Phloem-specific expression of a sucrose transporter enhances sucrose loading and improves rice yield.
Abstract
Yield in cereals is a function of grain number and size. Sucrose (Suc), the main carbohydrate product of photosynthesis in higher plants, is transported long distances from source leaves to sink organs such as seeds and roots. Here, we report that transgenic rice plants (Oryza sativa) expressing the Arabidopsis (Arabidopsis thaliana) phloem-specific Suc transporter (AtSUC2), which loads Suc into the phloem under control of the phloem protein2 promoter (pPP2), showed an increase in grain yield of up to 16% relative to wild-type plants in field trials. Compared with wild-type plants, pPP2::AtSUC2 plants had larger spikelet hulls and larger and heavier grains. Grain filling was accelerated in the transgenic plants, and more photoassimilate was transported from the leaves to the grain. In addition, microarray analyses revealed that carbohydrate, amino acid, and lipid metabolism was enhanced in the leaves and grain of pPP2::AtSUC2 plants. Thus, enhancing Suc loading represents a promising strategy to improve rice yield to feed the global population.
Rice (Oryza sativa) is a staple food for nearly one-half of the global population. Given the rapid growth of the world’s population, there is an urgent need to increase rice yield. Rice yield is a complex trait that is directly associated with grain size, panicle number, and the number of grains per panicle (Xing and Zhang, 2010). Increasing grain size is a prime breeding target, and several genes known to control rice grain size, such as GRAIN SIZE3 (GS3), GS5, GW2 QTL for rice grain width and weight (GW2), GW8, and rice seed width5, have been identified (Fan et al., 2006; Song et al., 2007; Shomura et al., 2008; Li et al., 2011a; Wang et al., 2012). However, our knowledge of the mechanisms that control rice yield is limited. Thus, further improving rice yield remains a challenge for breeders (Sakamoto and Matsuoka, 2008). Identifying and characterizing unique genes or targets that regulate yield traits would improve our understanding of the molecular mechanisms that regulate yield traits and facilitate the breeding of new rice varieties with higher yields.
The carbohydrates in rice grains originate from photosynthesis that is carried out predominantly in leaves (sources). Therefore, grain filling and rice yield depend on the efficient transport of carbohydrates from the leaves to seeds (sinks). In most plants, Suc is the main carbohydrate transported long distance in the veins to support the growth and development of roots, flowers, fruits, and seeds (Baker et al., 2012; Braun, 2012). Recently, the entire pathway for the export of Suc from leaves has been elucidated (Baker et al., 2012; Braun, 2012). Suc is synthesized in leaf mesophyll cells and diffuses from cell to cell through plasmodesmata until it reaches the phloem parenchyma cells (Slewinski and Braun, 2010). The SWEET transporters mediate Suc efflux from the phloem parenchyma cells into the apoplast, where Suc is subsequently loaded into the phloem sieve element-companion cell (SE/CC) complexes by Suc transporters (SUTs; Braun and Slewinski, 2009; Ayre, 2011; Chen et al., 2012). The resultant accumulation of Suc in sieve elements produces a hydrostatic pressure gradient that results in the bulk flow of Suc through a conduit of contiguous sieve elements, leading to its arrival and unloading in sink tissues (Lalonde et al., 2004; Baker et al., 2012).
Genetic evidence has demonstrated that apoplastic Suc phloem loading is critical for growth, development, and reproduction in Arabidopsis (Arabidopsis thaliana). AtSWEET11 and AtSWEET12 are localized to the plasma membrane of the phloem and are expressed in a subset of phloem parenchyma cells in minor veins. These transporters mediate Suc efflux from phloem parenchyma cells into the apoplast prior to Suc uptake by SE/CC (Chen et al., 2012). The atsweet11 or atsweet12 single mutants exhibit no aberrant phenotypes, possibly due to genetic redundancy. However, atsweet11;12 double mutants are mildly chlorotic and display slower growth and higher levels of starch and sugar accumulation in the leaves than do wild-type plants (Chen et al., 2012). Arabidopsis phloem-specific sucrose transporter (AtSUC2) is a phloem-specific SUT that is expressed specifically in companion cells (Stadler and Sauer, 1996). AtSUC2 plays an essential role in phloem Suc loading and is necessary for efficient Suc transport from source to sink tissues in Arabidopsis (Stadler and Sauer, 1996; Gottwald et al., 2000; Srivastava et al., 2008). The atsuc2 mutants show stunted growth, retarded development, and sterility. Furthermore, these mutants accumulate excess starch in the leaves and fail to transport sugar efficiently to the roots and inflorescences (Gottwald et al., 2000).
The proper control of carbohydrate partitioning is fundamental to crop yield (Braun, 2012). It has been reported that increasing sink grain strength by improving assimilate uptake capacity could be a promising approach toward obtaining higher yield. For example, seed-specific overexpression of a potato (Solanum tuberosum) SUT increased Suc uptake and growth rates of developing pea (Pisum sativum) cotyledons (Rosche et al., 2002). In addition, the Suc uptake capacity of grains and storage protein biosynthesis was increased in transgenic wheat (Triticum aestivum) plants expressing the barley (Hordeum vulgare) SUT HvSUT1 under the control of an endosperm-specific promoter (Weichert et al., 2010). Moreover, it was recently found that these transgenic wheat plants had a higher thousand grain weight and grain width and length, as well as a 28% increase in grain yield (Saalbach et al., 2014).
Since the carbohydrates in rice grains originate from photosynthesis in source leaves, and carbohydrate partitioning from source leaves to heterotrophic sinks (e.g. seeds) is mediated by Suc transport in plants (Lalonde et al., 2004; Ayre, 2011), enhancing the capacity for Suc transport from leaves to seeds theoretically could increase crop yield. However, until now, enhancing Suc transport from leaves to seeds has not been shown to improve yield (Ainsworth and Bush, 2011).
Here, we tested the hypothesis that enhancing Suc transport from leaves to seeds would increase rice yield. We expressed Arabidopsis SUC2 under control of the phloem protein2 promoter (pPP2) in rice and found that enhancing Suc loading did indeed increase rice yield. The pPP2::AtSUC2 plants produced larger grain than the wild type and showed grain yield increases of up to 16% in field trials. Our results suggest that manipulating phloem Suc transport is a useful strategy for increasing grain yield in rice and other cereal crops.
RESULTS
Generation of Transgenic Rice Plants Expressing AtSUC2
We prepared a sense gene construct from the full-length complementary DNA (cDNA) for the Arabidopsis phloem-specific Suc transporter AtSUC2 (Gottwald et al., 2000; Srivastava et al., 2008) in the pPP2 binary vector consisting of pCAMBIA1301 containing the strong companion cell-specific PP2-A1 promoter (Dinant et al., 2003; Supplemental Fig. S1). We expressed the resulting pPP2::AtSUC2 construct in rice by Agrobacterium tumefaciens-mediated gene transfer and selected four independent homozygous T3 lines (L6, L9, L26, and L51) displaying high levels of AtSUC2 mRNA (Supplemental Fig. S1) for further analysis. Immunoblot analysis using antiserum against AtSUC2 revealed the presence of AtSUC2 in the transgenic lines, whereas no such protein was detected in wild-type plants (Supplemental Fig. S1). Transmission electron microscopy showed that AtSUC2 was overexpressed in the companion cell (Supplemental Fig. S2) and was absent from the sieve element cells, from the phloem parenchyma cells, and from the mesophyll cell of the leaves (Supplemental Fig. S2). Reverse transcription (RT)-PCR analysis demonstrated that expression of AtSUC2 was highest in flag leaves, followed by stems, roots, and seeds (Supplemental Fig. S1), suggesting that expression of AtSUC2 is not restricted to the flag leaf companion cells, and the pPP2 promoters is also active in other organs, such as stems, roots, and seeds.
Heterologous Expression of AtSUC2 Enhances Phloem Loading and Transport
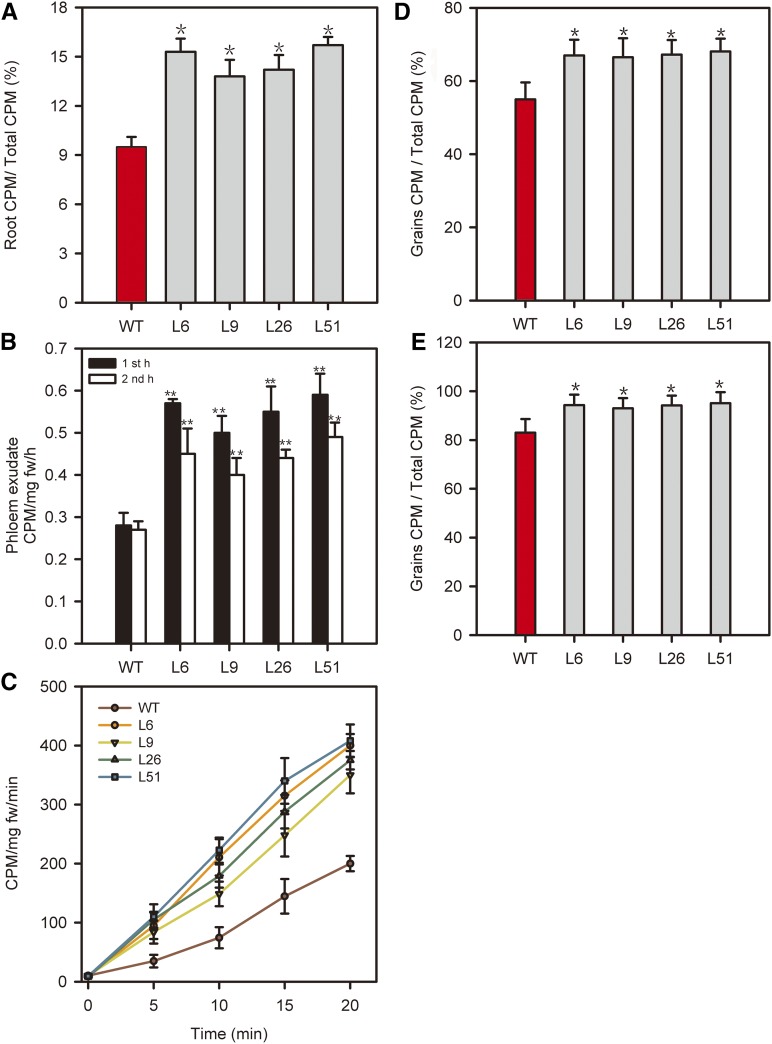
Since AtSUC2 functions in Suc loading from the apoplasm into SE/CC (Stadler and Sauer, 1996; Gottwald et al., 2000), we examined whether AtSUC2 expression would increase phloem transport from source leaves to sink organs (Fig. 1). The leaves of seedlings were photosynthetically labeled with [14C]CO2 for 20 min in the middle of the light period, and roots were harvested to determine transport of label by scintillation counting. The pPP2::AtSUC2 plants transported a higher proportion of the label to the root than did wild-type plants (Fig. 1A). This result suggests that more photoassimilate produced during the 20-min labeling period was loaded into the phloem and allocated to roots in the transgenic plants than in the wild-type plants.
Figure 1.
