A tropical legume requires the common symbiotic pathway to interact with a nitrogen-fixing bacterium devoid of the canonical nodulation genes.
Abstract
Nitrogen fixation in the legume-rhizobium symbiosis is a crucial area of research for more sustainable agriculture. Our knowledge of the plant cascade in response to the perception of bacterial Nod factors has increased in recent years. However, the discovery that Nod factors are not involved in the Aeschynomene-Bradyrhizobium spp. interaction suggests that alternative molecular dialogues may exist in the legume family. We evaluated the conservation of the signaling pathway common to other endosymbioses using three candidate genes: Ca2+/Calmodulin-Dependent Kinase (CCaMK), which plays a central role in cross signaling between nodule organogenesis and infection processes; and Symbiosis Receptor Kinase (SYMRK) and Histidine Kinase1 (HK1), which act upstream and downstream of CCaMK, respectively. We showed that CCaMK, SYMRK, and HK1 are required for efficient nodulation in Aeschynomene evenia. Our results demonstrate that CCaMK and SYMRK are recruited in Nod factor-independent symbiosis and, hence, may be conserved in all vascular plant endosymbioses described so far.
As sessile organisms, plants must be able to interpret and respond to their environment to grow and survive. Interactions with telluric microbes play a central role in these adaptive responses. An important example of plant-microorganism interactions is root endosymbiosis with fungi, which was established 400 million years ago and likely accompanied the rise of terrestrial flora (Remy et al., 1994). Today, more than 80% of land plants can establish an arbuscular mycorrhizal symbiosis.
Conversely, only plants belonging to the Rosid clade have the ability to form a nitrogen-fixing symbiosis with bacteria. The predisposition for nodulation was acquired around 100 million years ago by a common ancestor (Doyle, 2011). Nitrogen-fixing nodulation involves two distinct associations: the legume-rhizobium symbiosis and actinorhizal symbiosis, formed by members of the Fagales, Rosales, and Cucurbitales orders, which interact with Frankia spp. actinobacteria. The ability of such plants to symbiotically fix nitrogen has attracted attention because of their ecological and agronomic implications. It is now well established that nitrogen-fixing symbioses partially evolved from a preexisting genetic background present in the most ancient arbuscular mycorrhizal symbiosis (Markmann and Parniske, 2009; Kouchi el al., 2010).
This hypothesis has been strengthened by genetic screens in two temperate legumes, Medicago truncatula and Lotus japonicus, both used as models to study nitrogen-fixing symbioses, enabling major insights into the molecular components of the symbiotic signaling pathway (Oldroyd et al., 2011). One important advance was the finding in the early 1990s that plant-derived flavonoids stimulate rhizobia to produce lipochitooligosaccharide signal molecules called Nod factors (NFs; Lerouge et al., 1990). The perception of NFs by LysM-receptor-like kinase (LysM-RLK) receptors (L. japonicus Nod Factor Receptor1 [LjNFR1] /M. truncatula Lysin Motif Receptor-Like Kinase3 [MtLYK3] and L. japonicus Nod Factor Receptor5 [LjNFR5]/M. truncatula Nod Factor Perception [MtNFP]) is the keystone in the specific recognition of bacterial symbionts by the plant (Limpens et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006; Smit et al., 2007). This first recognition event is propagated by a set of proteins including a Leu-rich repeat receptor kinase (LjSYMRK/M. truncatula Does Not Make Infection2 [DMI2]), at least three nucleoporins (LjNUP85, LjNUP133, and NENA), and one or two cation channels (LjCASTOR and LjPOLLUX/MtDMI1), all required for the induction of Ca2+ spiking (Oldroyd and Downie, 2006). A Ca2+/calmodulin-dependent kinase (LjCCaMK/MtDMI3) is likely a decoder of calcium oscillations through the phosphorylation of target proteins (e.g. LjCYCLOPS/M. truncatula Interacting Protein of DMI3 [IPD3]; Messinese et al., 2007; Yano et al., 2008). The complex formed by CCaMK and its phosphorylated partner CYCLOPS triggers the expression of Nodule Inception (NIN; Singh et al., 2014), a transcription factor that eventually activates another round of specific transcription factors in the Gibberellic-Acid Insensitive (GAI)/Repressor of GAI/Scarecrow (GRAS) family (Kaló et al., 2005; Smit et al., 2005). This signaling cascade, together with a cytokinin signal detected by Lotus Histidine Kinase1 (LHK1)/M. truncatula Cytokinin Response1 (MtCRE1), enables nodule organogenesis (Murray et al., 2007; Tirichine et al., 2007).
An increasing number of genes primarily associated with rhizobial infection and nodule organogenesis (including the seven genes described above) have been shown to be also involved in arbuscular mycorrhizal symbiosis (for review, see Kouchi et al., 2010). Interestingly, two so-called common symbiotic genes, CgSYMRK/DgSYMRK and CgCCaMK, have been shown to be also necessary for the establishment of the symbiosis between actinorhizal plants (Casuarina glauca and Datisca glomerata) and Frankia spp. (Gherbi et al., 2008; Markmann et al., 2008; Svistoonoff et al., 2013).
Aeschynomene evenia recently emerged as a new model legume for the study of legume-rhizobium interactions (Arrighi et al., 2012). Most studies of rhizobium-legume symbioses up to now were those in temperate climates, which are far from representing the diversity of such interactions, particularly the range of infection processes. The best-known infection process, which is encountered in most temperate legumes, including L. japonicus and M. truncatula, involves the primary entry of bacteria into deformed root hairs through infection threads (ITs) and the induction of a nodule primordium in the root cortex (Gage, 2004). However, in approximately 25% of legumes found mainly in tropical and warm temperate areas, rhizobial invasion does not involve the formation of ITs (Sprent, 2007). Instead, invasion occurs through epidermal cracks, and the rhizobia are endocytosed directly by a cortical cell. These cortical cells divide repeatedly, resulting in uniformly infected tissue, with bacteria maintained in symbiosomes. Some Aeschynomene spp. display an NF-independent symbiotic process where the symbionts (photosynthetic Bradyrhizobium spp. strains) are able to induce nitrogen-fixing nodules in the absence of the canonical nodABC genes required for the synthesis of NFs (Giraud et al., 2000, 2007). In addition to the existence of alternative infection processes, results showing that common nod genes are absent in at least two photosynthetic Bradyrhizobium spp. strains naturally associated with Aeschynomene spp. show that alternative signaling pathways enabling nodulation exist even within the legume family.
In this context, it was important to investigate whether the nodulation signaling pathway, which is triggered by the recognition of NFs in legumes, is conserved in A. evenia. To this end, we analyzed the function of orthologs of symbiotic genes in A. evenia using RNA interference (RNAi) approaches. We selected CCaMK as the first candidate gene because it has been shown to play a central role in cross signaling between root nodule organogenesis and infection processes. The resulting data were extended to orthologs of LjSYMRK and LjHK1, which act upstream and downstream of CCaMK, respectively. The data presented here show that CCaMK, SYMRK, and HK1 are required for the efficient nodulation of A. evenia. Our results suggest that, beyond the perception of NFs, all nodulation genes are required in the legume NF-independent symbiosis and, hence, appear to be conserved either in legumes or in actinorhizal plants.
