A transcription factor functions as a negative feedback modulator of two mitogen-activated protein kinases and thereby acts as an early suppressor of herbivore-induced defenses in rice.
Abstract
The mechanisms by which herbivore-attacked plants activate their defenses are well studied. By contrast, little is known about the regulatory mechanisms that allow them to control their defensive investment and avoid a defensive overshoot. We characterized a rice (Oryza sativa) WRKY gene, OsWRKY53, whose expression is rapidly induced upon wounding and induced in a delayed fashion upon attack by the striped stem borer (SSB) Chilo suppressalis. The transcript levels of OsWRKY53 are independent of endogenous jasmonic acid but positively regulated by the mitogen-activated protein kinases OsMPK3/OsMPK6. OsWRKY53 physically interacts with OsMPK3/OsMPK6 and suppresses their activity in vitro. By consequence, it modulates the expression of defensive, MPK-regulated WRKYs and thereby reduces jasmonic acid, jasmonoyl-isoleucine, and ethylene induction. This phytohormonal reconfiguration is associated with a reduction in trypsin protease inhibitor activity and improved SSB performance. OsWRKY53 is also shown to be a negative regulator of plant growth. Taken together, these results show that OsWRKY53 functions as a negative feedback modulator of MPK3/MPK6 and thereby acts as an early suppressor of induced defenses. OsWRKY53 therefore enables rice plants to control the magnitude of their defensive investment during early signaling.
To effectively combat herbivores, plants have evolved sophisticated mechanisms that provide several layers of constitutive and inducible defense responses. Constitutive defenses are physical and chemical defensive traits that plants express regardless of the presence of herbivores. By contrast, inducible defenses are mounted only after plants are attacked by an herbivore (Wu and Baldwin, 2010). Induced defensive responses are the result of highly coordinated sequential changes at the cellular level, changes that activate multiple signaling pathways. These pathways mainly include mitogen-activated protein kinase (MPK) cascades and signaling pathways mediated by phytohormones, such as jasmonic acid (JA), jasmonoyl-isoleucine (JA-Ile), salicylic acid (SA), and ethylene (ET; van Loon et al., 2006; Bonaventure, 2012; Erb et al., 2012). Through cross talk, both synergistic and antagonistic interactions, this signaling network plays a central role in herbivore-induced defense responses by activating transcription factors (TFs) and regulating the transcript levels of many genes (van Loon et al., 2006; Bonaventure, 2012; Erb et al., 2012).
MPK cascades in all eukaryotes including plants generally consist of three components: MPK kinase kinases (MEKKs), MPK kinases (MEKs), and MPKs; these components are sequentially activated by phosphorylation (Rodriguez et al., 2010) to transfer information from sensors to responses and are involved in diverse physiological functions, including cell division, development, hormone synthesis and signaling, and response to abiotic and biotic stresses (Nakagami et al., 2005; Rodriguez et al., 2010; Liu, 2012). An MPK cascade consisting of MEKK1, MEK1/MEK2, and MPK4 (Qiu et al., 2008; Rodriguez et al., 2010), for instance, controls plant defenses by modulating defense-related signaling, WRKY TFs, and other genes. Furthermore, MPK3/MPK6 in Arabidopsis (Arabidopsis thaliana) mediate Flagellin-Sensitive2-N-terminal 22-amino-acid peptide of flagellin recognition and activate defense-related WRKYs (phosphorylation) as well as the biosynthesis of phytoalexins such as camalexin (Asai et al., 2002; Menke et al., 2004; Ren et al., 2008); they also modulate the ET-signaling pathway and plant resistance to pathogens (Kim et al., 2003; Kim and Zhang, 2004; Yoo et al., 2008; Han et al., 2010). In Nicotiana attenuata, wound-induced protein kinase (WIPK) and SA-induced protein kinase (SIPK; orthologs of AtMPK3 and AtMPK6) have been reported to regulate several WRKYs and to be involved in JA- and SA-signaling pathways and herbivore-induced defense responses (Wu et al., 2007).
WRKYs, which specifically bind W-box sequences (TTGACC/T) in the promoter region of target genes, are one of the largest families of TFs in plants (Rushton et al., 2010). In Arabidopsis and rice, there are more than 70 and 100 WRKYs, respectively (Wu et al., 2005; Xie et al., 2005; Eulgem and Somssich, 2007). According to the number of WRKY domains and the features of their zinc-finger motifs, WRKY TFs are divided into three groups (Rushton et al., 2010). In addition to playing an important role in plant growth and development as well as in shaping plant responses to abiotic stresses, WRKYs, by acting as positive or negative regulators of the target genes, also figure in the regulation of plant defense responses to pathogens (Eulgem and Somssich, 2007; Pandey and Somssich, 2009; Rushton et al., 2010). WRKYs can function at different regulatory levels: in addition to being phosphorylated by protein kinases as stated above, they can also act upstream and downstream of receptors and phytohormones as well as upstream of proteinase kinases (Ciolkowski et al., 2008; Bakshi and Oelmüller, 2014). In Arabidopsis, for example, small peptides encoded by Precursor Protein of Plant Elicitor Peptide (PROPEP) genes act as damage-associated molecular patterns that are perceived by two Leu-rich repeat receptor kinases, Plant Elicitor Peptide Receptor1(PEPR1) and PEPR2, to amplify defense responses. WRKY33 binds to the promoter of the PROPEP genes in a stimulus-dependent manner and regulates their expression (Logemann et al., 2013). AtWRKY33 has also been found to regulate redox homeostasis, SA signaling, ET/JA-mediated cross communication, and camalexin biosynthesis and to be essential for defense against the necrotrophic fungus Botrytis cinerea (Zheng et al., 2006; Birkenbihl et al., 2012). In rice, OsWRKY30, which may be phosphorylated by OsMPK3, positively regulates resistance to the rice sheath blight fungus Rhizoctonia solani and the blast fungus Magnaporthe grisea (Peng et al., 2012; Shen et al., 2012). In N. attenuata, NaWRKY3 and NaWRKY6 control the biosynthesis of herbivore-induced JA and JA-Ile/-Leu and, subsequently, herbivore-induced defenses (Skibbe et al., 2008). While the role of WRKYs as activators of plant defense against herbivores is established, the underlying molecular mechanisms remain unresolved. Furthermore, little is known about the potential of WRKYs to act as negative regulators of herbivory-induced defense responses.
Rice, one of the most important food crops worldwide, suffers heavily from insect pests (Cheng and He, 1996). The striped stem borer (SSB) Chilo suppressalis is one of the major lepidopteran pests of rice and causes severe yield losses in China (Chen et al., 2011). SSB larvae bore into and feed on rice stems, which results in dead heart and white heads symptoms at the vegetative and reproductive stage, respectively (Cheng and He, 1996). SSB attack in rice induces the biosynthesis of a variety of phytohormones, including JA, SA, and ET, which, in turn, regulate defense responses, such as the production of herbivore-induced volatiles and the accumulation of trypsin protease inhibitors (TrypPIs; Lou et al., 2005; Zhou et al., 2009; Lu et al., 2011; Qi et al., 2011; Li et al., 2013; Wang et al., 2013). Given the importance of WRKYs in mediating signaling pathways and defense responses, we isolated the rice group I WRKY TF OsWRKY53 and elucidated its roles in herbivore-induced defense responses. OsWRKY53 localizes to the nucleus, has specific binding activity toward W-box elements, and can be phosphorylated by the cascade OsMEK4-OsMPK3/OsMPK6 (Chujo et al., 2007, 2014; Yoo et al., 2014). OsWRKY53 has also been found to positively modulate resistance to pathogens, such as M. grisea (Chujo et al., 2007) and is strongly induced by herbivore infestation (Zhou et al., 2011). However, whether and how OsWRKY53 can regulate herbivore-induced defense in rice is unclear.
In this study, we reveal that OsWRKY53 is rapidly induced by mechanical wounding but only slowly induced by herbivore attack. Through silencing and overexpressing OsWRKY53, we show that it negatively regulates OsMPK3/OsMPK6 activity as well as the levels of herbivore-induced JA, JA-Ile, and ET, which subsequently mediate the activity of TrypPIs and resistance to SSB. Our study reveals that OsWRKY53 is an important herbivore-responsive component that functions as a negative feedback modulator of MPK3/MPK6, which allows rice plants to control the magnitude of defensive investment against a chewing herbivore during early signaling.
RESULTS
cDNA Cloning and Expression Analysis of OsWRKY53
We screened rice plants for herbivore-induced transcripts using rice microarrays and found that one WRKY TF, OsWRKY53, was up-regulated after SSB infestation (Zhou et al., 2011). Through reverse transcription PCR, we obtained the full-length complementary DNA (cDNA) of OsWRKY53, which includes an open reading frame of 1,464 bp (Supplemental Fig. S1). Phylogenetic analysis of the characterized group I-type WRKYs from different species revealed that OsWRKY53 is homologous to ZmWRKY33 in Zea mays (Li et al., 2013), TaWRKY53-a and TaWRKY53-b in Triticum aestivum (Van Eck et al., 2010), NaWRKY6 in N. attenuata (Skibbe et al., 2008), and AtWRKY33 in Arabidopsis (Supplemental Fig. S1; Zheng et al., 2006), which share 69%, 67%, 64%, 51%, and 51% amino acid sequence identity with OsWRKY53, respectively.
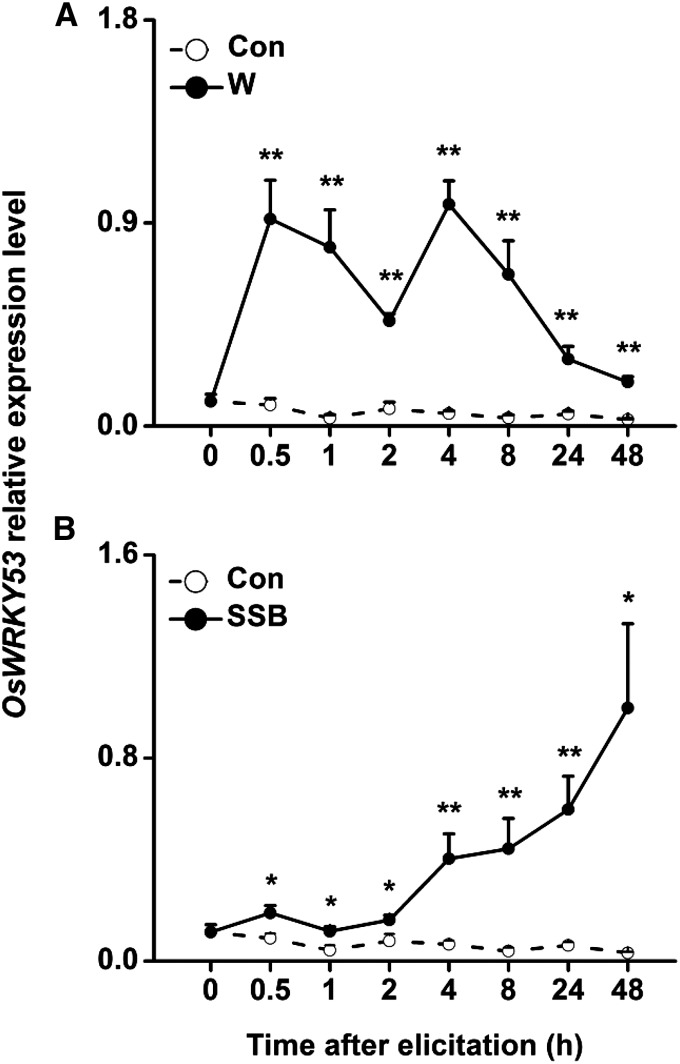
Quantitative real-time (qRT)-PCR analysis revealed that the OsWRKY53 gene is expressed at low levels in nonmanipulated wild-type plants, whereas mechanical wounding rapidly and strongly enhanced the mRNA levels of OsWRKY53 (Fig. 1). SSB larval feeding resulted in a slight increase in transcript levels in the stem after 1 and 2 h and a significant increase in OsWRKY53 transcript levels after 4 h (Fig. 1).