Suc loading and long-distance transport in wild-type (WT) and pPP2::AtSUC2 rice plants. A, Seedlings of the wild type and four independent lines of pPP2::AtSUC2 (L6, L9, L26, and L51) were photosynthetically labeled with [14C]CO2, and shoots and roots were analyzed separately by scintillation. The 14C transported to the roots is expressed as a percentage of total 14C incorporated. The data are means ± sd from four independent experiments. B, Phloem exudation from the flag leaves of wild-type and transgenic plants into EDTA-containing solution after photosynthetic labeling with [14C]CO2, expressed as counts per minute (CPM) per milligram of leaf fresh weight per hour (fm/h). The data are means ± sd from four independent experiments. C, Time course of uptake of [14C]Suc into flag leaves of wild-type and transgenic plants 10 d after fertilization, expressed as counts per minute per milligram of leaf fresh weight per minute. D, Partitioning of fed 14C in flag leaves of wild-type and pPP2::AtSUC2 rice plants. Labeling was performed on the flag leaves on the day of fertilization. The data are expressed as means ± sd of six plants. E, Partitioning of fed 14C in flag leaves of wild-type and pPP2::AtSUC2 rice plants. Labeling was performed on the flag leaves on the 10th day after fertilization. The data are expressed as means ± sd of six plants. For A, B, D, and E, significant differences from the wild type are based on Student’s t test (*, P < 0.05; and **, P < 0.01). For C, the differences between the wild-type and transgenic lines were significant at P < 0.01 at time points of 5, 10, 15, and 20 min based on Student’s t test.
We further examined whether the pPP2::AtSUC2 plants showed enhanced long-distance transport (Fig. 1B) by determining the rate at which 14C was exuded from cut leaves into EDTA solution, according to methods of Srivastava et al. (2008) and Dasgupta et al. (2014). The phloem exudation rate in pPP2::AtSUC2 plants was higher than that in wild-type plants, suggesting that the transgenic plants had more efficient loading and transport of carbon in the phloem.
The 14C allocation to roots and phloem exudation rate both measure long-distance transport, which is dependent on loading efficiency, but neither measures phloem loading directly (Turgeon and Wolf, 2009). To compare phloem loading between wild-type and transgenic plants, we measured 14C uptake into leaf veins. Leaf discs were infiltrated with a [14C]Suc solution for 20 min, washed thoroughly, and freeze dried, and phloem loading was assessed by scintillation counting. There was increased phloem loading in pPP2::AtSUC2 plants compared with wild-type plants (Fig. 1C).
We further investigated whether there was enhanced allocation of photosynthetic carbohydrates from leaves to grains in pPP2::AtSUC2 plants compared with wild-type plants. Flag leaves of wild-type and transgenic plants on the day of fertilization and the 10th day after fertilization were photosynthetically labeled with [14C]CO2 (Fig. 1, D and E). At maturity, transgenic plants transported more label to grains (as a percentage of total label in the plant) than did wild-type plants. This result indicates that more photosynthetic carbohydrate from leaves was allocated to grains in transgenic plants. Taken together, the above results show that phloem loading and long-distance transport of Suc is enhanced in rice plants expressing AtSUC2.
Heterologous Expression of AtSUC2 Leads to an Increase in Grain Size, Weight, and Yield
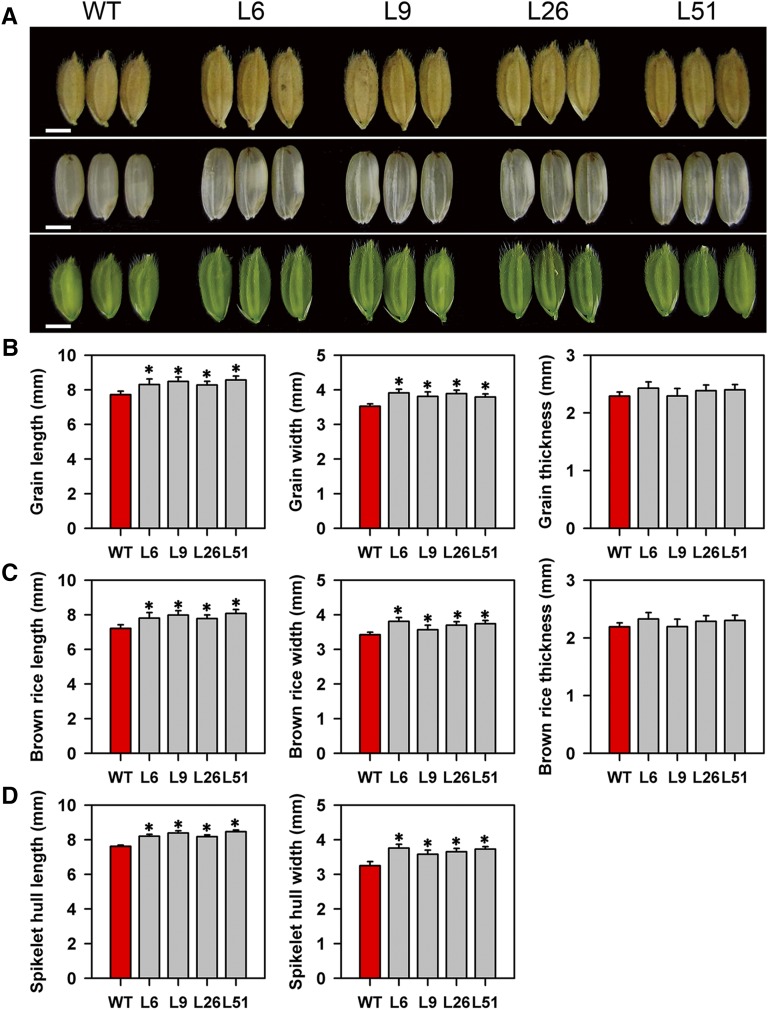
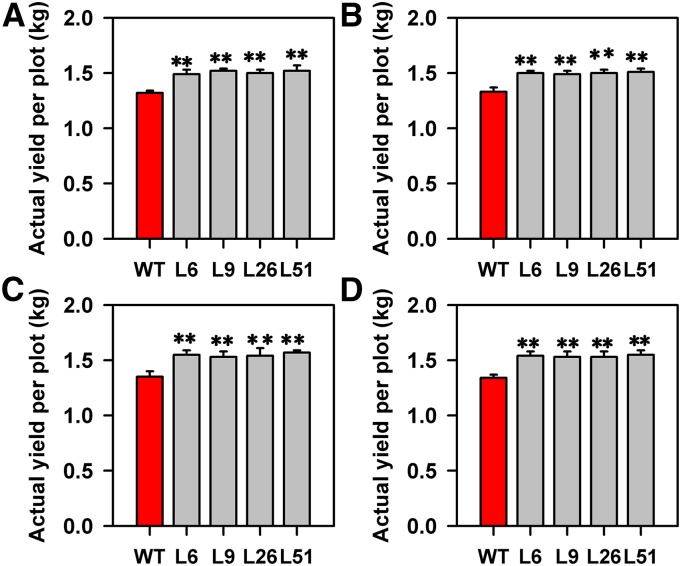
The grains, brown rice, and spikelet hulls were substantially wider and longer in the pPP2::AtSUC2 plants compared with wild-type plants (Fig. 2). We further examined the effects of AtSUC2 expression on the agricultural and grain yield traits (Supplemental Fig. S3), finding no significant differences in plant or panicle architecture between wild-type and transgenic plants. Although the number of grains per panicle was decreased in pPP2::AtSUC2 plants, the 1,000-grain weight and grain yield per plant were increased (Supplemental Fig. S3). Moreover, there was an increase of up to 16% in the plot grain yield over a 4-year period in pPP2::AtSUC2 plants as compared with wild-type plants in field trials (Fig. 3). The pPP2::AtSUC2 plants also showed a significant increase in biomass per plant. The increased biomass of aboveground parts in pPP2::AtSUC2 plants was associated with increased width of the leaves (Supplemental Fig. S4), which resulted from an increase in vein number (Supplemental Fig. S5).
Figure 2.
Characterization of grains in wild-type (WT) and pPP2::AtSUC2 rice plants. A, Comparisons of grains (top), brown rice (middle), and spikelets (bottom) in wild-type and four independent lines of pPP2::AtSUC2 (L6, L9, L26, and L51). Bars = 3 mm. B, Grain length, width, and thickness in wild-type and pPP2::AtSUC2 plants (n = 60 plants). C, Brown rice length, width, and thickness in wild-type and pPP2::AtSUC2 plants (n = 60 plants). D, Spikelet hull length and width just before heading in wild-type and pPP2::AtSUC2 plants (n = 60 plants). All data are given as means ± sd. Asterisks indicate significant difference from the wild type (*, P < 0.05; Student’s t test).
Figure 3.
Actual paddy yield of wild-type (WT) and pPP2::AtSUC2 rice plants. Plants were grown in a randomized complete block design with eight replications. Each block contained 60 plants. Student’s t test revealed a significant difference in grain yield between wild-type and pPP2::AtSUC2 plants, and this effect was consistent across 4 years. Data from 2010 (A), 2011 (B), 2012 (C), and 2013 (D). Data are given as means ± sd (n = 8). **, Statistically different from the wild type (P < 0.01). A Student’s t test was used to generate the P values.
Since increased rice grain size may have a negative effect on rice grain quality (Song et al., 2007; Wang et al., 2012), we investigated whether increased grain size in transgenic plants also affects grain quality (Supplemental Fig. S6). The grain quality mainly includes the following four aspects: cooking and eating quality, milling quality, appearance quality, and nutritional quality. A grain’s cooking and eating quality traits are determined mainly by amylose content and gel consistency (Tan et al., 2001). Amylose content and gel consistency did not differ significantly between wild-type and transgenic plants. There were also no significant differences in milling quality traits, brown rice percentage, and milled rice percentage between wild-type and transgenic plants. However, an appearance quality trait, chalky rice grain percentage, increased significantly in transgenic plants, whereas there were few differences with respect to head milled grain appearance between wild-type and transgenic plants. In addition, protein content, a nutritional quality, showed no significant difference between wild-type and transgenic plants. Overall, AtSUC2 expression in rice seems to enhance grain size and yield, but has little effect on appearance and no effect on cooking, eating, milling, or nutritional quality.
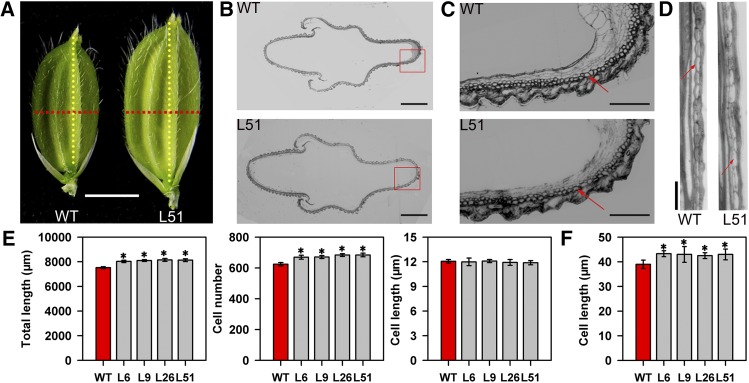
The Larger Spikelet Hulls of pPP2::AtSUC2 Plants Were Mainly Due to Increased Cell Number and Size
We examined cross and longitudinal sections of the spikelet hull in wild-type and transgenic plants before fertilization to investigate whether the increased size of spikelet hulls in transgenic plants was associated with an increase in cell number and/or size (Fig. 4). The cross sections showed that the outer parenchyma cell layers of transgenic plants were longer and contained more cells than those of the wild type, whereas there were no changes in cell length, indicating that the increased width of the spikelet hull in transgenic plants resulted mainly from an increase in cell number, but not cell size. The longitudinal sections revealed that there were no significant differences in cell number between the parenchyma cell layer of wild-type and pPP2::AtSUC2 plants, whereas cells were longer in transgenic plants than in wild-type plants, suggesting that the increased length of the spikelet hull in pPP2::AtSUC2 plants was mainly due to an increase in cell size, but not cell number.