RESULTS
Identification of CCaMK Orthologs in A. evenia ssp. serrulata
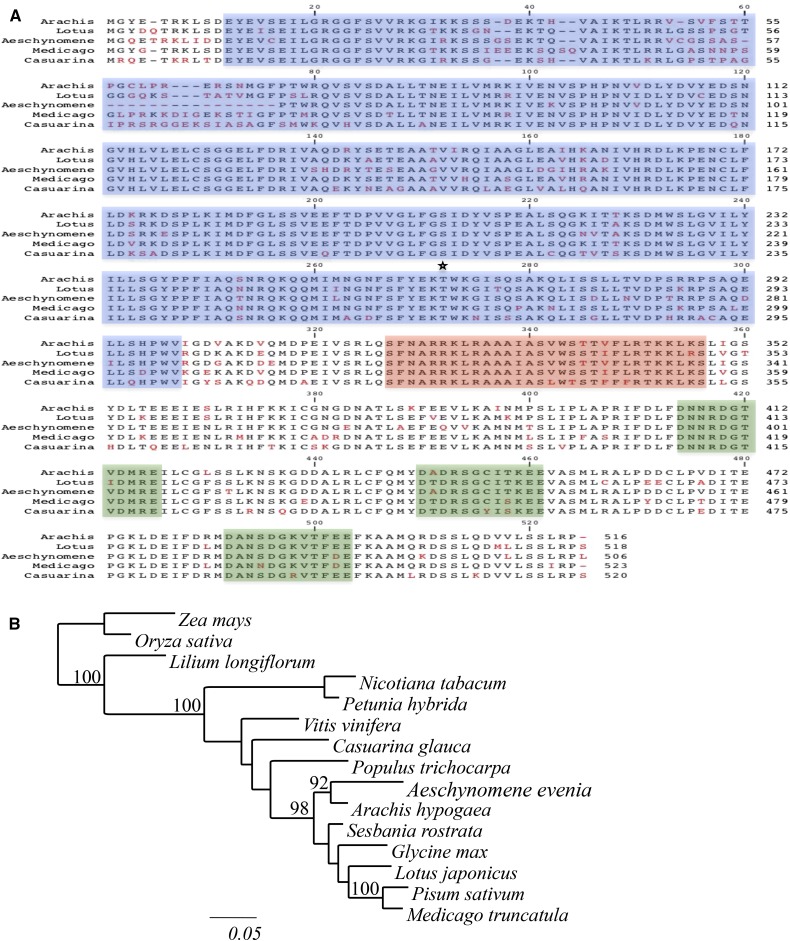
Screening of an EST database of A. evenia ssp. serrulata enabled the identification of a 1,686-bp EST containing the full-length CCaMK open reading frame (RNA was isolated from leaves, control roots, and roots sampled at different times after inoculation with Bradyrhizobium spp. ORS278; S. Fabre, D. Gully, A. Poitout, D. Patrel, J.-F. Arrighi, E. Giraud, P. Czernic, and F. Cartieaux, unpublished data). The AeCCaMK deduced protein was 506 amino acids long, and its alignment with available CCaMK sequences from legumes revealed strong conservation of protein domains in the Aeschynomene spp. ortholog (Fig. 1A). Phylogenetic analysis revealed that AeCCaMK clusters together with CCaMK from other legume species but forms an outgroup with A. hypogaea (87% amino acid identity), which belongs to the same Dalbergioid clade (Fig. 1B).
Figure 1.
Features of the CCaMK of A. evenia. A, Alignment of CCaMK from A. evenia, Arachis hypogea, L. japonicus, M. truncatula, and C. glauca. Sequences were aligned using ClustalW. Identical amino acids are in black, and different amino acids are in red. On the basis of in silico predictions, catalytic domain, calmodulin-binding domain, and EF-hands of the visinin-like domain are highlighted in blue, red, and green, respectively, and the autophosphorylation site is indicated by the star. B, Position of A. evenia CCaMK in a distance tree predicted based on available CCaMK protein sequences. The tree was generated based on a Clustal Omega alignment of 474 amino acids. Numbers at the nodes represent bootstrap values (percentage of 100 replicates; only values greater than 90% were retained). Accession numbers of the proteins are as follows: NP_001105906.1 (Zea mays), NP_001055895.1 (Oryza sativa), Q43531.1 (Lilium longiflorum), AAD52092.1 (Nicotiana tabacum), ABQ95545.1 (Petunia hybrida), XP_002273342.1 (Vitis vinifera), CCW43374.1 (C. glauca), XP_002315401.2 (Populus trichocarpa), ACB46142.1 (A. hypogaea), ACC94267.1 (Sesbania rostrata), XP_003531786.1 (Glycine max), A0AAR7.1 (L. japonicus), Q6RET6.2 (Pisum sativum), and XP_003628124.1 (M. truncatula).
Quantitative reverse transcription-PCR was performed to determine the steady-state levels of AeCCaMK transcripts in leaves, nodules, and roots during the nodulation process (Fig. 2A). AeCCaMK expression appeared to be restricted to the roots, since no expression was detected in leaves. Upon infection, a slight up-regulation of CCaMK was observed in nodules (Fig. 2A), which resembles the expression profiles of CCaMK in other legumes (Lévy et al., 2004; Sinharoy and DasGupta, 2009). To localize the expression of CCaMK, we isolated its promoter region by genome walking. A 564-bp promoter region was obtained corresponding to the sequence between the translation initiation site of CCaMK and an upstream transposon insertion site. This fragment, therefore, was considered to encompass the complete promoter region and was used to drive the transcription of the uidA reporter gene. This promoter fusion was introduced into A. evenia ssp. serrulata by transformation with A. rhizogenes strain ARqua1 (Bonaldi et al., 2010). GUS staining was observed in the root vasculature of uninoculated transgenic roots, and sectioning revealed GUS staining in cortical tissue (Fig. 2, B and C). The same expression pattern was observed over the 5-d period following inoculation with Bradyrhizobium spp. strain ORS278 (Fig. 2, D and E). At 10 dpi, the promoter was active in the infected cells of the nodule (Fig. 2, F and G). These observations revealed that the steady-state level of CCaMK expression in root is not induced during rhizobial invasion even though its expression pattern extends to the nodule.
Figure 2.
Expression analysis of AeCCaMK. A, Relative transcript abundance of AeCCaMK in roots, mature nodules (2 weeks after inoculation), and leaves. Relative transcript abundance was determined by quantitative reverse transcription-PCR and normalized against the A. evenia elongation factor EF1α, which was constitutively expressed in all tissues tested. B to G, Histochemical localization of GUS activity in transgenic roots expressing pAeCCaMK-GUS. B and C, Uninoculated transgenic roots showing faint AeCCaMK promoter activity in root vasculature and cortical tissue. D and E, In transgenic roots 5 days post inoculation (dpi) with Bradyrhizobium spp. ORS278, GUS staining is visible in the same tissues as in noninfected roots. F and G, In 10-d-old nodules, GUS activity is restricted to the infection zone. C, E, and G are longitudinal sections of roots and nodules. Bars = 100 µm.