Figure 1.
Expression of OsWRKY53 in rice stem after different treatments. Mean transcript levels (+SE, n = 5) of OsWRKY53 in rice stems that were mechanically wounded (A) and infested by rice SSB (B). Transcript levels were analyzed by qRT-PCR. Asterisks indicate significant differences in transcript levels between treatments and controls (*, P < 0.05; and **, P < 0.01, Student’s t tests). Con, Control plants; W, wounded plants.
Overexpression and RNAi of OsWRKY53
To investigate the function of OsWRKY53 in herbivore resistance, we obtained four T2 homozygous lines consisting of two OsWRKY53-silenced lines (ir-wrky lines: ir-14 and ir-29) and two OsWRKY53 overexpression lines (oe-WRKY lines: oe-5 and oe-6), all of which contain a single transfer DNA insertion (Supplemental Fig. S2). Transcription analysis showed that wound-induced transcript levels of OsWRKY53 in the ir-wrky lines were approximately 30% of those in wild-type plants at 1 h after wounding (Supplemental Fig. S3). By contrast, transcript levels were significantly increased in the oe-5 (13.8- and 9.5- to 11.6-fold) and oe-6 (14.5- and 9.8- to 14.9-fold) lines without or with SSB infestation compared with transcript levels in equally treated wild-type plants (Supplemental Fig. S3). In rice, genes whose nucleotide sequences have the highest similarity to OsWRKY53 are OsWRKY70 (69.96%, accession no. Os05g39720), OsWRKY35 (66.58%, Os04g39570), and OsWRKY24 (60.00%, Os01g61080; data not shown). Transcription analysis revealed that the RNA interference (RNAi) construct did not cosilence the transcript accumulation of these genes (Supplemental Fig. S4), suggesting that the specificity of the RNAi sequence is high. When grown in the greenhouse or the paddy, the overexpression lines consistently showed a semidwarf phenotype, and the root and stem lengths of oe-WRKY lines were almost one-half those of the wild-type plants (Supplemental Figs. S5 and S6). In addition, the oe-WRKY lines were darker green than the wild-type plants, owing to increased chlorophyll content (Supplemental Figs. S5 and S6). Conversely, in ir-wrky lines, root length was slightly longer than in the oe-WRKY lines, whereas stem length and chlorophyll content were identical to those of wild-type plants (Supplemental Figs. S5 and S6). Overexpressing plants showed a much higher leaf angle (Supplemental Fig. S7), delayed flowering time, and produced fewer filled pollen grains (data not shown).
OsWRKY53 Negatively Regulates MPK Activity
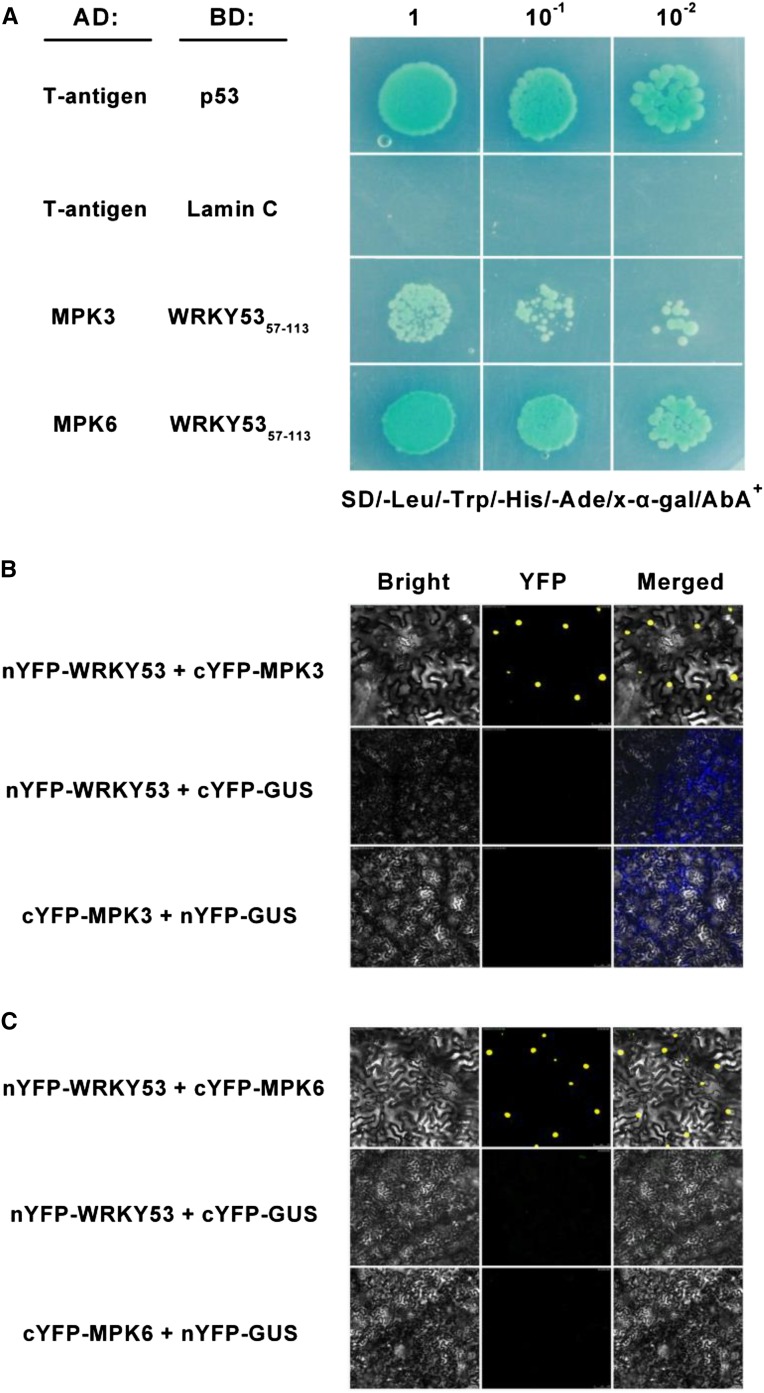
OsWRKY53 has been reported to be phosphorylated by the cascade OsMEK4-OsMPK3/OsMPK6, and phosphorylation enhances its transactivation activity (Chujo et al., 2014; Yoo et al., 2014). We here confirm that OsWRKY53 can physically interact with OsMPK3 or OsMPK6 in vitro and in vivo (Fig. 2; Supplemental Fig. S7). We investigated the interactions between OsWRKY53 and OsMPK3 or OsMPK6 in a yeast (Saccharomyces cerevisiae) two-hybrid (Y2H) assay system. Because the yeast transformed with full-length OsWRKY53 fused to the GAL4 DNA-binding domain showed autoactivation, we constructed the N-terminal region of OsWRKY53 (WRKY57–113), which contains the D domain and clustered proline-directed serines (SP cluster) and is sufficient for interaction with MPKs as bait based on the NbWRKY8 protein in tobacco (Nicotiana benthamiana; Ishihama et al., 2011). Positive interactions, revealed by β-galactosidase reporter activity (blue color) in the colonies, were observed only between one of the two MPKs and OsWRKY53, in addition to the positive control, which suggests that both OsMPK3 and OsMPK6 are capable of interacting with OsWRKY53 (Fig. 2A). To determine whether OsWRKY53 interacts with two MPKs in plant cells, bimolecular fluorescence complementation (BiFC) was performed in agroinfiltrated tobacco leaves. Pairwise expression of N-terminal part of yellow fluorescent protein (nYFP)-WRKY53/C-terminal part of yellow fluorescent protein (cYFP)-MPK3, cYFP-WRKY53/nYFP-MPK3, nYFP-WRKY53/cYFP-MPK6, and cYFP-WRKY53/nYFP-MPK6 resulted in a YFP fluorescence signal in the nucleus of agroinfiltrated cells at 72 h postinfiltration, whereas no fluorescence was detectable with combinations of nYFP-WRKY53/cYFP-GUS, cYFP-WRKY53/nYFP-GUS, nYFP-MPK3/cYFP-GUS, cYFP-MPK3/nYFP-GUS, nYFP-MPK6/cYFP-GUS, and cYFP-MPK6/nYFP-GUS (Fig. 2; Supplemental Fig. S7). From Supplemental Figure S8, it is clear that OsMPK3/OsMPK6-WRKY53 interactions occur in the nucleus. These results show that OsWRKY53 and OsMPK3/OsMPK6 are colocalized in nucleus and interact directly at the protein level in plant cells.
Figure 2.
OsWRKY53 interacts with OsMPK3/MPK6 in vitro and in vivo. A, Y2H analysis of the interaction between OsWRKY53 and MPK3/MPK6. Yeast was cotransformed with the constructs indicated, carrying a binding domain (BD) and an activation domain (AD), and was grown on synthetic dropout (SD)/-Leu/-Trp/-His/-Ade medium containing 5-Bromo-4-chloro-3-indoyl-α-galactosidel and 0.25 μg mL–1 aureobasidin A (AbA). T-antigen with p53 protein or with Lamin C served as positive and negative controls, respectively. B and C, BiFC visualization of WRKY53-MPK3 and WRKY53-MPK6 interactions. Tobacco leaves were cotransformed with the N-terminal part of YFP-fused WRKY53 or GUS (nYFP-WRKY53 and nYFP-GUS) and the C-terminal part of YFP-fused MPKs or GUS (cYFP-MPK3, cYFP-MPK6, and cYFP-GUS) by agroinfiltration.
