A nitrate transporter and a potassium channel act synergistically in nitrate-dependent root-to-shoot translocation of potassium in Arabidopsis.
Abstract
Root-to-shoot translocation and shoot homeostasis of potassium (K) determine nutrient balance, growth, and stress tolerance of vascular plants. To maintain the cation-anion balance, xylem loading of K+ in the roots relies on the concomitant loading of counteranions, like nitrate (NO3−). However, the coregulation of these loading steps is unclear. Here, we show that the bidirectional, low-affinity Nitrate Transporter1 (NRT1)/Peptide Transporter (PTR) family member NPF7.3/NRT1.5 is important for the NO3−-dependent K+ translocation in Arabidopsis (Arabidopsis thaliana). Lack of NPF7.3/NRT1.5 resulted in K deficiency in shoots under low NO3− nutrition, whereas the root elemental composition was unchanged. Gene expression data corroborated K deficiency in the nrt1.5-5 shoot, whereas the root responded with a differential expression of genes involved in cation-anion balance. A grafting experiment confirmed that the presence of NPF7.3/NRT1.5 in the root is a prerequisite for proper root-to-shoot translocation of K+ under low NO3− supply. Because the depolarization-activated Stelar K+ Outward Rectifier (SKOR) has previously been described as a major contributor for root-to-shoot translocation of K+ in Arabidopsis, we addressed the hypothesis that NPF7.3/NRT1.5-mediated NO3− translocation might affect xylem loading and root-to-shoot K+ translocation through SKOR. Indeed, growth of nrt1.5-5 and skor-2 single and double mutants under different K/NO3− regimes revealed that both proteins contribute to K+ translocation from root to shoot. SKOR activity dominates under high NO3− and low K+ supply, whereas NPF7.3/NRT1.5 is required under low NO3− availability. This study unravels nutritional conditions as a critical factor for the joint activity of SKOR and NPF7.3/NRT1.5 for shoot K homeostasis.
The macronutrient potassium (K) is essential for plant growth and development because of its crucial roles in various cellular processes (i.e. regulation of enzyme activities), stabilization of protein synthesis, and neutralization of negative charges. In addition, it is a major component of the cation-anion balance and osmoregulation in plants, thereby influencing cellular turgor, xylem and phloem transport, pH homeostasis, and the setting of membrane potentials (Maathuis, 2009; Marschner, 2012; Sharma et al., 2013). K+ uptake and distribution in Arabidopsis (Arabidopsis thaliana) are accomplished by a total of 71 membrane proteins that have been assigned to five gene families: the Shaker and Tandem-Pore K+ channels (now also including the inward-rectifier K-like (Kir-like) channels), the K+ uptake permeases (KUP/HAK/KT), the K+ transporter (HKT) family, and the cation proton antiporters (CPA; Gierth and Mäser, 2007; Gomez-Porras et al., 2012; Sharma et al., 2013).
Root xylem loading is a key step for the delivery of nutrients to the shoot (Poirier et al., 1991; Engels and Marschner, 1992a; Gaymard et al., 1998; Takano et al., 2002; Park et al., 2008). Root-to-shoot translocation of K+ is mediated by the voltage-dependent Shaker family K+ channel Stelar K+ Outward Rectifier (SKOR). The gene is primarily expressed in pericycle and root xylem parenchyma cells, and it is down-regulated upon K shortage and in response to treatments with the phytohormones abscisic acid, cytokinin, and auxin. Such gene expression changes are thought to control K+ secretion into the xylem sap and K+ reallocation through the phloem to adjust root K+ transport activity to K+ availability and shoot demand (Pilot et al., 2003). SKOR is activated upon membrane depolarization, and it is in a closed state when the driving force for K+ is inwardly directed. It elicits outward K+ currents, facilitating the release of the cation from the cells into the xylem. The voltage dependency of the channel is modulated by the external K+ concentration to minimize the risk of an undesired K+ influx under high K+ availability (Johansson et al., 2006). Root-to-shoot K+ transfer was strongly reduced in the knockout mutant skor-1, resulting in a decreased shoot K content, whereas the root K content remained unaffected (Gaymard et al., 1998).
Root xylem loading is subject to the maintenance of a cation-anion balance, and nitrate (NO3−) is the quantitatively most important anion counterbalancing xylem loading of K+ (Engels and Marschner, 1993). Members of the Nitrate Transporter1 (NRT1)/Peptide Transporter (PTR) transporter family (NPF) play a prominent role in NO3− uptake and allocation in Arabidopsis (summarized in Krouk et al., 2010; Wang et al., 2012; and Léran et al., 2014). Two of them have recently been reported to control xylem NO3− loading and unloading. The low-affinity, pH-dependent bidirectional NO3− transporter NPF7.3/NRT1.5 (subsequently termed NRT1.5) mediates NO3− efflux from pericycle cells to the xylem vessels, whereas the low-affinity influx protein NPF7.2/NRT1.8 removes NO3− from the xylem sap and transfers it into xylem parenchyma cells (Lin et al., 2008; Li et al., 2010; Chen et al., 2012). Accordingly, the expression of both genes is oppositely regulated under various stress conditions (Li et al., 2010). In nrt1.5 mutants, NRT1.8 expression is increased, which is thought to enhance NO3− reallocation to the root (Chen et al., 2012).
The NRT1.5 gene is mainly expressed in root pericycle cells close to the xylem, and the protein localizes to the plasma membrane. In nrt1.5 mutants, less NO3− is transported from the root to the shoot, and the NO3− concentration in the xylem sap is reduced. However, root-to-shoot NO3− transport is not completely abolished in these mutants, indicating the existence of additional xylem-loading activities for NO3− (Lin et al., 2008; Wang et al., 2012). The recent observation that NPF6.3/NRT1.1/CHL1 and NPF6.2/NRT1.4 are also capable of mediating bidirectional NO3− transport in Xenopus laevis oocytes might indicate that more NPF family members are contributing to xylem loading with NO3− (Léran et al., 2013).
Electrophysiological studies with NRT1.5-expressing X. laevis oocytes revealed that NO3− excited an inward current at pH 5.5, which would be expected for a proton-coupled nitrate transporter with a proton to nitrate ratio larger than one (Lin et al., 2008). The inward currents elicited by exposure to nitrate were pH dependent, and Lin et al. (2008) observed that NRT1.5 can also facilitate nitrate efflux when the oocytes were incubated at pH 7.4. Lin et al. (2008) concluded that NRT1.5 can transport nitrate in both directions, presumably through a proton-coupled mechanism. Interestingly, a K+ gradient was not sufficient to drive NRT1.5-mediated NO3− export. However, the determination of root and shoot cation concentrations in the nrt1.5-1 mutant revealed that the amount of K+ translocated to the shoot was reduced when NO3− but not NH4+ was supplied as the N source. Therefore, Lin et al. (2008) suggested a regulatory loop between NO3− and K+ at the xylem loading step.
A close relationship between these two nutrients concerning uptake, translocation, recycling, and reduction (of NO3−) has been described in physiological studies since the 1960s (e.g. Ben Zioni et al., 1971; Blevins et al., 1978; Barneix and Breteler, 1985), but only recently, common components in the NO3− and K+ uptake pathways were identified and led to the first ideas of how such a cross talk might be coordinated on the molecular level. The uptake activity of the K+ channel AKT1 as well as the affinity of the NO3− transporter NPF6.3/NRT1.1/CHL1 are both modulated by the activity of CALCINEURIN B-LIKE PROTEIN-INTERACTING PROTEIN KINASE23 (CIPK23), which itself is regulated by CALCINEURIN B-LIKE PROTEIN9 (CBL9) under both deficiencies (Xu et al., 2006; Ho et al., 2009). Yet, the details of this interaction in root K+ uptake, the (regulation of) xylem loading with K+ and NO3−, and the involvement of SKOR and NRT1.5 in this process are unknown.
In this study, we approached this problem by investigating the molecular and physiological responses of Arabidopsis wild-type (Columbia-0 [Col-0]), nrt1.5, and skor transfer DNA (T-DNA) insertion lines to varying NO3− and K+ regimes. The nrt1.5 mutant developed an early senescence phenotype under low NO3− nutrition, which could be attributed to a reduced K+ translocation to the shoot. The assessment of nrt1.5 and skor single- and double-knockout lines disclosed an interplay of the two proteins in the NO3−-dependent control of shoot K homeostasis. The presented data indicate that SKOR mediates K+ root-to-shoot translocation under high NO3− and low K+ availability, whereas NRT1.5 is important for K+ translocation under low NO3− availability, irrespective of the K+ supply.
RESULTS
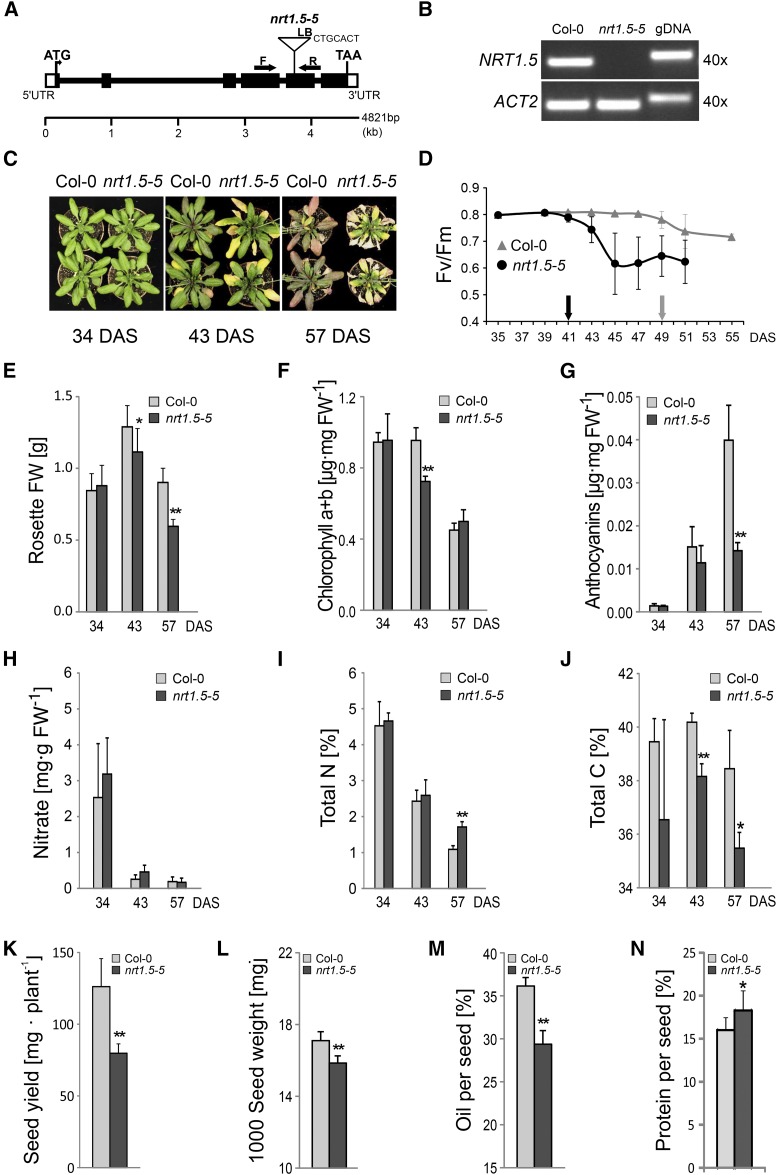
nrt1.5-5 Mutant Plants Exhibit a Pleiotropic Phenotype on Low-Fertilized Soil
Arabidopsis nrt1.5 mutant plants are phenotypically indistinguishable from wild-type plants under standard growth conditions (Lin et al., 2008; Chen et al., 2012). However, we found that, on low-fertilized soil, the three mutant lines nrt1.5-5, carrying a T-DNA insertion in exon 5 and lacking full-length NRT1.5 transcripts (Fig. 1, A and B), nrt1.5-2 (Lin et al., 2008), and nrt1.5-4 (Li et al., 2010) exhibited the same phenotype (Supplemental Fig. S1). We, therefore, focused subsequent analyses on line nrt1.5-5. Until bolting, the plants grew without any visible symptoms (Fig. 1C, 34 d after sowing [DAS]). In the following days, tips of old nrt1.5-5 leaves turned yellow and died, whereas the corresponding leaves of Col-0 plants developed a brown-reddish color and remained turgescent until 57 DAS (Fig. 1C, 43 and 57 DAS). The maximum quantum efficiency of PSII (maximum photochemical efficiency of PSII in the dark-adapted state [Fv/Fm]) was determined as an indicator for senescence initiation and progression in leaf number 8 (Fig. 1D). Fv/Fm declined earlier and faster in nrt1.5-5 than in Col-0. At 53 DAS, all leaves number 8 in the mutant were dead, whereas the wild-type leaves were still photosynthetically active. Determination of the rosette fresh weight revealed an earlier fresh weight loss of nrt1.5-5 plants, which coincided with the early senescence symptoms of the leaves (Fig. 1E). Leaf yellowing was reflected by decreased chlorophyll concentrations in the nrt1.5-5 whole rosettes at 43 DAS (Fig. 1F). At 57 DAS, mutant and wild-type plants contained similar amounts of the pigment. At this time point, the old nrt1.5-5 leaves were completely dried out, and only the inner part of the mutant rosette appeared light green and turgescent (Fig. 1C). The brown-reddish color of Col-0 rosettes at 57 DAS was the result of an increased anthocyanin production, which did not occur to such an extent in the mutant (Fig. 1G).
