S, Yu Y, Luo YC, Liu Q, Ju C, Zhang YC, Qu LH, Lucas WJ, Wang X. , et al. (2014) MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol J 12: 1132–1142 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhu ML, Wei ZM (2008) [Cotton laccase gene overexpression in transgenic Populus alba var. pyramidalis and its effects on the lignin biosynthesis in transgenic plants] (in Chinese). Fen Zi Xi Bao Sheng Wu Xue Bao 41: 11–18 [PubMed] [Google Scholar]
- Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T (2009) SQUAMOSA Promoter Binding Protein-Like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon EK, Yang JH, Lim J, Kim SH, Kim SK, Lee WS (2010) Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res 38: 1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Lin J, Zack DJ, Mendell JT, Qian J (2008) Analysis of regulatory network topology reveals functionally distinct classes of microRNAs. Nucleic Acids Res 36: 6494–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li L (2013) SQUAMOSA promoter binding protein-like7 regulated microRNA408 is required for vegetative development in Arabidopsis. Plant J 74: 98–109 [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA (2006) Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289: 3–16 [DOI] [PubMed] [Google Scholar]
- Zhao C, Johnson BJ, Kositsup B, Beers EP (2000) Exploiting secondary growth in Arabidopsis: construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiol 123: 1185–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Nakashima J, Chen F, Yin Y, Fu C, Yun J, Shao H, Wang X, Wang ZY, Dixon RA (2013) LACCASE is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25: 3976–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.