Knockdown of AeCCaMK Impairs Nodule Development
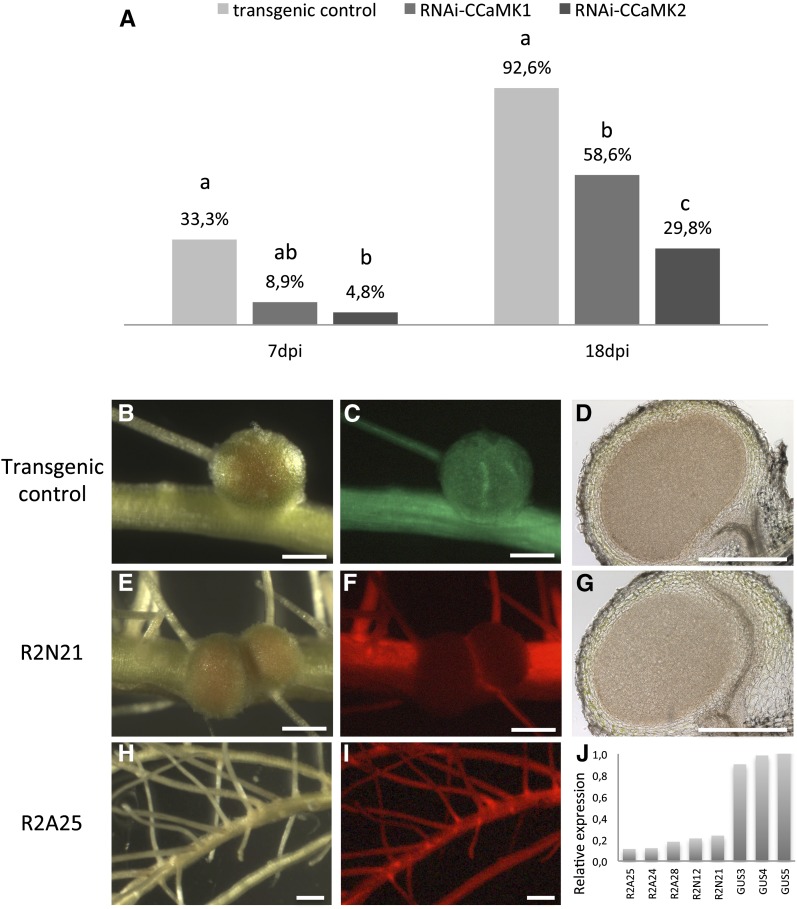
To investigate the function of AeCCaMK during nodulation, we reduced CCaMK transcript levels in transgenic A. evenia roots by RNAi. Two constructs were designed, one 410 bp long and one 326 bp long, targeting either the kinase (RNAiCCaMK1) or the visinin (RNAiCCaMK2) domain. These constructs were introduced into A. evenia ssp. serrulata via Agrobacterium rhizogenes-mediated transformation with DsRED as cotransformation marker. Roots transformed with the Gateway vector pK7WG2D carrying the uidA reporter gene under the control of the 35S promoter and GFP as cotransformation marker were analyzed as controls. Plants with transgenic roots, identified by green or red fluorescent protein (GFP or DsRED), were inoculated with Bradyrhizobium spp. ORS278, and the efficiency of nodulation was examined 1 week later (7 dpi) and again when the nodulation process was complete (18 dpi). We observed a clear impact of AeCCaMK silencing on nodulation efficiency: in the control transgenic lines (n = 27), more than 90% of transgenic roots had developed nodules at 18 dpi, whereas in lines transformed with the RNAiCCaMK1 knockdown construct (n = 84), only 58% of the transgenic roots had developed normal nodules at 18 dpi. In lines transformed with the RNAiCCaMK2 knockdown construct (n = 76), the phenotype was even more severe, as only 30% of the transgenic roots developed normal nodules. In the remaining 70% of RNAiCCaMK2 roots, absolutely no nodules developed (Fig. 3A). Besides a lower percentage of nodulated plants, the number of nodules was significantly reduced in the RNAiCCaMK2 lines: an average of only six nodules per plant was obtained compared with 11 in the control (Fisher’s lsd test), evidences that the formation of nodules is drastically impaired when the expression level of CCaMK is knocked down. Conversely, the nodules in the transgenic RNAi lines looked similar to those observed on control roots, and cytological analysis failed to reveal any particular disorders, pointing to normal organogenesis (Fig. 3, E–I). In addition, we observed no intermediate phenotypes, such as small nodules or bumps, on these transgenic RNAi roots, suggesting an all-or-nothing response in nodule organogenesis.
Figure 3.
Nodulation phenotypes of AeCCaMK knockdown mutants. A, Percentage of nodulated roots calculated at 7 and 18 dpi with the Bradyrhizobium spp. ORS278 strain (the control corresponds to transgenic roots transformed with an empty vector; transgenic control [n = 27], RNAiCCaMK1 [n = 84], and RNAiCCaMK2 [n = 76]). Different letters above the bars represent statistically significant differences (χ2 test, P < 0.05). B to D, Transgenic control nodule. E to I, RNAiCCaMK2 at 18 dpi. D to G are longitudinal sections (60 µm) of nodules. Bars = 500 µm. J, Quantification of AeCCaMK mRNA levels in RNAi plants determined by real-time quantitative PCR. Quantification was performed on five independent RNAiCCaMK2 lines. AeEF1α was used as a reference gene. Relative expression levels of AeCCaMK in three independent transgenic control roots are shown. Expression levels are relative to transgenic control line 5.
To evaluate the efficiency of AeCCaMK knockdown in RNAi roots, CCaMK transcript levels were determined using quantitative reverse transcription-PCR in a subset of the RNAiCCaMK1 and RNAiCCaMK2 lines (n = 32) as well as in control lines (n = 5). A link was found between the reduction in the transcript level and the severity of the phenotype (Fig. 3). Lines with the greatest reduction in expression (less than 20%; e.g. R2A25, R2A24, and R2A28) formed no nodules (Fig. 3, H and I), whereas those with greater than 20% expression, such as R2N21, formed normal nodules (five nodules in the R2N21 line; Fig. 3, E–G), suggesting that a threshold of 20% of CCaMK expression is critical to switch from an all-or-nothing phenotype in nodule organogenesis.
Expression of Deregulated Forms of AeCCaMK Leads to Spontaneous Nodule Inception
Ca2+ is the initial signal induced in response to symbiotic signals from rhizobia or arbuscular mycorrhizal fungi. A calmodulin-binding regulatory domain and a visinin-like Ca2+-binding domain including three canonical elongation factor EF-hand motifs are present on the C-terminal part of AeCCaMK. The N-terminal part of the protein contains a kinase domain with an autophosphorylation site (corresponding to the Thr in position 253 in AeCCaMK). Recent studies on M. truncatula CCaMK have shown that Ca2+ binding to the EF-hand domains promotes autophosphorylation, which negatively regulates CCaMK by stabilizing the inactive state of the protein. By contrast, calcium-dependent calmodulin binding overrides the effects of autophosphorylation and activates protein phosphorylation, which is responsible for the ability to accommodate rhizobia (Miller et al., 2013). Deregulated variants of CCaMK gain autonomous activity, leading to the development of spontaneous nodules in the absence of rhizobia (Gleason et al., 2006; Tirichine et al., 2006).
To further analyze the function of CCaMK in the NF-independent nodulation process, we constructed two gain-of-function CCaMK variants: a truncated and a mutated form of the protein. The truncated form consists of a 308-amino acid sequence corresponding to the kinase domain lacking the autoinhibition domain, while in the mutated form, the Thr at position 253 is replaced by an Asp (T253D), which mimics the autophosphorylation of Thr in response to the symbiotic Ca2+ signal. The complete coding sequence (CDS) of AeCCaMK and the two dominant active forms of the protein were introduced into A. evenia roots. Three weeks after transfer to liquid medium, nodule-like structures were observed in 46% (n = 11/24) and 39% (n = 12/31) of transgenic roots expressing the truncated and mutated variants of AeCCaMK, respectively. Spontaneous nodules were not observed either on control wild-type roots or on transgenic roots carrying the complete AeCCaMK CDS (n = 45). The spontaneous nodules were white and round and developed at the emergence of lateral roots, exactly where wild-type nodules are found (Fig. 4, A and B). Microscope analyses showed that spontaneous nodules lacked the typical central infection zone but initiated vascular bundles (Fig. 4, C–F), showing that the constitutive kinase activity of AeCCaMK is sufficient to induce nodule organogenesis in the absence of Bradyrhizobium spp.