Figure 1.
Pleiotropic phenotype of the T-DNA insertion mutant nrt1.5-5 on low-fertilized soil. A, NRT1.5 gene structure and T-DNA insertion site of the mutant line nrt1.5-5. LB, Left border. White boxes indicate untranslated regions (UTRs), and black boxes indicate exons. Arrows indicate positions of PCR primers used in B. B, RT-PCR analysis of NRT1.5 transcripts in rosette leaves of wild-type (Col-0) and homozygous nrt1.5-5 mutant plants. RT-PCR of ACTIN2 (ACT2; At3g18780) and PCR on genomic DNA (gDNA) served as controls. PCR cycle count was 40. C, Early leaf senescence phenotype of nrt1.5-5 on low-fertilized soil. Growth responses of Col-0 and nrt1.5-5 plants at 34, 43, and 57 DAS are shown. In three repeats of the experiment, plants developed the same phenotype. D, PSII maximum quantum efficiency decline in leaf number 8. Black and gray arrows indicate first declines in Fv/Fm in nrt1.5-5 and Col-0 plants, respectively, indicating senescence initiation (means ± sd; n = 8). E, Rosette fresh weight (FW) of Col-0 and nrt1.5-5 at 34, 43, and 57 DAS (means ± sd; n = 8). F, Chlorophyll content decreases at 43 DAS in nrt1.5-5 whole rosettes but not in Col-0 (means ± sd; n = 3). G, Reduced anthocyanin accumulation in nrt1.5-5 leaves (means ± sd; n = 3). H, Nitrate content in rosettes (means ± sd; n ≥ 5), I, Total N content in rosettes (means ± sd; n ≥ 4). J, Total C content in rosettes (means ± sd; n ≥ 4). K, Quantification of seed yield (means ± sd; n = 8). L, Thousand-seed weight (means ± sd; n = 8). M, Seed oil content (means ± sd; n = 8). N, Seed protein content (means ± sd; n = 8). Similar results were obtained in two independent experiments. *, Significant differences (Student’s t test) between nrt1.5-5 and Col-0 with P < 0.05; **, significant differences (Student’s t test) between nrt1.5-5 and Col-0 with P < 0.01.
Accelerated chlorophyll degradation is usually accompanied by N remobilization from chloroplasts (Hörtensteiner and Feller, 2002) and might be caused by N deficiency in nrt1.5-5. To address this issue, we determined NO3− and total N concentrations in the rosette material. The measurements revealed that the plants depleted their endogenous NO3− reserves in the leaves between 34 and 43 DAS to compensate for the reduced root supply on low-fertilized soil, but a significant difference in the NO3− concentrations was not observed between nrt1.5-5 and Col-0 (Fig. 1H). In contrast, mutant plants contained slightly more total N in their rosettes than the wild type at 57 DAS (Fig. 1I). In addition, the total C content was significantly decreased in nrt1.5-5 at 43 and 57 DAS (Fig. 1J). This observation was consistent with the early Fv/Fm decline and the lower photosynthetic activity of the mutant (Fig. 1D). The seed yield of nrt1.5-5 plants was reduced to only 63% relative to the wild type (Fig. 1K), and a significant seed weight reduction was observed (Fig. 1L). The reduced availability of carbon skeletons in nrt1.5-5 (Fig. 1J) might be the reason for the significantly lower seed oil content (Fig. 1M), whereas the total protein content in dry seeds was even slightly increased (Fig. 1N).
Supplementation of the low-fertilized soil with 10 mm KNO3 from 34 to 57 DAS diminished or even abolished symptom development of the nrt1.5-5 mutant (Supplemental Fig. S2). In both wild-type and mutant plants, additional KNO3 provision resulted in higher biomass production, later chlorophyll degradation, reduced anthocyanin accumulation, increased NO3− and total N concentrations at 43 and 57 DAS, and doubling of the seed yield. These results indicated that the pleiotropic phenotype of nrt1.5-5 was caused by the low KNO3 fertilization of the soil. NO3− and total N concentrations in nrt1.5-5 were close to wild-type levels under both cultivation conditions (Fig. 1, H and I; Supplemental Fig. S2, F and G), suggesting that K deficiency might be causal for the differential progression of leaf senescence in the mutant. Because Lin et al. (2008) did not report a similar phenotype for the nrt1.5 mutant plants when grown in hydroponic solutions containing high NO3− concentrations (4–12.5 mm), we hypothesized that the putative K deficiency symptoms in old leaves might depend on a limited NO3− supply.
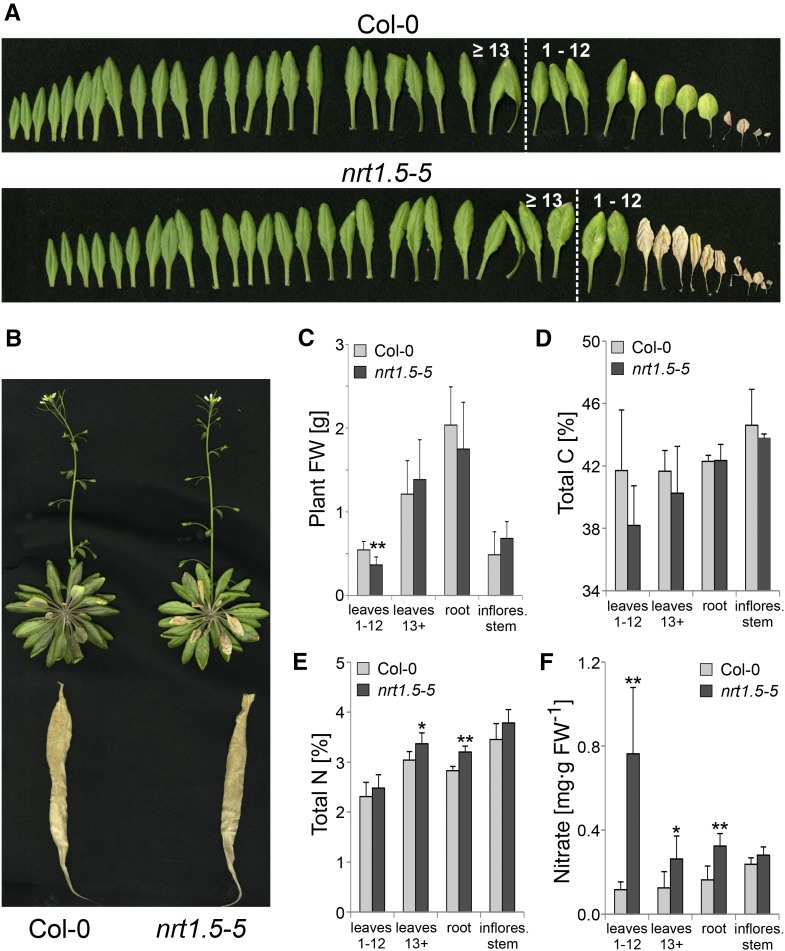
Limited NO3− Supply to nrt1.5-5 Plants Causes K Deficiency in Shoot Organs
By subjecting nrt1.5-5 mutant and Col-0 plants to limiting NO3− nutrition (0.1 mm) in hydroponic culture, we addressed the question if low NO3− fertilization causes K deficiency in the mutant shoot. As a result of the treatment, nrt1.5-5 leaves became earlier senescent than the wild type (Fig. 2, A and B), with the outcome of a significant fresh weight reduction in old leaves (Fig. 2C, leaves 1–12). Like in soil-grown mutants (Fig. 1J), total C was slightly (but not significantly) reduced in rosette leaves (Fig. 2D), and total N was elevated in young leaves (nos. ≥13) and roots (Fig. 2E). In comparison with Col-0 plants, all nrt1.5-5 organs, except the inflorescence stems, contained more NO3−, although the concentrations were generally close to the detection limit as a result of the limited NO3− supply (Fig. 2F).
Figure 2.
NO3− limitation-induced nrt1.5-5 phenotype in hydroponic culture. A, Individual rosette leaves of Col-0 and nrt1.5-5 plants grown for 5 weeks in hydroponic culture with 0.1 mm NH4NO3 in the medium. Left to right shows young (nos. ≥13) to old (nos. 1–12) leaves. B, Representative picture of Col-0 and nrt1.5-5 plants at the time of harvest showing the root, the abaxial side of the rosette, and the inflorescence stem. C, Fresh weight (FW) of harvested plant material used for further analyses: pooled old leaves numbers 1 to 12, pooled young leaves numbers ≥13, roots, and inflorescence stems (means ± sd; n ≥ 9). D, Total C content in said plant material (means ± sd; n ≥ 4). E, Total N content in said plant material (means ± sd; n ≥ 4). F, NO3− concentration in said plant material (means ± sd; n ≥ 5). *, Significant differences (Student’s t test) between nrt1.5-5 and Col-0 with P < 0.05; **, significant differences (Student’s t test) between nrt1.5-5 and Col-0 with P < 0.01.
When we determined the concentrations of mineral elements by inductively coupled plasma optical emission spectrometry (ICP-OES), most of the macroelements were increased in nrt1.5-5 leaves with the exception of K (Table I). K concentrations remained below 30% and 50% of the wild-type levels in leaves numbers 1 to 12 and ≥13, corresponding to <1% and <2% of the dry weight, respectively. Because the critical K concentration for plant growth is in the range of 0.5% to 2% (Leigh and Jones, 1984), nrt1.5-5 rosette leaves experienced K deficiency, whereas Col-0 rosettes with 3% to 4% K were well supplied (Table I). K deficiency is often accompanied by increased tissue concentrations of other cationic elements, which are partially compensating for the charge imbalances in the plant (Leigh and Jones, 1984). Indeed, we observed a remarkable increase in the two macronutrients Ca (1.5- to 3.0-fold) and Mg (1.3- to 2.0-fold) in rosette leaves and inflorescence stems of nrt1.5-5 plants. The micronutrients Mn, Zn, and Mo were also increased, whereas B was reduced in rosette leaves (Table I).
Table I. Elemental analysis in tissues of Arabidopsis nrt1.5-5 and Col-0 plants grown in hydroponic culture under supply of 0.1 mm NH4NO3 for 5 weeks.
The concentration of macro- and microelements was determined in homogenized material from leaf numbers 1 to 12, leaf numbers ≥13, roots, and inflorescence stems. Bold numbers indicate significantly lower or higher elemental concentration (Student's t test; *, P < 0.05; and **, P < 0.01) in nrt1.5-5 tissues when compared with Col-0.