Figure 4.
Histological analyses of spikelet hulls just before heading in wild-type (WT) and pPP2::AtSUC2 rice plants. A, Representative spikelets of wild-type and transgenic plants. Red and yellow dotted lines indicate positions of cross and longitudinal sections, respectively. Bars = 3 mm. B, Cross sections of spikelet hulls in wild-type and transgenic plants. Bars = 500 μm. C, Magnified views of spikelet hull cross sections boxed in B. Arrow shows the outer parenchyma cell layer. Bars = 150 μm. D, Magnified views of spikelet hull longitudinal sections in wild-type and transgenic plants. Arrow shows a parenchyma cell of the lemma. Bars = 50 μm. E, Comparison of total length, cell number, and mean cell length of the outer parenchyma cell layers of spikelet hulls in wild-type and transgenic plants (n = 10 spikelets). F, Comparison of parenchyma cell lengths in the lemma of spikelet hulls in wild-type and transgenic plants (n = 10 spikelets). All data are given as means ± sd. Asterisks indicate significant difference from the wild type (*, P < 0.05). A Student’s t test was used to generate the P values.
To investigate whether the increased grain size of transgenic plants is associated with altered regulation of genes controlling grain size and weight (Supplemental Fig. S7), we examined the transcriptional levels of genes/quantitative trait loci known to control grain size. There were no significant differences in the expression levels of these genes between wild-type and transgenic plants, suggesting that the increased grain size in transgenic plants was not associated with changes in expression of the genes that control grain size. The increase in cell number in the outer parenchyma cell layer of transgenic plants suggests that expression of AtSUC2 may influence the regulation of cell division (De Veylder et al., 2007; Berckmans and De Veylder, 2009). Indeed, we observed that the mRNA transcript levels of genes thought to be involved in the G1-to-S transition, such as cyclin-dependent kinase-activating kinase1 (CAK1), CAK1A, cyclin-dependent kinase A1 (CDKA1), cyclin D3 (CYCD3), CYCT1, and histone1 (H1), were much higher in pPP2::AtSUC2 plants than in wild-type plants (Supplemental Fig. S7), indicating that the expression of AtSUC2 regulates grain size by affecting the cell cycle machinery in rice.
Heterologous Expression of AtSUC2 Increases Grain Filling Rate
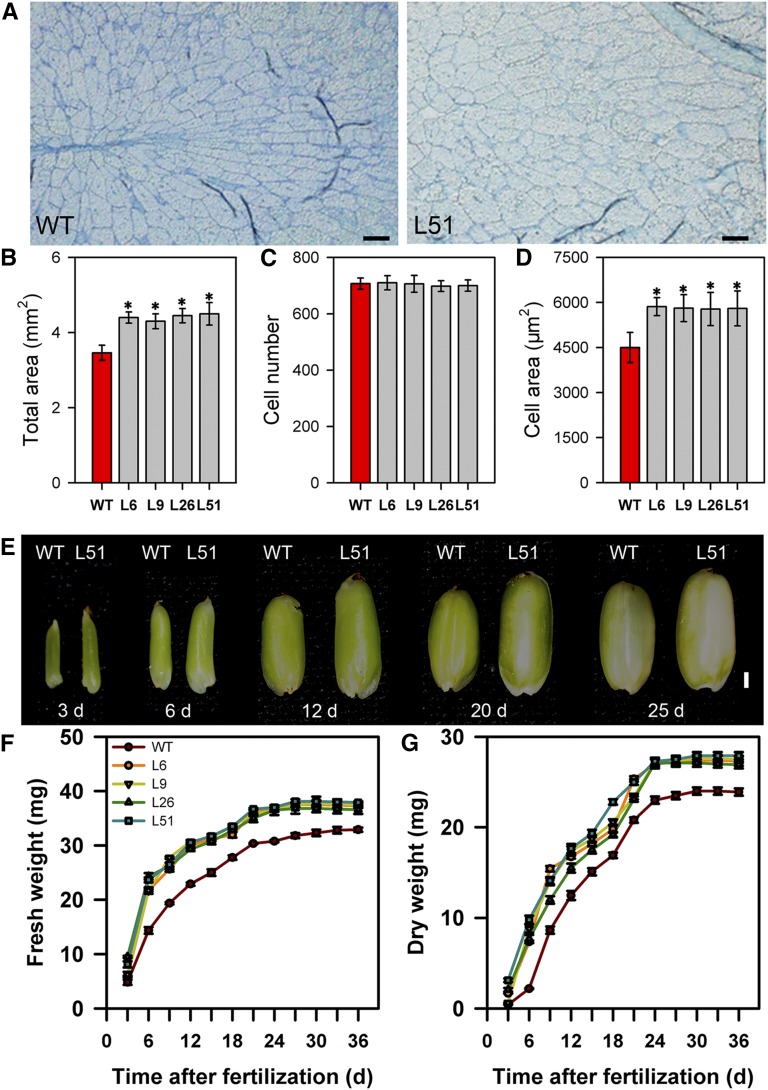
Since pPP2::AtSUC2 plants had a larger endosperm than wild-type plants (Fig. 2), we compared the endosperm cells in cross sections of mature grains in wild-type and transgenic plants (Fig. 5, A–D). Endosperm cells were larger in the pPP2::AtSUC2 plants than in wild-type plants, whereas there was no significant difference in endosperm cell number between wild-type and transgenic plants, suggesting that the increase in endosperm size in pPP2::AtSUC2 plants resulted mainly from an increase in cell size and not cell number.
Figure 5.
Histological analyses of endosperm at maturity and changes in endosperm size and grain filling after fertilization in wild-type (WT) and pPP2::AtSUC2 rice plants. A, Region between the dorsal and central points of the endosperm cross section in wild-type and pPP2::AtSUC2 plants. Bars = 100 μm. B, Comparison of total area of the endosperm cross section in wild-type and pPP2::AtSUC2 plants (n = 10 endosperms). C, Comparison of total cell number in the endosperm cross section of wild-type and pPP2::AtSUC2 plants (n = 20 endosperms). D, Comparison of mean cell area of the endosperm cross section of wild-type and pPP2::AtSUC2 plants (n = 20 endosperms). E, Representative endosperms after fertilization at 3, 6, 12, 20, and 25 d, respectively. Bars = 1 mm. F, Time course of endosperm fresh weight of wild-type and pPP2::AtSUC2 plants. Data are means ± sd (n = 20 plants). G, Time course of endosperm dry weight in wild-type and pPP2::AtSUC2 plants. Data are means ± sd (n = 20 plants). All data are given as means ± sd. Asterisks indicate significant difference from the wild type (*, P < 0.05; Student’s t test).
We then investigated grain filling in the wild-type and transgenic plants by comparing the endosperm weight after fertilization (Fig. 5, E–G). The endosperm fresh and dry weights were significantly higher in pPP2::AtSUC2 plants than in those of the wild type, starting 6 d after fertilization, and those differences almost reached a maximum 24 d after fertilization. These data suggest that the larger endosperms of pPP2::AtSUC2 plants were associated with a faster rate of accumulation of photosynthetic carbohydrates during grain filling.
An analysis of CO2 assimilation showed that there was no significant difference in CO2 assimilation between wild-type and transgenic plants (Supplemental Fig S8).
Gene Expression Analysis
We next investigated if the metabolic or phenotypic changes observed in pPP2::AtSUC2 plants could be correlated with changes in gene expression. We examined the expression of most of the genes in the rice genome using an Agilent rice microarray and RNA samples isolated from the flag leaves and developing seeds of wild-type and pPP2::AtSUC2 plants (line 51 [L51]) on the 10th day after fertilization. Statistical analysis revealed changes in gene expression in transgenic L51 plants that had P values <0.05 and fold changes >1.5 relative to wild-type plants. Only genes in this category were analyzed further. There were 211 and 333 genes that were differentially regulated in L51 flag leaves and seeds, respectively, compared with the wild type (Supplemental Tables S1 and S2). We selected genes that were up-regulated in flag leaves and developing seeds in wild-type and pPP2::AtSUC2 plants for gene ontology (GO) analysis. We conducted a Fisher’s test to calculate the significance of the percentage distribution of GO annotations for comparison with those in the whole genome (Supplemental Figs. S9 and S10).
Genes up-regulated by AtSUC2 expression in the flag leaves of transgenic plants had GO annotations highly associated with metabolic process, responses to stimulus, and biosynthetic process with a P value of 8.71 × 10−16, 2.98 × 10−13, and 2.03× 10−5, respectively (Supplemental Fig. S9), suggesting that genes related to metabolic and biosynthetic processes are overrepresented in the leaves of transgenic plants. Furthermore, genes up-regulated by AtSUC2 expression in the flag leaves of transgenic plants were highly related to primary metabolic process, nitrogen compound metabolic process, and carbohydrate metabolic process, with P values of 9.37 × 10−10, 4.67 × 10−5, and 1.52 × 10−3, respectively (Supplemental Fig. S9). In addition, genes up-regulated by AtSUC2 expression in the flag leaves of transgenic plants were highly related to starch metabolic process and polysaccharide metabolic process (Supplemental Fig. S9). We further identified a group of 71 genes with different functional categories, and expression of these 71 genes was confirmed by real-time PCR (Supplemental Table S3). Of these, many genes were involved in the metabolism of carbohydrates, amino acids, and lipids. These data suggest that AtSUC2 expression enhances carbohydrate, amino acid, and lipid metabolism in leaves.
Genes upregulated by AtSUC2 expression in developing seeds of transgenic plants had GO annotations highly associated with metabolic process, transport, and biosynthetic process, with a P value of 2.86 × 10−15, 7.83 × 10−7, and 3.51 × 10−4, respectively (Supplemental Fig. S10). Furthermore, genes up-regulated by AtSUC2 expression in developing seeds of transgenic plants were highly related to primary metabolic process, nitrogen compound metabolic process, and carbohydrate metabolic process, with P values of 5.25 × 10−9, 9.64 × 10−4, and 2.47 × 10−2, respectively (Supplemental Fig. S10). In addition, genes up-regulated by AtSUC2 expression in developing seeds of transgenic plants were highly related to anion transport and cation transport (Supplemental Fig. S10). We also further identified a group of 95 genes with different functional categories, and expression of these 95 genes was confirmed by real-time PCR (Supplemental Table S4). Of these, many genes were involved in the metabolism of carbohydrates, amino acids, and lipids. In addition, we found that the expression of many genes encoding sugar, amino acid, lipid, and metal transporters was up- or down-regulated in transgenic plants. These results suggest that enhanced Suc loading leads to enhanced metabolism of carbohydrates, amino acids, and lipid and increased translocation of nutrients during seed development. Interestingly, our microarray data also showed that the expression of genes involved in embryo and endosperm development (CycB1;1) and rice grain filling (rice prolamin box-binding factor [RPBF]) was significantly up-regulated in pPP2::AtSUC2 plants.