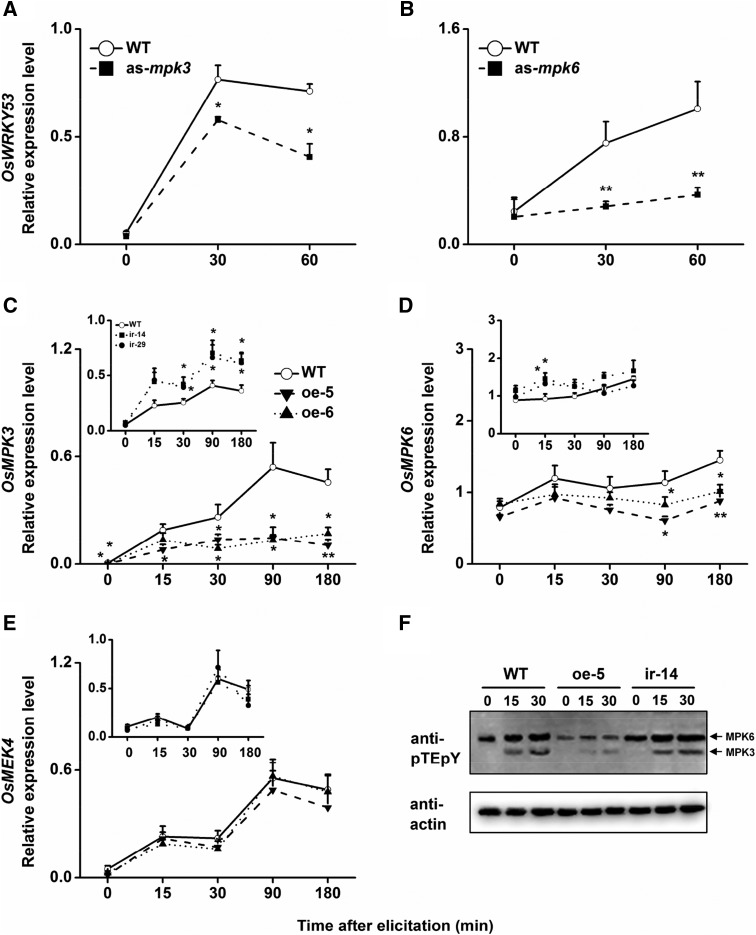
To examine if this interaction also influences transcript levels of OsWRKY53, we investigated the expression of OsWRKY53 in MPK mutants. We used the antisense expression lines OsMPK3 (as-mpk3) and OsMPK6 (as-mpk6), which had expression levels of 30% and 40% of OsMPK3 and OsMPK6 transcripts compared with wild-type plants (Lu et al., 2011; Wang et al., 2013). Transcript levels of OsWRKY53 were significantly reduced in as-mpk3 and as-mpk6 plants compared with wild-type plants measured 30 and 60 min after infestation with SSB larvae (Fig. 3).
Figure 3.
OsWRKY53 was downstream of MPK cascades but negatively regulated OsMPK3 and OsMPK6. A and B, Mean transcript levels (+se, n = 5) of OsWRKY53 in as-mpk3 (A) and as-mpk6 (B) lines and wild-type (WT) plants that were individually infested by a third-instar rice SSB larva. C to E, Mean transcript levels (+se, n = 5) of OsMPK3 (C), OsMPK6 (D), and OsMEK4 (E) in ir-wrky and oe-WRKY lines and wild-type plants that were individually infested by a third-instar SSB larva. F, SSB-elicited MPK activation in ir-wrky and oe-WRKY lines and wild-type plants. ir-wrky and oe-WRKY lines and wild-type plants were treated with or without SSB larva, and stems from five replicate plants were harvested at the indicated times. Immunoblotting was performed using either α-pTEpY antibody (top section) to detect phosphorylated MPKs or actin antibody (bottom section) as a loading control. This experiment was repeated three times. Asterisks indicate significant differences in ir-wrky, oe-WRKY, as-mpk3, and as-mpk6 lines compared with wild-type plants (*, P < 0.05; and **, P < 0.01, Student’s t tests).
We also measured transcription levels of OsMPK3, OsMPK6, and OsMEK4 in ir-wrky and oe-WRKY lines. Surprisingly, compared with wild-type plants, silencing OsWRKY53 increased the mRNA accumulation of OsMPK3 and OsMPK6, whereas overexpressing OsWRKY53 decreased their levels; moreover, the effect from oe-WRKY lines was bigger than that from ir-wrky lines, and the effect was stronger on OsMPK3 than on OsMPK6 (Fig. 3). To determine if this influence affects the activity of MPK3/MPK6, we used immunoblot analysis with an anti-phospho-extracellular signal-regulated kinase1 and 2 antibody to measure the activity of MPKs in wild-type and transgenic lines after SSB infestation. The result showed that SSB infestation quickly induced the activation of MPK3/MPK6 in wild-type plants. As the transcription results predicted, the activity of MPKs was lower in the oe-5 line and slightly higher in the ir-14 line than in wild-type plants (Fig. 3; Supplemental Fig. S9). These data show that OsWRKY53 functions as a repressor of MPK cascades.
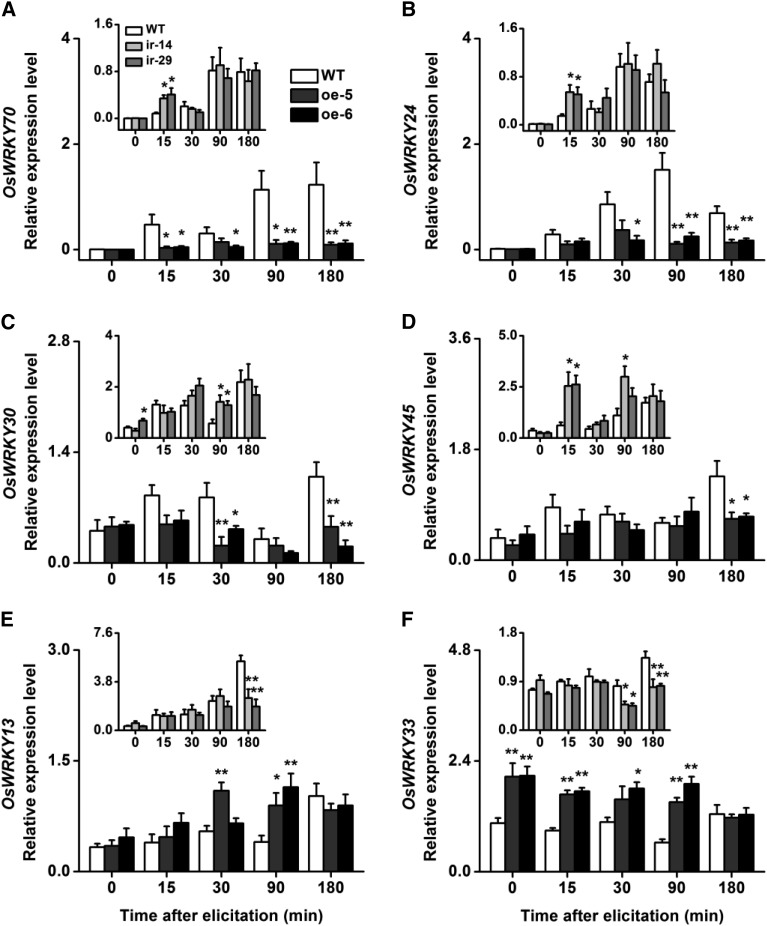
OsWRKY53 Regulates Other Defense-Related WRKYs
Autoregulation and cross regulation are common features of WRKY action (Ishihama and Yoshioka, 2012). Thus, we examined the transcript levels of OsWRKY70, OsWRKY24, OsWRKY30, OsWRKY45, OsWRKY13, WRKY35, and OsWRKY33, all of which have been reported to be involved in defense responses in rice (Qiu et al., 2007; Shimono et al., 2007; Koo et al., 2009; Li, 2012; Shen et al., 2012), in ir-wrky, oe-WRKY, and wild-type plants after SSB infestation. The results showed that silencing OsWRKY53 did not strongly change the elicited expression levels of the other WRKYs, whereas overexpression of OsWRKY53 altered WRKY mRNA levels, except the expression of OsWRKY35 (Fig. 4; Supplemental Fig. S4). Moreover, of the four WRKYs that were strongly influenced, OsWRKY33 was induced, but OsWRKY70, OsWRKY24, and OsWRKY30 were suppressed by overexpression of OsWRKY53 (Fig. 4).
Figure 4.
OsWRKY53 mediates the expression levels of defense-related OsWRKY genes. Mean transcript levels (+se, n = 5) of OsWRKY70 (A), OsWRKY24 (B), OsWRKY30 (C), OsWRKY45 (D), OsWRKY13 (E) and OsWRKY33 (F) in ir-wrky (insert) and oe-WRKY lines and wild-type (WT) plants that were individually infested by a third-instar rice SSB larva. Asterisks indicate significant differences in ir-wrky and oe-WRKY lines compared with wild-type plants (*, P < 0.05; and **, P < 0.01, Student’s t tests).