| Macro- and Microelements | Leaves 1-12 |
Leaves 13+ |
Roots |
Inflorescence Stem |
||||
|---|---|---|---|---|---|---|---|---|
| Col-0 | nrt1.5-5 | Col-0 | nrt1.5-5 | Col-0 | nrt1.5-5 | Col-0 | nrt1.5-5 | |
| Macroelements (mg g−1 dry wt) | ||||||||
| K | 30.7 ± 5.1 | 7.9 ± 1.0** | 43.8 ± 5.4 | 19.5 ± 1.5** | 56.5 ± 8.4 | 55.1 ± 8.0 | 32.3 ± 5.9 | 26.2 ± 3.3* |
| Ca | 17.4 ± 2.7 | 30.5 ± 3.6** | 7.5 ± 1.6 | 21.5 ± 2.2** | 0.5 ± 0.4 | 0.9 ± 0.8 | 1.7 ± 0.6 | 4.6 ± 0.6** |
| Mg | 15.8 ± 1.6 | 23.6 ± 2.9** | 8.1 ± 0.9 | 16.1 ± 1.6** | 1.9 ± 0.3 | 1.5 ± 0.3* | 3.0 ± 0.6 | 3.9 ± 0.4** |
| P | 7.8 ± 0.5 | 9.1 ± 1.6* | 8.0 ± 0.8 | 8.6 ± 0.8 | 9.5 ± 1.5 | 9.7 ± 1.3 | 8.2 ± 2.2 | 6.9 ± 0.6 |
| S | 6.0 ± 0.6 | 8.1 ± 1.5** | 8.4 ± 0.5 | 10.3 ± 1.0** | 10.1 ± 1.7 | 10.2 ± 1.4 | 8.0 ± 1.4 | 8.7 ± 0.9 |
| Microelements (μg g−1 dry wt) | ||||||||
| Fe | 102.8 ± 44.5 | 158.4 ± 164.8 | 81.8 ± 27.7 | 76.7 ± 20.2 | 5,375.6 ± 1,842.5 | 7,125.0 ± 1,586.9 | 61.4 ± 14.1 | 56.0 ± 5.1 |
| B | 166.8 ± 24.8 | 109.6 ± 12.5** | 85.9 ± 10.1 | 75.9 ± 8.2* | 15.6 ± 33.8 | 28.8 ± 66.7 | 28.6 ± 10.6 | 24.1 ± 1.7 |
| Mn | 507.9 ± 98.0 | 685.1 ± 131.4** | 348.1 ± 113.8 | 653.1 ± 107.6** | 258.0 ± 189.0 | 178.4 ± 74.9 | 120.1 ± 30.7 | 143.5 ± 16.5 |
| Zn | 83.7 ± 7.7 | 117.2 ± 18.9** | 79.1 ± 10.3 | 135.5 ± 14.5** | 200.7 ± 49.6 | 235.8 ± 50.9 | 53.8 ± 14.3 | 73.2 ± 7.2** |
| Mo | 24.1 ± 5.7 | 25.0 ± 5.3 | 9.9 ± 3.8 | 13.8 ± 1.8* | 130.8 ± 23.1 | 155.6 ± 22.8* | 0.4 ± 0.4 | 0.7 ± 0.1 |
| Cu | 6.0 ± 2.0 | 5.4 ± 1.6 | 8.7 ± 1.2 | 7.6 ± 1.3 | 50.1 ± 12.0 | 52.3 ± 14.6 | 4.4 ± 1.2 | 4.7 ± 1.3 |
Other than in shoots, the elemental composition in wild-type and nrt1.5-5 roots was almost the same (Table I). Apparently, mutant roots were able to absorb all nutrients in sufficient amounts but particularly failed to transfer K+ to the shoot. In turn, Ca2+ and Mg2+ were transported with increased efficiency to the aboveground tissues. These data largely resembled the phenotype of the skor-1 mutant. Gaymard et al. (1998) showed that the disruption of the SKOR gene had no effect on the K content in roots but resulted in an approximately 50% decrease in shoot K, which was essentially compensated for by an increase in Ca. Calculating the cationic charge balance further corroborated that Ca2+ and Mg2+, similarly as in skor-1, compensated for the decline in K+ in nrt1.5-5 shoots, whereas the root cation balance remained unchanged (Supplemental Fig. S3). These results showed that limited NO3− fertilization of nrt1.5-5 plants provoked the early senescence phenotype and eventually, resulted in severe K deficiency in the shoot. The comparison with skor-1 suggested that both NRT1.5 and SKOR might contribute to root-to-shoot translocation of K+, possibly influenced by the availability and uptake of NO3− in the roots.
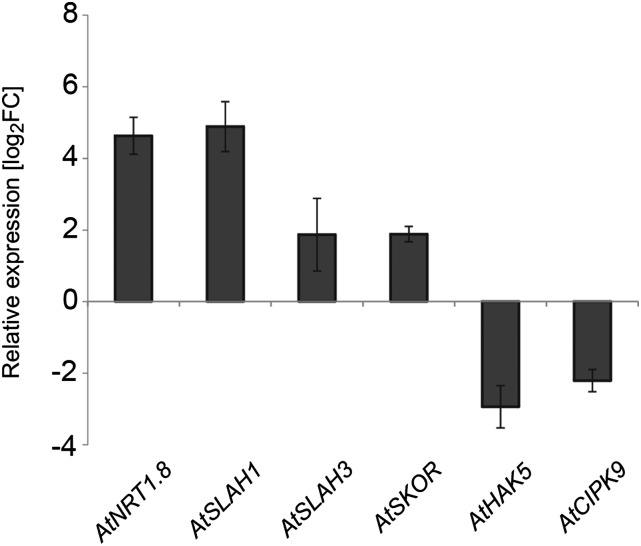
Transcription of Ion Homeostasis-Associated Genes Indicates K Deficiency in nrt1.5-5 Shoots
The K deficiency in shoots of nrt1.5-5 plants grown under low NO3− supply in hydroponics could be associated with altered expression of ion transporter or channel genes or selected NO3− and K+ signaling pathway genes in roots. Expression analysis of 48 genes belonging to NO3−, K+, Pi, or NH4+ transporter families and a small number of CIPK/CBL family members revealed significant expression changes of five genes that are important contributors in NO3−, K+, or anion transport and one gene encoding a protein kinase involved in the K+ signaling pathway (Fig. 3), whereas none of the other 42 genes were regulated (Supplemental Table S1). The NO3− transporter NRT1.8 was 32-fold up-regulated in nrt1.5-5 roots, corroborating earlier findings in nrt1.5-3 and nrt1.5-4 (Chen et al., 2012). In addition, expression of the membrane protein SLOW ANION CHANNEL-ASSOCIATED1 (SLAC1) HOMOLOG1 (SLAH1), which is likely involved in organic/inorganic anion homeostasis of plant cells (Negi et al., 2008), was strongly increased in nrt1.5-5 roots (Fig. 3). Also SLAH3, another member of the SLAC1/SLAH channel family, was significantly up-regulated. The protein was previously characterized as a preferentially NO3−-permeable channel, with activity that is regulated by NO3−, calcium, and an abscisic acid-sensitive phosphorylation step (Geiger et al., 2011).
Figure 3.
Differentially regulated genes in roots of nrt1.5-5 plants. Relative transcript levels of six regulated genes in roots of hydroponically grown plants were measured by qPCR and normalized to UBQ10 (means ± sd; n = 4). Plotted are the log2 fold expression changes (FCs) in nrt1.5-5 roots compared with Col-0 roots.
Although the shoot suffered from K deficiency, expression of the high-affinity K+ transporter HAK5 was 8-fold lower in mutant roots, and the 3.7-fold up-regulated expression of the voltage-dependent K+ channel SKOR did not compensate for the K deficit in the shoots (Fig. 3; Table I). Moreover, the CIPK9 gene, a critical K deprivation-inducible regulator of the low K response in Arabidopsis (Pandey et al., 2007; Liu et al., 2013), was 5-fold down-regulated in nrt1.5-5 roots, suggesting that the nrt1.5-5 root system was impaired in sensing or responding to K deficiency in the shoot. Considering that the elemental composition in nrt1.5-5 roots was almost the same as in the wild type (Table I), the root transcript patterns indicate an influence of NRT1.5 on the NO3− and K+ balance between roots and shoot.
Genes involved in biotic stress responses and jasmonic acid (JA) signaling have been described to play a prominent role in K-dependent gene regulation in Arabidopsis (Armengaud et al., 2004). We, therefore, surveyed by quantitative real-time PCR (qPCR) the expression of 20 genes related to JA metabolism in nrt1.5-5 shoots and another 14 genes related to calcium signaling and other plant stress response pathways, like defense, secondary metabolism, and reactive oxygen species production. Indeed, all 34 tested genes were more than 2-fold up-regulated in nrt1.5-5 leaves (Supplemental Fig. S4); 26 of the 34 tested genes were reported to be up-regulated in shoots under K starvation (Armengaud et al., 2004), supporting the notion that nrt1.5-5 mutant shoots sense and respond to K deficiency.
Overall, the transcription data are consistent with the altered K+ balance in the shoot and root system of nrt1.5-5 plants, defective shoot-to-root K deficiency signaling, and impaired root-to-shoot translocation of the cation. Our data suggest that NRT1.5 and the other alternatively regulated root genes are critical actors in these processes. In addition, the up-regulated expression of SKOR in nrt1.5-5 roots supports the idea of an interplay of the two proteins in the root-to-shoot translocation of K+.
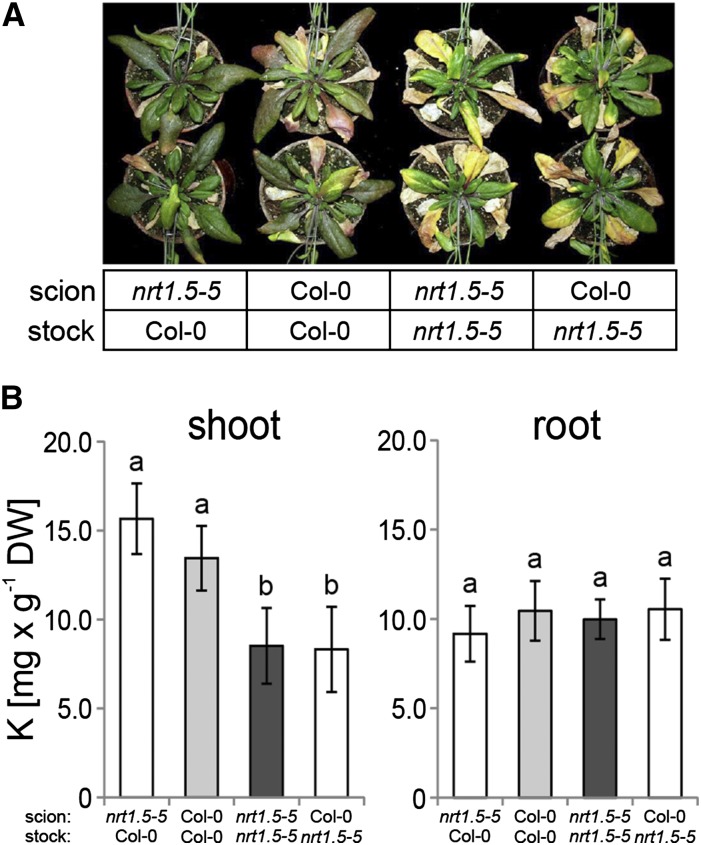
The Presence of NRT1.5 in Roots Is a Prerequisite for Proper Root-to-Shoot Translocation of K+ under Low NO3− Supply
To investigate whether the deficit in K+ translocation was caused by the root (loading of the xylem) or the shoot (unloading of the xylem), we generated homografted (Col-0 stock/Col-0 scion and nrt1.5-5 stock/nrt1.5-5 scion) and heterografted (Col-0 stock/nrt1.5-5 scion and nrt1.5-5 stock/Col-0 scion) plants and grew them on low-fertilized soil. The early leaf senescence phenotype of the mutant developed most severely on the nrt1.5-5 root stock (Fig. 4A), indicating that the absence of NRT1.5 function in the root was primarily responsible for symptom manifestation. This conclusion was corroborated by the observation of reduced shoot K concentrations (<1% of dry weight) on the nrt1.5-5 root stock, whereas root K concentrations were unchanged, irrespective of the grafting combination (Fig. 4B). Further support came from elevated Ca and Mg concentrations in shoots of grafted plants with an nrt1.5-5 root stock, suggesting that both cations contributed to compensating for the charge imbalances provoked by K deficiency (Supplemental Fig. S5).
Figure 4.
Rosette phenotype and K concentrations in shoots and roots of grafted Arabidopsis plants. A, Early leaf senescence phenotype as a result of the grafts as indicated in lower. Grafted plants were grown for 6 weeks on unfertilized type 0 soil supplemented for the first 2 weeks with 10 mm KNO3 to support plant growth and subsequently, 1 mm KNO3 to trigger development of the phenotype. B, K concentrations in shoots and roots of grafted plants. Data are means ± sd (n ≥ 6). The data were statistically analyzed by one-way ANOVA and subsequent multiple comparisons (Tukey’s range test). Different letters indicate a significant difference at P < 0.05. Vertical bars denote sds. DW, Dry weight.