Leaf Carbon Metabolism and Transport Processes
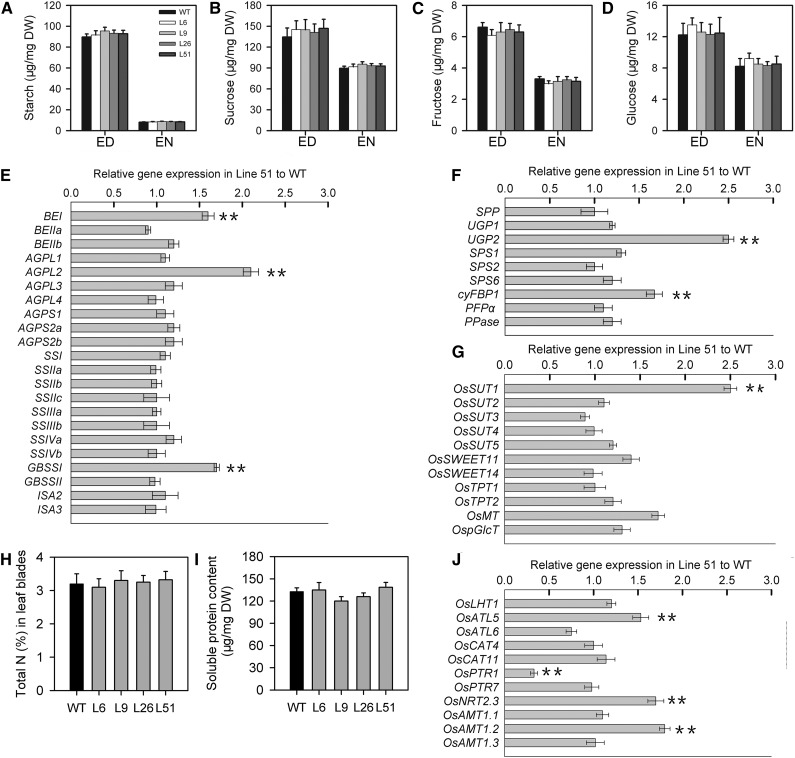
To assess whether primary carbon metabolites are altered in transgenic plants, we determined the levels of Suc, Glc, Fru, and starch in fully expanded leaves (i.e. source leaves) on the 10th day after fertilization, since grain filling was rapid at this time point. There were no significant differences in the levels of starch, Suc, Fru, and Glc in pPP2::AtSUC2 compared with wild-type plants (Fig. 6, A–D).
Figure 6.
Analyses of starch, sugar, nitrogen, and gene expression involved in carbon metabolism and carbon and nitrogen transport in flag leaves of wild-type (WT) and pPP2::AtSUC2 rice plants. A, Leaf starch levels (n = 5). B. Leaf Suc levels (n = 5). C, Leaf Fru levels (n = 5). D, Leaf Glc levels (n = 5). DW, Dry weight. E, Analysis of genes involved in starch biosynthesis in flag leaves. F, Analysis of genes involved in Suc biosynthesis in flag leaves. G, Analysis of genes involved in carbon transport in flag leaves. H, Total nitrogen (n = 6). I, Soluble protein content (n = 6). J, Analysis of genes involved in nitrogen transport in flag leaves. Analyses were performed on the flag leaves on the 10th day after fertilization. All data are given as the mean ± sd. Asterisks indicate significant difference from the wild type (**, P < 0.01). A Student’s t test was used to generate the P values. For the analysis of gene expression, real-time PCR was performed, and five independent experiments were done for each line using Actin (Os01g0376700) as control gene. Shown is the fold change in gene expression relative to the wild type. Starch and sugar were determined at the end of day (ED) and end of night (EN), respectively. BEI (Os06g0726400), BEIIa (Os02g0528200), and BEIIb (Os02g0528200), encoding for starch-branching enzymes; AGPL1 (Os05g0580000), AGPL2 (Os01g0633100), AGPL3 (Os03g0735000), AGPL4 (Os07g0243200), AGPS1 (Os09g0298200), AGPS2a (Os08g0345800), and AGPS2b (Os08g0345800), encoding for ADP-Glc pyrophosphorylase subunits; SSI (Os06g0160700), SSIIa (Os06g0229800), SSIIb (Os02g0744700), SSIIc (Os10g0437600), SSIIIa (Os08g0191500), SSIIIb (Os04g0624600), SSIVa (Os01g0720600), and SSIVb (Os05g0533600), encoding for starch synthase; GBSSI (Os06g0133000) and GBSSII (Os07g0412100), encoding for granule-bound starch synthases; ISA2 (Os05g0395300) and ISA3 (Os09g0468700), encoding for isoamylases; SPP (Os01g0376700), encoding for Suc-phosphatase; UGP1 (Os01g0264100) and UGP2 (Os02g0117700), encoding for UDP-Glc pyrophosphorylases; SPS1 (Os01g0702900), SPS2 (Os02g0184400), and SPS6 (Os06g0634800), encoding for Suc-phosphate synthases; cyFBP1 (Os01g0866400), encoding for Fru-1,6-bisphosphatase; PFPɑ (Os02g0714200), encoding for pyrophosphate-Fru 6-phosphate 1-phosphotransferase; PPase (Os01g0866500), encoding for pyrophosphatase.
The above microarray data suggest that AtSUC2 expression enhances expression of genes involved in carbohydrate and amino acid metabolism and transport in leaves (Supplemental Fig. S9; Supplemental Table S3); thus, we analyzed if the increased grain weight in pPP2::AtSUC2 plants is consistent with the expression of genes associated with carbon metabolism and transport in leaf cells, and we examined the RNA levels of genes involved in starch and Suc biosynthesis and carbon transport in mature leaves on the 10th day after fertilization by real-time PCR (Fig. 6, E–G). Among the genes that participate in starch biosynthesis in leaves, only the expression of those encoding branching enzyme I (BEI), ADP Glc pyrophosphorylase large subunit 2, and granule-bound starch synthase I (GBSSI) was increased. Among the genes involved in Suc biosynthesis in leaves, only the expression of those encoding UDP-Glc pyrophosphorylase 2 (UGP2) and cytosolic Fru-1,6-bisphosphatase 1 (cyFBP1) was increased in pPP2::AtSUC2 plants. We also examined the transcript levels of genes for Suc transporters and carbon metabolism-related plastidic translocators (Toyota et al., 2006; Braun and Slewinski, 2009; Ayre, 2011; Chen et al., 2012). Only the transcript level of the gene encoding Suc transporter OsSUT1 was increased in transgenic plants.
Because of the strong interaction between C and N metabolism (Foyer et al., 2006), we further examined differences in the expression of genes involved in N transport in mature leaves between wild-type and transgenic plants. There were no significant differences in total leaf N and soluble protein contents between wild-type and transgenic plants (Fig. 6, H and I). In addition, the transcript levels of genes encoding amino acid transporter-like5 (OsATL5), nitrate transporter (OsNRT2.3a), and ammonium transporter (OsAMT1.2), which are known to be involved in N transport (Sonoda et al., 2003; Yan et al., 2011; Tang et al., 2012), were up-regulated in transgenic plants (Fig. 6J). These results about expression of genes involved in carbon metabolism and carbon and nitrogen transport in leaves of wild-type and transgenic plants (Fig. 6) were consistent with microarray data observed in leaves of wild-type and transgenic plants (Supplemental Tables S1 and S3).
Seed Carbon Metabolism and Transport Processes
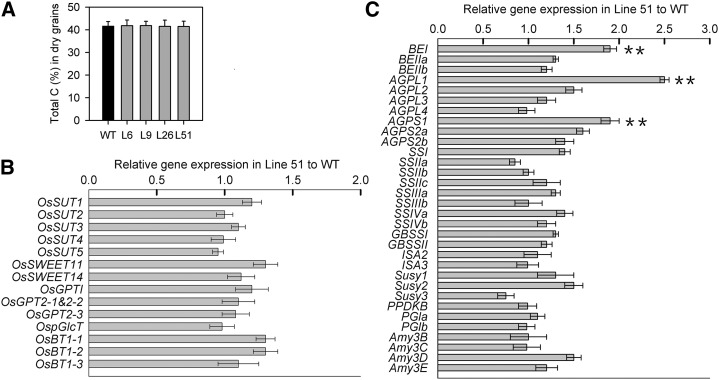
Since our microarray data also suggest that AtSUC2 expression enhances expression of genes involved in carbon and nitrogen metabolism and transport in developing seeds (Supplemental Figure S10; Supplemental Tables S2 and S4), we examined expression of genes involved in carbon and nitrogen transport, starch biosynthesis, and storage protein production in developing seeds in wild-type and transgenic plants by real-time PCR (Figs. 7 and 8). Percentage of carbon in the seeds of pPP2::AtSUC2 plants was comparable with those in wild-type plants (Fig. 7A). The transcript levels of genes for Suc transporters and starch metabolism-related plastidic translocators were not changed in the seeds of pPP2::AtSUC2 plants (Fig. 7B). However, the transcript levels of several genes involved in starch biosynthesis were increased, including those encoding BEI and ADP Glc pyrophosphorylase (AGPL1 and AGPS1; Fig. 7C).
Figure 7.
Analyses of total carbon and expression levels of genes involved in carbon metabolism and transport in seeds of wild-type (WT) and pPP2::AtSUC2 rice plants. A, Total carbon levels (percentage) in dry seeds (n = 5). B, Analysis of genes involved in carbon transport in developing seeds on the 10th day after fertilization. C, Analysis of genes involved in starch biosynthesis in developing seeds on the 10th day after fertilization. All data are given as the mean ± sd. Asterisks indicate significant difference from the wild type (**, P < 0.01). A Student’s t test was used to generate the P values. For the analysis of gene expression, real-time PCR was performed, and five independent experiments were done for each line using Actin (Os01g0376700) as control gene. Shown is the fold change in gene expression relative to the wild type. BEI (Os06g0726400), BEIIa (Os02g0528200), and BEIIb (Os02g0528200), encoding for starch-branching enzymes; AGPL1 (Os05g0580000), AGPL2 (Os01g0633100), AGPL3 (Os03g0735000), AGPL4 (Os07g0243200), AGPS1 (Os09g0298200), AGPS2a (Os08g0345800), and AGPS2b (Os08g0345800), encoding for ADP-Glc pyrophosphorylase subunits; SSI (Os06g0160700), SSIIa (Os06g0229800), SSIIb (Os02g0744700), SSIIc (Os10g0437600), SSIIIa (Os08g0191500), SSIIIb (Os04g0624600), SSIVa (Os01g0720600), and SSIVb (Os05g0533600), encoding for starch synthase; GBSSI (Os06g0133000) and GBSSII (Os07g0412100), encoding for granule-bound starch synthases; ISA2 (Os05g0395300) and ISA3 (Os09g0468700), encoding for isoamylases. Susy1 (Os06g0194900), Susy2 (Os03g0401300), and Susy3 (Os07g0616800), encoding for Suc synthases; PPDKB (Os05g0405000), encoding for pyruvate phosphate dikinase; PGIa (Os09g0465600) and PGIb (Os06g0256500), encoding for phosphoglucose isomerases; Amy3B (Os09g0457500), Amy3C (Os09g0457800), Amy3D (Os08g0473900), and Amy3E (Os08g0473600), encoding for alpha-amylase isozymes.
