A MADS-box transcription factor represses resistance to pathogenic microorganisms and water deficit, and its down-regulation results in improved biotic and abiotic stress tolerance of rice.
Abstract
Functional analyses of MADS-box transcription factors in plants have unraveled their role in major developmental programs (e.g. flowering and floral organ identity) as well as stress-related developmental processes, such as abscission, fruit ripening, and senescence. Overexpression of the rice (Oryza sativa) MADS26 gene in rice has revealed a possible function related to stress response. Here, we show that OsMADS26-down-regulated plants exhibit enhanced resistance against two major rice pathogens: Magnaporthe oryzae and Xanthomonas oryzae. Despite this enhanced resistance to biotic stresses, OsMADS26-down-regulated plants also displayed enhanced tolerance to water deficit. These phenotypes were observed in both controlled and field conditions. Interestingly, alteration of OsMADS26 expression does not have a strong impact on plant development. Gene expression profiling revealed that a majority of genes misregulated in overexpresser and down-regulated OsMADS26 lines compared with control plants are associated to biotic or abiotic stress response. Altogether, our data indicate that OsMADS26 acts as an upstream regulator of stress-associated genes and thereby, a hub to modulate the response to various stresses in the rice plant.
MADS-box transcription factors belong to a multigenic family and have been identified in yeasts, plants, insects, nematodes and lower vertebrates, and mammals, where they control different aspects of development and cell differentiation (Shore and Sharrocks, 1995). For example, the yeast (Saccharomyces cerevisiae) MINICHROMOSOME MAINTENANCE1 MADS-box transcription factor is involved in diverse regulatory mechanisms underlying cell viability, cell cycle control, mating, minichromosome maintenance, and recombination but also, osmotolerance (Messenguy and Dubois, 2003). The MADS-BOX PROTEIN REQUIRED FOR INFECTIOUS GROWTH1/RESISTANCE TO LEPTOSPHAERIA MACULANS1 MADS-box transcription factor is required for pathogenicity of the causal fungal agent of the rice (Oryza sativa) blast disease, Magnaporthe oryzae (Mehrabi et al., 2008). In plants, analyses of MADS-box transcription factors have mainly revealed a function in flower development, flowering induction, or fruit development (Theissen et al., 2000; Arora et al., 2007; Smaczniak et al., 2012). Expression of other MADS genes in pollen, endosperm, guard cells, roots, and trichomes suggests a function in the differentiation of these organs and tissues (Alvarez-Buylla et al., 2000; Parenicová et al., 2003; Puig et al., 2013). Some plant MADS-box transcription factors are involved in the control of stress-related developmental programs, such as abscission, fruit ripening, and senescence. For example, in Arabidopsis (Arabidopsis thaliana), overexpression (OX) of AGAMOUS-LIKE15 (AGL15) was found to delay flowering, senescence, fruit ripening, and floral organ abscission, suggesting that this MADS-box transcription factor is a negative regulator of these processes (Fernandez et al., 2000; Fang and Fernandez, 2002). Similarly, FOREVER YOUNG FLOWER represses floral organ senescence and abscission in Arabidopsis (Chen et al., 2011). SHATTERPROOF1 (SHP1) and SHP2 are involved in the cell specification of the dehiscence zone in Arabidopsis fruits, where they promote the lignification of cells adjacent to this zone (Liljegren et al., 2000). In tomato (Solanum lycopersicum), the MADS domain protein JOINTLESS is necessary to specify pedicel abscission zone MADS-RIPENING, and TOMATO AGL1 controls fruit ripening (Mao et al., 2000; Vrebalov et al., 2002, 2009; Itkin et al., 2009). Nevertheless, no MADS-box gene has been yet identified in plants to have a function related to biotic or abiotic stress response regulation.
The rice genome contains 75 genes encoding MADS-box transcription factors, but the function of only few of them has been determined. Most of the studied genes are involved in the control of development, including tillering, flower development, and flowering time (Arora et al., 2007; Guo et al., 2013). Some of them are involved in development by controlling stress-related processes, such as OsMADS3, which is involved in reactive oxygen species homeostasis during anther development, and OsMADS29, which controls cell degeneration during seed development (Hu et al., 2011; Yang et al., 2012). A possible specific involvement of rice MADS genes in stress response has been reported only for OsMADS26, the rice ortholog of AGL12 (Lee et al., 2008b, 2011). In Arabidopsis, AGL12 regulates cell proliferation in the root apical meristem as well as flowering transition and was suggested to control root secondary cell wall synthesis (Tapia-López et al., 2008; Chávez Montes et al., 2014). When overexpressed in Catharanthus roseus cell suspension, AGL12 promotes cell aggregation and stimulates expression of genes involved in the biosynthesis of terpene indole alkaloids (Montiel et al., 2007). In rice, OsMADS26 OX causes a severe stress phenotype that generally leads to plant death. Expression of OsMADS26 under the control of a dexamethasone-inducible promoter provokes the differential regulation of genes involved in jasmonic acid biosynthesis and reactive oxygen species production (Lee et al., 2008b).