Genetic Complementation of nrt1.5-5 by Expression of NRT1.5 under Control of the PHOSPHATE1 Promoter
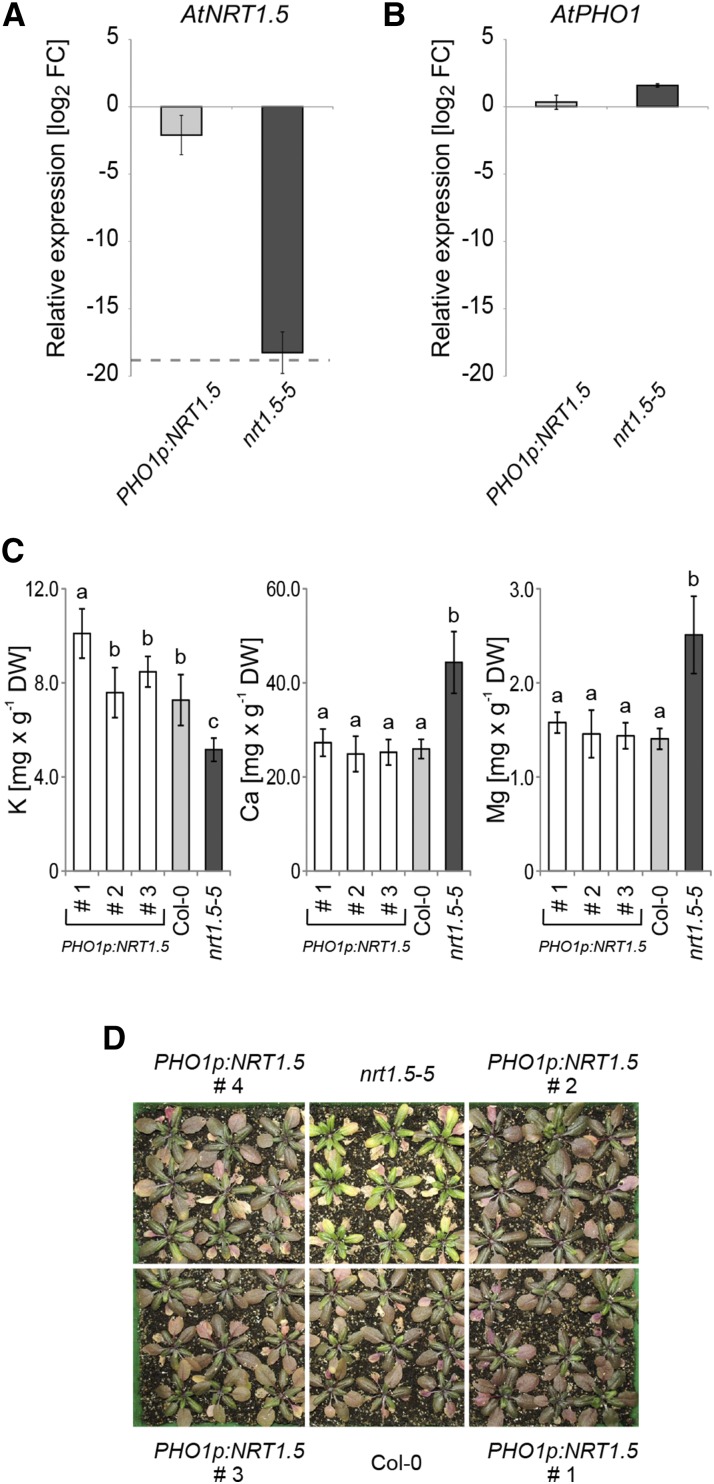
For complementation of the nrt1.5-5 mutant, we transformed the plants with the NRT1.5 coding region fused behind the promoter of the PHOSPHATE1 (PHO1) gene (At3g23430), with expression that is primarily directed to the root vasculature (Supplemental Note S1; Hamburger et al., 2002; Wege and Poirier, 2014). The transformants expressed approximately wild-type levels of NRT1.5 transcripts in the roots (Fig. 5A), and the endogenous PHO1 expression was unchanged (Fig. 5B). Determination of K, Ca, and Mg concentrations in three independent PHO1p:NRT1.5 transformed nrt1.5-5 lines (1–3) in comparison with Col-0 and nrt1.5-5 plants revealed a recovery of all three elements to wild-type concentrations in the shoot (Fig. 5C), which finally resulted in wild-type rosette phenotype of the complemented lines (Fig. 5D). This experiment showed that wild-type NRT1.5 expression in roots is needed to meet the K demand of the shoot.
Figure 5.
Complementation of the nrt1.5-5 mutant. A, Relative NRT1.5 transcript levels in seedling roots of PHO1p:NRT1.5-transformed and -nontransformed nrt1.5-5 plants compared with NRT1.5 transcript levels in Col-0 plants. PHO1 promoter-driven expression of NRT1.5 recovers almost wild-type transcript level in the nrt1.5-5 mutant background. The NRT1.5 signal in nontransformed nrt1.5-5 plants is at background noise level (gray dashed line). B, Relative PHO1 transcript levels in seedling roots of PHO1p:NRT1.5-transformed and -nontransformed nrt1.5-5 plants compared with PHO1 transcript levels in Col-0 plants. PHO1 promoter-driven expression of NRT1.5 does not alter the PHO1 transcript level in the nrt1.5-5 mutant background. Seedlings in A and B were raised in one-half-strength MS liquid culture. Relative transcript levels in A and B were measured by qPCR and normalized to UBQ10. Plotted are the log2 fold expression changes (FCs) compared with Col-0 seedling roots. Values are means ± sd of n = 6 independent transgenic PHO1p:NRT1.5 lines (including lines 1–4 shown in C and D) with eight pooled plants per sample and n = 3 nrt1.5-5 samples with eight pooled plants per sample. C, Potassium, Ca, and Mg concentrations (means ± sd; n = 9) in rosettes of Col-0, nrt1.5-5, and three independent PHO1p:NRT1.5-transformed nrt1.5-5 lines (1–3) grown on unfertilized type 0 soil and supplemented with a one-half-strength MS-based fertilization solution containing 1:1:10 mm N:K:P (Supplemental Table S2). The data were statistically analyzed by one-way ANOVA and subsequent multiple comparisons (Tukey’s honestly significant difference mean-separation test). In samples marked with different letters, concentrations differ significantly at P < 0.05. D, Rosette phenotype of nrt1.5-5, Col-0, and four independent PHO1p:NRT1.5-transformed nrt1.5-5 lines grown under the same condition as in C. DW, Dry weight.
NRT1.5 Enhances Hygromycin B Sensitivity in Yeast and Plants
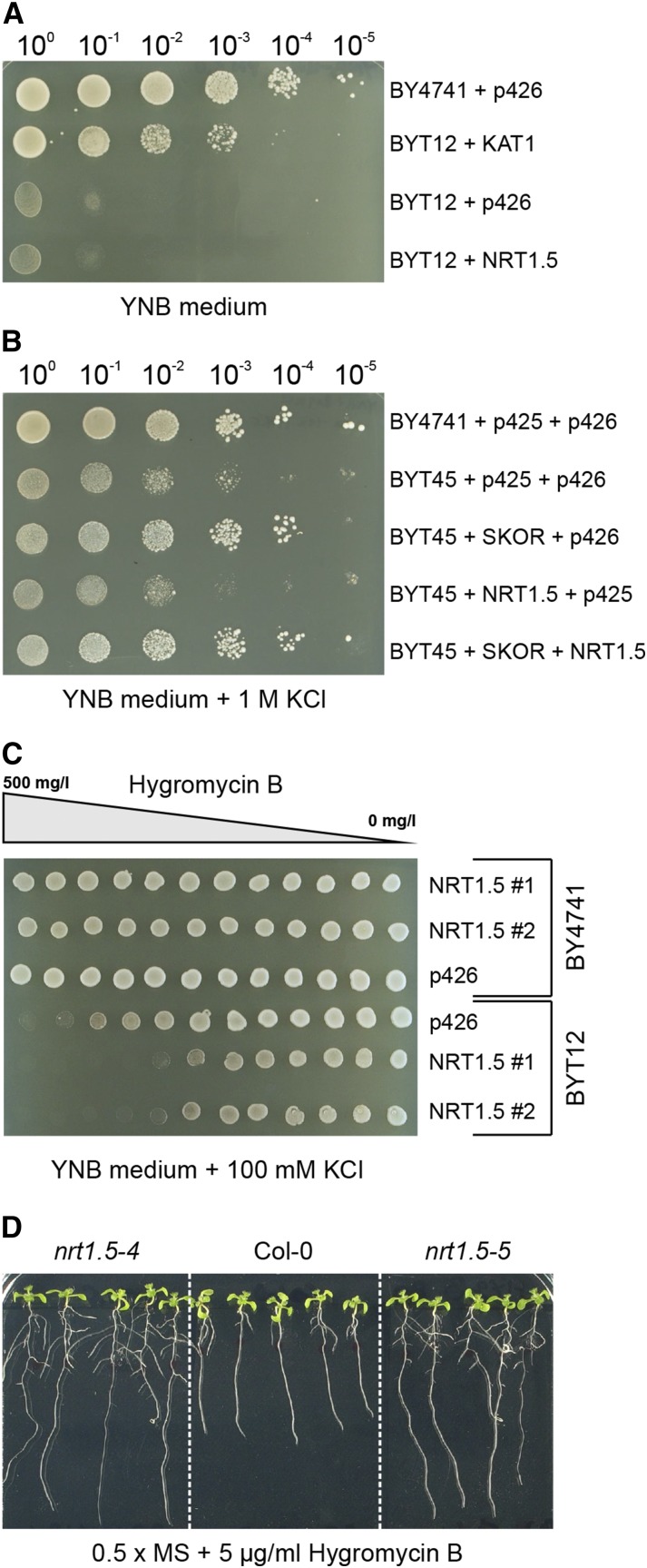
To functionally test NRT1.5 in a heterologous expression system for K+ transport activity, the yeast (Saccharomyces cerevisiae) mutants BYT12, lacking the main K+ uptake systems (trk1 trk2), and BYT45, lacking the main K+ efflux systems (nha1 ena1) were used (Zahrádka and Sychrová, 2012). The inward-rectifying Arabidopsis K+ channel KAT1 and the outward-rectifying K+ channel SKOR served as positive controls in the yeast assays. NRT1.5 was not able to complement the growth retardation of BYT12 under K deficiency (Fig. 6A) or counteract the accumulation of toxic levels of K+ in BYT45 cells (Fig. 6B). To test for an interaction of NRT1.5 and SKOR on K+ efflux, both proteins were coexpressed in BYT45 cells. Yeast growth of the double transformants was indistinguishable to the single transformants with SKOR, indicating no direct influence of NRT1.5 on SKOR activity under these conditions in yeast (Fig. 6B).
Figure 6.
Functional analysis of NRT1.5. A, Potassium uptake capacity was analyzed in yeast BY4741 and the potassium import mutant strain BYT12 (trk1Δ trk2Δ) and transformed with the expression constructs indicated on the right; 20-µL cell suspensions (OD600 from 1.0 to 10−5) were dropped on YNB (-Ura) agar plates containing 7 mm K+, which is growth limiting for BYT12 cells. p426 is the empty vector. KAT1 and NRT1.5 are coding sequences of the respective Arabidopsis genes cloned in p426. B, Potassium export capacity was analyzed in yeast BY4741 and the potassium export mutant strain BYT45 (ena1-5Δ nha1Δ) cells and transformed with the expression constructs indicated on the right; 20-µL cell suspensions (OD600 from 1.0 to 10−5) were dropped on YNB (-Ura -Leu) agar plates supplemented with 1 m KCl, which is growth suppressing for BYT45 cells. p425 and p426 are empty vectors; SKOR and NRT1.5 are coding sequences of the respective Arabidopsis genes cloned in p425 and p426, respectively. C, HygB sensitivity of yeast BY4741 and BYT12 cells transformed with the expression constructs indicated on the right on YNB (-Ura) agar plates containing 100 mm KCl to support growth of BYT12 and an HygB concentration gradient from 0 to 0.5 g L−1. Twelve 3-µL drops of each yeast cell suspension (OD600 = 1.0) were distributed along the HygB gradient on the plate; 1 and 2 indicate two independent yeast transformants. Representative pictures of three independent experiments are shown. D, Root sensitivity test of nrt1.5 mutants and Col-0 on HygB-containing medium. Seedlings pregerminated for 4 d on one-half-strength MS were transferred on one-half-strength MS containing 5 mg L−1 HygB and continued to grow vertically for 7 more d.
In yeast, sensitivity to toxic cationic drugs like Hygromycin B (HygB) is often linked to changes in the membrane potential, which can be provoked by alterations in K+ homeostasis (Barreto et al., 2011). Accordingly, the deletion of both TRK genes in BYT12 cells results in a higher sensitivity to toxic cations, even under nonlimiting K+ concentrations in the medium (Navarrete et al., 2010), because of a hyperpolarization of the plasma membrane. To investigate whether NRT1.5 has an influence on the plasma membrane potential, we expressed NRT1.5 in wild-type BY4741 cells and BYT12 cells and observed yeast growth on plates containing an HygB concentration gradient (Fig. 6C). Empty vector controls (p426) confirmed that BYT12 cells were more sensitive to the antibiotic, because they did not grow properly on HygB concentrations above 250 mg L−1. Transformation of BYT12 but not BY4741 cells with NRT1.5 resulted in strongly increased HygB sensitivity. These results suggest that expression of NRT1.5 causes hyperpolarization of the yeast plasma membrane in the K+ uptake-deficient mutant, but they do not provide support for K+ transport activity of NRT1.5.
