The hormone ethylene plays an important modulating role in the immune signaling network that regulates defense against microbial pathogens and insect herbivores.
Abstract
Ethylene (ET) is an important hormone in plant responses to microbial pathogens and herbivorous insects, and in the interaction of plants with beneficial microbes and insects. Early ET signaling events during these biotic interactions involve activities of mitogen-activated protein kinases and ETHYLENE RESPONSE FACTOR transcription factors. Rather than being the principal regulator, ET often modulates defense signaling pathways, including those regulated by jasmonic acid and salicylic acid. Hormonal signal integrations with ET steer the defense signaling network to activate specific defenses that can have direct effects on attackers, or systemically prime distant plant parts for enhanced defense against future attack. ET also regulates volatile signals that attract carnivorous enemies of herbivores or warn neighboring plants. Conversely, ET signaling can also be exploited by attackers to hijack the defense signaling network to suppress effective defenses. In this review, we summarize recent findings on the significant role of ET in the plants’ battle against their enemies.
Plants live in complex environments in which they are constantly exposed to a wide range of biotic interactors. Some of these interactors, such as mutualistic rhizosphere bacteria and fungi, are beneficial for the plant as they promote plant growth and protect the plant from attack by harmful interactors. Conversely, harmful interactors, such as herbivorous insects and microbial pathogens, reduce the fitness of the plant by retrieving energy-rich organic compounds without returning a net benefit to the plant (Pieterse et al., 2014b). To minimize the success of attack by other organisms, plants have evolved sophisticated defensive mechanisms that are either expressed constitutively or induced when an attacker is recognized. Active defense against biotrophic pathogens, which form a long-term relationship with living plant cells to derive nutrients, is mainly effectuated by programmed plant cell death (Glazebrook, 2005). Infection by necrotrophic pathogens, which first destroy host cells before feeding on the content, is usually repulsed by plant-produced antimicrobial compounds (Glazebrook, 2005). To defend themselves against insects, plants can activate both direct and indirect defenses. Direct defenses include plant traits that hinder the insect’s growth rate, adult size, and/or survival probability, such as trichomes or toxic secondary metabolites (Howe and Jander, 2008). Indirect defenses include plant traits that enhance the probability of attracting natural enemies of herbivorous insects, such as volatile organic compounds or extrafloral nectar (Dicke, 2015; Heil, 2015).
Inducible defenses are initiated after perception of microbial infection or insect infestation, for which plants possess a suite of pattern recognition receptors (PRRs). These PRRs specifically recognize general nonself molecules from attacking organisms and self molecules from already attacked plant cells (Cook et al., 2015). By detecting highly conserved structures of entire classes of microbes, so-called microbe-associated molecular patterns (MAMPs), plants can recognize attack by a wide variety of potential pathogens. Similarly, general elicitors that are present in the saliva of insects function as herbivore-associated molecular patterns. Moreover, enzymatic degradation of plant material by attacking microbes or insects generates endogenous elicitors, so-called damage-associated molecular patterns (Ferrari et al., 2013; Savatin et al., 2014; Acevedo et al., 2015). All of these different molecular patterns are detected through a general detection system consisting of PRRs and coreceptors, leading to the activation of pattern-triggered immunity (PTI; Jones and Dangl, 2006). Biosynthesis of the gaseous hormone ethylene (ET) is among the suite of immediate PTI responses that, together with the production of reactive oxygen species and the activation of mitogen-activated protein kinase (MAPK) signaling cascades, regulate the production of downstream defensive proteins and metabolites (Boller and Felix, 2009; Wu and Baldwin, 2010). A second layer of more specific perception of microbes and insects is accomplished by plant resistance (R) proteins. Successful pathogens are able to suppress or evade PTI by the production of attacker-specific effectors (Pel and Pieterse, 2013). In turn, R proteins in the plant have evolved to specifically recognize these effectors, initiating effector-triggered immunity (ETI) or R gene-mediated resistance (Jones and Dangl, 2006; Cui et al., 2015). ETI is accompanied by rapid ET production and a programmed cell death at the site of infection that prevents further ingress of the invading pathogen. Several R genes have been identified to confer resistance against insects (Broekgaarden et al., 2011), but whether they are involved in recognizing specific herbivore-derived elicitors is still unclear.
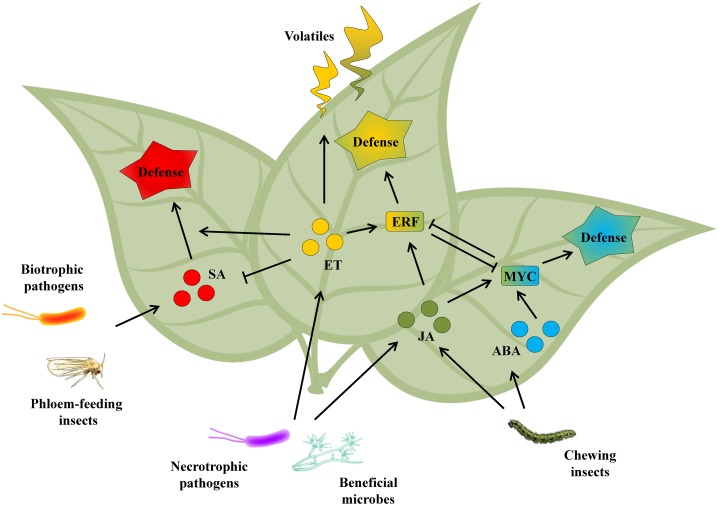
Subsequent to recognition of the attacker, a hormone-regulated cellular signaling network is triggered that orchestrates the production of defensive proteins and metabolites. Besides ET, several other plant hormones are implicated in this regulatory network, with jasmonic acid (JA), salicylic acid (SA), and abscisic acid (ABA) being major players (Fig. 1; Pieterse et al., 2012). Their abundant antagonistic or synergistic interactions provide the plant with an extensive regulatory potential that enables the activation of specific defenses, while minimizing fitness costs (Pieterse et al., 2012; Vos et al., 2013a). ET emerged as an important hormonal traffic controller on the hormone-regulated defense pathways that are activated in response to biotic stress. Here, we provide an update on the modulating role of ET in this process.
Figure 1.
Simplified schematic representation of plant defense signaling networks involving the hormones ET, SA, JA, and ABA. Necrotrophic pathogen and beneficial microbes induce or prime ET- and JA-dependent signaling pathways, whereas chewing insects induce JA- and ABA-dependent signaling pathways. The ET- and ABA-regulated branches of the JA pathway are mutually antagonistic. ET alone or together with JA plays a role in volatile signaling. Arrows and end-blocked lines indicate positive and negative regulation, respectively.