To precisely study the involvement of OsMADS26 in stress response in rice, we succeeded in generating viable plants overexpressing OsMADS26 and plants where OsMADS26 expression was down-regulated through RNA interference (RNAi). Our data showed that OsMADS26-down-regulated plants have no dramatic alteration of their development and were more resistant to M. oryzae and Xanthomonas oryzae pv oryzae, the main fungal and bacterial pathogens of rice. However, OsMADS26 OX increased moderately their susceptibility to these pathogens. Enhancement of recovery capacity after a severe water stress was also observed in OsMADS26-down-regulated plants. These phenotypes were further confirmed in the field, with OsMADS26 OX increasing M. oryzae susceptibility and OsMADS26 down-regulation (DR) promoting resistance against water deficit. A transcriptome analysis revealed that genes differentially regulated between control and overexpressing or down-regulated OsMADS26 plants were enriched with already known biotic and abiotic stress-related genes. Altogether, these results indicate that OsMADS26 is a major negative regulator of both biotic and abiotic stress responses in rice.
RESULTS
OsMADS26 Is Preferentially Expressed in Peripheral Tissues and Regulated by Biotic and Abiotic Stresses
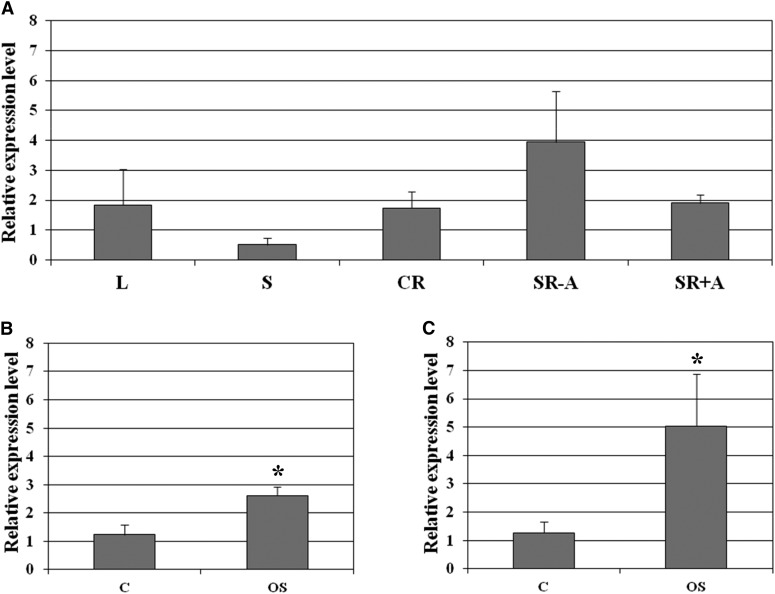
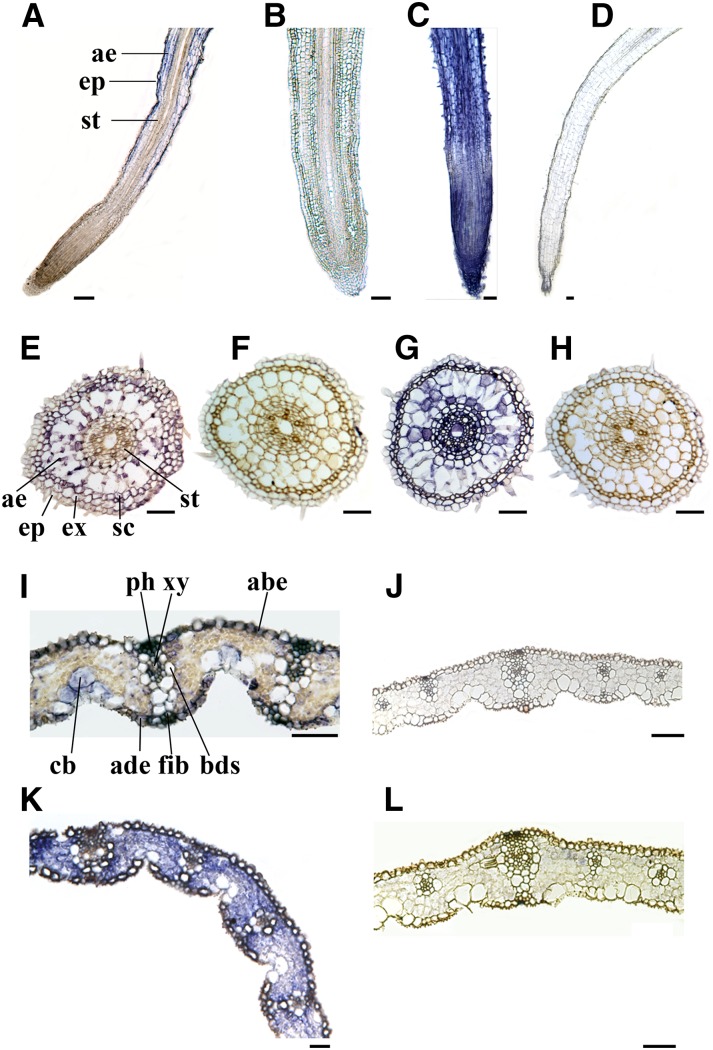
Accumulation of OsMADS26 transcripts in roots, leaves, and panicles has been previously reported (Shinozuka et al., 1999; Pelucchi et al., 2002; Arora et al., 2007) and was found to increase with organ aging (Lee et al., 2008b). To further precisely study the expression pattern of OsMADS26, we carried out quantitative reverse transcription (qRT)-PCR and in situ hybridization assays in the organs of 7-d-old rice seedlings. OsMADS26 was found to be expressed in all of the investigated organs (i.e. leaf blade, stem bases, and seminal and crown roots; Fig. 1A) in a consistent manner with regards to the available expression data (www.genevestigator.com with Os.4174.1.S1_at). In seminal roots, the expression of OsMADS26 in the 0.5-cm segment above the root tip was 2-fold higher than in the root tip itself (the 0.5-cm apical part of the seminal root; Fig. 1A). In situ hybridization specified