Remarkably, root growth of nrt1.5 mutants on one-half-strength Murashige and Skoog medium (MS) was markedly less impaired by HygB than in wild-type seedlings (Fig. 6D), which is in accordance with the result that overexpression of NRT1.5 caused yeast cells to be more susceptible to HygB. Because the membrane potential is the major proton-motive force that drives HygB uptake, we tested the development of nrt1.5 mutants, PHO1:NRT1.5 complementation lines, and Col-0 plants on one-half-strength MS with additional 50 mm KCl supply, because elevated K concentrations decrease the membrane potential (Hirsch et al., 1998). Indeed, the nrt1.5 mutants were more sensitive to 50 mm KCl than wild-type plants (Supplemental Fig. S6). Similar responses to HygB and high K concentrations in the medium were previously observed in plasma membrane proton atpase2 (aha2) mutants (Haruta et al., 2010). Moreover, the two independent complementation lines of nrt1.5-5 developed like the wild type on HygB or 50 mm KCl (Supplemental Fig. S6). Therefore, we speculate that, in nrt1.5 mutants, the membrane potential is similarly reduced as in aha2 mutants.
Both NRT1.5 and SKOR Contribute to Shoot K Homeostasis in an NO3− Supply-Dependent Manner
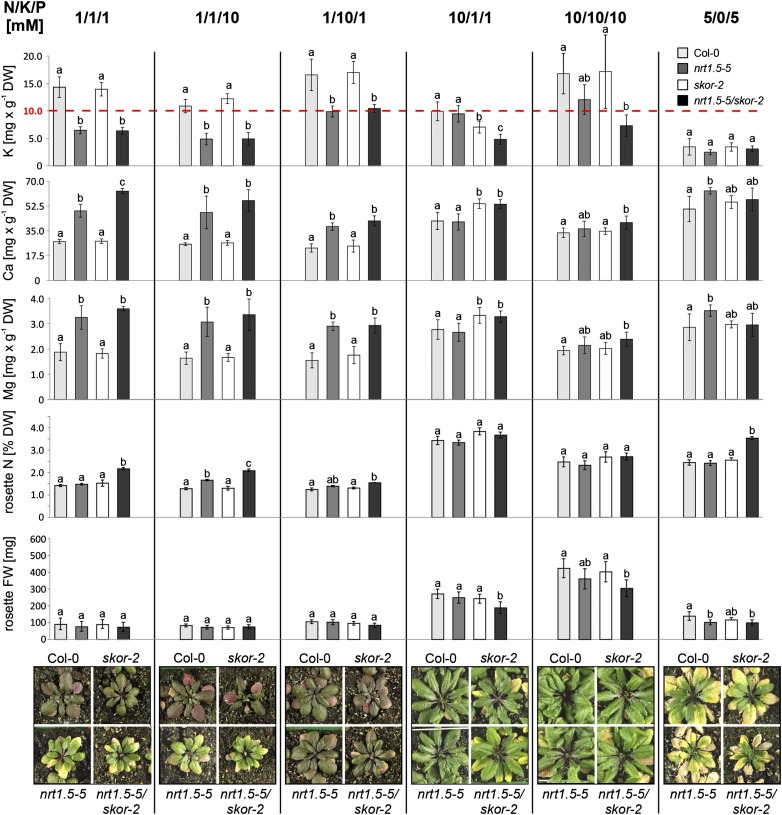
To investigate the interplay between NRT1.5 and SKOR on root-to-shoot translocation of K+ in planta, we isolated the two T-DNA insertion mutants skor-2 and skor-3 in the Col-0 background (Supplemental Fig. S7A). Both mutants exhibit the same phenotype (Supplemental Fig. S7C). We crossed skor-2, which carries the T-DNA insertion in the essential cyclic nucleotide binding domain (Dreyer et al., 2004), with nrt1.5-5 to generate a double mutant. Absence of gene-specific full-length transcripts in single and double mutants was verified by reverse transcription (RT)-PCR (Supplemental Fig. S7B). Wild-type Col-0, nrt1.5-5, skor-2, and nrt1.5-5/skor-2 plants were cultivated on unfertilized soil supplemented with modified one-half-strength MS solutions containing the macronutrients N (as NO3−):K:P in the concentrations 1:1:1, 1:1:10, 1:10:1, 10:1:1, 5:0:5, and 10:10:10 mm, respectively. When all plants were flowering, the inflorescence stems were removed, rosette phenotypes were documented, and the elemental composition was determined by ICP-OES (Fig. 7).
Figure 7.
Correlation of the rosette phenotype with the K, Ca, and Mg elemental composition in response to varying N, K, and P supply in Col-0, nrt1.5-5, skor-2, and nrt1.5-5/skor-2 plants. Each column shows the applied fertilization regime (N to K to P [mm]) along the top. The bar diagrams show the rosette K, Ca, Mg, and total N concentrations and fresh weight (FW) gain, respectively. On the bottom, the respective phenotypes of the plants are shown. The color codes of the bars are indicated in the top right corner. The dotted red line indicates a K concentration of 1% in the dry matter. The data were statistically analyzed by one-way ANOVA and subsequent multiple comparisons (Tukey’s honestly significant difference mean-separation test). Means (n ≥ 4) marked with different letters differ significantly at P < 0.05. Vertical bars denote sds. The experiment was performed three times independently with similar phenotypic growth responses. The elemental analysis by ICP-OES was performed for two of three independent experiments with similar results. DW, Dry weight.
In all four plant lines, early leaf senescence accompanied by yellow leaf tips and pale green inner rosettes occurred whenever the K concentration dropped below 1% dry weight (Fig. 7, red dotted line). At K concentrations of ≥1%, all plants regained the ability to accumulate anthocyanins and developed a red-brown leaf pigmentation that presumably indicated N deficiency (Fig. 7, treatments 1:1:1, 1:1:10, and 1:10:1 mm N:K:P). The concentrations of Ca and Mg in the rosettes were inversely correlated to that of K in all plant lines.
Interestingly, at 1 mm NO3− fertilization, the K concentrations in nrt1.5-5 and nrt1.5-5/skor-2 plants were reduced to approximately 50% of wild-type and skor-2 levels, respectively, and even supplying a 10-fold excess of K+ over NO3− could not revert them back to wild-type level (Fig. 7, treatment 1:10:1 mm N:K:P). Obviously, NRT1.5 made an important contribution to the shoot K status when NO3− supply is limited, whereas the skor-2 mutant behaved like the wild type.
Total N concentration in all lines grown at 1 mm NO3− supply was less than 2.5% (Fig. 7), which verified the low NO3− supply to the plants. Interestingly, when NO3− supply is limited, total N in nrt1.5-5/skor-2 mutant plants was higher than those of the wild type and single mutants. Fresh weight analysis further reflected the importance of NO3− supply to plant growth. Under 1 mm NO3− supply, the fresh weight gain of all lines was below 110 mg. In contrast, when 10 mm NO3− was supplied, fresh weight gain increased 2- to 3-fold.
Under a 10-fold excess of NO3− over K+, the nrt1.5-5 mutant contained similar K concentrations as wild-type plants (Fig. 7, treatment 10:1:1 mm N:K:P), which indicated that NRT1.5 was not primarily involved in the establishment of the shoot K status under these growth conditions. As expected, skor-2 plants had a significantly lower K concentration compared with Col-0 and nrt1.5-5. Lack of both NRT1.5 and SKOR decreased K levels even further. These results were confirmed by treating the skor-2 and nrt1.5-5/skor-2 plants with a 20-fold excess of NO3− over K+, which resulted in a similar reduction in shoot K concentration (Supplemental Fig. S8A). Under a high equimolar supply of NO3− and K+ (i.e. 10:10:10 and 10:10:1 mm N:K:P), both single mutants reached K levels close to Col-0 (Fig. 7; Supplemental Fig. S8B). However, even then the K concentrations in nrt1.5-5/skor-2 double mutant were significantly reduced. This highlighted the importance of both proteins for shoot K+ homeostasis under defined nutritional supply: SKOR under high NO3− and low K+ availability and NRT1.5 under low NO3− availability and independent of the K+ supply. Enhancement of K deficiency in the double mutant under a high NO3− to K+ ratio and a high equimolar supply further suggests an interdependency of NRT1.5 and SKOR in K+ root-to-shoot translocation under these conditions.
To investigate whether the regulation of SKOR and NRT1.5 in roots under different N to K regimes is consistent with this model, we analyzed their expression in roots of wild-type plants grown on unfertilized soil supplemented with 1:1:1, 10:1:1, 1:10:1, and 10:10:1 mm N:K:P. Expression of NRT1.5 was only very weakly regulated under the different NO3− to K+ ratios (Supplemental Fig. S9). Relative to equimolar supply (1:1:1 or 10:10:1 mm N:K:P), NRT1.5 was <1.5-fold up-regulated by high NO3− to K+ ratio (10:1:1 mm N:K:P) and < 1.5-fold down-regulated by low NO3− to K+ ratio (1:10:1 mm N:K:P). In contrast to NRT1.5, SKOR was, independent of the NO3− to K+ ratio, strongly up-regulated by high NO3− supply (10:1:1 and 10:10:1 mm N:K:P). These expression patterns of the two genes could partially explain the phenotypes of the mutant plants, but it is possible that also altered expression of other genes that was observed in nrt1.5-5 roots (Fig. 3) contributed to the phenotypes. Under low NO3− supply (1 mm), expression of NRT1.5 is nearly constant, whereas expression of SKOR is 5-fold lower than under high NO3− supply. Consistently, when grown with 1 mm NO3−, nrt1.5-5 but not skor-2 mutant plants had reduced K levels in the shoots (Fig. 7). In contrast, under high NO3− supply (10 mm), SKOR is strongly expressed in roots. It is conceivable that, under these conditions, SKOR can partially complement the lack of NRT1.5 in the nrt1.5 mutants and facilitate root-to-shoot translocation of K+. This view is concordant with the finding that, under high NO3− supply, the K concentrations in nrt1.5-5 and wild-type plants do not significantly differ (Fig. 7).
DISCUSSION
NRT1.5 Is Required to Maintain Root-to-Shoot Translocation of K+ under Low NO3− Supply
Lin et al. (2008) reported that the NPF member NRT1.5 functions in X. laevis oocytes as a low-affinity, pH-dependent bidirectional NO3− transporter and participates in plants in root xylem loading with NO3−, although in nrt1.5 mutants root-to-shoot translocation of NO3− was not completely blocked. In addition, Lin et al. (2008) observed a reduced K+ transport to the shoot when NO3− and not NH4+ was supplied to the mutants and therefore, suggested a homeostatic balance between the anion NO3− and the cation K+ in the xylem stream. Interestingly, we found that K deficiency in the shoot of nrt1.5-5 mutant plants was most severe under low NO3− supply (Figs. 4 and 7), and supplementation of soil with 10 mm NO3− (with or without additional supply of K+; Fig. 7; Supplemental Fig. S8) diminished or even completely abolished the K deficiency of nrt1.5-5 shoots. This indicated a much more subtle regulation of shoot K+ homeostasis, involving NRT1.5 and K+ transporters and channels.
Cation-anion fluxes in the xylem are strictly balanced, and the most important counteranion for K+ is NO3−. However, under conditions of low NO3− supply and without causing a concomitant shortage in K+, NO3− can be partially substituted by other anions (inorganic or organic), which then act as counteranions for the transport of K+ from roots to the shoot (Engels and Marschner, 1992b; Marschner et al., 1997). In this context, it was interesting to observe that the expression of SLAH1 was strongly up-regulated and that the expression of SLAH3 was weakly but still significantly up-regulated in nrt1.5-5 roots (Fig. 3). These slow anion channels belong to the SLAC/SLAH protein family, which consists in Arabidopsis of five members, SLAC1 and SLAH1 to SLAH4 (Barbier-Brygoo et al., 2011). Their up-regulation in nrt1.5-5 roots might indicate that the proteins can partially substitute the lack of NRT1.5 by facilitating the efflux of anions from the cells of the vasculature toward the xylem stream. This could possibly result in membrane depolarization and thereby, an activation of the xylem-loading K+ channel SKOR, which was indeed also transcriptionally up-regulated in nrt1.5-5 (Fig. 3). Although this model could explain the weaker phenotype of the mutant under high NO3− supply (Fig. 7; Supplemental Fig. S8), it fails under limited NO3− supply, where increased SLAH1 expression in nrt1.5-5 roots did not prevent K deficiency in the shoot. Our results suggest that NRT1.5 is important for a continuous root-to-shoot translocation of K+ under low NO3− supply, whereas its absence is less critical for the plant when ample NO3− is available.
The nrt1.5-5 mutant has, even more pronounced under low NO3− supply, a slightly higher content of nitrogenous compounds (Figs. 1, H and I and 2, E and F). Interestingly, this coincides with an accumulation of nearly all macro- and microelements in the mutant shoot, whereas the elemental composition in the roots was only marginally altered (Table I). The increased shoot Ca and Mg concentrations could be explained by charge compensation for the reduced amount of K (Leigh and Jones, 1984). The higher N content and the accumulation of other elements, like P and S, might result from impaired remobilization capability of the nrt1.5-5 shoot, possibly because of the K deficiency. Cycling and recycling of phloem-mobile mineral nutrients, like phosphate, sulfate, and NO3−, is also charge balanced by cations mostly in the form of K+, and the deficiency can therefore directly affect phloem loading and transport (Marschner et al., 1996, 1997).