Figure 4.
Induction of spontaneous nodules in A. evenia roots expressing gain-of-function CCaMK variants. A, C, and E, Nodules induced by the Bradyrhizobium spp. ORS278 strain on wild-type roots. B, D, and F, Spontaneous nodules. A, Wild-type nodule 3 weeks after Bradyrhizobium spp. ORS278 inoculation. B, Spontaneous nodules on hairy roots formed 4 weeks after transfer to hydroponics. C and D, Semithin sections (60 µm) of nodules observed with a confocal microscope. The wild-type nodule displays a typical central infection zone with infected cells filled with Bradyrhizobium spp. ORS278 expressing GFP (C), while the spontaneous nodule lacks the typical infection zone but is starting to form vascular bundles (D; arrows). E and F, Closeups of C and D, respectively. In the wild-type nodule, bacteria have differentiated into spherical bacteroids. The spontaneous nodule is devoid of bacteria. Bars = 500 µm (A and B), 100 µm (C and D), and 10 µm (E and F).
To What Extent Is the Nod Signaling Pathway Conserved in A. evenia ssp. serrulata?
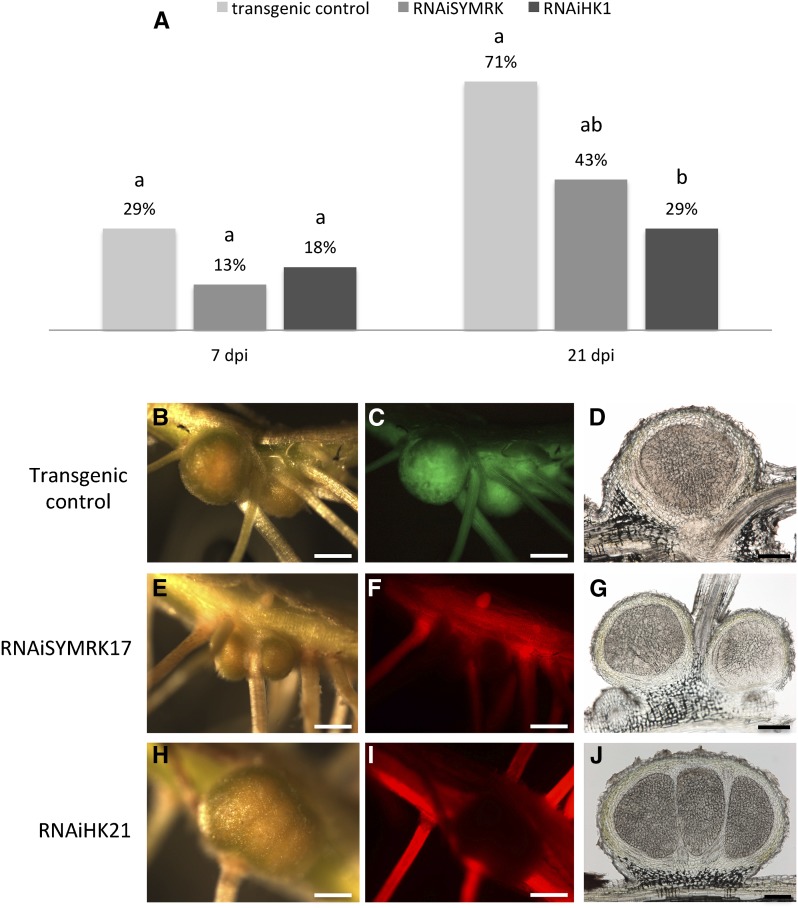
To answer this question, we performed RNAi assays on two additional symbiotic genes: AeSYMRK, which encodes the Leu-rich repeat receptor kinase acting upstream from CCaMK in the common symbiosis pathway, and the cytokinin receptor AeHK1 involved in cell divisions downstream from CCaMK. We identified AeSYMRK and AeHK1 by screening the EST database of A. evenia ssp. serrulata. Phylogenetic analyses were performed to unambiguously identify AeSYMRK and AeHK1 orthologs among their respective gene families (Supplemental Figs. S1 and S2). Two RNAi constructs were made: one with a 297-bp fragment of the AeSYMRK coding sequence encompassing the transmembrane domain of the protein, and the other with a 277-bp fragment of AeHK1 corresponding to part of the receiver domain. In total, 70 RNAiSYMRK and 68 RNAiHK1 roots were selected for analysis. Chimeric plants with transgenic roots were inoculated with Bradyrhizobium spp. ORS278, and nodulation was scored 1 week later and when the nodulation process was completed (7 and 21 dpi). At 21 dpi, 72% of control lines exhibited N2-fixing nodules (n = 24), and RNAiSYMRK and RNAiHK1 lines displayed 43% and 29% of nodulated roots, respectively (Fig. 5). In addition to reduced nodulation, we also observed a decrease in the number of nodules. The average number of nodules per root was 1.6 in RNAiSYMRK and 1.3 in RNAiHK1 lines, compared with 5.8 in the control transgenic lines. A Fisher’s test revealed that the number of nodules in RNAiSYMRK and RNAiHK1 lines was significantly lower than in the transgenic control lines (P < 0.05).
Figure 5.
Nodulation phenotypes of SYMRK and HK1 knockdown in A. evenia. A, Percentage of nodulated roots calculated at 7 and 21 dpi with the Bradyrhizobium spp. ORS278 strain (the control corresponds to transgenic roots transformed with an empty vector; transgenic control [n = 24], RNAiSYMRK [n = 70], and RNAiHK1 [n = 68]). Different letters above the bars represent statistically significant differences (χ2 test, P < 0,05). B to D, Transgenic control nodules. E to G and H to J, Wild-type phenotypes of nodules obtained on roots silenced for SYMRK and HK1, respectively. B, E, and F are bright-field views of mature nodules obtained 3 weeks after Bradyrhizobium spp. ORS278 inoculation. C, F, and I are UV light views of the same images (the transgenic line contains the GFP marker gene, and the RNAi lines express the red fluorescent protein). D, E, and J are longitudinal sections (60 µm) of corresponding nodules. Bars = 500 µm (B, C, E, F, H, and I) and 200 µm (D, G, and J).
RNAi nodules displayed the classical histological organization of aeschynomenoid nodules, in which the core infected zone is not interspersed with uninfected cells (Fig. 5, E–J).
AeSYMRK and AeHK1 extinction levels were quantified by quantitative reverse transcription-PCR in a subset of 15 and 18 RNAi roots displaying a major reduction in the number of nodules. A 39% to 89% reduction in AeSYMRK mRNA levels and a 45% to 80% reduction in AeHK1 mRNA levels were observed, demonstrating the impact of silencing on nodulation.
DISCUSSION
The discovery that NFs are not required for the interaction between Aeschynomene and Bradyrhizobium spp. challenged the long-held view that NFs are essential for all legume-rhizobia associations (Giraud et al., 2007). In this study, we assessed whether the widely studied components of the symbiotic signaling pathway are necessary for this NF-independent mode of nodulation. In all vascular plant endosymbioses described so far (i.e. nodulation and mycorrhization), calcium oscillations play an essential role as a secondary messenger. The symbiotic calcium- and calmodulin-dependent protein kinase CCaMK is required for the interpretation of this Ca2+ signature (Oldroyd and Downie, 2006; Madsen et al., 2010; Singh et al., 2014). Mutations in CCaMK or CCaMK RNAi prevent root infections by microsymbionts through the inhibition of either the formation of ITs or the endocytosis of rhizobia inside the nodules, and they block or, in the case of RNAi lines, reduce nodule organogenesis in legumes (Catoira et al., 2000; Lévy et al., 2004; Mitra et al., 2004; Capoen et al., 2009; Sinharoy and DasGupta, 2009) and actinorhizal plants (Svistoonoff et al., 2013).