qRT-PCR data, showing that OsMADS26 transcripts accumulate in the differentiated epidermis, exodermis, sclerenchyma, and cortical aerenchyma layers but do not accumulate in the meristematic zone of the root or the root cap (Fig. 2, A–H). OsMADS26 mRNA was not detected in the stele tissues (Fig. 2, A and E). In leaves, OsMADS26 was expressed in the epidermal cells, bulliform cells, phloem, and xylem-associated parenchyma cells (Fig. 2, I–L).
Figure 1.
OsMADS26 is expressed in shoots and roots and induced by osmotic stress. A, Expression of OsMADS26 in different organs of 7-d-old rice seedlings cultivated in standard condition (MS). CR, Crown root; L, leaf; S, stem base; SR+A, seminal root apex; SR−A, seminal root without apex. B and C, Expression patterns of OsMADS26 in root (B) and shoot (C) in standard condition (C) or under osmotic stress (OS; MS + 100 mm mannitol). Means and se were calculated from two independent experiments consisting of three technical replicates each. A Student’s t test was used to compare the relative expression level observed in standard and stress conditions. *, Significant difference with P < 0.05.
Figure 2.
OsMADS26 is expressed in differentiated peripheral tissues. In situ hybridizations were revealed with the VectorBlue Kit III. Antisense (A, E, and I) and sense (B, F, and J) OsMADS26 probe hybridizations on a longitudinal section of the root tip (A and B), a transverse section in the seminal root (E and F), and a transverse section in the third leaf (I and J) of 7-d-old rice seedling. Hybridization with antisense (C, G, and K) and sense (D, H, and L) 18S ribonucleic RNA probes, which were used as positive and negative controls, respectively. abe, Abaxial epidermis; ade, adaxial epidermis; ae, aerenchyma; bds, bundle sheath; cb, bulliform cells; ep, epidermis; ex, exodermis; fib, fiber; ph, phloem; sc, sclerenchyma; st, stele; xy, xylem. Bars = 70 µm.
To determine whether OsMADS26 expression is influenced by osmotic stress, rice seedlings were grown on culture media supplemented with 100 mm mannitol. Under these conditions, the seedling growth is reduced but not abolished (data not shown). Mannitol treatment induced the expression level of OsMADS26 in both shoot and root tissues (Fig. 1, B and C).