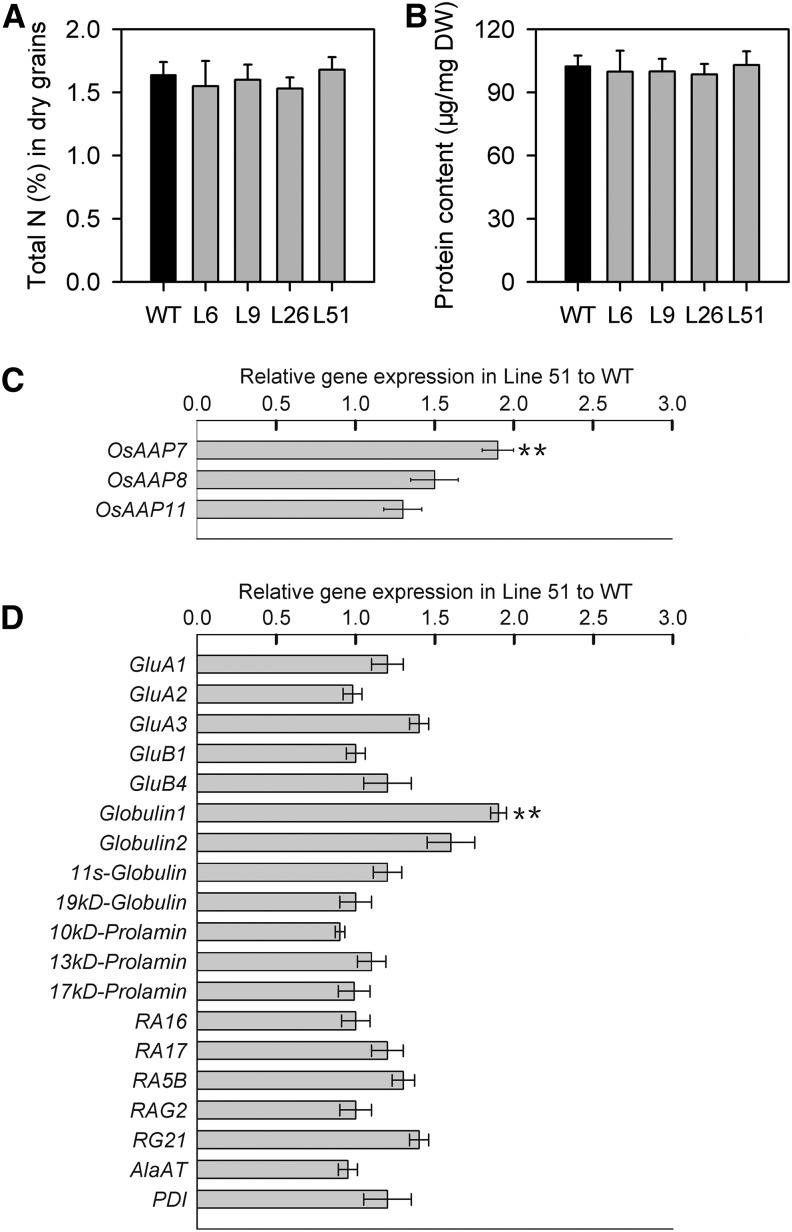
Figure 8.
Nitrogen levels and expression of genes involved in nitrogen transport and production of storage protein in seeds of wild-type (WT) and pPP2::AtSUC2 rice plants. All data are given as the mean ± sd. Asterisks indicate significant difference from the wild type (**, P < 0.01). A Student’s t test was used to generate the P values. A, Total nitrogen in dry seeds (n = 5). B, Protein content in dry seeds (n = 5). C, Real-time PCR analysis of genes involved in nitrogen transport in developing seeds on the 10th day after fertilization. Five independent experiments were done for each line using Actin (Os01g0376700) as control gene. Shown is the fold change in gene expression relative to the wild type. (D) Real-time PCR analysis of genes involved in production of storage protein in developing seeds on the 10th day after fertilization. Five independent experiments were done for each line using Actin (Os01g0376700) as control gene. Shown is the fold change in gene expression relative to the wild type. OsAAP7 (Os05g0424000), OsAAP8 (Os01g0882800), and OsAAP11 (Os11g0195600), encoding for amino acid transporters 7, 8, and 11, respectively; GluA1 (Os01g0762500), GluA2 (Os10g0400200), GluA3 (Os03g0427300), GluB1 (Os02g0249800), and GluB4 (Os02g0242600), encoding for glutelin A1, A2, A3, B1, and B4, respectively; Globulin1 (Os03g0663800), Globulin2 (Os03g0793700), 11S-globulin (Os05g0116000), and 19 kD globulin (Os05g0499100), encoding for different globulin species; 10-, 13-, and 17-kD prolamin (Os03g0766100, Os07g0219400, Os06g0507100, respectively), encoding for prolamins with different size; RA16 (Os07g0214600), RA17 (Os03g0791200), RA5B (Os07g0215500), RAG2 (Os07g0214300), and RG21 (Os02g0268100), encoding for species of rice allergenic proteins; AlaAT (Os10g0390500), encoding for Ala aminotransferase; PDI (Os11g0199200), encoding for protein disulfate isomerase.
There were no significant changes in the total N and protein amounts between wild-type and pPP2::AtSUC2 plants (Fig. 8, A and B). We further compared the expression of genes possibly involved in the transfer of amino acids to the seeds of wild-type and pPP2::AtSUC2 plants (Zhang et al., 2010; Zhao et al., 2012). Our results show that the expression of only encoding rice amino acid transporter7 (OsAAP7) was up-regulated in transgenic plants (Fig. 8C). We also examined the expression of genes involved in seed storage protein biosynthesis in wild-type and transgenic plants. The expression of only Globulin1 was up-regulated in pPP2::AtSUC2 plants (Fig. 8C). These results of seed carbon and nitrogen metabolism and transport processes (Figs. 7 and 8) further confirm microarray data observed in developing seeds of wild-type and transgenic plants (Supplemental Tables S2 and S4).
DISCUSSION
In this study, we successfully generated transgenic rice plants that express AtSUC2 (Supplemental Fig. S1). The transgenic plants showed enhanced phloem loading and long-distance transport of Suc, and more photoassimilate from leaves was allocated to grains (Fig. 1). Importantly, we found that this increased Suc loading was accompanied by substantially increased rice yield (Fig. 3). Thus, our data support the conclusion that enhancing Suc loading by genetic engineering promoted rice yield.
The increased yield resulted primarily from increased grain size, which is associated with an increase in spikelet hull size and endosperm size (Fig. 2). Seed development depends on the accumulation of storage starch and storage protein during grain filling. We found that the expression of BEI, AGPL1, and AGPS1, which are involved in storage starch production, was significantly induced in transgenic plants (Supplemental Table S4). AGPase is a key regulatory step in the starch biosynthesis pathway, and its activity strongly affects rice grain yield (Smidansky et al., 2002, 2003; Kawagoe et al., 2005; Li et al., 2011b; Hannah et al., 2012). BEI has a specific role in determining the amylopectin fine structure in rice endosperm (Satoh et al., 2003). Induction of AGPase and BEI in seeds of transgenic plants likely enhanced these activities and contributed to the increase in grain weight. In addition, CycB1;1 and CycD5;1 were up-regulated in the seeds of transgenic plants (Supplemental Table S2). OsCycB1;1 is critical for endosperm formation in rice and plays an important role in coordinating embryo and endosperm development. Therefore, up-regulating OsCycB1;1 expression in the seeds of transgenic plants may be a helpful strategy to promote endosperm development and increase grain weight (Guo et al., 2010). Interestingly, our microarray data showed that the expression of the gene for RPBF was significantly induced in the transgenic plants (Supplemental Table S4). RPBF is a transcriptional activator of rice storage protein genes in vivo, and most rice seed storage protein genes are transactivated by RPBF (Yamamoto et al., 2006). RPBF plays an important role during grain filling in rice (Kawakatsu et al., 2009). Our finding that the RPBF gene was induced in transgenic plants suggests that the regulation of RPBF might play an important role in grain filling. Taken together, our results suggest that enhanced Suc loading leads to increased expression of genes involved in the accumulation of storage starch and storage protein and endosperm development in developing seeds, which contributes to increased endosperm size and grain weight in the transgenic plants.
Seed development also depends on the amount of Suc in source leaves that can be transported into seeds. The lack of significant difference in the level of Suc and CO2 assimilation in leaves between wild-type and transgenic plants (Supplemental Fig. S8; Fig. 6) suggests that Suc fluxes, rather than steady-state levels, are affected in transgenic plants. Furthermore, we observed that the pPP2::AtSUC2 plants had wider leaves and thus larger leaf areas (Supplemental Fig. S4). The increased leaf area in the transgenic plants likely improves total photosynthetic performance and may increase carbon partitioning from source to sink organs, resulting in improved seed development. In addition, the pPP2::AtSUC2 plants showed a significant increase in the expression of genes, such as cyFBP1 and UGP2, that are involved in Suc biosynthesis in leaves (Fig. 6F; Kleczkowski et al., 2004; Lee et al., 2008). These results suggest that enhanced Suc loading also promotes processes involved in Suc biosynthesis in rice leaves to maintain the steady-state level of Suc in the face of increased flux.
Although AtSUC2 expression did not increase the sugar content in the mature leaves of the transgenic rice plants, we were surprised to observe that the expression of OsSUT1 was significantly up-regulated in those leaves (Fig. 6G). OsSUT1 primarily functions in phloem loading of Suc retrieved from the apoplasm along the transport pathway to maintain the supply of assimilate to the filling grain (Scofield et al., 2002, 2007). Thus, the increased Suc loading in the pPP2::AtSUC2 plants may trigger induction of OsSUT1, which may serve to enhance Suc loading from the apoplasm and further promote assimilate transport to the filling grain. In addition, the expression of several transporter genes (OsATL5, OsNRT2.3, and OsAMT1.2) involved in the remobilization of nitrogen was significantly up-regulated in the mature leaves of pPP2::AtSUC2 plants (Sonoda et al., 2003; Yan et al., 2011; Tang et al., 2012; Fig. 6J). Thus, the enhanced Suc loading appears to coordinate nitrogen remobilization in the mature leaves of the transgenic plants. Our microarray data show that several classes of nutrient transport-related genes were regulated in the seeds of the transgenic plants (Supplemental Table S4). These genes include one encoding an amino acid transporter, two encoding sorbitol transporters, two encoding phosphate transporters, two encoding ATP-binding cassette transporters, one encoding a potassium transporter, one encoding a heavy metal transporter, one encoding a lipid transfer protein, and one encoding a member of the α-tocopherol transport family. These results suggest that enhanced Suc loading may also influence nutrient remobilization to seeds during grain filling.
SUTs can be divided into three clades: type I, II, and III according to phylogenetic analysis (Aoki et al., 2003; Reinders et al., 2012). Type I SUTs are exclusive to eudicots, whereas type II and III SUTs exist in all plants. AtSUC2 and OsSUT1 are in different phylogenetic clades and belong to type I and II SUTs, respectively (Reinders et al., 2012). In eudicot species, type I SUTs, such as AtSUC2 in Arabidopsis, are required for loading Suc into the phloem (Gottwald et al., 2000). Monocot species such as rice lack type I SUTs (e.g. AtSUC2), and they use type II SUTs for Suc phloem loading (Slewinski et al., 2009). There are differences in substrate affinity and specificity between type I and II SUTs (Reinders et al., 2012). Transport activities of OsSUT1 and AtSUC2 were already analyzed. AtSUC2 was found to have a higher apparent affinity for Suc (1.44 mm) compared with OsSUT1 (7.5 mm; Chandran et al., 2003; Sun et al., 2010). In addition, differences in substrate specificity between type I SUTs such as AtSUC2 and type II SUTs such as OsSUT1 were identified. AtSUC2 appeared to be more selective than OsSUT1 (Sun et al., 2010; Reinders et al., 2012), suggesting that the substrate specificity of phloem loading in eudicots is different from that in monocots. However, OsSUT1 can complement the growth reduction caused by the loss of AtSUC2 in Arabidopsis (Sun and Ward, 2012), indicating that difference in substrate specificity between type I and II SUTs may not reflect a significant difference in physiological function (e.g. Suc phloem loading). Thus, the difference in activity between AtSUC2 and OsSUT1 may be important to achieve the increase in yield in rice. Since rice does not have type I SUTs, AtSUC2 expression in rice may be helpful to have more Suc phloem loading and more Suc transported to grains, leading to increased yield in rice.