OsWRKY53 Is a Regulator of SSB-Elicited JA, JA-Ile, SA, and ET
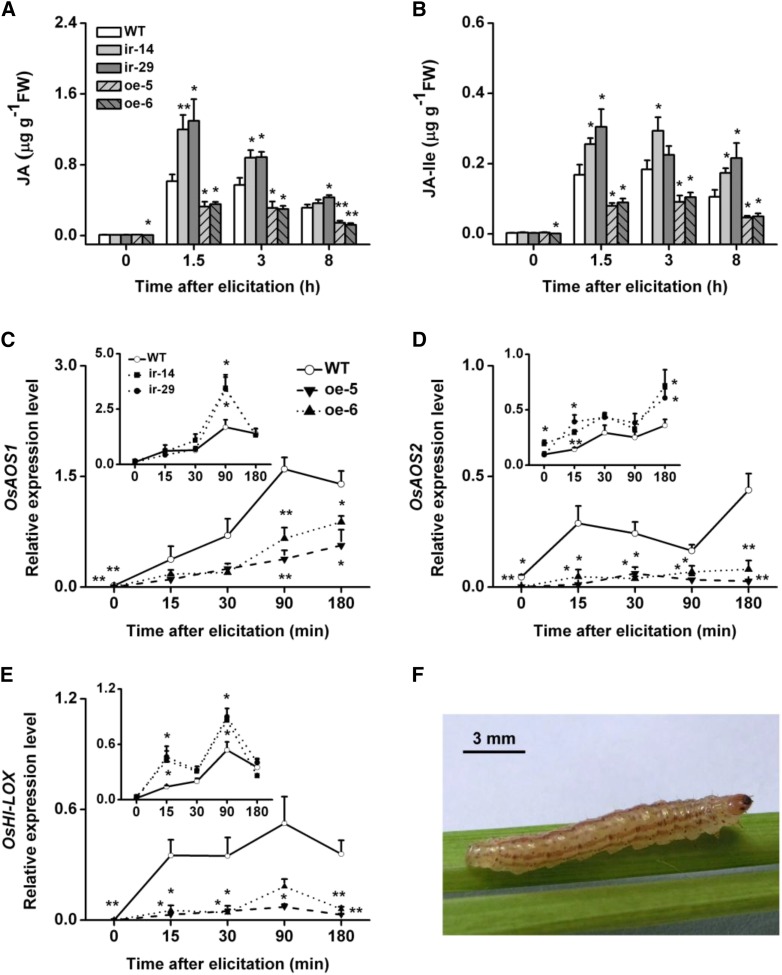
Plant hormones play major roles in plant defense (Ahuja et al., 2012; Erb et al., 2012; Nomura et al., 2012). The importance of JA, SA, and ET in rice defense against herbivores has also been reported previously (Zhou et al., 2009; Lu et al., 2011; Li et al., 2013; Wang et al., 2013). To evaluate whether the altered expression of OsWRKY53 affected the production of JA, JA-Ile, SA, and ET, levels of these phytohormones were quantified in ir-wrky, oe-WRKY, and wild-type plants after SSB infestation. Basal JA and JA-Ile levels were similar between the ir-wrky lines and wild-type plants, whereas JA and JA-Ile levels in the ir-wrky lines were significantly increased (by approximately 95%–110% and 52%–82% at 1.5 h after SSB infestation), compared with those of wild-type plants in response to SSB attack. In agreement with this finding, overexpression lines showed significantly decreased constitutive (in one line oe-6) and SSB-induced JA and JA-Ile levels (reduced by 42%–61% and 43%–56%, respectively; Fig. 5). Consistent with the JA and JA-Ile levels, the transcript levels of JA biosynthesis-related genes, an herbivore-induced 13-lipoxygenase gene (OsHI)-LOX (Zhou et al., 2009), and two putative allene oxide synthase genes, OsAOS1 and OsAOS2 (Supplemental Fig. S10), were decreased in oe-WRKY lines and slightly enhanced in ir-wrky lines (Fig. 5).
Figure 5.
OsWRKY53 negatively mediates rice SSB-induced JA and JA-Ile biosynthesis. A and B, Mean levels (+se, n = 5) of JA (A) and JA-Ile (B) in ir-wrky and oe-WRKY lines and wild-type (WT) plants that were individually infested by a third-instar rice SSB larva. C to E, Mean transcript levels (+se, n = 5) of JA biosynthesis-related genes OsAOS1 (C), OsAOS2 (D), and OsHI-LOX (E) in ir-wrky and oe-WRKY lines and wild-type plants that were individually infested by a third-instar SSB larva. Asterisks indicate significant differences in ir-wrky and oe-WRKY lines compared with wild-type plants (*, P < 0.05; and **, P < 0.01, Student’s t tests). F, An SSB larva. FW, Fresh weight.
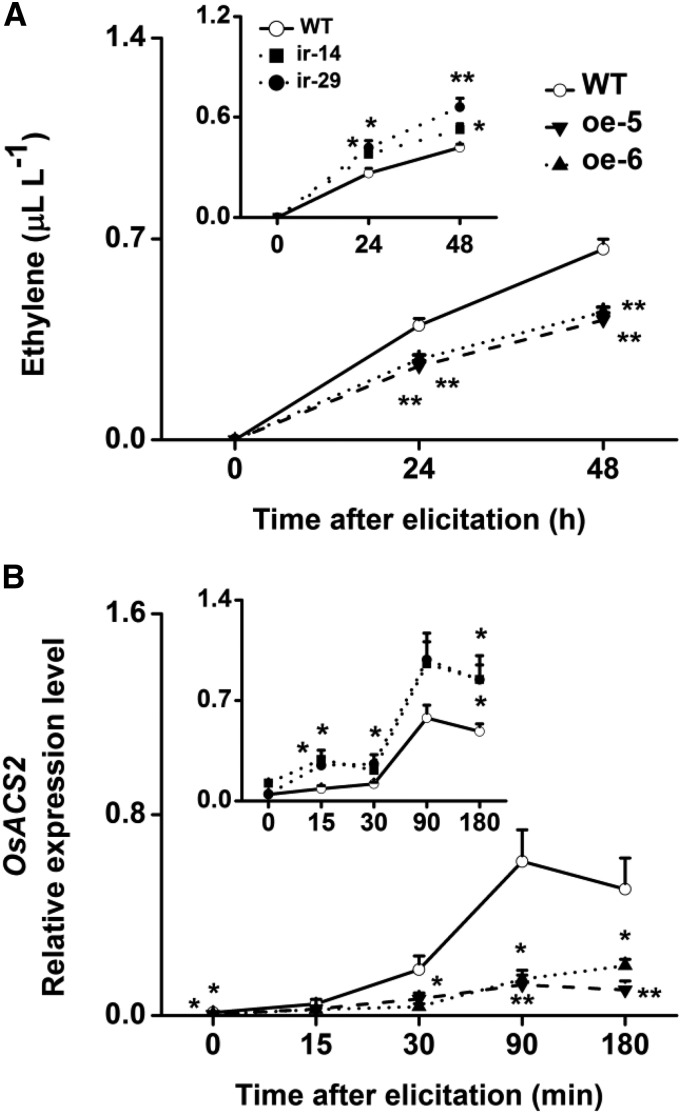
The wild-type plants and transgenic lines (ir-wrky and oe-WRKY lines) showed similar constitutive SA levels, whereas the SA levels were increased in oe-WRKY lines and decreased in ir-wrky lines after SSB infestation, although SSB infestation did not induce the biosynthesis of SA in wild-type plants (Supplemental Fig. S11). The transcript levels of an isochorismate synthase gene OsICS1 that is involved in herbivore-induced SA biosynthesis in rice (Wang, 2012) were also positively regulated by OsWRKY53 (Supplemental Fig. S11). A significantly lower accumulation of ET in the oe-WRKY lines and higher production in the ir-wrky lines compared with wild-type plants were observed at 24 and 48 h after infestation with SSB larvae (Fig. 6). The different levels of ET accumulation in transgenic plants compared with in wild-type plants correlate with distinct transcript levels of the OsACS2 gene, which encodes the ET biosynthetic enzyme 1-aminocyclopropane-1-carboxylic acid synthase (ACS; Fig. 6; Lu et al., 2014).
Figure 6.
OsWRKY53 mediates rice SSB-induced ET accumulation. A, Mean levels (+se, n = 5) of ET in ir-wrky (insert) and oe-WRKY lines and wild-type (WT) plants that were individually infested by a third-instar SSB larva. B, Mean transcript levels (+se, n = 5) of OsACS2 in ir-wrky (insert) and oe-WRKY lines and wild-type plants that were individually infested by a third-instar SSB larva. Asterisks indicate significant differences in ir-wrky and oe-WRKY lines compared with wild-type plants (*P < 0.05; and **P < 0.01, Student’s t tests).
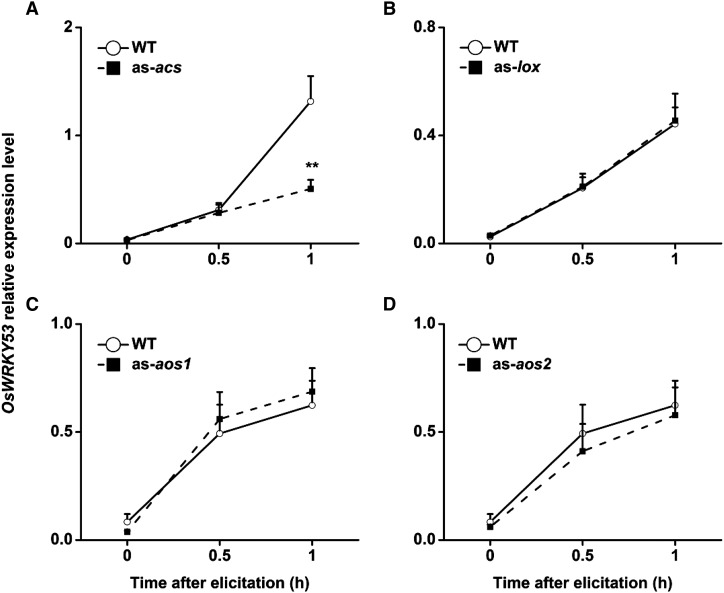
To explore the notion that OsWRKY53 may be an upstream component that regulates the biosynthesis of these signals, we investigated the expression of OsWRKY53 in transgenic plants with impaired JA or ET biosynthesis. We used our previous transgenic lines with antisense expression of OsHI-LOX (as-lox; Zhou et al., 2009), OsAOS1 (as-aos1), and OsAOS2 (as-aos2; Supplemental Fig. S10), all of which produced remarkably lower JA levels compared with those found in wild-type plants when infested by SSB larvae, as well as with antisense expression of OsACS (as-acs), which produced significantly less SSB-elicited ET than was found in wild-type plants (Lu et al., 2014). The levels of constitutive and induced OsWRKY53 transcripts in as-lox, as-aos1, and as-aos2 plants were identical to those in wild-type plants, whereas levels of induced OsWRKY53 transcripts in as-acs plants were significantly lower than in wild-type plants (Fig. 7). These results indicate that OsWRKY53 is induced upstream of the JA pathway but may form a negative feedback loop with the ET pathway.
Figure 7.
OsWRKY53 transcripts in JA and ET biosynthesis mutants. Mean transcript levels (+se, n = 5) of OsWRKY53 in as-acs (A), as-lox (B), as-aos1 (C), and as-aos2 (D) lines and wild-type (WT) plants that were individually infested by a third-instar SSB larva. Asterisks indicate significant differences in as-acs, as-lox, as-aos1, and as-aos2 lines compared with wild-type plants (*, P < 0.05; and **, P < 0.01, Student’s t tests).