An increased expression of JA-responsive and stress-related genes was previously described as a response of Arabidopsis plants suffering from K deficiency (Armengaud et al., 2004). Thus, the altered transcription pattern of selected JA and stress pathway genes in nrt1.5 shoots likely is a consequence of the reduced amount of K+ translocated to the shoot. The transcription changes of ion homeostasis-associated genes in roots did not reflect K deficiency. Decreased expression of CIPK9, a protein kinase gene with increased expression under K deprivation (Pandey et al., 2007), and HAK5, a transporter involved in high-affinity K+ uptake at external concentration below approximately 100 μm (Rubio et al., 2008) that is up-regulated in wild-type roots during K+ starvation (Armengaud et al., 2004), rather indicated that the K deficiency status of the shoot was not transmitted to the outer root cell layers, which are responsible for the uptake of the cation from the soil solution (Drew et al., 1990). This assumption is corroborated by earlier results from Engels and Marschner (1992a), who suggested that xylem loading of K+ is regulated separately from K+ uptake into the root cells and that the adjustment of root-to-shoot translocation of K+ to the demand of the shoot is coupled with an altered xylem loading capacity in the root. The increased expression of SLAH1, SLAH3, SKOR, and NRT1.8 in nrt1.5-5 roots would fit such a regulatory adjustment of xylem loading capacities (Fig. 3). SLAH1, SKOR, and NRT1.8 are primarily expressed in the root vascular cylinder, and SKOR and NRT1.8 were described to have xylem loading or unloading properties, respectively (Gaymard et al., 1998; Negi et al., 2008; Li et al., 2010). The grafting experiment further suggested that the presence of NRT1.5 in the root vasculature is a prerequisite for a proper root-to-shoot translocation of K+ under NO3− limitation and that the shoot itself has no or only a minor influence on the K concentrations in the shoot or the entire root system under those conditions (Fig. 4, A and B). Accordingly, NRT1.5 may be a molecular regulator of the root capacity for xylem loading with K+ and therefore, responsible for the maintenance of the root-to-shoot translocation of K+ under limited NO3− supply.
NRT1.5: Transporter or Trigger for Root-to-Shoot Translocation of K+?
A plausible explanation for the physiological consequences observed in nrt1.5-5 plants would be a K+ transport function of NRT1.5. However, the complementation studies using yeast trk1 trk2 mutants did not provide evidence for a direct K+ import or export function of the protein. Functional characterization of plant K+ channels with this mutant was sometimes successful (e.g. Anderson et al., 1992; Sentenac et al., 1992; Obata et al., 2007) but failed in other cases, possibly because of an incompatibility of the heterologous expression system (Dreyer et al., 1999) or the dependence of transport activity on a regulatory network involving protein-protein interactions and posttranslational modifications as described (e.g. for SLAC1 and AKT1; Honsbein et al., 2009; Maierhofer et al., 2014). The lack of such modifying activities or appropriate regulators in yeast could explain the negative results obtained with NRT1.5. Alternatively, NRT1.5 might only modulate or influence the activity of other K+-transporting membrane proteins as a result of its own transport function or subsequent signaling events or through direct protein-protein interaction.
Because NO3− also fulfils an important function as a signaling molecule (Krouk et al., 2010), it is conceivable that the transport of only a few NO3− molecules through NRT1.5 modifies the K+ flux through the transporter itself or associated K+ channel(s). In K+ uptake-deficient yeast cells with hyperpolarized plasma membranes, NRT1.5 expression resulted in increased membrane hyperpolarization illustrated by the increased HygB sensitivity of the cells (Fig. 6). If NRT1.5 expression in plant roots is also involved in regulating the plasma membrane potential, plasma membrane polarization changes of pericycle and xylem parenchyma cells in response to fluctuating NO3− supply might explain the NRT1.5-triggered alterations in root-to-shoot K+ transport. This hypothesis is indirectly corroborated by the enhanced HygB tolerance and susceptibility of nrt1.5-5 mutant plants to high K concentrations. However, it is currently unclear how plasma membrane potential differences in the vascular tissue of nrt1.5 mutants might be transferred to the outer root cell layers, which are responsible for the uptake of substances (e.g. HygB).
The activity of ion channels can be influenced by membrane polarization. In Arabidopsis, six and two members of the Shaker K+ channel family are activated by hyperpolarization and depolarization, respectively (for review, see Lebaudy et al., 2007). It is thus tempting to speculate that NRT1.5 modulates translocation of K+ by indirectly regulating the activity of voltage-dependent potassium channel proteins.
Job Sharing between NRT1.5 and SKOR in NO3−-Dependent K+ Translocation
SKOR is presently known as the main xylem loader for root-to-shoot translocation of K+ (summarized, for example, in Sharma et al., 2013), a conclusion based on the finding that its disruption strongly reduced the K+ content in the shoot, whereas the K+ content in the roots remained unaffected (Gaymard et al., 1998). The nrt1.5-5 mutant displayed a similar phenotype under NO3−-limiting conditions, although the expression of SKOR was increased in the roots. Thus, an NO3−-dependent job sharing between SKOR and NRT1.5 in the root-to-shoot translocation of K+ is conceivable.
The interplay between NRT1.5 and SKOR was further elucidated in a fertilization experiment utilizing single and double T-DNA insertion lines (Fig. 7; Supplemental Fig. S8). Independent of the chlorosis phenotype, under low NO3− (1 mm) conditions, the K concentration in nrt1.5-5 mutants was always lower than in the wild type and skor-2. Even at a 10-fold excess of K+ over NO3−, K levels in nrt1.5-5 rosettes remained lower than in Col-0 and skor-2, indicating that, irrespective of the K+ supply, primarily NRT1.5 but not SKOR is involved in root-to-shoot translocation of K+ in a low-NO3− environment. Interestingly, the skor-1 mutant phenotype in the Wassilewskija background differs from skor-2 in Col-0 (Supplemental Fig. S10). Therefore, the results by Gaymard et al. (1998)—reduced K+ content in the shoot of skor-1 plants (grown on substrate with unknown NO3− to K+ ratio)—preclude a direct comparison of the two skor mutants.
SKOR activity is influenced by a variety of triggers, including membrane depolarization (Gaymard et al., 1998), pH (Lacombe et al., 2000), intracellular and external K+ status (Johansson et al., 2006; Liu et al., 2006), and reactive oxygen species in the form of hydrogen peroxide (Garcia-Mata et al., 2010). Here, we show that lack of SKOR results in reduced shoot K concentrations when the mutant grows on soil with a high NO3−:K+ ratio (10:1 or 20:1). It thus seems that SKOR is most important for the cation-anion balance in the xylem and consequently, the root-to-shoot translocation of K+ when the transport of NO3− toward the xylem is not restricted, whereas the supply with the counterion K+ is limited.
Finally, the importance of both proteins for root-to-shoot translocation of K+ is illustrated by the observation that, under high equimolar NO3− and K+ supplies (10:10:10 and 10:10:1 mm N:K:P), both single mutants reached K shoot concentrations close to wild-type plants, whereas the nrt1.5-5/skor-2 double mutant failed to do so. Accordingly, NRT1.5 and SKOR cooperate on K+ root-to-shoot translocation under certain nutritional conditions, and this cooperation is important for K homeostasis in the shoot. However, root-to-shoot translocation of K+ was not completely blocked in the double mutant, indicating the activity of additional K+ transport systems in the vasculature.
CONCLUSION
The transporter NRT1.5 and the channel protein SKOR are both involved in root-to-shoot translocation of K+ and regulators of shoot K homeostasis. SKOR activity is responsible for K+ translocation when plants are supplied with ample NO3− but low K+, whereas the presence of NRT1.5 in the root vasculature is needed for K+ transport to the shoot when little NO3− is available to the plant, irrespective of the K+ supply. The two proteins act synergistically, which is indicated by reduced shoot K levels in the double mutant but not the single mutants. Although the molecular mechanism determining the interplay between SKOR and NRT1.5 activities remains to be uncovered, this study defines the physiological conditions under which these transporters cooperate. Because in yeast and plants, NRT1.5 seems to induce plasma membrane hyperpolarization, it is conceivable that the NO3− concentration in pericycle and xylem parenchyma cells regulates NRT1.5 activity in K+ translocation through membrane polarization changes. The ability of a plant to adapt to varying NO3− and K+ concentrations in the soil is important for its survival. Our results suggest that NRT1.5 and SKOR synergistically contribute to this ability.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center. The nrt1.5-2 (SALK_005099), nrt1.5-4 (SALK_063393), nrt1.5-5 (GABI_347B03), skor-2 (GABI_391G12), and skor-3 (SALK_097435) lines are in the Col-0 background, and the skor-1 T-DNA insertion line (N3816) is in the Wassilewskija background. Homozygous mutant plants were identified by PCR (primers are listed in Supplemental Table S3). The T-DNA insertion site in skor-2 was located in exon 8 at codon number 462 by sequencing the PCR product generated with the GABI T-DNA left border primer and the skor-2 reverse primer (Supplemental Table S3). Plants were grown on either a low-fertilized 1:1 (v/v) mixture of the commercial soil types P and 0 (Einheitserde) or on the unfertilized type 0 soil supplemented with nutrient solutions as indicated. Soil composition is given in Supplemental Table S4. Plants were cultivated in a growth chamber under long-day conditions (16-h/8-h light-dark cycle and 21°C/18°C day-night cycle) with a light intensity of 120 µmol m−2 s−1 and a relative humidity of 55% to 70%. For the fertilization experiment in Supplemental Figure S2, plants were supplied from 35 to 55 DAS with 10 mm KNO3 every 3 to 5 d depending on soil humidity. Plant material was harvested 2 to 3 d after fertilization treatments. For the fertilization experiments in Figure 7 and Supplemental Figures S8 and S10, plants were germinated on fertilized soil (type P) after 10- to 14-d seedlings were singled out on unfertilized soil (type 0) in trays and supplied with the respective fertilization solutions (Supplemental Table S2). Plants were irrigated every 5 to 7 d depending on soil humidity. At harvest, inflorescence stems were removed, and the rosette material was frozen in liquid nitrogen. For the root assay, seeds of Col-0, nrt1.5-4, and nrt1.5-5 were surface sterilized, distributed on one-half-strength MS plates containing 1% (w/v) Suc and 0.3% (w/v) Gelrite (pH 5.8), and stratified in darkness for 2 d at 4°C. After 4 d of growth in long-day conditions (16-h/8-h light-dark cycle, 21°C/18°C day-night cycle, and 120 µmol m−2 s−1), seedlings were transferred on one-half-strength MS (1% [w/v] Suc and 0.6% [w/v] Gelrite, pH 5.8) containing HygB or KCl and grown vertically for 7 more d.
Plant Growth in Hydroponic Culture
Plants were grown hydroponically under nonsterile conditions in a growth cabinet. Arabidopsis seeds were germinated under short-day conditions and precultured on rockwool moistened with tap water for 1 week. Tap water was substituted in the second and third weeks by one-half-strength nutrient solution containing 0.5 mm KH2PO4, 0.5 mm MgSO4, 125 μm K2SO4, 125 μm CaCl2, 50 μm Na-Fe-EDTA, 50 μm KCl, 30 μm H3BO3, 5 μm MnSO4, 1 μm ZnSO4, 1 μm CuSO4, and 0.7 μm NaMoO4 (pH 5.8) with KOH. Nitrogen was supplied as 0.5 mm KNO3. In the fourth week, plants were supplied with full-strength hydroponic growth medium (1 mm KH2PO4, 1 mm MgSO4, 250 μm K2SO4, 250 μm CaCl2, 100 μm Na-Fe-EDTA, 50 μm KCl, 30 μm H3BO3, 5 μm MnSO4, 1 μm ZnSO4, 1 μm CuSO4, and 0.7 μm NaMoO4 [pH 5.8] with KOH) containing 1 mm KNO3. After 4 weeks, plants were transferred into long-day conditions until harvest (week 9), and the plants were further cultivated in the full-strength growth medium, but nitrogen was supplied as 0.1 mm NH4NO3. The nutrient solution was renewed once a week during the first 3 weeks, twice in the fourth week, and every second to third day for the following weeks until harvest. Conditions in the growth chambers were as follows: 10-h/14-h light-dark cycle for short-day conditions and 16-h/8-h light-dark cycle for long-day conditions, light intensity 240 μmol m−2 s−1, 22°C/18°C day-night temperature cycle, and 70% relative humidity.
RNA Isolation
Total RNA was extracted from frozen plant material using the hot-phenol extraction method (Verwoerd et al., 1989).