ATTACKER-INDUCED ET SIGNALING
During induced plant defenses in general, activation of ET-mediated defenses starts with the recognition of the attacker, after which calcium fluxes and MAPK cascades are initiated that in turn stimulate ET biosynthesis. The amount and timing of ET production is then further regulated in an attacker-specific manner, which modulates the plant’s defense response (Hu et al., 2011; Van der Ent and Pieterse, 2012; Groen and Whiteman, 2014; Rehrig et al., 2014). In addition to its role in defense, ET is also known to be an important regulator of developmental processes, and accordingly, ET-dependent defense patterns change during development. For instance, treating Nicotiana attenuata leaves of various ages with saliva from the Manduca sexta caterpillar showed that induced ET emission decreased with leaf and plant age (Diezel et al., 2011).
Recognition of Attack Leading to ET Signaling
A classic example of attacker perception leading to the activation of ET signaling is the recognition of MAMPs such as flagellin and elongation factor (EF)-TU (Boller and Felix, 2009). Upon perception of these MAMPs by their cognate PRRs, MAPK signaling is activated, leading to rapid production of ET, which results in effective downstream PTI responses. In the case of flagellin perception, ET signaling regulates accumulation of the cognate PRR receptor FLAGELLIN SENSITIVE2, and is required for the oxidative burst that contributes to flagellin-triggered plant immunity (Mersmann et al., 2010). Furthermore, plant elicitor peptide ZmPep3 in maize (Zea mays) serves as an endogenous signal to stimulate ET production and increase the expression of genes involved in direct and indirect defense, leading to enhanced resistance against Spodoptera exigua caterpillars (Wu and Baldwin, 2010; Huffaker et al., 2013). A recent example of effector-triggered activation of ET signaling comes from the interaction between the bacterial pathogen Erwinia amylovora and Arabidopsis (Arabidopsis thaliana). The E. amylovora-derived elicitor HrpNEa activates ET-mediated expression of the Arabidopsis transcription factor MYB44, which in turn enhances the expression of ETHYLENE INSENSITIVE2 (EIN2), an essential gene in ET signaling (Liu et al., 2011a).
Molecular Players in Signal Transduction from Recognition to ET Biosynthesis
Recently, several unique components involved in the steps between pathogen/insect recognition and the activation of ET signaling have been reported. For example, during infection of Arabidopsis by the necrotrophic pathogen Botrytis cinerea, MITOGEN-ACTIVATED PROTEIN KINASE3 (MPK3) and MPK6 phosphorylate the ET biosynthesis proteins 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID (ACC) SYNTHASE2 (ACS2) and ACS6, resulting in rapid ET production (Han et al., 2010). Additionally, silencing SALICYLIC ACID-INDUCED PROTEIN KINASE (SIPK), encoding the N. attenuata ortholog of Arabidopsis MPK6, abolished insect-induced ET accumulation in N. attenuata (Wu et al., 2007; Mase et al., 2012). Moreover, SIPK- and WOUND INDUCED PROTEIN KINASE-silenced Nicotiana umbratica plants showed disrupted ET accumulation and compromised necrosis upon application of the pathogen-derived Alternaria alternata sp. lycopersici toxin (Mase et al., 2012). Taken together, these results highlight the importance of MAPK signaling cascades in the regulation of ET signaling and its downstream defense responses. Furthermore, the Ca2+-binding protein Calreticulin 3a of Nicotiana benthamiana was shown to be involved in the induced production of ET upon recognition of MAMPs derived from the pathogen Phytophthora infestans. Induced ET signaling was further demonstrated to be required for the production of antimicrobial phytoalexins, which is critical for the resistance of N. benthamiana to P. infestans (Matsukawa et al., 2013).
ET-Mediated Downstream Responses
Downstream in the ET signaling cascade, transcription factors of the ETHYLENE RESPONSE FACTOR (ERF) family play a dominant role in the regulation of defenses. For example, ERF3 was identified in rice (Oryza sativa) as a gene that positively affects expression of trypsin proteinase inhibitors and to mediate resistance toward Chilo suppressalis caterpillars (Lu et al., 2011). Well-characterized ERF transcription factors from Arabidopsis with a role in ET-dependent defense are ERF1 (Lorenzo et al., 2003) and OCTADECANOID-RESPONSIVE ARABIDOPSIS APETALA2/ ETHYLENE RESPONSE FACTOR DOMAIN PROTEIN59 (ORA59; Pré et al., 2008). Overexpressing or silencing these ERF transcription factors in Arabidopsis leads to enhanced or reduced resistance to several fungi, respectively (Memelink, 2009). Overall, ERFs are conserved among different plant species (Groen and Whiteman, 2014) and mediate resistance to especially necrotrophic pathogens (Anderson et al., 2010; Liu et al., 2011b; Zhu et al., 2014).
The ETI response that is triggered in Arabidopsis upon infection by avirulent Pseudomonas syringae pv tomato avrRpm1 was reported to be accompanied by a biphasic accumulation of ET (Mur et al., 2009). This ETI-associated ET production is dependent on ETHYLENE RECEPTOR1 (ETR1) and ET signaling component EIN2 and accelerates the hypersensitive cell death response, confirming previous findings that ET plays a role in lesion expansion during the hypersensitive response (Van Loon et al., 2006).
COMMUNICATION WITH OTHER HORMONES
A plant’s response to insect or pathogen attack is based on a complex network of interactions between different hormonal signals. Cross communication between the initiated hormone signaling pathways contributes to the activation of attacker-specific defenses (Robert-Seilaniantz et al., 2011; Pieterse et al., 2012; León et al., 2014; Caarls et al., 2015; Fig. 1). JA and SA are dominant players in the regulation of the immune signaling network, where JA is generally effective against necrotrophic pathogens and insects while SA is primarily effective against biotrophic pathogens (Vos et al., 2013a). Accumulating evidence indicates that ET can interact both positively and negatively with SA, depending on the plant-attacker interaction (Van der Ent and Pieterse, 2012). For example, ET acts positively on the level of SA-mediated resistance against the pathogen Leptosphaeria maculans in Brassica napus (Sašek et al., 2012), while it acts negatively on SA-mediated defense against the pathogen P. syringae in Arabidopsis (Chen et al., 2009). The majority of studies on the modulating role of ET report on its effect on the JA signaling pathway and on the interplay between the JA and SA pathways. Therefore, we focus on these ET-hormone interactions in more detail below.