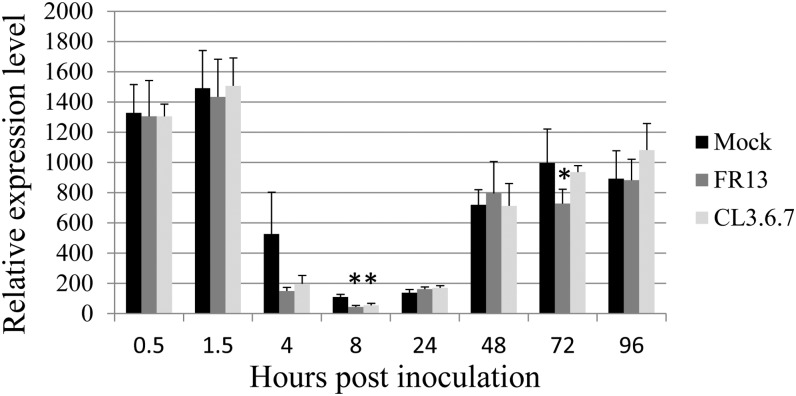
Because available microarray data indicate that OsMADS26 is slightly down-regulated late after infection (48 h postinoculation [hpi]) by the FR13 virulent isolate of the blast fungus M. oryzae (Ribot et al., 2008; Gene Expression Omnibus accession no. GSE7256), we further investigated its expression time course after inoculation with virulent and avirulent isolates (FR13 and CL3.6.7, respectively; Delteil et al., 2012) of M. oryzae (Fig. 3). We confirmed that OsMADS26 transcription is slightly repressed late after inoculation (72 hpi) with the virulent isolate FR13 but not the avirulent isolate CL3.6.7. More strikingly, OsMADS26 was strongly repressed in an early phase of infection by both isolates (4 and 8 hpi) before the fungus had penetrated into the leaf (Fig. 3).
Figure 3.
OsMADS26 expression is regulated by M. oryzae infection. Three-week-old rice seedlings of cv Nipponbare were challenged with two isolates of M. oryzae virulent FR13 and avirulent CL3.6.7 or mock treated. The expression of each gene was normalized using the actin gene as control. The mean and sd were calculated from three independent experiments. A Student’s t test was done to establish whether the relative expression level in inoculated condition was different from mock treated. *, P < 0.05; **, P < 0.01.
OsMADS26 Misregulation Does Not Strongly Affect Plant Development
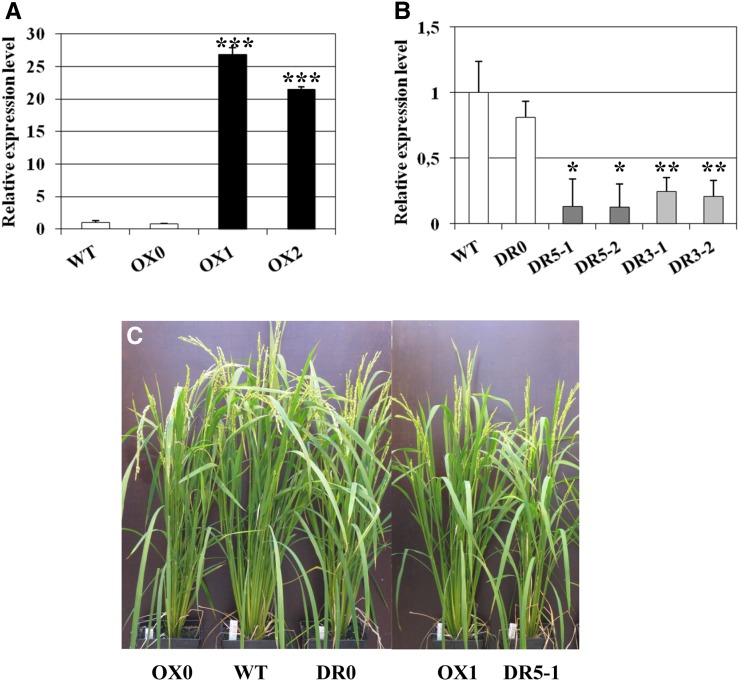
To precisely study the function of OsMADS26, we investigated the effect of its OX and its RNAi-mediated DR in rice plants. For OX, the OsMADS26 complementary DNA (cDNA) was placed under the control of the maize (Zea mays) ubiquitin1 promoter that allows high-level, constitutive expression in rice (Cornejo et al., 1993). We selected two independent, homozygous single-transfer DNA (T-DNA) copy events, OX1 and OX2, accumulating OsMADS26 transcripts at 30- and 20-fold higher levels than the control, respectively (Fig. 4A). OsMADS26 OX remained stable in further generations (Supplemental Fig. S1A). For constitutive RNAi-mediated DR of OsMADS26, two constructs specifically targeting either its 5′ untranslated region (UTR; DR5) or the 3′ UTR (DR3) region were prepared. Two independent, homozygous, single-T-DNA copy events were randomly selected for each construct (DR5-1 and DR5-2 and DR3-1 and DR3-2). A wild-type line regenerated from untransformed callus used for the transformation experiment was kept as control (the wild type). In addition, one line transformed with the empty OX T-DNA (OX0) and one line obtained by transformation with the empty RNAi T-DNA (DR0) were used as additional controls. Plantlets of these three control lines accumulated OsMADS26 transcripts at a similar level (Fig. 4, A and B). In all of the RNAi lines, OsMADS26 expression was reduced strongly and stably over the subsequent generations (Fig. 4B; Supplemental Fig. S1B) and did not respond to osmotic stress (Supplemental Fig. S1C).