To address the question of the role of CCaMK in the Aeschynomene-Bradyrhizobium spp. symbiotic couple through reverse genetic approaches, we isolated and characterized the CCaMK gene from A. evenia spp. serrulata. The AeCCaMK protein has all the features of other plant CCaMKs (i.e. a kinase domain and an autoinhibition domain overlapping a calmodulin-binding domain, such that autoinhibition is released in the presence of calmodulin). Deletion of this autoinhibition domain or a phosphomimetic version of CCaMK leads to the formation of spontaneous nodules in the absence of symbionts in legumes (Gleason et al., 2006; Tirichine et al., 2006; Takeda et al., 2012; Miller et al., 2013), in C. glauca (Svistoonoff et al., 2013), and in Parasponia andersonii, the only nonlegume able to form a nitrogen-fixing symbiosis with rhizobia (Op den Camp et al., 2011).
Our results showed that in A. evenia, two similar gain-of-function AeCCaMK variants also led to the inception of spontaneous nodules at the emergence of lateral roots, where nodules normally develop in legumes infected via a crack-entry process. In parallel with the gain-of-function strategy, we down-regulated the expression of AeCCaMK using an RNAi strategy. The silencing of AeCCaMK led to a significant decrease in the percentage of roots that developed nodules and also in the average number of nodules per root. Taken together, the data obtained using gain-of-function and RNAi approaches demonstrate the central role of CCaMK in nodule inception in a legume NF-independent context.
The next step was to determine if other components of the nodulation signaling pathway were conserved in A. evenia. To this end, we performed RNAi assays against the Leu-rich repeat receptor kinase gene SYMRK, which acts upstream from CCaMK, and against the cytokinin receptor (LHK1/MtCRE1), which is involved in cell divisions after Ca2+ spiking perception. In model legumes, mutations in SYMRK almost completely block epidermal responses to rhizobia, including root hair curling and the formation of ITs (Catoira et al., 2000; Endre et al., 2002; Stracke et al., 2002), while mutations in HK1 allow ITs to form but alter nodule development and prohibit the endocytosis of bacteria in primordium cells (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Plet et al., 2011). Observations made on S. rostrata, a tropical aquatic legume like A. evenia, demonstrated that the epidermis lock observed in symrk or ccamk mutants in model legumes can be bypassed (Capoen et al., 2010). In S. rostrata, SrSYMRK and SrCCaMK RNAi lines are still able to form bumps or small nonfunctional nodules at lateral root bases. Those permissive phenotypes are correlated with the nodulation features displayed by S. rostrata. On hydroponic roots, the microsymbiont of S. rostrata starts to invade the cortex intercellularly via cracks at lateral root bases exactly like Bradyrhizobium spp. on A. evenia roots. Thus, rhizobial invasion involves the formation of intercellular infection pockets and the subsequent formation of ITs that spread toward the nodule primordium (Ndoye et al., 1994; Goormachtig et al., 2004). SrSYMRK and SrCCaMK RNAi roots still form infection pockets and nodules, but these are not functional because the ITs are no longer able to release bacteria. This study (1) demonstrated that during the crack-entry infection process, the epidermal stages of nodulation are independent of SYMRK and CCaMK activities and (2) confirmed that SYMRK and CCaMK play an additional role at later stages of nodule development (i.e. bacterial release into the primordium cells; Capoen et al., 2005, 2009). The role of SYMRK and CCaMK in the endocytosis of bacteria inside nodules is shared with HK1 (Murray et al., 2007; Plet et al., 2011).
In A. evenia, silencing of SYMRK or HK1 led to a significant reduction in the number of nodules. Taken together, our data show that AeCCaMK, AeSYMRK, and AeHK1 are required for functional NF-independent symbiosis in Aeschynomene spp.
Another interesting observation was that for each RNAi construct, nodulation was achieved with no impact on the morphology of the nodules. Contrary to what was described in S. rostrata (Capoen et al., 2005, 2009), we observed no transitional phenotypes such as small, nonfixing nodule-like structures. The absence of transitional phenotypes might be related to the more primitive intercellular infection process at play in A. evenia, which not only bypasses epidermal responses to rhizobia (no ITs) but does not involve the distant induction of a primordium. In Aeschynomene spp., rhizobia are directly endocytosed in cortical cells from intercellular spaces (Bonaldi et al., 2011). The infected cortical cells, so-called founder cells, then divide repeatedly to form the typical aeschynomenoid nodule characterized by the absence of uninfected cells in the central symbiotic tissue. As a consequence, we hypothesize that, in the RNAi lines in which the residual amount of the targeted gene transcripts is necessary and sufficient to allow at least one rhizobial endocytosis event inside a founder cell, nodule organogenesis can be triggered. This could explain the drastic differences in the symbiotic phenotype, ranging from the complete absence of nodules to the formation of a few perfectly functional nodules, depending on the transgenic line concerned.
The fact that AeCCaMK, AeSYMRK, and AeHK1 are required for functional NF-independent symbiosis strengthens the well-established evolutionary hypothesis that a core set of common symbiotic genes is conserved in all vascular plant endosymbioses described so far (Markmann and Parniske, 2009). Our results are consistent with the development of functional nodules in L. japonicus mutants (affected in the perception of the NFs and able to form spontaneous nodules) upon infection with a Nod− bacterial mutant (Madsen et al., 2010). In other words, nodulation can occur even in the absence of NFs, as long as the common symbiosis signal transduction pathway is switched on: the so-called ground state described by Madsen et al. (2010). Our Aeschynomene-Bradyrhizobium spp. pair probably represents a natural case of such a ground state infection process.
Similar symbiotic cascade requirements were also found in the actinorhizal plant C. glauca, in which both SYMRK (Gherbi et al., 2008) and CCaMK (Svistoonoff et al., 2013) were shown to be required for nodulation. It is noteworthy that a microorganism lacking nod genes activates such a signaling pathway. Indeed, the sequencing of several Frankia spp. genomes revealed the absence of the common nod genes (Normand et al., 2007), as is the case for Bradyrhizobium spp. ORS278 (Giraud et al., 2007). A simple explanation could be the existence of original signal molecule(s) originating from these symbiotic partners. In the case of Frankia spp., the only published data concern a root hair-deforming factor whose biochemical properties differ from those of NFs (Cérémonie et al., 1999). In Bradyrhizobium spp., a mutant screen was conducted on the ORS278 strain, but neither complete Nod− mutants nor mutants affected in the synthesis of a putative signal molecule were isolated (Bonaldi et al., 2011). However, several mutants were found to be affected in the synthesis of purine, suggesting a role for bacterial cytokinins as a signal. This hypothesis has since been evaluated, and although bacterial cytokinins contribute positively to nodule development in Aeschynomene spp. plants, they are not the key signal that triggers nodule formation during NF-independent symbiosis (Podlešáková et al., 2013). The absence of nod mutants could be due to either functional redundancy or the need for bacterial viability. A putative candidate could be the bicyclic sugar bradyrhizose, which is the O-antigen-specific component of Bradyrhizobium spp. lipopolysaccharide (Silipo et al., 2011). Notably, bradyrhizose does not trigger an innate immune response in different plant families (Silipo et al., 2011). It remains to be seen if bradyrhizose is able to induce the Aeschynomene spp. symbiotic pathway and the expression of symbiotic genes. Another argument in favor of a structural component of the bacteria as a signal comes from the specific environment of the symbiotic interaction in our model. Aeschynomene spp. are tropical flooded legumes, and a diffusible signal such as the NF would be highly diluted. Alternatively, a diffusible signal may be involved but at a later stage of infection, when the bradyrhizobia colonize the subepidermal layers of the root. It is worth noting that such behavior is also observed in the Sesbania spp. infection process upon flooding (Goormachtig et al., 2004). Identifying Aeschynomene spp. genes as markers of the early steps in the symbiotic interaction would be useful in the search for the Bradyrhizobium spp. ORS278 signal.