Interaction between ET and JA Signaling
Although the JA pathway has a dominant role in induced plant defenses (Wasternack, 2015), ET plays an important part in fine tuning these responses. The JA signaling pathway comprises two separate branches: (1) the so-called ERF branch, which in Arabidopsis is regulated by ERF-type transcription factors such as ERF1 and ORA59 (Lorenzo et al., 2003; McGrath et al., 2005; Pré et al., 2008), and (2) the so-called MYC branch, which in Arabidopsis is regulated by MYC-type transcription factors such as MYC2, 3, and 4 (Boter et al., 2004; Dombrecht et al., 2007; Fernández-Calvo et al., 2011). ET synergizes the ERF branch to activate defense-related genes, such as PLANT DEFENSIN1.2 (PDF1.2), leading to effective defense against necrotrophic pathogens. The MYC branch, on the other hand, is synergized by ABA to activate defense-related genes, such as VEGETATIVE STORAGE PROTEIN2, resulting in defense against chewing insects (Fig. 1). The ERF and MYC branches of the JA pathway act as communicating vessels in which the relative balance is dependent on the relative strength of the concomitantly activated ET and ABA pathways (Lorenzo et al., 2004; Verhage et al., 2011; Vos et al., 2013b). This is exemplified by the fact that ABA-deficient mutants are more resistant to necrotrophic pathogens and more susceptible to certain insects, whereas ET-deficient mutants are more susceptible to necrotrophs and more resistant to certain insects (Berrocal-Lobo et al., 2002; Lorenzo et al., 2003, 2004; Nickstadt et al., 2004; van Loon et al., 2006; Bodenhausen and Reymond, 2007; Kazan and Manners, 2012; Dinh et al., 2013). However, it should be noted that there are exceptions; for example, an ET-insensitive mutant of N. attenuata displayed reduced JA-mediated defenses against M. sexta caterpillars (Onkokesung et al., 2010).
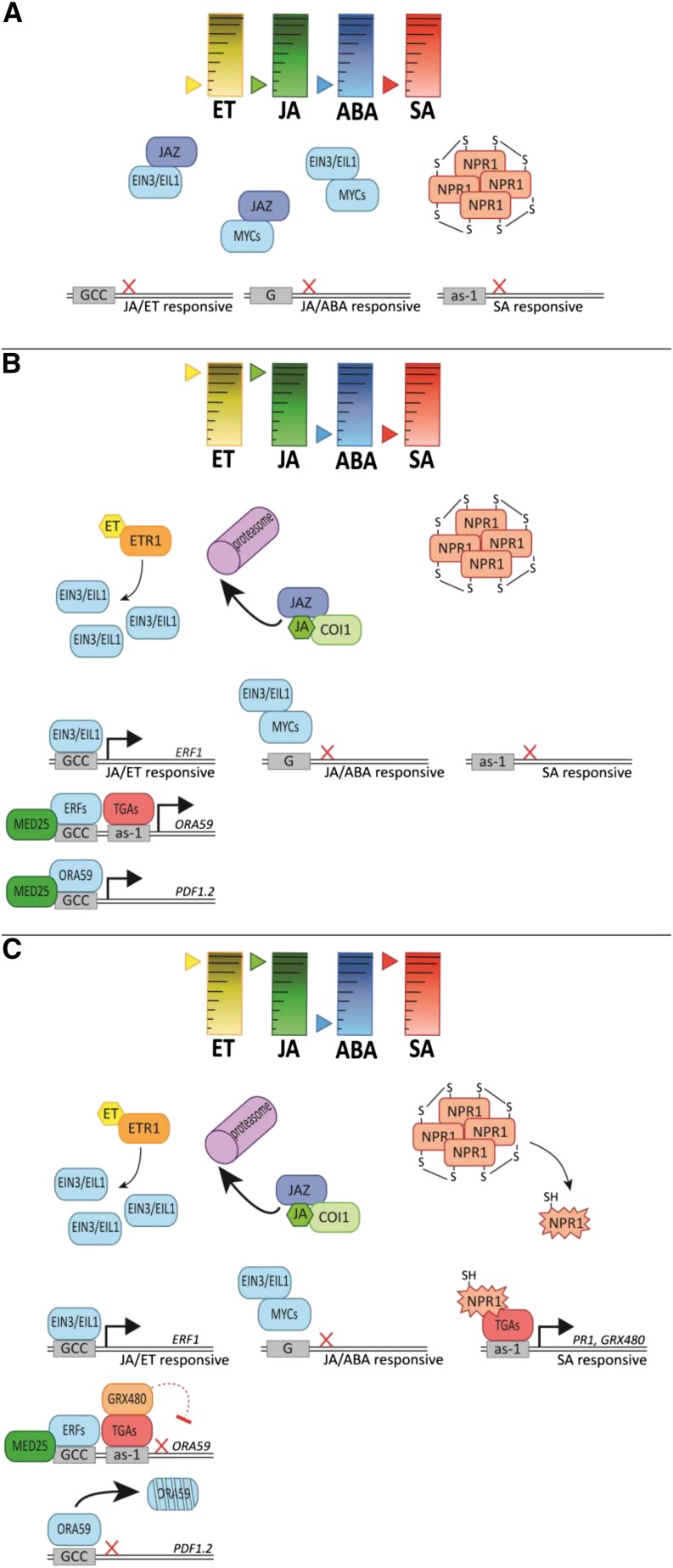
Recent studies provide evidence that the balance between the ERF and MYC branch of the JA pathway is modulated through interactions between JA/ABA-activated MYC2/3/4 and ET-stabilized EIN3 and ETHYLENE INSENSITIVE3-LIKE1 (EIL1; Fig. 2). Zhang et al. (2014) showed that MYC2 represses EIN3 by inducing expression of the EIN3-repressor ETHYLENE INSENSITIVE3 BINDING F-BOX PROTEIN1 as well as by physically interacting with EIN3 to inhibit its DNA binding activity. Additionally, Song et al. (2014) showed that this antagonistic effect of MYC2 on EIN3 also involves EIL1 and attenuates resistance to B. cinerea. Furthermore, they demonstrated that the interaction between MYC2 and EIN3 is mutually antagonistic. EIN3 and EIL1 interact with and repress MYC2, MYC3, and MYC4, resulting in attenuated JA-regulated defense against Spodoptera littoralis and S. exigua caterpillars. Important modulators of the activity of the MYC2/3/4 and EIN3/EIL1 transcription factors are JAZ repressor proteins that physically bind to them (Fig. 2B; Chini et al., 2007; Fernández-Calvo et al., 2011; Niu et al., 2011b; Zhu et al., 2011). Another modulator of the activity of the MYC and ERF branches of the JA pathway is the mediator complex subunit MEDIATOR25 (MED25; Çevik et al., 2012). In eukaryotes, the multiprotein mediator complex connects transcription factors to the core transcriptional machinery. MED25 binds to and is required for the transcriptional activation of the MYC and ERF branch transcription factors MYC2, ERF1, and ORA59 (Fig. 2B). Consequently, med25 mutants are impaired in ET- and JA-dependent defenses against pathogens and insects (Çevik et al., 2012).