It was recently reported that the expression of Suc transporter cDNAs specifically in companion cells results in increased phloem loading and long-distance transport of Suc, but inhibition of growth in Arabidopsis, due to phosphate limitation (Dasgupta et al., 2014). These results suggest that enhancing sugar transport may disrupt carbon and phosphate homeostasis in Arabidopsis. It is also possible that the inhibition of growth in these plants is due to decreased photosynthesis (Dasgupta et al., 2014). By contrast, our results demonstrate that the expression of the Suc transporter AtSUC2 specifically in companion cells of rice led to increased phloem loading and long-distance transport of Suc as well as an increase in leaf growth and grain yield in rice. One possible explanation is that our study focused the grain-filling process in rice, which is predominantly Suc derived, whereas the study by Dasgupta et al. (2014) focused on vegetative growth, which may require a different balance of phloem-transported nutrients.
Phloem loading of Suc is the first step of long-distance transport of photoassimilate from source leaves to sink organs. Three phloem loading strategies have been identified: two active mechanisms (i.e. apoplastic phloem loading via Suc transporters and symplastic polymer trapping) and one passive mechanism (i.e. the symplastic phloem loading pathway; Eom et al., 2012). Although the phloem loading mechanisms in rice have not been elucidated, both apoplastic and symplastic phloem loading pathways could exist, based on observations of the ultrastructure of vascular bundles (Kaneko et al., 1980; Chonan et al., 1984). It has been proposed that rice uses a passive symplastic pathway to translocate Suc through plasmodesmata into the phloem of the minor vein (Eom et al., 2012). However, the results of our study suggest that phloem loading of Suc through the apoplastic phloem loading pathway may also play an important role in the long-distance transport of photoassimilate from source leaves to grains in rice.
In conclusion, our results show that enhancing phloem Suc loading increases grain size and substantially enhances grain yield in rice. This work establishes that Suc loading controls seed size and grain yield. Our results demonstrate that manipulating phloem Suc loading could be useful for increasing grain yields in rice and other cereal crops.
MATERIALS AND METHODS
Generation of Transgenic Rice Plants
AtSUC2 cDNA (1,539 bp; accession no. At1g22710) was fused to the companion cell-specific PP2-A1 promoter, pPP2 (1,500-bp fragment carrying the promoter region of AtPP2-A1; accession no. At4g19840) and the nopaline synthase (NOS) terminator (260 bp; accession no. DQ005463). The pPP2::AtSUC2::NOS construct was cloned into the pCAMBIA1301 binary vector. The resultant construct was introduced into Agrobacterium tumefaciens strain GV3101. For transformation, calli were induced from mature seeds and transformed via Agrobacterium tumefaciens-mediated transformation (Hiei et al., 1994). Individual transgenic plants were selected on the basis of resistance to 75 mg L−1 hygromycin in one-half-strength Murashige and Skoog medium. The resistant lines were transplanted into soil and grown in a greenhouse (28°C). T1 plants were backcrossed to the wild type. The homozygous T3 seeds were used for further experiments.
Plant Materials, Growth Conditions, and Measurements of Agronomic Traits
The wild-type rice (Oryza sativa) used in this study was the japonica variety, ‘Zhonghua 11.’ The wild-type and transgenic plants were grown in a field station in Beijing, China, from May through October 2010 to 2013. Each line was grown in eight replicates in paddy fields laid out in a randomized complete block design, with plots of 60 plants arranged in four rows of 15 plants each. The distance between the plants in a row was 16.5 cm, and the distance between rows was 26 cm. The plants were maintained under routine management practices. The grain length, width, and thickness; the number of primary and secondary branches; and the number of effective grains were determined when the seeds were harvested. Plant height and tiller number were determined just after flowering. The biomass and seed weight measurements were carried out after harvesting, and dry mature plants and seeds were kept at 42°C for 1 week. For each line, data from 60 individual plants were obtained and subjected to statistical analyses.
Determination of Rice Grain Quality
The contents of amylose and gel consistency were measured as described previously (Song et al., 2007). The protein content was measured by the Kjeldahl method (Kjeltec System 1002, Tecator). The brown rice percentage, milled rice percentage, and chalky rice grain percentage were measured according to previously reported methods (Tan et al., 2001).
Determination of Suc, Glc, Fru, Starch, and Total Carbon and Nitrogen
Suc, Glc, and Fru were analyzed by an HPLC system (Agilent Technologies 1200 series) as previously described (Eom et al., 2011). Starch was measured enzymatically (Lee et al., 2005). Total carbon and nitrogen in dried rice flour and leaves were determined using a TOC/TNb analyzer (Elementar Analysensysteme GmbH), and protein content was calculated by multiplying the percentage of total nitrogen by a conversion factor of 5.26 for rice grains and leaves (Fujihara et al., 2008).
CO2 Assimilation
CO2 assimilation was measured in attached leaves using a portable gas-exchange system (Li-6400, Li-Cor) at a sequentially changed photon flux density, leaf temperature of 25°C, and reference CO2 concentration of 350 μmol mol−1.
Immunoblot and SDS-PAGE Analysis
Immunoblot and SDS-PAGE analyses were performed according to our previous studies (Peng et al., 2006; Liu et al., 2012) using AtSUC2 antibody. For immunoblot analysis, total proteins were prepared and quantified as previously described (Ouyang et al., 2011).
Immunogold Electron Microscopy
Immunoelectron microscopy experiments were carried out as previously described (Saito et al., 2009; Li et al., 2011c). In brief, nickel grids carrying ultrathin leaf sections were sequentially floated in 0.01 m sodium phosphate buffer (PBS, pH 7.2) containing 5% (w/v) bovine serum albumin for 5 min, and then for 1 h at 37°C in PBS containing diluted anti-AtSUC2 antibody (1:10,000). After several washes in PBS, sections were incubated for 1 h at 37°C in PBS containing goat anti-rabbit IgG antibody conjugated to 10 nm colloidal gold (1:40, Sigma-Aldrich). After five washes with PBS, ultrathin sections were washed with distilled water, air dried, and counterstained with 2% (w/v) uranyl acetate. Images were observed at 80 kV with a JEOL 1400 transmission electron microscope and captured with a Fastscan F214 digital camera (TVIPS).
Histological Analysis
Spikelet hulls just before heading, mature rice grain, and leaf blades were fixed in formaldehyde-acetic acid solution (60% [v/v] ethanol, 5% [v/v] formaldehyde, and 5% [v/v] acetic acid) for 24 h, dehydrated in an ethanol series, and finally embedded in Epon812 resin (Shell Chemical, USA). Tissue sections (4–8 μm thick) were cut with a rotary microtome, mounted, and stained with toluidine blue. The sections were photographed under a light microscope (Olympus BX51) coupled to a DP70 CCD camera. The length of the cells was determined using ImageJ (National Institutes of Health).
Radiolabeling and EDTA Exudate Analysis
Partitioning of 14CO2 fed in flag leaves of rice grown under field conditions was determined according to Yang et al. (2001). Flag leaves from six main stems of wild-type and transgenic plants were labeled with 14CO2 on the day of fertilization and on the 10th day after fertilization. Labeling was performed between 9 am and 11 am on a clear day with photosynthetically active radiation at the top of the canopy ranging from 1,100 to 1,200 μmol m−2 s−1. The whole flag leaf was placed in a polyethylene chamber (25-cm length and 4-cm diameter) and sealed with tape and plasticine to obtain a gas-tight seal. Six milliliters of air in the chamber was drawn out, and the same volume of gas was injected into the chamber containing 0.01 mol L−1 CO2 at a specific radioactivity of 14C at 1.48 MBq L−1. The chamber was removed after 0.5 h.
The labeled plants were sampled at maturity. Each plant was divided into two parts: grains and the remaining aboveground portion. Samples were dried at 80°C to a constant weight, ground into powder, and then extracted by shaking in 630 g L−1 boiling ethanol. The 14C radioactivity in the extracted aliquots was then measured by scintillation counting. Radioactivity distribution in grains was expressed as a percentage of the total radioactivity of the aboveground portion of the plant.
To investigate phloem transport, an EDTA exudation method (Srivastava et al., 2008; Dasgupta et al., 2014) was used to collect phloem sap from the leaf blades of wild-type and transgenic plants after photosynthetic labeling with [14C]CO2 under field conditions, as described above. After labeling in the presence of [14C]CO2 for 30 min, the chamber was removed, and leaf photosynthesis continued for 15 min in air. The leaf blades were cut at the base of flag leaves, and then the end of the leaf blades were cut again under EDTA solution. Exudates from the first 20 min were discarded, because there may be contamination from cut cells, and subsequent exudates from each of two 1-h periods were collected. The 14C-radioactivity in exudates was then measured by scintillation counting.
To analyze 14C transport to roots in wild-type and transgenic plants, rice seedlings were grown using the hydroponic culture method with one-half-strength concentrated Murashige and Skoog medium through which seedling roots could be collected easily. Seedlings were grown in a 14-h-light/10-h-dark cycle (260 μmol m−2 s−1) at a constant temperature of 29°C for 10 d. The whole leaves of each seedling were labeled 6 to 8 h into a 16-h-light cycle. The labeling procedure was the same as described above for flag leaves. The distribution of 14C in roots and shoots of individual seedlings was determined by scintillation counting.
Suc uptake was determined as described by Leterrier et al. (2003) and Dasgupta et al. (2014). Leaf blade discs were excised with a cork borer from flag leaves on the 10th day after fertilization. The discs were incubated in incubation medium (20 mm MES[ pH 5.8], 175 mm mannitol, 0.5 mm CaCl2, 0.5 mm K2SO4) for a 20-min period. The incubation medium was then replaced with the same medium supplemented with [14C]Suc (1 mm Suc, 10 MBq mL−1). At the end of the incubation period, leaf discs were rinsed three times for 3 min each with the incubation medium. Then, the discs were digested in a solution of perchloric acid (65% [w/w]) and hydrogen peroxide (33% [w/w]) at 55°C for 16 h. Radioactivity was counted by scintillation counting.
Scintillation counting was performed using a liquid scintillation counter (Packard Tricarb 1900TR, Packard Instruments).
RNA Isolation, cDNA Synthesis, RT-PCR, and Quantitative Real-Time RT-PCR
Procedures for the purification of total RNAs for cDNA synthesis, RT-PCR, and quantitative real-time RT-PCR (for primers used, see Supplemental Table S5) were carried out according to our previous study (Chi et al., 2008). Amplified Actin was used as an internal control for normalization.