Another consequence of the existence of a Bradyrhizobium spp. ORS278 signal concerns its perception. Until recently, NFs and plant NF receptors (LysM-RLK) were considered to be specific to nitrogen-fixing root nodule symbioses. However, this border has been crossed through the identification of the mycorrhizal lipochitooligosaccharide, whose structure appears very close to that of the NFs (Maillet et al., 2011), and the discovery in P. andersonii of a LysM-RLK receptor involved in both rhizobial and mycorrhizal recognition (Op den Camp et al., 2011). Another member of the LysM-RLK family, Chitin Elicitor Receptor Kinase1 (CERK1), was recently shown to be involved in the perception of pathogenic and symbiotic microbial patterns (Miyata et al., 2014). These data support the hypothesis of a role for LysM-RLKs in the perception of a broad range of microbial patterns, and our next goal is to identify their role in legume-rhizobium NF-independent symbioses. Exploring such avenues supports the current goal of circumventing the nodulation lock in nonnodulating plants by redirecting the existing symbiosis pathway to enable the creation of artificial plant-bacteria associations/symbioses.
MATERIALS AND METHODS
Plant and Bacterial Strains
Aeschynomene evenia seeds used in this study came from line IRFL6945 (U.S. Department of Agriculture; Arrighi et al., 2012). Seeds were scarified with concentrated (95% [v/v]) sulfuric acid for 40 min. The seeds were then abundantly rinsed with sterile deionized water, surface sterilized with sodium hypochlorite (3% [w/v]) for 5 min, and again rinsed abundantly. They were then incubated overnight in sterile water at room temperature, transferred onto 0.8% (w/v) water agar plates, and put upside down for another night at 34°C to induce germination.
Agrobacterium rhizogenes strain ARqua1 (Quandt et al., 1993) was used for hairy root transformation of A. evenia (Bonaldi et al., 2010). Bradyrhizobium spp. strain ORS278 (Molouba et al., 1999) was used for nodulation assays (Giraud et al., 2000).
Isolation of AeCCaMK, AeSYMRK, and AeHK1 Sequences
A full-length AeCCaMK fragment was amplified from nodule complementary DNA (cDNA) using CCaMK(CDS)-F (5′-CACCATGGGACAAGAAACCAGAAAG-3′) and CCaMK(CDS)-R (5′-CTATAGTGGGCGAAGAGAAGAAA-3′) primers designed from an A. evenia EST library homologous to the CCaMK gene. The complete AeCCaMK CDS was then cloned in the pENTR/D-TOPO vector (Invitrogen). The AeCCaMK promoter (564 bp) was cloned using the Universal Genome Walker kit (Clontech) applied to genomic DNA following the manufacturer’s recommendations. An amplified fragment was cloned into pGEM-T vector (Promega) and sequenced. Screening of the A. evenia EST database enabled the identification of two ESTs corresponding to SYMRK and HK1 gene transcripts. For phylogenetic analyses, CCaMK sequences (GenBank accession numbers in parentheses) from Zea mays (162459834), Oryza sativa (115464591), Lilium longiflorum (71152362), Nicotiana tabacum (5814023), Petunia hybrida (148607974), Vitis vinifera (225465367), Casuarina glauca (511630596), Populus trichocarpa (224110060), Arachis hypogaea (195542475), Sesbania rostrata (186909461), Glycine max (356526360), Pisum sativum (71152364), Medicago truncatula (357515671), and Lotus japonicus (122250440) were retrieved from GenBank. Sequences similar to AeSYMRK and AeHK1 were also recovered from GenBank.
A. rhizogenes-Mediated Root Transformation
This procedure was described by Arrighi and associates (2012). Four-day-old seedlings were inoculated by direct application of A. rhizogenes on freshly sectioned radicles. One week after transformation with A. rhizogenes, the plantlets were subcultured twice at 7-d intervals on solid one-half-strength Murashige and Skoog medium containing cefotaxim (300 mg L−1). Chimeric root systems obtained after A. rhizogenes transformation using binary vectors derived from pK7WG2D or pK7GWIWG2D(II)-RedRoot Gateway vectors were dissected in sterile conditions to select one cotransformed root expressing GFP or DsRED. Dissected plants were transferred to Falcon tubes containing liquid buffered nodulation media supplemented with 1 mm KNO3, which were covered with aluminum to protect the root system from light. The plants were grown in a growth chamber at 28°C with a 16-h-light/8-h-dark regime and 70% humidity.
For studies of nodulation, plants were inoculated with 1 mL of a 5-d-old Bradyrhizobium spp. ORS278 culture grown in yeast mold modified medium. Plants were regularly inspected for nodule formation, and nodulation was quantified 1 week after inoculation and when the nodulation process was completed (corresponding to the beginning of the second infection wave when new young nodules started to appear, usually between 18 and 21 dpi). The significance of the difference between treatments was determined with a Fisher’s test.
Transgenic Roots
RNAiCCaMK1 and RNAiCCaMK2 constructs corresponding to 410 bp of the AeCCaMK kinase domain and 326 bp of the visinin domain, respectively, were generated by amplification from AeCCaMK cDNA template with the following primers: CCaMKRNAi1-F (5′-CACCTGTGAGAAAAGGCATCAGGA-3′), CCaMKRNAi1-R (5′-CAGGAACAAGCAATTCTCAGG-3′), CCaMKRNAi2-F (5′-CACCATGTGGAAATGGGGAGAATG-3′), and CCaMKRNAi2-R (5′-TCTCATCCAACTTCCCAGGT-3′). RNAiSYMRK and RNAiHK1 constructs were generated by amplification from A. evenia nodule cDNA template with the Gateway-modified primers TOPO-AeSYMRKfor (5′-CACCAATAACTTTCCAGAACAAGCAATC-3′), TOPO-AeSYMRKrev (5′-GTAGACCGAACCGAACCC-3′), CRE1(RNAi)-F (5′-CACCACGGTGATCATGAAGCCTCT-3′), and CRE1(RNAi)-R (5′-CCATGAAGCAAGCATCAAAA-3′). The specificity of all the fragments used for silencing was checked by a BLAST analysis against the Aeschynomene spp. EST database. Amplified fragments were cloned into pENTR/D-TOPO (Invitrogen) and then into binary vector pK7GWIWG2D(II)-RedRoot (Vlaams Instituut voor Biotechnologie-Ghent University) by Gateway technology.
The AeCCaMK promoter was identified with the Universal Genome Walker kit (Clontech) applied to genomic DNA, and the 564-bp region upstream from the start codon was amplified with primers PromAeCCaMK.fwd (5′-CACCCTAGTACATGTTGCATATCCT-3′) and PromAeCCaMK.rev2 (5′-CATGGTGGGGTGTGGGAATGA-3′) and recombined into pENTR/D-TOPO (Invitrogen) and then into binary vector pKGWFS7 (Karimi et al., 2002).