Figure 2.
Simplified model of the molecular machinery involved in the transcriptional regulation of ET-modulated JA- and SA-dependent defense responses. A, In an unstressed plant, several mechanisms repress activation of ET-, JA-, and SA-dependent defense pathways. Binding of jasmonic acid-Zim domain (JAZ) repressor proteins to transcriptional activators such as EIN3/EIL1 and the MYC transcription factors suppresses their activity and, thus, downstream defense-related gene transcription. Furthermore, EIN3/EIL1 and MYC transcription factors bind to each other, which also represses their activity. In the SA pathway, oligomerization of the regulatory protein NPR1 prevents NPR1 from going into the nucleus, thereby preventing downstream SA-dependent defense responses. B, Infection with a necrotrophic pathogen induces JA and ET levels in the plant. JA binds to the JA receptor CORONATINE INSENSITIVE1 (COI1), which results in the degradation of JAZ repressor proteins by the proteasome and subsequent release of activating transcription factors. ET binds to the receptor ETR1, eventually resulting in the stabilization of the EIN3/EIL1 transcription factors. Accumulation of both JA and ET activates the ERF branch of the JA defense pathway, resulting in activation of JA/ET-responsive genes such as the PDF1.2 marker gene, leading to defense against necrotrophic pathogens. At the same time, activation of the JA and ET pathways suppresses the MYC branch of the JA pathway via interaction between EIN3/EIL1 and MYCs. Upon insect herbivory, JA acts together with ABA to activate the MYC branch of the JA pathway and to simultaneously suppress of the ERF branch (details not shown here). C, When SA levels are increased in the plant in addition to elevated JA and ET levels, NPR1 monomers are released, and NPR1 can translocate to the nucleus to activate downstream SA responses. Activation of the SA pathway leads to antagonism of the JA/ET-dependent transcription, likely via suppression of ORA59 transcription by TGAs and GLUTAREDOXIN480 (GRX480), and degradation of ORA59 protein. See text for details on the molecular processes underlying the transcriptional control. Solid lines indicate established activities and dashed lines indicate hypothesized activities, where black arrows specify activation and red lines suppression. Red crosses indicate arrested gene transcription. GCC box, Binding site of ERF transcription factors; G box, binding site for MYC transcription factors; as-1 motif, binding site for TGA transcription factors.
Effect of ET on SA-JA Cross Talk
Among the best-studied hormone signal interactions is the antagonistic relationship between the SA and JA signaling pathways, often referred to as SA-JA cross talk (Pieterse et al., 2012). A number of important regulators of the interaction between the SA and JA pathways have been identified, including the redox sensitive transcriptional coregulator NONEXPRESSOR OF PATHOGENESIS-RELATED PROTEINS1 (NPR1; Spoel et al., 2003). Interestingly, timing and concentration of ET production modulate the strength of the antagonistic effect of SA on the JA pathway and the dependency on NPR1. Leon-Reyes et al. (2009) demonstrated that simultaneous stimulation of the ET and JA pathways in Arabidopsis strongly suppressed the NPR1 dependency of SA-JA cross talk. Additionally, full activation of ET and JA signaling (e.g. during infection with a necrotrophic pathogen) resulted in insensitivity to future SA-mediated suppression of JA/ET-dependent defenses. Hence, ET helps the plant to prioritize the JA/ET pathway over the SA/NPR1 pathway during multiattacker interactions (Leon-Reyes et al., 2010). Also, in other plant species, a link between ET and NPR1-dependent regulation of defense has been reported. For instance, antisense expression of NPR1 in rice increased JA and ET levels, resulting in increased levels of antiherbivore compounds and reduced performance of rice striped corn borer (Li et al., 2013).
Besides NPR1, other SA-regulated signaling components emerged as important regulators of the interplay between the SA and JA/ET pathways (Fig. 2C). In Arabidopsis, TGA transcription factors are primarily known as regulators of SA-mediated transcription, but they control activity of the ET/JA-dependent ERF branch as well. TGA2, TGA5, and TGA6 are required for B. cinerea-induced expression of PDF1.2 to confer resistance against this necrotrophic fungus (Zander et al., 2010). However, in the presence of SA, the same TGAs have a role in SA-mediated antagonism of ERF branch signaling. The TGAs can directly target the activation sequence-1 (as-1) promoter sequence in the ERF transcription factor gene ORA59, thereby regulating its expression (Zander et al., 2014). Possibly through SA-induced glutaredoxins such as GLUTAREDOXIN480 (GRX480) that interact with TGA transcription factors, expression of ORA59 is suppressed (Zander et al., 2012; Caarls et al., 2015). Together with the observation that SA affects ORA59 protein accumulation (Van der Does et al., 2013), these findings demonstrate that ORA59 plays a central role in the interaction between the SA and JA/ET pathways.
SPATIAL CONTROL OF ET-DEPENDENT DEFENSES
Plants activate defenses not only locally at the site of attack, but also systemically throughout the plant to protect the still-healthy tissue against future attack. Pathogen-induced systemic acquired resistance (SAR), beneficial microbe-induced systemic resistance (ISR), and herbivore/wound-induced systemic resistance are three well-characterized types of systemic defense responses (Wu and Baldwin, 2010; Pieterse et al., 2014b). Although ET seems to have only a minor role in systemic acquired resistance (Fu and Dong, 2013) and herbivore/wound-induced systemic resistance (Vos et al., 2013b), its essential role in the regulation of ISR is widely accepted (Pieterse et al., 2014b). Interestingly, ET is also implicated in volatile signaling, either by serving as a volatile itself or regulating the production of other volatiles to communicate with distant plant tissues or neighboring plants about pathogen or insect attack. As part of an indirect defense mechanism, volatiles also function to repel insects or attract the insect’s natural enemies (Scala et al., 2013).