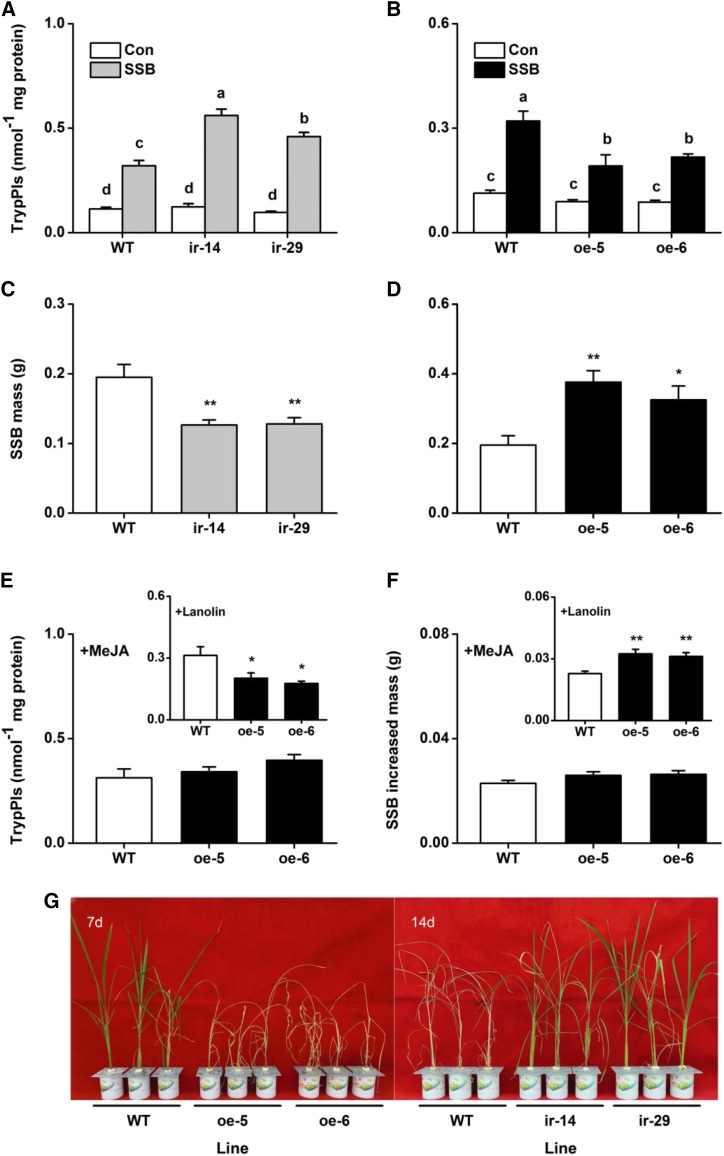
OsWRKY53 Lowers Levels of TrypPIs and Resistance to SSB
TrypPIs are important direct defense proteins that help plants resist herbivores that chew on rice, and their activity was regulated by JA- and ET-mediated signaling pathways (Zhou et al., 2009; Lu et al., 2014). Therefore, we investigated TrypPI activity and SSB performance on transgenic lines and on wild-type plants. SSB-induced TrypPI activity was enhanced in the ir-wrky lines and suppressed in the oe-wrky lines unlike in wild-type plants (Fig. 8). Consistent with the TrypPI activity, larvae of SSB gained less mass on the ir-wrky lines than on wild-type plants. By day 12, the mass of larvae that fed on the ir-wrky lines was about 65% of larvae that fed on wild-type plants (Fig. 8). By contrast, the mass of SSB larvae that fed on the oe-wrky lines oe-5 and oe-6 was 1.93- and 1.67-fold higher than the mass of SSB larvae that fed on wild-type plants (Fig. 8). Moreover, the oe-WRKY lines were more severely damaged by SSB larvae than were the wild-type plants, whereas the ir-wrky lines were less damaged (Fig. 8).
Figure 8.
OsWRKY53 negatively regulates TrypPI production and resistance of rice to the SSB. A and B, Mean TrypPI activities (+se, n = 5) in ir-wrky and oe-WRKY lines and wild-type (WT) plants that were individually infested by a third-instar SSB larva for 3 d. C and D, Mean larval mass (+se, n = 60) of SSB that fed on ir-wrky and oe-WRKY lines or wild-type plants for 12 d. E, Mean activities (+se, n = 5) of TrypPIs in oe-WRKY lines and wild-type plants that were individually treated with 100 μg of MeJA in 20 μL of lanolin paste (MeJA) or with 20 μL of pure lanolin (insert), followed by a SSB larva feeding for 3 d. F, Mean increased larval mass (+se, n = 60) of SSB larvae 12 d after they fed on oe-WRKY lines and wild-type plants that were individually treated with 100 μg of MeJA in 20 μL of lanolin paste (MeJA) or with 20 μL of pure lanolin (insert). G, Damaged phenotypes of ir-wrky and oe-WRKY lines and wild-type plants that were individually infested by a third-instar SSB larva for 14 or 7 d (n = 20). Letters indicate significant differences between lines (P < 0.05, Duncan’s multiple range test). Asterisks indicate significant differences in ir-wrky and oe-WRKY lines compared with wild-type plants (*, P < 0.05; and **, P < 0.01, Student’s t tests). Con, Control plants.
To determine whether impaired resistance to herbivores and compromised defense responses in oe-WRKY plants could be due to lower JA and JA-Ile levels, we treated the overexpression lines with 100 μg of methyl jasmonate (MeJA). This direct JA complementation restored TrypPI activity in oe-WRKY plants to the levels observed in wild-type plants (Fig. 8). Larvae of SSB that fed on MeJA-treated oe-WRKY plants showed the same low growth rate as larvae that fed on wild-type plants (Fig. 8). These results show that the attenuated TrypPI accumulation and resistance to SSB of the oe-WRKY lines is probably largely caused by defective jasmonate signaling, which is negatively mediated by OsWRKY53.
DISCUSSION
In this study, we elucidate the mechanism by which OsWRKY53 acts as a negative regulator of rice defenses and growth. Several lines of evidence point to a key role of OsWRKY53 in controlling induced rice defense responses against SSB. First, the expression levels of OsWRKY53 are induced when plants are wounded or infested with a chewing herbivore (Fig. 1). Second, OsWRKY53 interacts directly with the MPK proteins OsMPK3 and OsMPK6 (Fig. 2) in a feedback loop (Fig. 3). Third, altering expression of OsWRKY53 affects the elicited accumulation of JA, JA-Ile, SA, and ET and the expression of their biosynthesis genes (Figs. 5 and 6; Supplemental Fig. S11). Fourth, mutants with impaired JA pathway do not influence the levels of OsWRKY53 transcripts, but the ET biosynthesis mutant decreases the expression of OsWRKY53 (Fig. 7). Finally, OsWRKY53 regulates the production of defense compounds, such as TrypPIs, and resistance in rice to SSB (Fig. 8).
OsWRKY53 Functions as a Negative Feedback Modulator of MPK3/MPK6-Mediated Plant Defense Responses
WRKYs can act as positive or negative regulators of the target genes and function at different regulatory levels (Ciolkowski et al., 2008; Rushton et al., 2010; Bakshi and Oelmüller, 2014), and MPKs can mediate the activity of WRKYs via transcriptional and translational regulation (Ishihama et al., 2011; Li et al., 2012). Both OsMPK3 and OsMPK6 have been reported to phosphorylate OsWRKY53 (Yoo et al., 2014). Here, we found that OsWRKY53 negatively influenced the activity of OsMPK3 and OsMPK6 in turn. OsWRKY53 overexpression in particular strongly suppressed MPK activity (Fig. 3F). The relatively weak influence of OsWRKY53 silencing on MPK activities, which is also reflected in weaker phytohormone and gene expression patterns, may be caused by functional redundancy with other homologous WRKY genes or noncomplete silencing of OsWRKY53. Our results suggest that OsWRKY53 and OsMPK3/OsMPK6 form an interactive loop: OsMPK3 and OsMPK6 elicit the activity of OsWRKY53, whereas the activated OsWRKY53 suppresses the activity of MPK3 and MPK6, acting as a negative feedback regulator. It has been reported that WIPK and SIPK in N. attenuata, the homologs of MPK3 and MPK6 in rice, can regulate each other at the transcriptional level (Wu et al., 2007). Thus, it is possible that OsWRKY53 directly suppresses the activity of one of the two MPKs and then influences the activity of the other indirectly by the interaction between the two MPKs. The mechanism on how OsWRKY53 inhibits MPK3/MPK6 activities might be related to OsWRKY53 regulation of MPK3/MPK6 phosphorylation: by interacting physically with MPK3/MPK6, OsWRKY53 may prevent access of mitogen-activated protein kinase phosphatases to the MPKs. Further experiments will be required to test these hypotheses.
Given the fact that MPK3 and MPK6 play an important role in plant defense responses by regulating defense-related signaling pathways, such as JA, SA, and ET (Schweighofer et al., 2007; Li et al., 2012; Tsuda et al., 2013; Wang et al., 2013), and that herbivore infestation induced the expression of OsWRKY53 at later time points (Fig. 1), we propose that OsWRKY53 may function mainly as a regulator for herbivore-induced defense responses and may allow plants to control the strength of their defense response and investment during early signaling. SSB infestation elicits a MPK3-dependent JA burst (Wang et al., 2013) that reaches a maximum at 3 h after infestation and subsides to control levels at 8 h (Zhou et al., 2011). The early expression pattern of OsWRKY53 upon SSB attack fits its role as a negative regulator that contributes to bringing JA signaling down after the initial burst (Fig. 1B). In rice, other negative modulators of herbivore-induced defenses, such as 9-lipoxygenase (Osr9)-LOX1 (Zhou et al., 2014) and OsNPR1 (Li et al., 2013), have been described. In other plants, SA signaling and jasmonate catabolism have been shown to be involved in attenuating herbivore defenses (Pieterse and Van Loon, 2004; Campos et al., 2014). This suggests that plants possess a set of mechanisms to control the magnitude of herbivore-induced defenses in space and time. Because of its involvement upstream of phytohormone signaling, OsWRKY53 is among the earliest modulators described so far in this context. Interestingly, we also found that the expression level of OsWRKY53 was continuously up-regulated by SSB infestation up to 48 h (Fig. 1B). Because low JA levels impair resistance of rice to SSB (Zhou et al., 2009), this phenomenon opens questions that need to be elucidated in the future. Especially, the role of OsWRKY53 at later stages of SSB infestation should be addressed.