Microarray Hybridization and Data Analysis
For the microarray experiment, Agilent 4*44K rice oligoarrays containing 43,803 features were used. Twelve chips were hybridized for microarray experiments. There were two sample types (flag leaves and seeds 10 d after fertilization), two genotypes (the wild type and the transgenic line, L51), and three biological replicates per genotype. Total RNA was prepared using TRIzol reagent (Invitrogen), and the mRNA was isolated from total RNAs using an RNAeasy Mini Kit (Qiagen). Microarray hybridization was carried out in a Hybridization Chamber (Agilent, G2534A) according to procedures provided by the company. The microarrays were scanned with an Agilent instrument (G2565BA), and the quality of the chip data was analyzed using R statistical language and the Limma package of the Bioconductor project (http://www.bioconductor.org/). Acquired signals were normalized internally and across all arrays as described by Smyth and Speed (2003), and the average signals of replicates were used for analysis. Linear models and empirical Bayes methods were applied to identify the differentially expressed genes (Smyth, 2004). The genes that were up- or down-regulated >1.5-fold with a P value <0.05 (without applying correction) in transgenic plants as compared with the wild type were identified. Last, GO terms enriched in up-regulated genes were identified with GO::TermFinder (Boyle et al., 2004). Raw P values of GO term enrichment were corrected for multiple tests using false discovery rate (Benjamini and Hochberg, 1995). Expression data are available from the National Center for Biotechnology Gene Expression Omnibus repository (www.ncbi.nlm.nih.gov/geo/) under accession number GSE74024.
Antiserum Production
To produce polyclonal antibodies against AtSUC2, two synthetic peptides, ETQTGELDQPERLRK (amino acids 17–31) and QAENHRRDHGGAKTG (amino acids 384–398), were conjugated with keyhole limpet hemocyanin and used to immunize rabbits. Antibodies were affinity purified using a cyanogen bromide-activated Sepharose 4B column (Sigma-Aldrich) conjugated with the peptides. The dilution ratio for antibody against AtSUC2 in the immunoblots was 1:10,000.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: AtSUC2 (AT1G22710), GS3 (Os01g0239200), GS5 (Os05g0158500), GW2 (Os02g0244100), GW5 (Os09g0465600), GW8 (Os08g0531600), GRAIN INCOMPLETE FILLING1 (GIF1, Os04g0413500), DENSE AND ERECT PANICLE1 (DEP1, Os09g0441900), SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE4 (OsSPL4, Os09g0457800), SEED ROUND AND SMALL1 (SRS1, Os07g0616000), SEED ROUND AND SMALL3 (SRS3, Os01g0376700), DENSE AND ERECT PANICLE3 (DEP3, Os02g0260200), SHORT GRAIN1 (SG1, Os09g0459200), CAK1 (Os06g0171700), CAK1A (Os06g0334400), cell division cycle protein20 (CDC20, Os04g0599800), CDKA1 (Os03g0118400), CDKA2 (Os02g0123100), CDKB (Os08g0512600), cell division cycle protein (CDT2, Os03g0699100), CYCA2.1 (Os12g0581800), CYCA2.2 (Os12g0502300), CYCA2.3 (Os01g0233500), CYCB2.1 (Os04g0563700), CYCB2.2 (Os06g0726800), CYCD3 (Os11g0706801), CYCD4 (Os09g0466100), cyclin IdZm (CYCIdZm, Os01g0805600), cyclin T (CYCT1, Os02g0438200), transcription factor (E2F2, Os12g0158800), H1 (Os04g0253000), syntaxin-related protein KNOLLE (KN, Os03g0736500), mitotic spindle checkpoint protein (MAD2, Os04g40940), lysine and histidine transporter (OsLHT1, Os08g0127100), OsATL5 (Os06g0633800), OsATL6 (Os02g0191300), cationic amino acid transporter (OsCAT4, Os03g0654400), OsCAT11 (Os12g0623500), peptide transporter (OsPTR1, Os07g0100600), OsPTR7 (Os01g0142600), OsNRT2.3 (Os06g0194900), OsAMT1.1 (Os08g0191500), OsAMT1.2 (Os06g0229800), OsAMT1.3 (Os06g0229800), OsAAP7 (Os05g0424000), OsAAP8 (Os06g0194900), OsAAP11 (Os03g0793700), OsSUT1 (Os03g0170900), OsSUT2 (Os12g0641400), OsSUT3 (Os10g0404500), OsSUT4 (Os02g0827200), OsSUT5 (Os02g0576600), OsSWEET11 (Os08g0535200), OsSWEET14 (Os11g0508600), OsTPT1 (triose phosphate/phosphate translocator, Os01g0239200), OsTPT2 (Os05g0241200), maltose translocator (OsMT, 0s04g06020400), plastidic glucose translocator (OspGLcT, Os01g0133400), glucose 6-phosphate/phosphate translocator (OsGPT1, Os08g0187800), OsGPT2-1 (LOC_Os07g34010), OsGPT2-2 (LOC_Os07g33960), OsGPT2-3 (Os07g0523400), BRITTLE1 (OsBT1-1, Os02g0202400), OsBT1-2 (Os05g0171300), OsBT1-3 (Os06g0602700), BEI (Os06g0726400), BEIIa (Os02g0528200), BEIIb (Os02g0528200), AGPL1 (Os05g0580000), AGPL2 (Os01g0633100), AGPL3 (Os03g0735000), AGPL4 (Os07g0243200), AGPS1 (Os09g0298200), AGPS2a (Os08g0345800), AGPS2b (Os08g0345800), SSI (Os06g0160700), SSIIa (Os06g0229800), SSIIb (Os02g0744700), SSIIc (Os10g0437600), SSIIIa (Os08g0191500), SSIIIb (Os04g0624600), SSIVa (Os01g0720600), SSIVb (Os05g0533600), GBSSI (Os06g0133000), GBSSII (Os07g0412100), ISA2 (Os05g0395300), ISA3 (Os09g0468700), SPP (Os01g0376700), UGP1 (Os01g0264100), UGP2 (Os02g0117700), SPS1 (Os01g0702900), SPS2 (Os02g0184400), SPS6 (Os06g0634800), cyFBP1 (Os01g0866400), PFPɑ (Os02g0714200), PPase (Os01g0866500), Susy1 (Os06g0194900), Susy2 (Os03g0401300), Susy3 (Os07g0616800), PPDKB (Os05g0405000), PGIa (Os09g0465600), PGIb (Os06g0256500), Amy3B (Os09g0457500), Amy3C (Os09g0457800), Amy3D (Os08g0473900), Amy3E (Os08g0473600), GluA1 (Os01g0762500), GluA2 (Os10g0400200), GluA3 (Os03g0427300), GluB1 (Os02g0249800), GluB4 (Os02g0242600), Globulin1 (Os03g0663800), Globulin2 (Os03g0793700), 11s-globulin (Os05g0116000), 19 kD globulin (Os05g0499100), 10-kD prolamin (Os03g0766100), 13-kD prolamin (Os07g0219400), 17-kD prolamin (Os06g0507100), RA16 (Os07g0214600), RA17 (Os03g0791200), RA5B (Os07g0215500), RAG2 (Os07g0214300), RG21 (Os02g0268100), AlaAT (Os10g0390500), PDI (Os11g0199200), and Actin (Os01g0376700).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization of pPP2::AtSUC2 rice plants.
Supplemental Figure S2. Localization of AtSUC2 in leaves of wild-type and pPP2::AtSUC2 rice plants (line 51).
Supplemental Figure S3. Phenotype and grain yield of wild-type and pPP2::AtSUC2 rice plants.
Supplemental Figure S4. Leaf length and width and biomass of wild-type and pPP2::AtSUC2 rice plants.
Supplemental Figure S5. Histological analyses of flag leaves on the 10th day after fertilization in wild-type and pPP2::AtSUC2 rice plants.
Supplemental Figure S6. Comparisons of grain quality traits between wild-type and pPP2::AtSUC2 rice plants.
Supplemental Figure S7. Comparisons of transcripts of genes determining for grain size and genes involved in cell cycle between wild-type and pPP2::AtSUC2 rice plants.
Supplemental Figure S8. CO2 assimilation in leaves of wild-type and pPP2::AtSUC2 rice plants.
Supplemental Figure S9. Global analysis of gene expression profiles in flag leaves of pPP2::AtSUC2 rice plants (line 51).
Supplemental Figure S10. Global analysis of gene expression profiles in developing seeds of pPP2::AtSUC2 rice plants (line 51).
Supplemental Table S1. Genes found to be significantly up-regulated (≥1.5) or down-regulated (≤1.5) in flag leaves of pPP2::AtSUC2 rice plants (line 51).
Supplemental Table S2. Genes found to be significantly up-regulated (≥1.5) or down-regulated (≤1.5) in developing seeds of pPP2::AtSUC2 rice plants (line 51).
Supplemental Table S3. Statistical analyses of microarray and quantitative RT-PCR data for genes in flag leaves of pPP2::AtSUC2 rice plants (line 51) on the 10th day after fertilization.
Supplemental Table S4. Statistical analyses of microarray and quantitative RT-PCR data for genes in developing seeds of pPP2::AtSUC2 rice plants (line 51) on the 10th day after fertilization.
Supplemental Table S5. Primer sequences used for PCR and quantitative real-time RT-PCR.
Supplementary Material
Glossary
- SE/CC
sieve element-companion cell
- cDNA
complementary DNA
- RT
reverse transcription
- GO
gene ontology
- PBS
sodium phosphate buffer
Footnotes
This work was supported by the State Key Basic Research and Development Plan of China (grant no. 2015CB150105) and the Ministry of Agriculture of China (grant no. 2014ZX08009–003–005).
Articles can be viewed without a subscription.