The Role of ET in ISR
Plants produce exudates and lysates at their root surface, resulting in the attraction of a large number of beneficial microorganisms (Berendsen et al., 2012). Besides stimulating plant growth and outcompeting soil-borne pathogens, some beneficial microbes are capable of activating ISR in above- and belowground plant tissues, a phenomenon that is effective against a broad spectrum of pathogens and insects (Pineda et al., 2010; Pieterse et al., 2014b; Zamioudis et al., 2015). In many cases, ET has been demonstrated to play an important role in the modulation of host immune responses by beneficial microbes (Zamioudis et al., 2012). At the root-microbe interface, the MAMP flagellin of the ISR-inducing rhizobacterium Pseudomonas fluorescens WCS417 (recently renamed as Pseudomonas simiae WCS417; Berendsen et al., 2015) initially triggers an ET-dependent immune response in Arabidopsis roots, but this becomes readily suppressed, probably to facilitate accommodation of the microbe on the root surface (Millet et al., 2010). In contrast, activation of ET signaling is essential in the initial stages of colonization of barley (Hordeum vulgare) roots by the mutualistic fungus Piriformospora indica (Khatabi et al., 2012). In the interaction between Populus trichocarpa and the ectomycorrhizal fungus Laccaria bicolor, ET acts together with JA in constraining fungal growth, possibly to maintain an economic balance between costs and benefits in this mutualistic plant-fungus interaction (Plett et al., 2014).
To develop ISR, plants need an intact ET response, as was demonstrated by the impaired ability of ET signaling mutants to develop ISR (Van Wees et al., 2008; Niu et al., 2011a; Salas-Marina et al., 2011; Fracetto et al., 2013; Pieterse et al., 2014b). In tomato (Solanum lycopersicum), the ET dependency of ISR triggered by a mutualistic Methylobacterium spp. strain is related to the suppression of ET accumulation induced by the pathogen Xanthomonas campestris, resulting in a reduction of disease severity (Yim et al., 2014). Hence, both locally in the root and systemically in the leaves, ET modulates immune responses, either to accommodate mutualists or to counteract enemies.
The Role of ET in Volatile Signaling
Besides playing a vital role as endogenous plant hormone, ET is also involved in volatile signaling, especially during plant-insect interactions. ET is not only volatile by itself but also positively regulates the accumulation of volatile organic compounds, often together with JA (Fig. 1; Pierik et al., 2014). For example, exogenous application of the ET precursor ACC on lima bean (Phaseolus lunatus) leaves enhanced the production of three JA-mediated volatiles [i.e. (E)- and (Z)-β-ocimene and (Z)-3-hexenyl acetate], leading to the enhanced attraction of the carnivorous mite Phytoseiulus persimilis (Horiuchi et al., 2001). ET-mediated volatile production can alter insect behavior in an insect species-dependent manner. For example, silencing of ET biosynthesis in rice reduced the volatile release upon infestation by C. suppressalis caterpillars, leading to reduced plant resistance. In contrast, infestation of ET-silenced rice plants by the phloem-feeding brown plant hopper (Nilaparvata lugens) increased the emission of volatiles that repelled this insect (Lu et al., 2014). Besides altering insect behavior directly, ET-mediated volatile production can also play a role in indirect defense through the attraction of carnivorous enemies of the insect (Scala et al., 2013). For example, the S. exigua caterpillar-induced production of JA/ET-mediated volatile terpenes and benzoxazinoids in maize enhanced the attraction of the parasitoid Cotesia merginiventris, which is a natural enemy of S. exigua (Huffaker et al., 2013).
Finally, ET also plays a role in plant-plant communication, in which an attacked plant transfers volatile information to its neighboring plants, which in turn tailor their defenses (Holopainen and Blande, 2012). For example, exposure of intact maize plants to (Z)-3-hezen-1-ol, a volatile emitted by green plants upon mechanical damage, induced the emission of a volatile blend that is typically emitted after caterpillar feeding and attracts natural enemies of the herbivores. The volatile emission increased when ET was added, indicating a synergistic role for ET in plant-plant signaling mediated by (Z)-3-hezen-1-ol (Ruther and Kleier, 2005). Although there is a clear role for ET in plant volatile signaling, knowledge of the function of ET in plant-insect and plant-plant communication is still limited.
ATTACKERS HIJACK ET SIGNALING TO SUPPRESS PLANT DEFENSE
ET controls multiple aspects of plant defense by connecting different hormonal signaling pathways and often decisively modulating their relative output. This central role of ET makes it a valid target for attackers in their battle against induced plant defenses. Certain pathogens and insects activate diverse hormone signal integration mechanisms to manipulate the plant’s defense system, resulting in the circumvention or suppression of effectual defenses (Walling, 2008; Robert-Seilaniantz et al., 2011; Pieterse et al., 2012; Kazan and Lyons, 2014). ET plays a role in this decoy strategy of several attackers, resulting in increased plant susceptibility. For example, the soil-borne pathogen Fusarium oxysporum produces ET itself, leading to expression of the ET receptor gene ETR1 in Arabidopsis, which in turn leads to suppression of effective SA-dependent defenses, and thus promotion of Fusarium spp. disease (Pantelides et al., 2013). Additionally, saliva of Pieris rapae caterpillars activated the ERF branch of the JA signaling pathway in Arabidopsis, leading to suppression of effective defense responses that are mediated by the MYC branch (Verhage et al., 2011). Moreover, saliva of S. exigua caterpillars increased cellular oxidative stress in an ET-dependent manner in leaves of Medicago truncatula, which led to SA/NPR1-mediated suppression of JA-related defensive proteins (Paudel and Bede, 2015). This suggests that S. exigua engages the plant’s ET signaling pathway to activate SA-mediated antagonism of JA signaling for its own benefit. The role of ET in plant defense manipulation by attackers is still poorly understood, and future research in this direction may provide us with novel insight in regulatory components of hormone signal integration.
FUTURE PERSPECTIVES
In past decades, the pathways of ET biosynthesis and signaling have been elucidated in detail. Moreover, ET emerged as an important modulator of other hormonal signaling pathways that play a role in the regulation of plant growth and adaptive responses to biotic and abiotic stresses (Kazan, 2015; Vos et al., 2015). More and more molecular players active at the crossroad of hormone signaling pathways that regulate both growth and defense were identified (Pieterse et al., 2014a; Caarls et al., 2015). Knowledge of master regulators and their gene regulatory networks provides important tools to investigate how the plant's stress signaling network functions during different environmental conditions. In natural and agricultural settings, plants often have to cope with multiple biotic and abiotic stresses at the same time. So how do ET and other hormones steer the plant stress signaling network under these conditions of combinatorial stress? And how can we utilize this knowledge to improve our crops? To obtain a deep understanding of the dynamics of the hormone signaling network, it will be essential to investigate the dynamics of the transcriptome and proteome during single and multistress interactions. Computational analysis of high-density time series of transcriptome changes during ET-inducing stress conditions, such as leaf senescence (Breeze et al., 2011) or infection by the necrotrophic pathogen B. cinerea (Windram et al., 2012), or during cross talk between defense hormones (Van Verk et al., 2013) has been shown to be highly instrumental in the prediction and validation of unique players in the network. Unique technologies such as ChIP-seq (Chang et al., 2013) and protein interactome mapping (Braun et al., 2013) will further integrate ET signaling proteins and transcription factors in the dynamic hormone-mediated signaling network. These systems’ approaches will ultimately lead to a better understanding of how the plant growth regulator ET evolved as a traffic controller on the hormonal crossroads to defense.