OsWRKY53 and Its Regulation on Other WRKYs and Phytohormones
Increasing evidence shows that both MPKs and WRKYs can modulate the biosynthesis of JA, JA-Ile, SA, and ET by directly regulating the activity of related enzymes (Li et al., 2006; Wu et al., 2007; Skibbe et al., 2008; Birkenbihl et al., 2012; Li et al., 2012). In Arabidopsis, for example, AtMPK6 can directly phosphorylate AtACS2 and AtACS6, which subsequently elevates ACS activities and the production of ET (Liu and Zhang, 2004); WRKY33 modulates the expression of ACS2 and ACS6 by binding to the W-boxes in the promoters of the two genes (Liu and Zhang, 2004; Li et al., 2012). We found that OsWRKY53 negatively modulated the production of elicited JA, JA-Ile, and ET as well as the transcript levels of JA and ET biosynthesis-related genes, such as OsHI-LOX and OsACS2 (Figs. 5 and 6), whereas it positively influenced the accumulation of SA after SSB infestation, including the transcript level of a SA biosynthesis-related gene ICS1 (Supplemental Fig. S11). Because SSB infestation did not elicit the production of SA in wild-type plants, the latter suggests that OSWRKY53 plays a role in SA homeostasis. Moreover, OsWRKY53 also affected transcript levels of other WRKYs (Fig. 4). In rice, OsMPK3/OsMPK6 and these OsWRKYs are known to be involved in regulating signaling pathways and defense responses, and it seems that OsWRKY53 negatively mediates the components activating JA and ET pathways but positively regulates the components activating the SA pathway. OsWRKY13 and OsWRKY33 (Qiu et al., 2007; Koo et al., 2009), for instance, both of which suppress the JA-dependent pathway but activate the SA-dependent pathway by regulating the transcript levels of JA biosynthesis- or SA biosynthesis-related genes, such as AOS2, LOX, and ICS1, were positively modulated by OsWRKY53. OsMPK3 (Wang et al., 2013), OsWRKY30 (Peng et al., 2012), and OsWRKY70 and OsWRKY24 (Li, 2012), all of which have been reported to positively regulate the JA and ET pathways, were negatively regulated by OsWRKY53. Given the fact that MPKs can modulate the activity of WRKYs as stated above and that WRKYs can regulate each other (Xu et al., 2006; Chen et al., 2009; Besseau et al., 2012; Chi et al., 2013), the influence of OsWRKY53 on these WRKYs and on phytohormone biosynthesis might occur via its direct and indirect (by mediating MPKs and other WRKYs) regulation. Here, we observed some synchronized changes between OsMPK3/OsMPK6 and some WRKYs, such as OsWRKY70 and OsWRKY30, both of which have been reported to be positively regulated by these MPKs (Li, 2012; Shen et al., 2012). Therefore, the indirect regulation of OsWRKY53, i.e., its functioning as a negative feedback regulator of OsMPK3/OsMPK6 as stated above, may also play an important role in regulating the biosynthesis of phytohormones. Further research should investigate the direct target genes of OsWRKY53 and elucidate which OsWRKYs and/or OsMPKs can directly mediate the activity of phytohormone biosynthesis-related enzymes.
In addition, we also found that altering OsWRKY53 expression influenced the growth phenotype of plants, especially oe-WRKY lines (Supplemental Figs. S5 and S6). In Arabidopsis, WRKY53 regulates leaf senescence and leaf development (Zentgraf et al., 2010; Xie et al., 2014). Moreover, in rice, the homologs of OsWRKY53, OsWRKY70 (Li, 2012), and OsWRKY24 (Zhang et al., 2009) negatively mediate the biosynthesis of GAs and/or abscisic acid and their signaling. Thus, the effect of OsWRKY53 on plant growth may be related to its influence on these phytohormones. Interestingly, the characteristics of the effect of OsWRKY53 on plant growth we observed here contradict what Chujo et al. (2007) found. This difference might be related to different levels of OsWRKY53 transcripts in mutants and the different genetic backgrounds. It has been reported that different transcription levels of a target gene caused different growth phenotypes (Kang et al., 2006). The mechanism of OsWRKY53 underlying rice morphological alterations is worthy of elucidation in the future.
CONCLUSION
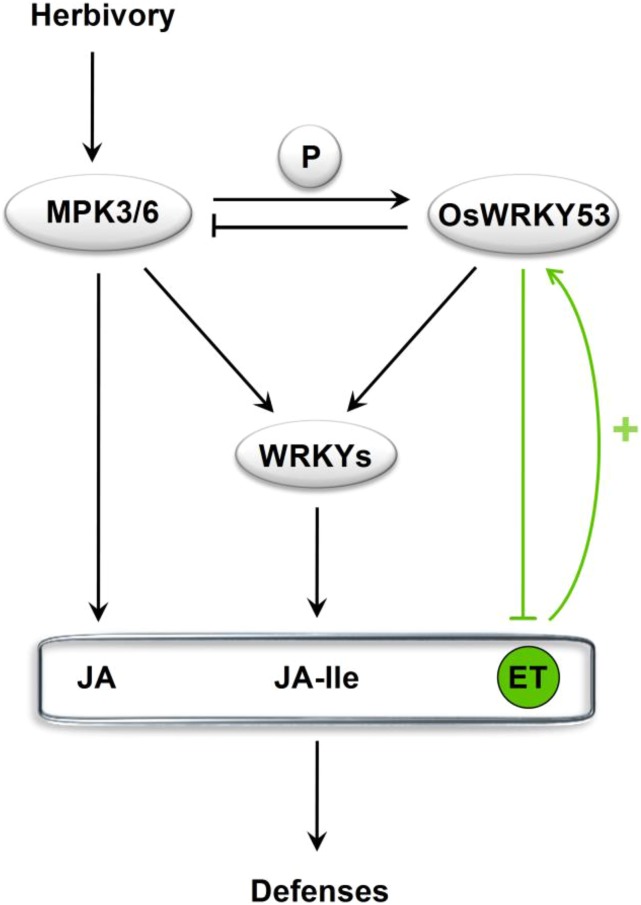
In summary, our results demonstrate that OsWRKY53 is a regulator of herbivore-induced defense responses in rice (Fig. 9). When infested by an herbivore, rice plants perceive the signals from the herbivore and immediately activate MPKs, such as OsMPK3 and OsMPK6; these subsequently increase the activity of some OsWRKYs, except for OsWRKY53, such as OsWRKY70. The activated MPKs and WRKYs then regulate the biosynthesis of defense-related signal molecules, including JA, JA-Ile, and ET. Moreover, the activated OsMPK3 and OsMPK6 also gradually activate OsWRKY53 and then enhance its transcript level, which, in turn, inhibits OsMPK3 and OsMPK6 directly and indirectly by the interaction of the two MPKs and thereby controls the magnitude of the plant’s defense response. This system likely enables plants to fine-tune the activity of their defensive investment in space and time in a highly coordinated fashion.
Figure 9.
Preliminary model summarizing how OsWRKY53 regulates herbivore-induced signaling pathways and defenses. Plants recognize signals from wounding and herbivore infestation and quickly transduce these to MPK cascades, which leads to the activation of OsMPK3/OsMPK6. Active OsMPK3/OsMPK6 activates some WRKYs, and thus both OsMPK3/OsMPK6 and WRKYs regulate the biosynthesis of defense-related signals, such as JA, JA-Ile, and ET. The activated OsMPK3/OsMPK6 gradually elicits OsWRKY53 by phosphorylating it. Moreover, the ET pathway may also positively mediate the activity of OsWRKY53. OsWRKY53 can inhibit the activity of OsMPK3/OsMPK6 directly and indirectly by the interaction of the two MPKs and may mediate other WRKYs with an unknown way, which keeps the defense response at an appropriate level. Arrows represent regulation negatively or positively; barred lines represent negative regulation; and arrows with a plus symbol represent positive regulation. Lines in green represent the OsWRKY53-ET regulation loop.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The rice (Oryza sativa) genotypes used in this study were cv Xiushui 11 wild-type and transgenic lines of ir-wrky, oe-WRKY, as-acs (Lu et al., 2014), as-aos1, as-aos2 (Supplemental Fig. S10), as-mpk3, as-mpk6 (Wang et al., 2013), and as-lox (Zhou et al., 2009). Pregerminated seeds of the different lines were cultured in plastic bottles (diameter, 8 cm; height, 10 cm) in a greenhouse (28°C ± 2°C, 14-h light, 10-h dark). Ten-day-old seedlings were transferred to 20-L hydroponic boxes with a rice nutrient solution (Yoshida et al., 1976). After 40 d, seedlings were transferred to individual 500-mL hydroponic plastic pots. Plants were used for experiments 4 to 5 d after transplanting.
Insects
An SSB colony was originally obtained from rice fields in Hangzhou, China, and maintained on rice seedlings of TN1, a rice variety that is susceptible to infestation by SSB. All of the plants were kept in a controlled climate chamber at 26°C ± 2°C, with a 12-h photoperiod and 80% relative humidity.
Isolation and Characterization of OsWRKY53 cDNA
The full-length cDNA of OsWRKY53 was PCR amplified. The primers WRKY-F (5′-CGTTCTCGTCTCCGATCACT-3′) and WRKY-R (5′-ATACGGCGAGGCGAAAATAC-3′) were designed based on the sequence of rice OsWRKY53. The PCR products were cloned into the pMD19-T vector (TaKaRa) and sequenced.
Phylogenetic Analysis
For the phylogenetic analysis, the program MEGA 6.0 (Tamura et al., 2013) was used. The protein sequences aligned using the ClustalW method in MEGA 6.0 (pairwise alignment: gap opening penalty 10, gap extension penalty 0.1; multiple alignment: gap opening penalty 10, gap extension penalty 0.2, protein weight matrix using Gonnet). The residue-specific and hydrophilic penalties were on, and the end gap separation and the use negative separation matrix were off. Gap separation distance was 4, and the delay divergence cutoff (percentage) was at 30. This alignment (available as Supplemental Data Set S1) was then used to generate an unrooted tree with statistical tests (parameters for phylogeny reconstruction were neighbor-joining method [Saitou and Nei, 1987] and bootstrap [Felsenstein, 1985], n = 1,000, amino acid, Poisson model, rate among sites: uniform rates gaps/missing, data treatment: complete deletion, traditional tree without modification for graphics) using MEGA 6.0.
qRT-PCR
For qRT-PCR analysis, five independent biological samples were used. Total RNA was isolated using the SV Total RNA Isolation System (Promega) following the manufacturer’s instructions. One microgram of each total RNA sample was reverse transcribed using the PrimeScript RT-PCR Kit (TaKaRa). The qRT-PCR assay was performed on CFX96 Real-Time system (Bio-RAD) using the SsoFast Probes Supermix (Bio-RAD). A linear standard curve, threshold cycle number versus log (designated transcript level), was constructed using a series dilution of a specific cDNA standard, and the relative levels of the transcript of the target gene in all unknown samples were determined according to the standard curve. A rice actin gene OsACT (accession no. Os03g50885) was used as an internal standard to normalize cDNA concentrations. The primers and probes used for qRT-PCR for all tested genes are listed in Supplemental Table S1.