References
- Ainsworth EA, Bush DR (2011) Carbohydrate export from the leaf: a highly regulated process and target to enhance photosynthesis and productivity. Plant Physiol 155: 64–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki N, Hirose T, Scofield GN, Whitfeld PR, Furbank RT (2003) The sucrose transporter gene family in rice. Plant Cell Physiol 44: 223–232 [DOI] [PubMed] [Google Scholar]
- Ayre BG. (2011) Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol Plant 4: 377–394 [DOI] [PubMed] [Google Scholar]
- Baker RF, Leach KA, Braun DM (2012) SWEET as sugar: new sucrose effluxers in plants. Mol Plant 5: 766–768 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc, B 57: 289–300 [Google Scholar]
- Berckmans B, De Veylder L (2009) Transcriptional control of the cell cycle. Curr Opin Plant Biol 12: 599–605 [DOI] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G (2004) GO:TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DM. (2012) Plant science. SWEET! The pathway is complete. Science 335: 173–174 [DOI] [PubMed] [Google Scholar]
- Braun DM, Slewinski TL (2009) Genetic control of carbon partitioning in grasses: roles of sucrose transporters and tie-dyed loci in phloem loading. Plant Physiol 149: 71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran D, Reinders A, Ward JM (2003) Substrate specificity of the Arabidopsis thaliana sucrose transporter AtSUC2. J Biol Chem 278: 44320–44325 [DOI] [PubMed] [Google Scholar]
- Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB (2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207–211 [DOI] [PubMed] [Google Scholar]
- Chi W, Ma J, Zhang D, Guo J, Chen F, Lu C, Zhang L (2008) The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol 147: 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chonan N, Kawahara H, Matsuda T (1984) Ultrastructure of vascular bundles and fundamental parenchyma in relation to movement of photosynthate in leaf sheath of rice. Jpn J Crop Sci 53: 435–444 [Google Scholar]
- Dasgupta K, Khadilkar AS, Sulpice R, Pant B, Scheible W-R, Fisahn J, Stitt M, Ayre BG (2014) Expression of sucrose transporter cDNAs specifically in companion cells enhances phloem loading and long-distance transport of sucrose but leads to an inhibition of growth and the perception of a phosphate limitation. Plant Physiol 165: 715–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inzé D (2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8: 655–665 [DOI] [PubMed] [Google Scholar]
- Dinant S, Clark AM, Zhu Y, Vilaine F, Palauqui JC, Kusiak C, Thompson GA (2003) Diversity of the superfamily of phloem lectins (phloem protein 2) in angiosperms. Plant Physiol 131: 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom JS, Cho JI, Reinders A, Lee SW, Yoo Y, Tuan PQ, Choi SB, Bang G, Park YI, Cho MH, et al. (2011) Impaired function of the tonoplast-localized sucrose transporter in rice, OsSUT2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol 157: 109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom JS, Choi SB, Ward JM, Jeon JS (2012) The mechanism of phloem loading in rice (Oryza sativa). Mol Cells 33: 431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet 112: 1164–1171 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G, Verrier P (2006) Photosynthetic carbon-nitrogen interactions: Modeling inter-pathway control and signaling. In Plaxton WC, McManus MT, eds, Control of Primary Metabolism in Plants. Blackwell Publishing, Oxford, pp 325–347 [Google Scholar]
- Fujihara S, Sasaki H, Aoyagi Y, Sugahara T (2008) Nitrogen-to-protein conversion factors for some cereal products in Japan. J Food Sci 73: C204–C209 [DOI] [PubMed] [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR (2000) Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc Natl Acad Sci USA 97: 13979–13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wang F, Song J, Sun W, Zhang XS (2010) The expression of Orysa;CycB1;1 is essential for endosperm formation and causes embryo enlargement in rice. Planta 231: 293–303 [DOI] [PubMed] [Google Scholar]
- Hannah LC, Futch B, Bing J, Shaw JR, Boehlein S, Stewart JD, Beiriger R, Georgelis N, Greene T (2012) A shrunken-2 transgene increases maize yield by acting in maternal tissues to increase the frequency of seed development. Plant Cell 24: 2352–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Kaneko M, Chonan N, Matsuda T (1980) Ultrastructure of the small vascular bundles and transfer pathways for photosynthate in the leaves of rice plant. Jpn J Crop Sci 49: 42–50 [Google Scholar]
- Kawagoe Y, Kubo A, Satoh H, Takaiwa F, Nakamura Y (2005) Roles of isoamylase and ADP-glucose pyrophosphorylase in starch granule synthesis in rice endosperm. Plant J 42: 164–174 [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Yamamoto MP, Touno SM, Yasuda H, Takaiwa F (2009) Compensation and interaction between RISBZ1 and RPBF during grain filling in rice. Plant J 59: 908–920 [DOI] [PubMed] [Google Scholar]
- Kleczkowski LA, Geisler M, Ciereszko I, Johansson H (2004) UDP-glucose pyrophosphorylase: an old protein with new tricks. Plant Physiol 134: 912–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S, Wipf D, Frommer WB (2004) Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu Rev Plant Biol 55: 341–372 [DOI] [PubMed] [Google Scholar]
- Lee JW, Lee DS, Bhoo SH, Jeon JS, Lee YH, Hahn TR (2005) Transgenic Arabidopsis plants expressing Escherichia coli pyrophosphatase display both altered carbon partitioning in their source leaves and reduced photosynthetic activity. Plant Cell Rep 24: 374–382 [DOI] [PubMed] [Google Scholar]
- Lee SK, Jeon JS, Börnke F, Voll L, Cho JI, Goh CH, Jeong SW, Park YI, Kim SJ, Choi SB, et al. (2008) Loss of cytosolic fructose-1,6-bisphosphatase limits photosynthetic sucrose synthesis and causes severe growth retardations in rice (Oryza sativa). Plant Cell Environ 31: 1851–1863 [DOI] [PubMed] [Google Scholar]
- Leterrier M, Atanassova R, Laquitaine L, Gaillard C, Coutos-Thévenot P, Delrot S (2003) Expression of a putative grapevine hexose transporter in tobacco alters morphogenesis and assimilate partitioning. J Exp Bot 54: 1193–1204 [DOI] [PubMed] [Google Scholar]
- Li C, Wang Y, Liu L, Hu Y, Zhang F, Mergen S, Wang G, Schläppi MR, Chu C (2011c) A rice plastidial nucleotide sugar epimerase is involved in galactolipid biosynthesis and improves photosynthetic efficiency. PLoS Genet 7: e1002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang S, Zhao Y, Li B, Zhang J (2011b) Over-expression of AGPase genes enhances seed weight and starch content in transgenic maize. Planta 233: 241–250 [DOI] [PubMed] [Google Scholar]
- Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J. , et al. (2011a) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet 43: 1266–1269 [DOI] [PubMed] [Google Scholar]
- Liu J, Yang H, Lu Q, Wen X, Chen F, Peng L, Zhang L, Lu C (2012) PsbP-domain protein1, a nuclear-encoded thylakoid lumenal protein, is essential for photosystem I assembly in Arabidopsis. Plant Cell 24: 4992–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang M, Li X, Ma J, Chi W, Xiao J, Zou M, Chen F, Lu C, Zhang L (2011) LTD is a protein required for sorting light-harvesting chlorophyll-binding proteins to the chloroplast SRP pathway. Nat Commun 2: 277. [DOI] [PubMed] [Google Scholar]
- Peng L, Ma J, Chi W, Guo J, Zhu S, Lu Q, Lu C, Zhang L (2006) LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders A, Sivitz AB, Ward JM (2012) Evolution of plant sucrose uptake transporters. Front Plant Sci 3: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche E, Blackmore D, Tegeder M, Richardson T, Schroeder H, Higgins TJ, Frommer WB, Offler CE, Patrick JW (2002) Seed-specific overexpression of a potato sucrose transporter increases sucrose uptake and growth rates of developing pea cotyledons. Plant J 30: 165–175 [DOI] [PubMed] [Google Scholar]
- Saalbach I, Mora-Ramírez I, Weichert N, Andersch F, Guild G, Wieser H, Koehler P, Stangoulis J, Kumlehn J, Weschke W. , et al. (2014) Increased grain yield and micronutrient concentration in transgenic winter wheat by ectopic expression of a barley sucrose transporter. J Cereal Sci 60: 75–81 [Google Scholar]
- Saito Y, Kishida K, Takata K, Takahashi H, Shimada T, Tanaka K, Morita S, Satoh S, Masumura T (2009) A green fluorescent protein fused to rice prolamin forms protein body-like structures in transgenic rice. J Exp Bot 60: 615–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Matsuoka M (2008) Identifying and exploiting grain yield genes in rice. Curr Opin Plant Biol 11: 209–214 [DOI] [PubMed] [Google Scholar]
- Satoh H, Nishi A, Yamashita K, Takemoto Y, Tanaka Y, Hosaka Y, Sakurai A, Fujita N, Nakamura Y (2003) Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol 133: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield GN, Hirose T, Aoki N, Furbank RT (2007) Involvement of the sucrose transporter, OsSUT1, in the long-distance pathway for assimilate transport in rice. J Exp Bot 58: 3155–3169 [DOI] [PubMed] [Google Scholar]
- Scofield GN, Hirose T, Gaudron JA, Upadhyaya NM, Ohsugi R, Furbank RT (2002) Antisense suppression of the rice sucrose transporter gene, OsSUT1, leads to impaired grain filling and germination but does not affect photosynthesis. Funct Plant Biol 29: 815–826 [DOI] [PubMed] [Google Scholar]
- Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M (2008) Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet 40: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Slewinski TL, Braun DM (2010) Current perspectives on the regulation of whole-plant carbohydrate partitioning. Plant Sci 178: 341–349 [Google Scholar]
- Slewinski TL, Meeley R, Braun DM (2009) Sucrose transporter1 functions in phloem loading in maize leaves. J Exp Bot 60: 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidansky ED, Clancy M, Meyer FD, Lanning SP, Blake NK, Talbert LE, Giroux MJ (2002) Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc Natl Acad Sci USA 99: 1724–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidansky ED, Martin JM, Hannah LC, Fischer AM, Giroux MJ (2003) Seed yield and plant biomass increases in rice are conferred by deregulation of endosperm ADP-glucose pyrophosphorylase. Planta 216: 656–664 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T (2003) Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet 39: 623–630 [DOI] [PubMed] [Google Scholar]
- Sonoda Y, Ikeda A, Saiki S, von Wirén N, Yamaya T, Yamaguchi J (2003) Distinct expression and function of three ammonium transporter genes (OsAMT1;1-1;3) in rice. Plant Cell Physiol 44: 726–734 [DOI] [PubMed] [Google Scholar]
- Srivastava AC, Ganesan S, Ismail IO, Ayre BG (2008) Functional characterization of the Arabidopsis AtSUC2 Sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol 148: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Sauer N (1996) The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot Acta 109: 299–306 [Google Scholar]
- Sun Y, Reinders A, LaFleur KR, Mori T, Ward JM (2010) Transport activity of rice sucrose transporters OsSUT1 and OsSUT5. Plant Cell Physiol 51: 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Ward JM (2012) Arg188 in rice sucrose transporter OsSUT1 is crucial for substrate transport. BMC Biochem 13: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Sun M, Xing Y, Hua J, Sun X, Zhang Q, Corke H (2001) Mapping quantitative trait loci for milling quality, protein content and color characteristics of rice using a recombinant inbred line population derived from an elite rice hybrid. Theor Appl Genet 103: 1037–1045 [Google Scholar]
- Tang Z, Fan X, Li Q, Feng H, Miller AJ, Shen Q, Xu G (2012) Knockdown of a rice stelar nitrate transporter alters long-distance translocation but not root influx. Plant Physiol 160: 2052–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota K, Tamura M, Ohdan T, Nakamura Y (2006) Expression profiling of starch metabolism-related plastidic translocator genes in rice. Planta 223: 248–257 [DOI] [PubMed] [Google Scholar]
- Turgeon R, Wolf S (2009) Phloem transport: cellular pathways and molecular trafficking. Annu Rev Plant Biol 60: 207–221 [DOI] [PubMed] [Google Scholar]
- Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q. , et al. (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44: 950–954 [DOI] [PubMed] [Google Scholar]
- Weichert N, Saalbach I, Weichert H, Kohl S, Erban A, Kopka J, Hause B, Varshney A, Sreenivasulu N, Strickert M, et al. (2010) Increasing sucrose uptake capacity of wheat grains stimulates storage protein synthesis. Plant Physiol 152: 698–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Zhang Q (2010) Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61: 421–442 [DOI] [PubMed] [Google Scholar]
- Yamamoto MP, Onodera Y, Touno SM, Takaiwa F (2006) Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiol 141: 1694–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Fan X, Feng H, Miller AJ, Shen Q, Xu G (2011) Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ 34: 1360–1372 [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang J, Wang Z, Zhu Q, Wang W (2001) Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol 127: 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Tan Q, Lee R, Trethewy A, Lee Y-H, Tegeder M (2010) Altered xylem-phloem transfer of amino acids affects metabolism and leads to increased seed yield and oil content in Arabidopsis. Plant Cell 22: 3603–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Ma H, Yu L, Wang X, Zhao J (2012) Genome-wide survey and expression analysis of amino acid transporter gene family in rice (Oryza sativa L.). PLoS One 7: e49210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.