Acknowledgments
We thank Roeland Berendsen, Silvia Proietti, and two anonymous reviewers for valuable comments on the article.
Glossary
- PRR
pattern recognition receptor
- MAMP
microbe-associated molecular pattern
- PTI
pattern-triggered immunity
- ET
ethylene
- MAPK
mitogen-activated protein kinase
- ETI
effector-triggered immunity
- JA
jasmonic acid
- SA
salicylic acid
- ABA
abscisic acid
- JAZ
jasmonic acid-Zim domain
- ISR
microbe-induced systemic resistance
Footnotes
This work was supported by the Dutch Technology Foundation STW, part of the Netherlands Organization of Scientific Research (VENI grant no. 13087 to C.B. and VIDI grant no. 11281 to S.C.M.V.W.), and the European Research Council (ERC advanced grant no. 269072 to C.M.J.P.).
References
- Acevedo FE, Rivera-Vega LJ, Chung SH, Ray S, Felton GW (2015) Cues from chewing insects - the intersection of DAMPs, HAMPs, MAMPs and effectors. Curr Opin Plant Biol 26: 80–86 [DOI] [PubMed] [Google Scholar]
- Anderson JP, Lichtenzveig J, Gleason C, Oliver RP, Singh KB (2010) The B-3 ethylene response factor MtERF1-1 mediates resistance to a subset of root pathogens in Medicago truncatula without adversely affecting symbiosis with rhizobia. Plant Physiol 154: 861–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17: 478–486 [DOI] [PubMed] [Google Scholar]
- Berendsen RL, Van Verk MC, Stringlis IA, Zamioudis C, Tommassen J, Pieterse CMJ, Bakker PAHM (2015) Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genomics 16: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Bodenhausen N, Reymond P (2007) Signaling pathways controlling induced resistance to insect herbivores in Arabidopsis. Mol Plant Microbe Interact 20: 1406–1420 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Boter M, Ruíz-Rivero O, Abdeen A, Prat S (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun P, Aubourg S, Van Leene J, De Jaeger G, Lurin C (2013) Plant protein interactomes. Annu Rev Plant Biol 64: 161–187 [DOI] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekgaarden C, Snoeren TAL, Dicke M, Vosman B (2011) Exploiting natural variation to identify insect-resistance genes. Plant Biotechnol J 9: 819–825 [DOI] [PubMed] [Google Scholar]
- Caarls L, Pieterse CMJ, Van Wees SCM (2015) How salicylic acid takes transcriptional control over jasmonic acid signaling. Front Plant Sci 6: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çevik V, Kidd BN, Zhang P, Hill C, Kiddle S, Denby KJ, Holub EB, Cahill DM, Manners JM, Schenk PM, et al. (2012) MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol 160: 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KN, Zhong S, Weirauch MT, Hon G, Pelizzola M, Li H, Huang SSC, Schmitz RJ, Urich MA, Kuo D, et al. (2013) Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2: e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, Cai R, Zuo J, Tang X, Li X, et al. (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Cook DE, Mesarich CH, Thomma BPHJ (2015) Understanding plant immunity as a surveillance system to detect invasion. Annu Rev Phytopathol 53: 541–563 [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol 66: 487–511 [DOI] [PubMed] [Google Scholar]
- Dicke M. (2015) Herbivore-induced plant volatiles as a rich source of information for arthropod predators: Fundamental and applied aspects. J Indian Inst Sci 95: 35–42 [Google Scholar]
- Diezel C, Allmann S, Baldwin IT (2011) Mechanisms of optimal defense patterns in Nicotiana attenuata: flowering attenuates herbivory-elicited ethylene and jasmonate signaling. J Integr Plant Biol 53: 971–983 [DOI] [PubMed] [Google Scholar]
- Dinh ST, Baldwin IT, Galis I (2013) The HERBIVORE ELICITOR-REGULATED1 gene enhances abscisic acid levels and defenses against herbivores in Nicotiana attenuata plants. Plant Physiol 162: 2106–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Savatin DV, Sicilia F, Gramegna G, Cervone F, Lorenzo GD (2013) Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front Plant Sci 4: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fracetto GGM, Peres LEP, Mehdy MC, Lambais MR (2013) Tomato ethylene mutants exhibit differences in arbuscular mycorrhiza development and levels of plant defense-related transcripts. Symbiosis 60: 155–167 [Google Scholar]
- Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64: 839–863 [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Groen SC, Whiteman NK (2014) The evolution of ethylene signaling in plant chemical ecology. J Chem Ecol 40: 700–716 [DOI] [PubMed] [Google Scholar]
- Han L, Li GJ, Yang KY, Mao G, Wang R, Liu Y, Zhang S (2010) Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J 64: 114–127 [DOI] [PubMed] [Google Scholar]
- Heil M. (2015) Extrafloral nectar at the plant-insect interface: a spotlight on chemical ecology, phenotypic plasticity, and food webs. Annu Rev Entomol 60: 213–232 [DOI] [PubMed] [Google Scholar]
- Holopainen JK, Blande JD (2012) Molecular plant volatile communication. Adv Exp Med Biol 739: 17–31 [DOI] [PubMed] [Google Scholar]
- Horiuchi J, Arimura G, Ozawa R, Shimoda T, Takabayashi J, Nishioka T (2001) Exogenous ACC enhances volatiles production mediated by jasmonic acid in lima bean leaves. FEBS Lett 509: 332–336 [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Hu J, Zhou J, Peng X, Xu H, Liu C, Du B, Yuan H, Zhu L, He G (2011) The Bphi008a gene interacts with the ethylene pathway and transcriptionally regulates MAPK genes in the response of rice to brown planthopper feeding. Plant Physiol 156: 856–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Veyrat N, Erb M, Turlings TCJ, Sartor R, Shen Z, Briggs SP, Vaughan MM, Alborn HT, et al. (2013) Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proc Natl Acad Sci USA 110: 5707–5712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kazan K. (2015) Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci 20: 219–229 [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM (2012) JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci 17: 22–31 [DOI] [PubMed] [Google Scholar]