The gain-of-function CCaMK constructs were generated by PCR with the following primers, CCaMK(CDS)-F (5′-CACCATGGGACAAGAAACCAGAAAG-3′), CCaMK(truncated)-R (5′-CTAGCTTTGCAGCCTCGACAC-3′), CCaMK-T253D-F (5′-GCTTCTATGAGAAGGACTGGAAGGGCATTTC-3′), and CCaMK-T253D-R (5′-GAAATGCCCTTCCAGTCCTTCTCATAGAAGC-3′), using AeCCaMK cDNA cloned into the Gateway entry vector pENTR/D-TOPO (Invitrogen) as template. The PCR products were recombined into the destination vector pK7WG2D (Karimi et al., 2002).
As a transgenic control construct, we used the PENTR-GUS positive control provided with the Gateway LR Clonase II enzyme mix (Invitrogen) cloned in pK7WG2D vector (Karimi et al., 2002). All binary vectors were introduced into A. rhizogenes strain ARqua1 by the freeze-thaw method as described by Höfgen and Willmitzer (1988).
Expression Analysis of AeCCaMK, AeSYMRK, and AeHK1
Total RNA was extracted from roots using the SV Total RNA Isolation System (Promega) and quantified using a NanoDrop ND-1000 spectrophotometer. Two hundred nanograms per sample was reverse transcribed using SuperScriptII reverse transcriptase (Invitrogen) and oligo(dT)12–18. Real-time quantitative PCR was performed using the Brilliant II SYBR Green QPCR Master Mix (Agilent Technologies). The primers used were qPCRAeCCaMKfwd (5′-CCTTTCATTGCTCAGACTAATCGC-3′), qPCRAeCCaMKrev (5′-TTGGCTCCATCACCTCTCACC-3′), AeSYMRKqPCRfor (5′-AATGTGGAGAGTGGTGGAAGTAGC-3′), AeSYMRKqPCRrev (5′-TGATGATGGAGTAGCGGTTGGAC-3′), AeCre1qPCRfor (5′-AGCAAGTCAAATGGGCAAGTCAC-3′), AeCre1qPCRrev (5′-GTACCACCGTAGTTCCGAGAGG-3′), qPCRAeEF1.for (5′-AATGGTGATGCTGGTATGGTTAAG-3′), and qPCRAeEF1.rev (5′-TCTTCTTCTGTGCTGCCTTGG-3′). The real-time SYBR Green cycling PCR program on a Stratagene MX3005P (Agilent Technologies) was as follows: one cycle at 95°C for 10 min; 40 cycles at 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s; and then one cycle at 95°C for 1 min, 55°C for 30 s, and 95°C for 30 s. Reactions were performed in triplicate, and the efficiency-corrected comparative quantification method was used to quantify AeCCaMK, AeSYMRK, and AeHK1 expression (Pfaffl 2001). The results were standardized with AeEF1α expression levels.
Histochemical Analysis and Microscopy
Observations were made with a fluorescence MACROscope (Nikon AZ100), a microscope (Olympus PROVIS), or a confocal laser-scanning microscope (Carl Zeiss LSM 700). Semithin nodule sections (40–60 µm thick) were prepared using a vibratome (VT1000S; Leica). For confocal microscopy observations, the nodule sections were incubated for 20 min in live/dead staining solution (5 µm SYTO 9 and 30 µm propidium iodide in 50 mm Tris-HCl, pH 7 buffer; Live/Dead BacLight [Invitrogen]). Sections were then removed and incubated for 15 min more in 10 mm phosphate saline buffer containing Calcofluor White M2R (Sigma) at a final concentration of 0.01% (w/v) to stain the plant cell wall (Nagata and Takebe, 1970). After washing with phosphate saline buffer, the sections were mounted on microscope slides in phosphate saline buffer containing glycerol at a final concentration of 50% (v/v). Calcofluor was excited at 405 nm and detected with a 460- to 500-nm emission filter. Excitation wavelengths of 488 and 555 nm were used for SYTO 9 and propidium iodide with emission signal collection at 490 to 522 nm and 600 to 700 nm, respectively. Images were obtained using ZEN 2008 software (Zeiss). GUS activity was localized by incubating roots or sectioned nodules for 1 h at −20°C in 90% methanol, and samples were rinsed in 0.2 m phosphate buffer (pH 7) and finally incubated in a GUS assay buffer at 37°C for 12 h in the dark. The substrate mixture (adapted from Mendel et al., 1989) contained 100 µm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide in 50 mm potassium/sodium phosphate buffer (pH 8), 1% Triton X-100, 1 mm potassium ferricyanide and 1 mm potassium ferrocyanide.
The sequence data cited in this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AeCCaMK (KT248236), AeSYMRK (KT248237), and AeHK1 (KT248238).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of the SymRK ortholog of A. evenia.
Supplemental Figure S2. The A. evenia Histidine Kinase protein family.
Glossary
- NF
Nod factor
- ITs
infection threads
- RNAi
RNA interference
- dpi
days post inoculation
- CDS
coding sequence
- cDNA
complementary DNA
Footnotes
This work was supported by the French National Research Agency (grant no. ANR–SESAM–2010–BLAN–170801).
References
- Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Ghérardi M, Huguet T, et al. (2006) The Medicago truncatula lysin motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi JF, Cartieaux F, Brown SC, Rodier-Goud M, Boursot M, Fardoux J, Patrel D, Gully D, Fabre S, Chaintreuil C, et al. (2012) Aeschynomene evenia, a model plant for studying the molecular genetics of the nod-independent rhizobium-legume symbiosis. Mol Plant Microbe Interact 25: 851–861 [DOI] [PubMed] [Google Scholar]
- Bonaldi K, Gargani D, Prin Y, Fardoux J, Gully D, Nouwen N, Goormachtig S, Giraud E (2011) Nodulation of Aeschynomene afraspera and A. indica by photosynthetic Bradyrhizobium sp. strain ORS285: the nod-dependent versus the nod-independent symbiotic interaction. Mol Plant Microbe Interact 24: 1359–1371 [DOI] [PubMed] [Google Scholar]
- Bonaldi K, Gherbi H, Franche C, Bastien G, Fardoux J, Barker D, Giraud E, Cartieaux F (2010) The Nod factor-independent symbiotic signaling pathway: development of Agrobacterium rhizogenes-mediated transformation for the legume Aeschynomene indica. Mol Plant Microbe Interact 23: 1537–1544 [DOI] [PubMed] [Google Scholar]
- Capoen W, Den Herder J, Sun J, Verplancke C, De Keyser A, De Rycke R, Goormachtig S, Oldroyd G, Holsters M (2009) Calcium spiking patterns and the role of the calcium/calmodulin-dependent kinase CCaMK in lateral root base nodulation of Sesbania rostrata. Plant Cell 21: 1526–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoen W, Goormachtig S, De Rycke R, Schroeyers K, Holsters M (2005) SrSymRK, a plant receptor essential for symbiosome formation. Proc Natl Acad Sci USA 102: 10369–10374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoen W, Oldroyd G, Goormachtig S, Holsters M (2010) Sesbania rostrata: a case study of natural variation in legume nodulation. New Phytol 186: 340–345 [DOI] [PubMed] [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet EP, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J (2000) Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12: 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cérémonie H, Debellé F, Fernandez MP (1999) Structural and functional comparison of Frankia root hair deforming factor and rhizobia Nod factor. Can J Bot 77: 1293–1301 [Google Scholar]
- Doyle JJ. (2011) Phylogenetic perspectives on the origins of nodulation. Mol Plant Microbe Interact 24: 1289–1295 [DOI] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kaló P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Gage DJ. (2004) Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev 68: 280–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherbi H, Markmann K, Svistoonoff S, Estevan J, Autran D, Giczey G, Auguy F, Péret B, Laplaze L, Franche C, et al. (2008) SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankia bacteria. Proc Natl Acad Sci USA 105: 4928–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Hannibal L, Fardoux J, Verméglio A, Dreyfus B (2000) Effect of Bradyrhizobium photosynthesis on stem nodulation of Aeschynomene sensitiva. Proc Natl Acad Sci USA 97: 14795–14800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Moulin L, Vallenet D, Barbe V, Cytryn E, Avarre JC, Jaubert M, Simon D, Cartieaux F, Prin Y, et al. (2007) Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science 316: 1307–1312 [DOI] [PubMed] [Google Scholar]