Semiquantitative RT-PCR
Semiquantitative RT-PCR was performed on complementary DNA corresponding to 50 to 200 ng of reverse-transcribed total RNA as described previously (Rausch et al., 2004). Gene-specific primer pairs are listed in Supplemental Table S3. PCR products were visualized by agarose gel electrophoresis.
qPCR
Total RNA was DNase I treated according to the manufacturer’s instructions (Fermentas). Absence of genomic DNA was subsequently tested by qPCR using gene At5g65080 intron-specific primers. First-strand complementary DNA was synthesized from 2 μg of total RNA using either SuperScript III Reverse Transcriptase (Life Technologies) or RevertAid H Minus Reverse Transcriptase (Life Technologies) and quality controlled. Unless otherwise noted, expression analyses were performed with tissue from separately grown plants in at least three independent replicates. qPCR reactions were conducted on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems) using the Power SYBR Green PCR Master Mix (Applied Biosystems) following the thermal profile: 1 time (95°C for 10 min) and 40 times (95°C for 15 s and 60°C for 1 min). SDS 2.2.1 software (Applied Biosystems) was used for data analysis. Expression values for each gene were normalized to the reference gene At4g05320 (UBIQUITIN10 [UBQ10]), and relative expression levels were determined according to the work by Czechowski et al. (2005). Primer sequences are listed in Supplemental Table S5.
Complementation of nrt1.5-5 with PHO1:NRT1.5
A 1.6-kb genomic fragment upstream of the PHO1 initiation codon was PCR amplified on genomic Arabidopsis DNA using primers listed in Supplemental Table S3. The amplification product was cloned upstream of the NRT1.5 coding region in the binary vector pTkan3. The construct was transformed in nrt1.5-5 plants by the floral dip method (Clough and Bent, 1998). For gene expression analysis of PHO1p:NRT1.5 plants, seeds of six independent transgenic lines (wild-type and nrt1.5-5 plants) were germinated on one-half-strength MS plates. After 1 week, eight seedlings per line were transferred into Erlenmeyer flasks containing 10 mL of one-half-strength MS. Plants were cultivated shaking in a growth chamber under long-day conditions (16-h/8-h light-dark cycle and 21°C/18°C day-night cycle) with a light intensity of 120 µmol m−2 s−1. One-half-strength MS was exchanged weekly. At 20 DAS, root material was harvested for RNA extraction with TRIsure Reagent (Bioline). Primers for qPCR are listed in Supplemental Table S5. Growth analysis of three transgenic lines in comparison with Col-0 and nrt1.5-5 was performed on unfertilized soil (Supplemental Table S4) supplemented with the nutrient solution 1:1:10 mm N:K:P (Supplemental Table S2).
Pulse-Amplitude Modulated Fluorometry
Chlorophyll fluorescence was measured with a FluorCam 800MF (Photon Systems Instruments). After a 20-min dark adaptation of the plants, the minimum fluorescence in dark-adapted state was captured. The maximum yield of chlorophyll fluorescence in the dark-adapted state was induced by an 800-ms pulse of saturating white light (2,100 μmol m−2 s−1). The maximum quantum yield of PSII photochemistry (Fv/Fm) was calculated.
Chlorophyll Measurements
Thirty milligrams of frozen and homogenized plant material (whole rosettes including dry leaves) was extracted in 1 mL of 80% (v/v) buffered aqueous acetone containing 2.5 mm sodium phosphate buffer (pH 7.8) for 30 min in the dark at 4°C under gentle shaking. After centrifugation for 10 min at 15,000g and 4°C, the supernatant was collected and stored at 4°C until the second extraction of the pellet with 500 µL of buffered aqueous acetone was performed. Both supernatants were combined, and the chlorophyll content was measured spectrophotometrically and calculated according to Porra et al., 1989.
Anthocyanin Measurements
Anthocyanin levels were determined basically according to Neff and Chory, 1998; 50 mg of frozen and homogenized plant material was incubated overnight at 4°C under gentle shaking in 300 μL of methanol acidified with 1% (w/v) HCl. After the addition of 200 μL of deionized water, anthocyanins were separated from chlorophylls by addition of 500 μL of chloroform. After centrifugation for 2 min at 15,000g, 400 µL of aqueous phase was diluted with 400 µL of acidified methanol, and the anthocyanin concentration was determined by measuring the A530 and A657 with a spectrophotometer. Anthocyanin concentrations were calculated by subtracting A657 from A530.
Nitrate Measurements
Nitrate was determined colorimetrically according to the protocol by Cataldo et al. (1975). Briefly, 30 to 50 mg of frozen and homogenized plant material (whole rosettes including dry leaves or separated young and old leaves) was mixed with 1.0 mL of deionized water and centrifuged for 5 min at 4°C and 12,000g; 40 µL of supernatant was mixed with 160 µL of 1% (w/v) salicylic acid in concentrated sulfuric acid. For background measurement, 40 µL of supernatant were mixed with 160 µL of deionized water. After a 20-min incubation on ice, 1.8 mL of cooled 4 m NaOH was added and mixed carefully. When the samples reached room temperature, the A410 was determined with a microplate reader (BioTek), and the values were compared to a nitrate standard curve (0–8.0 mm KNO3).
Determination of the Total N and C Contents
C and N analysis was performed on a EuroEA3000-Single Elemental Analyzer (www.eurovector.it) according to the manufacturer’s instructions; 1.5 to 2.5 mg of homogenized and oven-dried (2 d at 85°C) plant material (whole rosettes including dry leaves or separated young and old leaves) was used for each measurement. Data were analyzed with the Callidus 5.1 software.
Seed Lipid and Protein Analyses
Determination of seed lipid and protein content was basically performed according to Reiser et al., 2004; 40 mg of homogenized dry seeds were mixed in a mortar with 1.5 mL of isopropanol. The suspension was transferred to a reaction tube and incubated for 20 h at 8°C on a laboratory shaker. Samples were then centrifuged for 10 min at 12,000g, and the supernatants transferred into preweighted reaction tubes. Isopropanol was finally evaporated, and the total lipid content quantified gravimetrically.
For seed protein quantification, 10 mg of dry seeds were homogenized in a mortar with 0.5 mL of extraction buffer (50 mm HEPES, 5 mm MgCl2, pH 7.5, 1% [v/v] Triton X-100, 15% [v/v] glycerol, 2% [w/v] SDS, 1 mm EDTA, and 1% [w/v] phenylmethylsulfonyl fluoride). After 10 min of centrifugation at 12,000g, proteins in the supernatant were precipitated with acetone to remove bicinchoninic acid assay interfering substances: 25 µL of supernatant was diluted with 175 µL of deionized water and incubated with 6 volumes of ice-cold acetone for 30 min at −20°C. After 10 min of centrifugation at 16,000g and 4°C, protein pellets were dissolved in 200 µL of deionized water. The protein concentration was determined by BCA-Protein Assay (Pierce).
Yeast Growth Assays
The protein coding sequences of KAT1, NRT1.5, and SKOR were PCR amplified with primers containing restriction sites. SKOR was cloned in the yeast (Saccharomyces cerevisiae) expression vector p425 (Ura3 marker), and NRT1.5 and KAT1 were cloned in p426 (Leu-2 marker; Mumberg et al., 1995) behind the constitutive Translational Elongation Factor EF-1α promoter. Primer combinations are listed in Supplemental Table S3. The constructs were transformed in the yeast strain BY4741 (Brachmann et al., 1998), BYT12 (trk1Δ trk2Δ; Petrezselyova et al., 2010), or BYT45 (ena1-5Δ nha1Δ; Navarrete et al., 2010) by the lithium acetate method (Ito et al., 1983). For each combination of two plasmids, two independent double transformants were generated to confirm identical growth properties. For potassium uptake experiments with BYT12 and export experiments with BYT45, transformed yeast cells were pregrown overnight in liquid yeast nitrogen base (YNB) medium supplemented with or without 100 mm KCl, respectively. The next day, the cells were washed with deionized water and resuspended to an optical density at 600 nm wavelength (OD600) value of 1, and 10-fold serial dilutions from OD600 of 1 to 10−5 were prepared; 20 µL of each dilution was dropped on YNB agar plates supplemented with or without 1 m KCl for export or import experiments, respectively. The HygB sensitivity test was performed according to Petrezselyova et al., 2010; 3 µL of cell suspension (OD600 = 1) was spotted on HygB gradient plates consisting of YNB medium supplemented with 100 mm KCl to support growth of mutant BYT12.
Elemental Analyses
The plant material was dried for 48 h at 75°C, weighed into polytetrafluorethylene digestion tubes, and digested in HNO3 under pressure using a microwave digester (Ultraclave 4; MLS GmbH). The concentrations of macro- and microelements were determined by ICP-OES using an iCAP 6500 Dual OES Spectrometer (Thermo Fischer Scientific). The certified Standard Reference Material 1515 Apple Leaves (National Institute of Standards and Technology) was used for quality control. Letters highlight significantly lower or higher elemental concentration in nrt1.5-5 tissues when compared with Col-0. Data are means ± sd (n ≥ 9). Asterisks indicate statistically significant differences (Student’s t test) between nrt1.5-5 and Col-0: *, P < 0.05; **, P < 0.01, respectively.
Grafting
Grafts between nrt1.5-5 and Col-0 plants were performed according to the root-shoot grafts protocol by Bainbridge et al. (2006).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Genomic structure and phenotypes of mutant lines nrt1.5-2, nrt1.5-4, and nrt1.5-5.
Supplemental Figure S2. Supplementation with 10 mm KNO3 suppressed the pleiotrophic nrt1.5-5 phenotype on low-fertilized soil.
Supplemental Figure S3. Altered cation distribution in nrt1.5-5 plants.
Supplemental Figure S4. Expression analysis of JA metabolism-related, Ca-related, and other genes in Col-0 and nrt1.5-5 leaves.
Supplemental Figure S5. Ca and Mg concentrations in shoots and roots of grafted Arabidopsis plants.
Supplemental Figure S6. Root growth phenotypes of nrt1.5 mutants in response to HygB and 50 mm KCl.
Supplemental Figure S7. Identification of the T-DNA insertion mutant skor-2 in the Col-0 background and confirmation of the absence of full-length transcripts in skor-2 and the double mutant nrt1.5-5/skor-2.
Supplemental Figure S8. Rosette phenotype and K, Ca, and Mg concentrations in nrt1.5-5, skor-2, nrt1.5-5/skor-2, and Col-0 plants after 20:1:1 and 10:10:1 mm N:K:P supply, respectively.
Supplemental Figure S9. Expression of NRT1.5 and SKOR under the different NO3− to K+ ratios.
Supplemental Figure S10. Phenotypic differences between skor mutants in Arabidopsis accessions Col-0 and Wassilewskija.
Supplemental Table S1. Ion transporter and signaling genes with similar expression levels in roots of nrt1.5-5 and Col-0 plants grown in hydroponic culture.
Supplemental Table S2. Composition of N, K, and P fertilization solutions.
Supplemental Table S3. Primer sequences for T-DNA insertion lines, RT-PCR, cloning, and complementation analysis.
Supplemental Table S4. Results of soil composition analysis performed by a certified analytical laboratory.
Supplemental Table S5. qPCR primer sequences.
Supplemental Note S1. Choice of the PHO1 promoter for complementation.
Supplementary Material
Acknowledgments
We thank Eva Häffner for help with the statistical analysis (ANOVA), Susanne Reiner for technical assistance with the ICP-OES measurements, Manfred Forstreuter and Sabine Artelt for technical assistance with the C:N analyzer, Hana Sychrova for providing the yeast strains BYT12 and BYT45, and Guillaume Pilot for the binary vectors pUTkan3 and pTkan3.
Glossary
- Col-0
Columbia-0
- DAS
days after sowing
- Fv/Fm
maximum photochemical efficiency of PSII in the dark-adapted state
- HygB
Hygromycin B
- ICP-OES
inductively coupled plasma optical emission spectrometry
- JA
jasmonic acid
- MS
Murashige and Skoog medium
- qPCR
quantitative real-time PCR
- RT
reverse transcription
- T-DNA
transfer DNA
- YNB
yeast nitrogen base
Footnotes
This work was supported by the Studienstiftung des Deutschen Volkes (N.D.), the China Scholarship Council (Y.Z.), and the Deutsche Forschungsgemeinschaft (Forschergruppe FOR 948 grant nos. WI1728/14–2 to N.v.W. and KU715/10–2 to R.K. and C.R.).
Articles can be viewed without a subscription.