- Kazan K, Lyons R (2014) Intervention of phytohormone pathways by pathogen effectors. Plant Cell 26: 2285–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatabi B, Molitor A, Lindermayr C, Pfiffi S, Durner J, von Wettstein D, Kogel KH, Schäfer P (2012) Ethylene supports colonization of plant roots by the mutualistic fungus Piriformospora indica. PLoS One 7: e35502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Reyes A, Du Y, Koornneef A, Proietti S, Körbes AP, Memelink J, Pieterse CMJ, Ritsema T (2010) Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic Acid. Mol Plant Microbe Interact 23: 187–197 [DOI] [PubMed] [Google Scholar]
- Leon-Reyes A, Spoel SH, De Lange ES, Abe H, Kobayashi M, Tsuda S, Millenaar FF, Welschen RAM, Ritsema T, Pieterse CMJ (2009) Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol 149: 1797–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León J, Castillo MC, Coego A, Lozano-Juste J, Mir R (2014) Diverse functional interactions between nitric oxide and abscisic acid in plant development and responses to stress. J Exp Bot 65: 907–921 [DOI] [PubMed] [Google Scholar]
- Li R, Afsheen S, Xin Z, Han X, Lou Y (2013) OsNPR1 negatively regulates herbivore-induced JA and ethylene signaling and plant resistance to a chewing herbivore in rice. Physiol Plant 147: 340–351 [DOI] [PubMed] [Google Scholar]
- Liu R, Chen L, Jia Z, Lü B, Shi H, Shao W, Dong H (2011a) Transcription factor AtMYB44 regulates induced expression of the ETHYLENE INSENSITIVE2 gene in Arabidopsis responding to a harpin protein. Mol Plant Microbe Interact 24: 377–389 [DOI] [PubMed] [Google Scholar]
- Liu WY, Chiou SJ, Ko CY, Lin TY (2011b) Functional characterization of three ethylene response factor genes from Bupleurum kaoi indicates that BkERFs mediate resistance to Botrytis cinerea. J Plant Physiol 168: 375–381 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Ju H, Zhou G, Zhu C, Erb M, Wang X, Wang P, Lou Y (2011) An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J 68: 583–596 [DOI] [PubMed] [Google Scholar]
- Lu J, Li J, Ju H, Liu X, Erb M, Wang X, Lou Y (2014) Contrasting effects of ethylene biosynthesis on induced plant resistance against a chewing and a piercing-sucking herbivore in rice. Mol Plant 7: 1670–1682 [DOI] [PubMed] [Google Scholar]
- Mase K, Mizuno T, Ishihama N, Fujii T, Mori H, Kodama M, Yoshioka H (2012) Ethylene signaling pathway and MAPK cascades are required for AAL toxin-induced programmed cell death. Mol Plant Microbe Interact 25: 1015–1025 [DOI] [PubMed] [Google Scholar]
- Matsukawa M, Shibata Y, Ohtsu M, Mizutani A, Mori H, Wang P, Ojika M, Kawakita K, Takemoto D (2013) Nicotiana benthamiana calreticulin 3a is required for the ethylene-mediated production of phytoalexins and disease resistance against oomycete pathogen Phytophthora infestans. Mol Plant Microbe Interact 26: 880–892 [DOI] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memelink J. (2009) Regulation of gene expression by jasmonate hormones. Phytochemistry 70: 1560–1570 [DOI] [PubMed] [Google Scholar]
- Mersmann S, Bourdais G, Rietz S, Robatzek S (2010) Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol 154: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, Ausubel FM (2010) Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22: 973–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LAJ, Lloyd AJ, Cristescu SM, Harren FJM, Hall MA, Smith AR (2009) Biphasic ethylene production during the hypersensitive response in Arabidopsis: a window into defense priming mechanisms? Plant Signal Behav 4: 610–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickstadt A, Thomma BPHJ, Feussner I, Kangasjärvi J, Zeier J, Loeffler C, Scheel D, Berger S (2004) The jasmonate-insensitive mutant jin1 shows increased resistance to biotrophic as well as necrotrophic pathogens. Mol Plant Pathol 5: 425–434 [DOI] [PubMed] [Google Scholar]
- Niu DD, Liu HX, Jiang CH, Wang YP, Wang QY, Jin HL, Guo JH (2011a) The plant growth-promoting rhizobacterium Bacillus cereus AR156 induces systemic resistance in Arabidopsis thaliana by simultaneously activating salicylate- and jasmonate/ethylene-dependent signaling pathways. Mol Plant Microbe Interact 24: 533–542 [DOI] [PubMed] [Google Scholar]
- Niu Y, Figueroa P, Browse J (2011b) Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onkokesung N, Baldwin IT, Gális I (2010) The role of jasmonic acid and ethylene crosstalk in direct defense of Nicotiana attenuata plants against chewing herbivores. Plant Signal Behav 5: 1305–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantelides IS, Tjamos SE, Pappa S, Kargakis M, Paplomatas EJ (2013) The ethylene receptor ETR1 is required for Fusarium oxysporum pathogenicity. Plant Pathol 62: 1302–1309 [Google Scholar]
- Paudel JR, Bede JC (2015) Ethylene signaling modulates herbivore-induced defense responses in the model legume Medicago truncatula. Mol Plant Microbe Interact 28: 569–579 [DOI] [PubMed] [Google Scholar]
- Pel MJC, Pieterse CMJ (2013) Microbial recognition and evasion of host immunity. J Exp Bot 64: 1237–1248 [DOI] [PubMed] [Google Scholar]
- Pierik R, Ballaré CL, Dicke M (2014) Ecology of plant volatiles: taking a plant community perspective. Plant Cell Environ 37: 1845–1853 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Pierik R, Van Wees SCM (2014a) Different shades of JAZ during plant growth and defense. New Phytol 204: 261–264 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM (2014b) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52: 347–375 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Pineda A, Zheng S-J, van Loon JJA, Pieterse CMJ, Dicke M (2010) Helping plants to deal with insects: the role of beneficial soil-borne microbes. Trends Plant Sci 15: 507–514 [DOI] [PubMed] [Google Scholar]
- Plett JM, Khachane A, Ouassou M, Sundberg B, Kohler A, Martin F (2014) Ethylene and jasmonic acid act as negative modulators during mutualistic symbiosis between Laccaria bicolor and Populus roots. New Phytol 202: 270–286 [DOI] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CMJ, Memelink J (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehrig EM, Appel HM, Jones AD, Schultz JC (2014) Roles for jasmonate- and ethylene-induced transcription factors in the ability of Arabidopsis to respond differentially to damage caused by two insect herbivores. Front Plant Sci 5: 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JDG (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Ruther J, Kleier S (2005) Plant-plant signaling: ethylene synergizes volatile emission in Zea mays induced by exposure to (Z)-3-hexen-1-ol. J Chem Ecol 31: 2217–2222 [DOI] [PubMed] [Google Scholar]