- Gleason C, Chaudhuri S, Yang T, Muñoz A, Poovaiah BW, Oldroyd GED (2006) Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18: 2680–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goormachtig S, Capoen W, James EK, Holsters M (2004) Switch from intracellular to intercellular invasion during water stress-tolerant legume nodulation. Proc Natl Acad Sci USA 101: 6303–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16: 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P, Gleason C, Edwards A, Marsh J, Mitra RM, Hirsch S, Jakab J, Sims S, Long SR, Rogers J, et al. (2005) Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, Suganuma N, Kawaguchi M (2010) How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol 51: 1381–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784 [DOI] [PubMed] [Google Scholar]
- Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ané JM, Lauber E, Bisseling T, et al. (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J (2010) The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, et al. (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- Markmann K, Giczey G, Parniske M (2008) Functional adaptation of a plant receptor-kinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biol 6: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmann K, Parniske M (2009) Evolution of root endosymbiosis with bacteria: how novel are nodules? Trends Plant Sci 14: 77–86 [DOI] [PubMed] [Google Scholar]
- Mendel RR, Müller B, Schulze J, Kolesnikov V, Zelenin A (1989) Delivery of foreign genes to intact barley cells by high-velocity microprojectiles. Theor Appl Genet 78: 31–34 [DOI] [PubMed] [Google Scholar]
- Messinese E, Mun JH, Yeun LH, Jayaraman D, Rougé P, Barre A, Lougnon G, Schornack S, Bono JJ, Cook DR, et al. (2007) A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol Plant Microbe Interact 20: 912–921 [DOI] [PubMed] [Google Scholar]
- Miller JB, Pratap A, Miyahara A, Zhou L, Bornemann S, Morris RJ, Oldroyd GED (2013) Calcium/calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell 25: 5053–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Gleason CA, Edwards A, Hadfield J, Downie JA, Oldroyd GE, Long SR (2004) A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc Natl Acad Sci USA 101: 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata K, Kozaki T, Kouzai Y, Ozawa K, Ishii K, Asamizu E, Okabe Y, Umehara Y, Miyamoto A, Kobae Y, et al. (2014) The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol 55: 1864–1872 [DOI] [PubMed] [Google Scholar]
- Molouba F, Lorquin J, Willems A, Hoste B, Giraud E, Dreyfus B, Gillis M, de Lajudie P, Masson-Boivin C (1999) Photosynthetic bradyrhizobia from Aeschynomene spp. are specific to stem-nodulated species and form a separate 16S ribosomal DNA restriction fragment length polymorphism group. Appl Environ Microbiol 65: 3084–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K (2007) A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104 [DOI] [PubMed] [Google Scholar]
- Nagata T, Takebe I (1970) Cell wall regeneration and cell division in isolated tobacco mesophyll protoplasts. Planta 92: 301–308 [DOI] [PubMed] [Google Scholar]
- Ndoye I, de Billy F, Vasse J, Dreyfus B, Truchet G (1994) Root nodulation of Sesbania rostrata. J Bacteriol 176: 1060–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand P, Lapierre P, Tisa LS, Gogarten JP, Alloisio N, Bagnarol E, Bassi CA, Berry AM, Bickhart DM, Choisne N, et al. (2007) Genome characteristics of facultatively symbiotic Frankia sp. strains reflect host range and host plant biogeography. Genome Res 17: 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA (2006) Nuclear calcium changes at the core of symbiosis signalling. Curr Opin Plant Biol 9: 351–357 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144 [DOI] [PubMed] [Google Scholar]
- Op den Camp R, Streng A, De Mita S, Cao Q, Polone E, Liu W, Ammiraju JSS, Kudrna D, Wing R, Untergasser A, et al. (2011) LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science 331: 909–912 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plet J, Wasson A, Ariel F, Le Signor C, Baker D, Mathesius U, Crespi M, Frugier F (2011) MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J 65: 622–633 [DOI] [PubMed] [Google Scholar]
- Podlešáková K, Fardoux J, Patrel D, Bonaldi K, Novák O, Strnad M, Giraud E, Spíchal L, Nouwen N (2013) Rhizobial synthesized cytokinins contribute to but are not essential for the symbiotic interaction between photosynthetic Bradyrhizobia and Aeschynomene legumes. Mol Plant Microbe Interact 26: 1232–1238 [DOI] [PubMed] [Google Scholar]
- Quandt HJ, Pühler A, Broer I (1993) Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 6: 699–706 [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Remy W, Taylor TN, Hass H, Kerp H (1994) Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA 91: 11841–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silipo A, Leone MR, Erbs G, Lanzetta R, Parrilli M, Chang WS, Newman MA, Molinaro A (2011) A unique bicyclic monosaccharide from the Bradyrhizobium lipopolysaccharide and its role in the molecular interaction with plants. Angew Chem Int Ed Engl 50: 12610–12612 [DOI] [PubMed] [Google Scholar]
- Singh S, Katzer K, Lambert J, Cerri M, Parniske M (2014) CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15: 139–152 [DOI] [PubMed] [Google Scholar]
- Sinharoy S, DasGupta M (2009) RNA interference highlights the role of CCaMK in dissemination of endosymbionts in the Aeschynomeneae legume Arachis. Mol Plant Microbe Interact 22: 1466–1475 [DOI] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Raedts J, Portyanko V, Debellé F, Gough C, Bisseling T, Geurts R (2005) NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Sprent JI. (2007) Evolving ideas of legume evolution and diversity: a taxonomic perspective on the occurrence of nodulation. New Phytol 174: 11–25 [DOI] [PubMed] [Google Scholar]
- Stracke S, Kistner C, Yoshida S, Mulder L, Sato S, Kaneko T, Tabata S, Sandal N, Stougaard J, Szczyglowski K, et al. (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Svistoonoff S, Benabdoun FM, Nambiar-Veetil M, Imanishi L, Vaissayre V, Cesari S, Diagne N, Hocher V, de Billy F, Bonneau J, et al. (2013) The independent acquisition of plant root nitrogen-fixing symbiosis in Fabids recruited the same genetic pathway for nodule organogenesis. PLoS One 8: e64515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, Maekawa T, Hayashi M (2012) Nuclear-localized and deregulated calcium- and calmodulin-dependent protein kinase activates rhizobial and mycorrhizal responses in Lotus japonicus. Plant Cell 24: 810–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L, Imaizumi-Anraku H, Yoshida S, Murakami Y, Madsen LH, Miwa H, Nakagawa T, Sandal N, Albrektsen AS, Kawaguchi M, et al. (2006) Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156 [DOI] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, Radutoiu S, Albrektsen AS, Sato S, Asamizu E, Tabata S, Stougaard J (2007) A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315: 104–107 [DOI] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Müller J, Singh S, Banba M, Vickers K, Markmann K, White C, Schuller B, Sato S, et al. (2008) CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc Natl Acad Sci USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]