References
- Anderson JA, Huprikar SS, Kochian LV, Lucas WJ, Gaber RF (1992) Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 89: 3736–3740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud P, Breitling R, Amtmann A (2004) The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol 136: 2556–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge K, Bennett T, Turnbull C, Leyser O (2006) Grafting. Methods Mol Biol 323: 39–44 [DOI] [PubMed] [Google Scholar]
- Barbier-Brygoo H, De Angeli A, Filleur S, Frachisse JM, Gambale F, Thomine S, Wege S (2011) Anion channels/transporters in plants: from molecular bases to regulatory networks. Annu Rev Plant Biol 62: 25–51 [DOI] [PubMed] [Google Scholar]
- Barneix AJ, Breteler H (1985) Effect of cations on uptake, translocation and reduction of nitrate in wheat seedlings. New Phytol 99: 367–379 [Google Scholar]
- Barreto L, Canadell D, Petrezsélyová S, Navarrete C, Maresová L, Peréz-Valle J, Herrera R, Olier I, Giraldo J, Sychrová H, et al. (2011) A genomewide screen for tolerance to cationic drugs reveals genes important for potassium homeostasis in Saccharomyces cerevisiae. Eukaryot Cell 10: 1241–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Zioni A, Vaadia Y, Herman Lips S (1971) Nitrate uptake by roots as regulated by nitrate reduction products of shoot. Physiol Plant 24: 288–290 [Google Scholar]
- Blevins DG, Barnett NM, Frost WB (1978) Role of potassium and malate in nitrate uptake and translocation by wheat seedlings. Plant Physiol 62: 784–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Cataldo DA, Haroon M, Schrader LE, Youngs VL (1975) Rapid colorimetric determination of nitrate in plant-tissue by nitration of salicylic-acid. Commun Soil Sci Plant Anal 6: 71–80 [Google Scholar]
- Chen CZ, Lv XF, Li JY, Yi HY, Gong JM (2012) Arabidopsis NRT1.5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiol 159: 1582–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC, Webb J, Saker LR (1990) Regulation of K+ uptake and transport to the xylem in barley roots: K+ distribution determined by electron-probe x-ray-microanalysis of frozen-hydrated cells. J Exp Bot 41: 815–825 [Google Scholar]
- Dreyer I, Horeau C, Lemaillet G, Zimmermann S, Bush DR, Rodriguez-Navarro A, Schachtman DP, Spalding EP, Sentenac H, Gaber RF (1999) Identification and characterization of plant transporters using heterologous expression systems. J Exp Bot 50: 1073–1087 [Google Scholar]
- Dreyer I, Porée F, Schneider A, Mittelstädt J, Bertl A, Sentenac H, Thibaud JB, Mueller-Roeber B (2004) Assembly of plant Shaker-like K(out) channels requires two distinct sites of the channel alpha-subunit. Biophys J 87: 858–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels C, Marschner H (1992a) Adaptation of potassium translocation into the shoot of maize (Zea mays) to shoot demand: evidence for xylem loading as a regulating step. Physiol Plant 86: 263–268 [Google Scholar]
- Engels C, Marschner H (1992b) Root to shoot translocation of macronutrients in relation to shoot demand in maize (Zea mays L) grown at different root zone temperatures. Zeitschrift für Pflanzenernährung und Bodenkunde 155: 121–128 [Google Scholar]
- Engels C, Marschner H (1993) Influence of the form of nitrogen supply on root uptake and translocation of cations in the xylem exudate of maize (Zea Mays L.). J Exp Bot 44: 1695–1701 [Google Scholar]
- Garcia-Mata C, Wang J, Gajdanowicz P, Gonzalez W, Hills A, Donald N, Riedelsberger J, Amtmann A, Dreyer I, Blatt MR (2010) A minimal cysteine motif required to activate the SKOR K+ channel of Arabidopsis by the reactive oxygen species H2O2. J Biol Chem 285: 29286–29294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, Boucherez J, Michaux-Ferrière N, Thibaud JB, Sentenac H (1998) Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94: 647–655 [DOI] [PubMed] [Google Scholar]
- Geiger D, Maierhofer T, Al-Rasheid KA, Scherzer S, Mumm P, Liese A, Ache P, Wellmann C, Marten I, Grill E, et al. (2011) Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci Signal 4: ra32. [DOI] [PubMed] [Google Scholar]
- Gierth M, Mäser P (2007) Potassium transporters in plants: involvement in K+ acquisition, redistribution and homeostasis. FEBS Lett 581: 2348–2356 [DOI] [PubMed] [Google Scholar]
- Gomez-Porras JL, Riaño-Pachón DM, Benito B, Haro R, Sklodowski K, Rodríguez-Navarro A, Dreyer I (2012) Phylogenetic analysis of k(+) transporters in bryophytes, lycophytes, and flowering plants indicates a specialization of vascular plants. Front Plant Sci 3: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Burch HL, Nelson RB, Barrett-Wilt G, Kline KG, Mohsin SB, Young JC, Otegui MS, Sussman MR (2010) Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J Biol Chem 285: 17918–17929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch RE, Lewis BD, Spalding EP, Sussman MR (1998) A role for the AKT1 potassium channel in plant nutrition. Science 280: 918–921 [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Honsbein A, Sokolovski S, Grefen C, Campanoni P, Pratelli R, Paneque M, Chen Z, Johansson I, Blatt MR (2009) A tripartite SNARE-K+ channel complex mediates in channel-dependent K+ nutrition in Arabidopsis. Plant Cell 21: 2859–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S, Feller U (2002) Nitrogen metabolism and remobilization during senescence. J Exp Bot 53: 927–937 [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson I, Wulfetange K, Porée F, Michard E, Gajdanowicz P, Lacombe B, Sentenac H, Thibaud JB, Mueller-Roeber B, Blatt MR, et al. (2006) External K+ modulates the activity of the Arabidopsis potassium channel SKOR via an unusual mechanism. Plant J 46: 269–281 [DOI] [PubMed] [Google Scholar]
- Krouk G, Crawford NM, Coruzzi GM, Tsay YF (2010) Nitrate signaling: adaptation to fluctuating environments. Curr Opin Plant Biol 13: 266–273 [DOI] [PubMed] [Google Scholar]
- Lacombe B, Pilot G, Gaymard F, Sentenac H, Thibaud JB (2000) pH control of the plant outwardly-rectifying potassium channel SKOR. FEBS Lett 466: 351–354 [DOI] [PubMed] [Google Scholar]
- Lebaudy A, Véry AA, Sentenac H (2007) K+ channel activity in plants: genes, regulations and functions. FEBS Lett 581: 2357–2366 [DOI] [PubMed] [Google Scholar]
- Leigh RA, Jones RGW (1984) A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol 97: 1–13 [Google Scholar]
- Léran S, Muños S, Brachet C, Tillard P, Gojon A, Lacombe B (2013) Arabidopsis NRT1.1 is a bidirectional transporter involved in root-to-shoot nitrate translocation. Mol Plant 6: 1984–1987 [DOI] [PubMed] [Google Scholar]
- Léran S, Varala K, Boyer JC, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, et al. (2014) A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Li JY, Fu YL, Pike SM, Bao J, Tian W, Zhang Y, Chen CZ, Zhang Y, Li HM, Huang J, et al. (2010) The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22: 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SH, Kuo HF, Canivenc G, Lin CS, Lepetit M, Hsu PK, Tillard P, Lin HL, Wang YY, Tsai CB, et al. (2008) Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20: 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Li L, Luan S (2006) Intracellular K+ sensing of SKOR, a Shaker-type K+ channel from Arabidopsis. Plant J 46: 260–268 [DOI] [PubMed] [Google Scholar]
- Liu LL, Ren HM, Chen LQ, Wang Y, Wu WH (2013) A protein kinase, Calcineurin B-Like Protein-Interacting Protein Kinase9, interacts with calcium sensor Calcineurin B-Like Protein3 and regulates potassium homeostasis under low-potassium stress in Arabidopsis. Plant Physiol 161: 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJM. (2009) Physiological functions of mineral macronutrients. Curr Opin Plant Biol 12: 250–258 [DOI] [PubMed] [Google Scholar]
- Maierhofer T, Diekmann M, Offenborn JN, Lind C, Bauer H, Hashimoto K, S Al-Rasheid KA, Luan S, Kudla J, Geiger D, et al. (2014) Site- and kinase-specific phosphorylation-mediated activation of SLAC1, a guard cell anion channel stimulated by abscisic acid. Sci Signal 7: ra86. [DOI] [PubMed] [Google Scholar]
- Marschner H, Kirkby EA, Cakmak I (1996) Effect of mineral nutritional status on shoot-root partitioning of photoassimilates and cycling of mineral nutrients. J Exp Bot 47: 1255–1263 [DOI] [PubMed] [Google Scholar]
- Marschner H, Kirkby EA, Engels C (1997) Importance of cycling and recycling of mineral nutrients within plants for growth and development. Bot Acta 110: 265–273 [Google Scholar]
- Marschner P. (2012) Marschner’s Mineral Nutrition of Higher Plants. Academic Press, London
- Mumberg D, Müller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Navarrete C, Petrezsélyová S, Barreto L, Martínez JL, Zahrádka J, Ariño J, Sychrová H, Ramos J (2010) Lack of main K+ uptake systems in Saccharomyces cerevisiae cells affects yeast performance in both potassium-sufficient and potassium-limiting conditions. FEMS Yeast Res 10: 508–517 [DOI] [PubMed] [Google Scholar]
- Neff MM, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi J, Matsuda O, Nagasawa T, Oba Y, Takahashi H, Kawai-Yamada M, Uchimiya H, Hashimoto M, Iba K (2008) CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452: 483–486 [DOI] [PubMed] [Google Scholar]
- Obata T, Kitamoto HK, Nakamura A, Fukuda A, Tanaka Y (2007) Rice shaker potassium channel OsKAT1 confers tolerance to salinity stress on yeast and rice cells. Plant Physiol 144: 1978–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Cheong YH, Kim BG, Grant JJ, Li L, Luan S (2007) CIPK9: a calcium sensor-interacting protein kinase required for low-potassium tolerance in Arabidopsis. Cell Res 17: 411–421 [DOI] [PubMed] [Google Scholar]
- Park J, Kim YY, Martinoia E, Lee Y (2008) Long-distance transporters of inorganic nutrients in plants. J Plant Biol 51: 240–247 [Google Scholar]
- Petrezselyova S, Zahradka J, Sychrova H (2010) Saccharomyces cerevisiae BY4741 and W303-1A laboratory strains differ in salt tolerance. Fungal Biol 114: 144–150 [DOI] [PubMed] [Google Scholar]
- Pilot G, Gaymard F, Mouline K, Chérel I, Sentenac H (2003) Regulated expression of Arabidopsis shaker K+ channel genes involved in K+ uptake and distribution in the plant. Plant Mol Biol 51: 773–787 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J (1991) Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta-Bioenergetics 975: 384–394 [Google Scholar]
- Rausch C, Zimmermann P, Amrhein N, Bucher M (2004) Expression analysis suggests novel roles for the plastidic phosphate transporter Pht2;1 in auto- and heterotrophic tissues in potato and Arabidopsis. Plant J 39: 13–28 [DOI] [PubMed] [Google Scholar]
- Reiser J, Linka N, Lemke L, Jeblick W, Neuhaus HE (2004) Molecular physiological analysis of the two plastidic ATP/ADP transporters from Arabidopsis. Plant Physiol 136: 3524–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio F, Nieves-Cordones M, Alemán F, Martínez V (2008) Relative contribution of AtHAK5 and AtAKT1 to K+ uptake in the high-affinity range of concentrations. Physiol Plant 134: 598–608 [DOI] [PubMed] [Google Scholar]
- Sentenac H, Bonneaud N, Minet M, Lacroute F, Salmon JM, Gaymard F, Grignon C (1992) Cloning and expression in yeast of a plant potassium ion transport system. Science 256: 663–665 [DOI] [PubMed] [Google Scholar]
- Sharma T, Dreyer I, Riedelsberger J (2013) The role of K(+) channels in uptake and redistribution of potassium in the model plant Arabidopsis thaliana. Front Plant Sci 4: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Noguchi K, Yasumori M, Kobayashi M, Gajdos Z, Miwa K, Hayashi H, Yoneyama T, Fujiwara T (2002) Arabidopsis boron transporter for xylem loading. Nature 420: 337–340 [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Hsu PK, Tsay YF (2012) Uptake, allocation and signaling of nitrate. Trends Plant Sci 17: 458–467 [DOI] [PubMed] [Google Scholar]
- Wege S, Poirier Y (2014) Expression of the mammalian Xenotropic Polytropic Virus Receptor 1 (XPR1) in tobacco leaves leads to phosphate export. FEBS Lett 588: 482–489 [DOI] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360 [DOI] [PubMed] [Google Scholar]
- Zahrádka J, Sychrová H (2012) Plasma-membrane hyperpolarization diminishes the cation efflux via Nha1 antiporter and Ena ATPase under potassium-limiting conditions. FEMS Yeast Res 12: 439–446 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.