- Salas-Marina MA, Silva-Flores MA, Uresti-Rivera EE, Castro-Longoria E, Herrera-Estrella A, Casas-Flores S (2011) Colonization of Arabidopsis roots by Trichoderma atroviride promotes growth and enhances systemic disease resistance through jasmonic acid/ethylene and salicylic acid pathways. Eur J Plant Pathol 131: 15–26 [Google Scholar]
- Sašek V, Nováková M, Jindřichová B, Bóka K, Valentová O, Burketová L (2012) Recognition of avirulence gene AvrLm1 from hemibiotrophic ascomycete Leptosphaeria maculans triggers salicylic acid and ethylene signaling in Brassica napus. Mol Plant Microbe Interact 25: 1238–1250 [DOI] [PubMed] [Google Scholar]
- Savatin DV, Gramegna G, Modesti V, Cervone F (2014) Wounding in the plant tissue: the defense of a dangerous passage. Front Plant Sci 5: 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala A, Allmann S, Mirabella R, Haring MA, Schuurink RC (2013) Green leaf volatiles: a plant’s multifunctional weapon against herbivores and pathogens. Int J Mol Sci 14: 17781–17811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Huang H, Gao H, Wang J, Wu D, Liu X, Yang S, Zhai Q, Li C, Qi T, et al. (2014) Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SMC, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux JP, Brown R, Kazan K, et al. (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Does D, Leon-Reyes A, Koornneef A, Van Verk MC, Rodenburg N, Pauwels L, Goossens A, Körbes AP, Memelink J, Ritsema T, et al. (2013) Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25: 744–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ent S, Pieterse CMJ (2012) Ethylene: multi-tasker in plant–attacker interactions. Annu Plant Rev 44: 343–377 [Google Scholar]
- Van Loon LC, Geraats BP, Linthorst HJ (2006) Ethylene as a modulator of disease resistance in plants. Trends Plant Sci 11: 184–191 [DOI] [PubMed] [Google Scholar]
- Van Verk MC, Hickman R, Pieterse CMJ, Van Wees SCM (2013) RNA-Seq: revelation of the messengers. Trends Plant Sci 18: 175–179 [DOI] [PubMed] [Google Scholar]
- Van Wees SCM, Van der Ent S, Pieterse CMJ (2008) Plant immune responses triggered by beneficial microbes. Curr Opin Plant Biol 11: 443–448 [DOI] [PubMed] [Google Scholar]
- Verhage A, Vlaardingerbroek I, Raaymakers C, Van Dam NM, Dicke M, Van Wees SCM, Pieterse CMJ (2011) Rewiring of the jasmonate signaling pathway in Arabidopsis during insect herbivory. Front Plant Sci 2: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos IA, Moritz L, Pieterse CMJ, Van Wees SCM (2015) Impact of hormonal crosstalk on plant resistance and fitness under multi-attacker conditions. Front Plant Sci 6: 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos IA, Pieterse CMJ, Van Wees SCM (2013a) Costs and benefits of hormone-regulated plant defences. Plant Pathol 62: 43–55 [Google Scholar]
- Vos IA, Verhage A, Schuurink RC, Watt LG, Pieterse CMJ, Van Wees SCM (2013b) Onset of herbivore-induced resistance in systemic tissue primed for jasmonate-dependent defenses is activated by abscisic acid. Front Plant Sci 4: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling LL. (2008) Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol 146: 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C. (2015). How jasmonates earned their laurels: past and present. J Plant Growth Regul 34: 761–794 [Google Scholar]
- Windram O, Madhou P, McHattie S, Hill C, Hickman R, Cooke E, Jenkins DJ, Penfold CA, Baxter L, Breeze E, et al. (2012) Arabidopsis defense against Botrytis cinerea: chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell 24: 3530-3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Baldwin IT (2010) New insights into plant responses to the attack from insect herbivores. Annu Rev Genet 44: 1–24 [DOI] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Meldau S, Baldwin IT (2007) Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19: 1096–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim WJ, Kim KY, Lee YW, Sundaram SP, Lee Y, Sa TM (2014) Real time expression of ACC oxidase and PR-protein genes mediated by Methylobacterium spp. in tomato plants challenged with Xanthomonas campestris pv. vesicatoria. J Plant Physiol 171: 1064–1075 [DOI] [PubMed] [Google Scholar]
- Zamioudis C, Korteland J, Van Pelt JA, Van Hamersveld M, Dombrowski N, Bai Y, Hanson J, Van Verk MC, Ling HQ, Schulze-Lefert P, et al. (2015) Rhizobacterial volatiles and photosynthesis-related signals coordinate MYB72 in Arabidopsis roots during onset of induced systemic resistance and iron deficiency responses. Plant J 84: 308–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamioudis C, Pieterse CMJ (2012) Modulation of host immunity by beneficial microbes. Mol Plant-Microbe Interact 25: 139–150 [DOI] [PubMed] [Google Scholar]
- Zander M, Chen S, Imkampe J, Thurow C, Gatz C (2012) Repression of the Arabidopsis thaliana jasmonic acid/ethylene-induced defense pathway by TGA-interacting glutaredoxins depends on their C-terminal ALWL motif. Mol Plant 5: 831–840 [DOI] [PubMed] [Google Scholar]
- Zander M, La Camera S, Lamotte O, Métraux JP, Gatz C (2010) Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J 61: 200–210 [DOI] [PubMed] [Google Scholar]
- Zander M, Thurow C, Gatz C (2014) TGA transcription factors activate the salicylic acid-suppressible branch of the ethylene-induced defense program by regulating ORA59 expression. Plant Physiol 165: 1671–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhu Z, An F, Hao D, Li P, Song J, Yi C, Guo H (2014) Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 26: 1105–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Qi L, Liu X, Cai S, Xu H, Huang R, Li J, Wei X, Zhang Z (2014) The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol 164: 1499–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, An F, Feng Y, Li P, Xue L, A M, Jiang Z, Kim J-M, To TK, Li W, et al. (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA 108: 12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]