Generation and Characterization of Transgenic Plants
The full-length cDNA sequence and a 333-bp fragment of OsWRKY53 were inserted into the pCAMBIA-1301 transformation vector to yield an overexpression and an RNAi construct, respectively (Supplemental Fig. S12). Both vectors were inserted into the rice variety Xiushui 11 using Agrobacterium tumefaciens-mediated transformation. The transformation of rice, the screening of the homozygous T2 plants, and the identification of the number of insertions followed the same method as described in Zhou et al. (2009). Two T2 homozygous lines (ir-14 and ir-29) of ir-wrky and two lines (oe-5 and oe-6) of oe-WRKY, each harboring a single insertion (Supplemental Fig. S2), were used in subsequent experiments.
Plant Treatments
For mechanical wounding, the lower portion of plant stems (approximately 2 cm long) was individually pierced 200 times with a needle. Control plants were not pierced. For SSB treatment, plants were individually infested by a third-instar SSB larva that had been starved for 2 h. Control plants were not infested. For MeJA treatment, plant stems were individually treated with 100 μg of MeJA in 20 μL of lanolin paste. Controls (lanolin) were similarly treated with 20 μL of pure lanolin.
Y2H Assay
The OsWRKY5357-113 cDNA fragment was cloned into the pGBKT7 vector in-frame with the GAL4-binding domain. Full-length OsMPK3 and OsMPK6 were cloned into the pGADT7 vector, in the in-frame next to the activation domain (Clontech). The combinations of bait and prey plasmids (Fig. 2) were cotransformed into yeast (Saccharomyces cerevisiae) Y2H Gold (Clontech). The interactions were tested on selective medium lacking Leu, Trp, Ade, and His (SD-Leu-Trp-His-Ade) and containing 5-Bromo-4-chloro-3-indoyl-α-galactoside and 0.25 μg mL–1 aureobasidin A, according to the Matchmaker Gold Yeast Two-Hybrid System User Manual (Clontech). Serial 1:10 dilutions were prepared in water, and 4 μL of each dilution was used to yield one spot. Plates were incubated at 30°C for about 72 h before the scoring and capturing of photographs took place. SV40 T-antigen with p53 or Lamin C (Clontech) was used as the positive and negative control, respectively.
BiFC Assay
For BiFC studies, full-length OsWRKY53, OsMPK3, OsMPK6, and GUS were cloned into the pCV-nYFP or pCV-cYFP vector (Lu et al., 2011) to produce fused N- or C-terminal half of YFP, i.e., pCV-nYFP-WRKY53, pCV-nYFP-MPK3, pCV-nYFP-MPK6, pCV-nYFP-GUS, pCV-cYFP-WRKY53, pCV-cYFP-MPK3, pCV-cYFP-MPK6, and pCV-cYFP-GUS, respectively. Constructed plasmids were separately transformed into A. tumefaciens EHA105. The plasmid-containing A. tumefaciens was coinfiltrated into tobacco (Nicotiana benthamiana) leaves at optical density at 600 nm of 0.5:0.5 (see combinations in Fig. 2 and Supplemental Fig. S7). Small living pieces of tobacco leaves were cut from the infected area at 72 h after infiltration and mounted in water for microscopic observation. YFP fluorescence was observed and photographed by using a Leica TCS SP5 confocal laser scanning microscope (Leica Microsystems) with an argon laser.
Detection of MPK Activity
To detect MPK activities, 1-month-old plants of different genotypes were randomly assigned to SSB treatment. Plant stems were harvested at 0, 15, and 30 min after treatment. Five replicates at each time point were pooled together, and total proteins were extracted using the method described by Wu et al. (2007). Forty micrograms of total proteins was separated by SDS-PAGE and transferred onto Bio Trace pure nitrocellulose blotting membrane (PALL). Immunoblotting was performed using rabbit anti-pTEpY (Cell Signaling Technologies) or plant-actin rabbit polyclonal antibody (EarthOx). Chemiluminescence-based detection (Thermo Scientific) was performed using horseradish peroxidase-conjugated goat anti-rabbit antibody (Sigma).
JA, JA-Ile, SA, and ET Analysis
Plants of the different genotypes were randomly assigned to SSB and control treatments. For JA, JA-Ile, or SA analysis, plant stems were harvested at 0, 1.5, 3, and 8 h after the start of SSB infestation. Samples were ground in liquid nitrogen, and JA and JA-Ile were extracted with ethyl acetate spiked with labeled internal standards (13C2-JA and 13C6-JA-Ile, each with 100 ng) and analyzed with HPLC/mass spectrometry/mass spectrometry system following the method as described in Lu et al. (2015). SA levels were analyzed by gas chromatography-mass spectrometry using labeled internal standards as described previously (Lou and Baldwin, 2003). For ET analysis, infested and control plants were covered with sealed glass cylinders (diameter, 4 cm; height, 50 cm). ET production was determined using the method described by Lu et al. (2006). Each treatment at each time interval was replicated five times.
Analysis of TrypPI Activity
The stems of wild-type plants and transgenic lines (ir-wrky and oe-WRKY; 0.12–0.15 g sample–1) were harvested with or without SSB treatment for 3 d. The TrypPI concentrations were measured using a radial diffusion assay as described by van Dam et al. (2001). Each treatment at each time interval was replicated five times.
Herbivore Resistance Experiments
The performance of SSB larvae on different genotypes (ir-14, ir-29, oe-5, and oe-6) and wild-type plants was determined by infesting with freshly hatched larvae. For testing the effect of MeJA on SSB larval performance, the second-instar SSB larvae, which had been weighed and starved for 2 h, were placed individually on each transgenic (oe-5 and oe-6) plant that had been treated with MeJA (20 μL of lanolin containing 100 μg of MeJA). Sixty replicate plants from each line and treatment were used. Larval mass (to an accuracy of 0.1 mg) was measured 12 d after the start of the experiment. For the effect of MeJA, the increased percentage of larval mass on each line or treatment was calculated.
To determine differences in the tolerance of plants to herbivore attack, the different genotypes were individually infested with individual SSB third-instar larvae. The damage levels of plants were checked and photographs were taken.
Data Analysis
Differences in herbivore performance, expression levels of genes, and levels of herbivore-induced JA, JA-Ile, SA, and ET in different treatments, lines, or treatment times were determined by ANOVA (or Student’s t test for comparing two treatments). All tests were carried out with Statistica (SAS Institute).
Sequence data from this article can be found in the Rice Annotation Project under accession numbers OsWRKY53 (Os05g27730), OsWRKY70 (Os05g39720), OsWRKY45 (Os05g25770), OsWRKY35 (Os04g39570), OsWRKY33 (Os03g33012), OsWRKY30 (Os08g38990), OsWRKY24 (Os01g61080), OsWRKY13 (Os01g54600), OsMEK4 (Os2g54600), OsMPK3 (Os03g17700), OsMPK6 (Os06g06090), OsHI-LOX (Os08g39840), OsAOS1 (Os03g55800), OsAOS2 (Os03g12500), OsICS1 (Os09g19734), OsACS2 (Os04g48850), and OsACTIN (Os03g50885).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequences of nucleotides and deduced amino acids of OsWRKY53 and phylogenetic analysis of group I-type WRKYs from different plant species.
Supplemental Figure S2. DNA gel-blot analysis of ir-wrky and oe-WRKY lines, and wild-type plants.
Supplemental Figure S3. OsWRKY53 expression levels of ir-wrky and oe-WRKY lines, and wild-type plants.
Supplemental Figure S4. Expression levels of OsWRKY24, OsWRKY35, and OsWRKY70 in ir-wrky, oe-WRKY lines and wild-type plants.
Supplemental Figure S5. Growth phenotypes of ir-wrky and oe-WRKY lines and wild-type plants.
Supplemental Figure S6. OsWRKY53 influences the phenotype of rice plants.
Supplemental Figure S7. OsWRKY53 interacts with MPK3/MPK6 in vivo.
Supplemental Figure S8. High-resolution photographs of interactions between OsWRKY53 and MPK3/MPK6 in the nucleus.
Supplemental Figure S9. Activity of OsMPK3 and OsMPK6 in ir-wrky and oe-WRKY lines and wild-type plants.
Supplemental Figure S10. OsAOS1 and OsAOS2 mediate herbivore-induced JA biosynthesis in rice.
Supplemental Figure S11. OsWRKY53 mediates SA accumulation in rice after infestation with the SSB.
Supplemental Figure S12. Transformation vectors were used in this study.
Supplemental Table S1. Primers and probes used for qRT-PCR of target genes.
Supplemental Data Set S1. Text file of alignments used for the phylogenetic analysis in Supplemental Figure S1B.
Acknowledgments
We thank Emily Wheeler and Matthias Erb for editorial assistance and Matthias Erb for valuable scientific suggestions.
Glossary
- SSB
striped stem borer
- JA
jasmonic acid
- JA-Ile
jasmonoyl-isoleucine
- SA
salicylic acid
- ET
ethylene
- TF
transcription factor
- TrypPI
trypsin protease inhibitor
- cDNA
complementary DNA
- qRT
quantitative real-time
- RNAi
RNA interference
- Y2H
yeast two-hybrid
- BiFC
bimolecular fluorescence complementation
- MeJA
methyl jasmonate
Footnotes
This work was supported by the National Natural Science Foundation of China (grant no. 31330065), the Innovation Research Team Program of the National Natural Science Foundation of China (grant no. 31321063), the Special Fund for Agroscientific Research in the Public Interest (grant no. 201403030), and the China Agriculture Research System (grant no. CARS–01–21).
References
- Ahuja I, Kissen R, Bones AM (2012) Phytoalexins in defense against pathogens. Trends Plant Sci 17: 73–90 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Bakshi M, Oelmüller R (2014) WRKY transcription factors: jack of many trades in plants. Plant Signal Behav 9: e27700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S, Li J, Palva ET (2012) WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J Exp Bot 63: 2667–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Diezel C, Somssich IE (2012) Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol 159: 266–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventure G. (2012) Perception of insect feeding by plants. Plant Biol (Stuttg) 14: 872–880 [DOI] [PubMed] [Google Scholar]
- Campos ML, Kang JH, Howe GA (2014) Jasmonate-triggered plant immunity. J Chem Ecol 40: 657–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Shelton A, Ye GY (2011) Insect-resistant genetically modified rice in China: from research to commercialization. Annu Rev Entomol 56: 81–101 [DOI] [PubMed] [Google Scholar]
- Chen YF, Li LQ, Xu Q, Kong YH, Wang H, Wu WH (2009) The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21: 3554–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, He J (1996) Rice Insect Pests. China Agricultural Press, Beijing [Google Scholar]
- Chi Y, Yang Y, Zhou Y, Zhou J, Fan B, Yu JQ, Chen Z (2013) Protein-protein interactions in the regulation of WRKY transcription factors. Mol Plant 6: 287–300 [DOI] [PubMed] [Google Scholar]
- Chujo T, Miyamoto K, Ogawa S, Masuda Y, Shimizu T, Kishi-Kaboshi M, Takahashi A, Nishizawa Y, Minami E, Nojiri H, et al. (2014) Overexpression of phosphomimic mutated OsWRKY53 leads to enhanced blast resistance in rice. PLoS One 9: e98737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Takai R, Akimoto-Tomiyama C, Ando S, Minami E, Nagamura Y, Kaku H, Shibuya N, Yasuda M, Nakashita H, et al. (2007) Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim Biophys Acta 1769: 497–505 [DOI] [PubMed] [Google Scholar]
- Ciolkowski I, Wanke D, Birkenbihl RP, Somssich IE (2008) Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol Biol 68: 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M, Meldau S, Howe GA (2012) Role of phytohormones in insect-specific plant reactions. Trends Plant Sci 17: 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Han L, Li GJ, Yang KY, Mao G, Wang R, Liu Y, Zhang S (2010) Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J 64: 114–127 [DOI] [PubMed] [Google Scholar]
- Ishihama N, Yamada R, Yoshioka M, Katou S, Yoshioka H (2011) Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell 23: 1153–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama N, Yoshioka H (2012) Post-translational regulation of WRKY transcription factors in plant immunity. Curr Opin Plant Biol 15: 431–437 [DOI] [PubMed] [Google Scholar]
- Kang JH, Wang L, Giri A, Baldwin IT (2006) Silencing threonine deaminase and JAR4 in Nicotiana attenuata impairs jasmonic acid-isoleucine-mediated defenses against Manduca sexta. Plant Cell 18: 3303–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CY, Liu Y, Thorne ET, Yang H, Fukushige H, Gassmann W, Hildebrand D, Sharp RE, Zhang S (2003) Activation of a stress-responsive mitogen-activated protein kinase cascade induces the biosynthesis of ethylene in plants. Plant Cell 15: 2707–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CY, Zhang S (2004) Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. Plant J 38: 142–151 [DOI] [PubMed] [Google Scholar]
- Koo SC, Moon BC, Kim JK, Kim CY, Sung SJ, Kim MC, Cho MJ, Cheong YH (2009) OsBWMK1 mediates SA-dependent defense responses by activating the transcription factor OsWRKY33. Biochem Biophys Res Commun 387: 365–370 [DOI] [PubMed] [Google Scholar]
- Li G, Meng X, Wang R, Mao G, Han L, Liu Y, Zhang S (2012) Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet 8: e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brader G, Kariola T, Palva ET (2006) WRKY70 modulates the selection of signaling pathways in plant defense. Plant J 46: 477–491 [DOI] [PubMed] [Google Scholar]
- Li R (2012) Function characterization of herbivore resistance-related genes OsWRKY24, OsWRKY70 and OsNPR1 in rice. PhD thesis. Zhejiang University, Hangzhou, China [Google Scholar]
- Li R, Afsheen S, Xin Z, Han X, Lou Y (2013) OsNPR1 negatively regulates herbivore-induced JA and ethylene signaling and plant resistance to a chewing herbivore in rice. Physiol Plant 147: 340–351 [DOI] [PubMed] [Google Scholar]
- Liu Y. (2012) Roles of mitogen-activated protein kinase cascades in ABA signaling. Plant Cell Rep 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Birkenbihl RP, Rawat V, Schneeberger K, Schmelzer E, Somssich IE (2013) Functional dissection of the PROPEP2 and PROPEP3 promoters reveals the importance of WRKY factors in mediating microbe-associated molecular pattern-induced expression. New Phytol 198: 1165–1177 [DOI] [PubMed] [Google Scholar]
- Lou Y, Baldwin IT (2003) Manduca sexta recognition and resistance among allopolyploid Nicotiana host plants. Proc Natl Acad Sci USA 100(Suppl 2): 14581–14586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou YG, Du MH, Turlings TCJ, Cheng JA, Shan WF (2005) Exogenous application of jasmonic acid induces volatile emissions in rice and enhances parasitism of Nilaparvata lugens eggs by the parasitoid Anagrus nilaparvatae. J Chem Ecol 31: 1985–2002 [DOI] [PubMed] [Google Scholar]
- Lu J, Ju H, Zhou G, Zhu C, Erb M, Wang X, Wang P, Lou Y (2011) An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J 68: 583–596 [DOI] [PubMed] [Google Scholar]
- Lu J, Li J, Ju H, Liu X, Erb M, Wang X, Lou Y (2014) Contrasting effects of ethylene biosynthesis on induced plant resistance against a chewing and a piercing-sucking herbivore in rice. Mol Plant 7: 1670–1682 [DOI] [PubMed] [Google Scholar]
- Lu J, Robert CAM, Riemann M, Cosme M, Mène-Saffrané L, Massana J, Stout MJ, Lou Y, Gershenzon J, Erb M (2015) Induced jasmonate signaling leads to contrasting effects on root damage and herbivore performance. Plant Physiol 167: 1100–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wang X, Lou Y, Cheng J (2006) Role of ethylene signaling in the production of rice volatiles induced by the rice brown planthopper Nilaparvata lugens. Chin Sci Bull 51: 2457–2465 [Google Scholar]
- Menke FL, van Pelt JA, Pieterse CM, Klessig DF (2004) Silencing of the mitogen-activated protein kinase MPK6 compromises disease resistance in Arabidopsis. Plant Cell 16: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H (2005) Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci 10: 339–346 [DOI] [PubMed] [Google Scholar]
- Nomura H, Komori T, Uemura S, Kanda Y, Shimotani K, Nakai K, Furuichi T, Takebayashi K, Sugimoto T, Sano S, et al. (2012) Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat Commun 3: 926. [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Hu Y, Tang X, Zhou P, Deng X, Wang H, Guo Z (2012) Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 236: 1485–1498 [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Van Loon LC (2004) NPR1: the spider in the web of induced resistance signaling pathways. Curr Opin Plant Biol 7: 456–464 [DOI] [PubMed] [Google Scholar]
- Qi J, Zhou G, Yang L, Erb M, Lu Y, Sun X, Cheng J, Lou Y (2011) The chloroplast-localized phospholipases D α4 and α5 regulate herbivore-induced direct and indirect defenses in rice. Plant Physiol 157: 1987–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S (2007) OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol Plant Microbe Interact 20: 492–499 [DOI] [PubMed] [Google Scholar]
- Qiu JL, Fiil BK, Petersen K, Nielsen HB, Botanga CJ, Thorgrimsen S, Palma K, Suarez-Rodriguez MC, Sandbech-Clausen S, Lichota J, et al. (2008) Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J 27: 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Liu Y, Yang KY, Han L, Mao G, Glazebrook J, Zhang S (2008) A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 105: 5638–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61: 621–649 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Kazanaviciute V, Scheikl E, Teige M, Doczi R, Hirt H, Schwanninger M, Kant M, Schuurink R, Mauch F, et al. (2007) The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19: 2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Liu C, Zhang Y, Meng X, Zhou X, Chu C, Wang X (2012) OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol Biol 80: 241–253 [DOI] [PubMed] [Google Scholar]
- Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, Takatsuji H (2007) Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19: 2064–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbe M, Qu N, Galis I, Baldwin IT (2008) Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 20: 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Mine A, Bethke G, Igarashi D, Botanga CJ, Tsuda Y, Glazebrook J, Sato M, Katagiri F (2013) Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genet 9: e1004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam NM, Horn M, Mares M, Baldwin IT (2001) Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. J Chem Ecol 27: 547–568 [DOI] [PubMed] [Google Scholar]
- Van Eck L, Schultz T, Leach JE, Scofield SR, Peairs FB, Botha AM, Lapitan NL (2010) Virus-induced gene silencing of WRKY53 and an inducible phenylalanine ammonia-lyase in wheat reduces aphid resistance. Plant Biotechnol J 8: 1023–1032 [DOI] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44: 135–162 [DOI] [PubMed] [Google Scholar]
- Wang B (2012) Functional analysis of herbivore resistance-related genes OsICS and OsHPL3 in rice. PhD thesis. Zhejiang University, Hangzhou, China [Google Scholar]
- Wang Q, Li J, Hu L, Zhang T, Zhang G, Lou Y (2013) OsMPK3 positively regulates the JA signaling pathway and plant resistance to a chewing herbivore in rice. Plant Cell Rep 32: 1075–1084 [DOI] [PubMed] [Google Scholar]
- Wu J, Baldwin IT (2010) New insights into plant responses to the attack from insect herbivores. Annu Rev Genet 44: 1–24 [DOI] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Meldau S, Baldwin IT (2007) Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19: 1096–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KL, Guo ZJ, Wang HH, Li J (2005) The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res 12: 9–26 [DOI] [PubMed] [Google Scholar]
- Xie Y, Huhn K, Brandt R, Potschin M, Bieker S, Straub D, Doll J, Drechsler T, Zentgraf U, Wenkel S (2014) REVOLUTA and WRKY53 connect early and late leaf development in Arabidopsis. Development 141: 4772–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, Shen QJ (2005) Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 137: 176–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J (2008) Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SJ, Kim SH, Kim MJ, Ryu CM, Kim YC, Cho BH, Yang KY (2014) Involvement of the OsMKK4-OsMPK1 cascade and its downstream transcription factor OsWRKY53 in the wounding response in rice. Plant Pathol J 30: 168–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory Manual for Physiological Studies of Rice, Ed 3. International Rice Research Institute, Manila, Philippines [Google Scholar]
- Zentgraf U, Laun T, Miao Y (2010) The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur J Cell Biol 89: 133–137 [DOI] [PubMed] [Google Scholar]
- Zhang ZL, Shin M, Zou X, Huang J, Ho TH, Shen QJ (2009) A negative regulator encoded by a rice WRKY gene represses both abscisic acid and gibberellins signaling in aleurone cells. Plant Mol Biol 70: 139–151 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48: 592–605 [DOI] [PubMed] [Google Scholar]
- Zhou G, Qi J, Ren N, Cheng J, Erb M, Mao B, Lou Y (2009) Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J 60: 638–648 [DOI] [PubMed] [Google Scholar]
- Zhou G, Ren N, Qi J, Lu J, Xiang C, Ju H, Cheng J, Lou Y (2014) The 9-lipoxygenase Osr9-LOX1 interacts with the 13-lipoxygenase-mediated pathway to regulate resistance to chewing and piercing-sucking herbivores in rice. Physiol Plant 152: 59–69 [DOI] [PubMed] [Google Scholar]
- Zhou G, Wang X, Yan F, Wang X, Li R, Cheng J, Lou Y (2011) Genome-wide transcriptional changes and defence-related chemical profiling of rice in response to infestation by the rice striped stem borer Chilo suppressalis. Physiol Plant 143: 21–40 [DOI] [PubMed] [Google Scholar]