Figure 4.
OX and DR of OsMADS26 do not interfere with overall plant development. A, OsMADS26 relative expression levels in 3-week-old T2 overexpressing (OX1 and OX2; black bars) and control (the wild type [WT] and OX0; white bars) plants cultivated in the greenhouse. B, OsMADS26 expression levels in RNA down-regulated (DR5-1, DR5-2, DR3-1, and DR3-2; gray bars) and control (the wild type and DR0; white bars) plants cultivated in the greenhouse. Means and se were obtained from two individual plants of each line. C, Control and transgenic OsMADS26 T2 plants cultivated in the greenhouse observed at flowering stage. A Student’s t test was done to establish whether the relative expression level in the transgenic line was different from the corresponding null segregant line. *, Significant difference with P < 0.05; **, significant difference with P < 0.01; ***, significant difference with P < 0.001.
To further establish the influence of OsMADS26 on rice development, the phenology of the transformed lines was investigated. The height of 7-d-old seedlings grown in vitro was scored. All control lines (the wild type, OX0, and DR0) exhibited similar development, whereas the heights of the OX1, OX2, DR5, and DR3 lines were significantly reduced (Table I). DR5 and DR3 plantlets were the most affected. However, 2 months after transfer in pots in the greenhouse (76 d after germination [DAG]), the average heights of OX1, OX2, DR5, and DR3 lines were similar to those of control lines, except the DR5-1 line, which still exhibited a reduced size (Table I). At the same time, all of the down-regulated lines displayed a reduction in tiller number (Table I; Fig. 4C). This was particularly significant for the DR5-2 line, which displayed a 45% reduction in the number of tillers compared with its control (DR0; Table I). The dry weights of the aerial parts of the DR plants, especially the two DR5 lines, were lower than those of the control and OX plants (Table I). The two DR3 lines also exhibited significant delay of 3 to 4 d in flowering (Table I). No significant difference for these two traits was observed among the rest of the lines. Total weight and 1,000-seed weight of the main panicle were comparable in all of the lines studied (Table I). In summary, although the overexpressing and down-regulated OsMADS26 lines exhibited a retarded growth at early stages of development after germination, further transfer and growth in the greenhouse allowed them to recover and exhibit a performance generally similar or close to that of control plants. The weak impact of constitutive OsMADS26 OX or DR on plant development was confirmed in the field, where we observed only a reduced height for the OX2 line and a higher biomass and yield for the DR3-1 line compared with their relative controls (Supplemental Fig. S2).
Table I. Plant phenotype of control and transgenic OsMADS26 lines after 7 d of in vitro culture (MS), 72 DAG in greenhouse, and from flowering to harvest.
Results shown are from one of two independent biological repetitions that produced similar results. Reported values are the means and se for three individual plants of each line. A Student’s t test was done to establish whether the parameter measured in transgenic lines was different from corresponding control lines. BEG, Flowering beginning; DW, plant dry weight after seed harvesting; FD, flowering date; HTG_7, plant height measured at 7 DAG; HTG_76, plant height measured at 76 DAG; P1,000:, weight of 1,000 seeds; PW, panicle weight; TIL_76, number of tillers counted at 76 DAG. Significant difference: *, P < 0.05, **, P < 0.01; and ***, P < 0.001.
| Line name | HTG_7 | HTG_76 | TIL_76 | BEG | FD | DW | PW | P1,000 |
|---|---|---|---|---|---|---|---|---|
| cm | DAG | g | ||||||
| Wild type | 6.06 ± 1.51 | 97.53 ± 0.59 | 12.33 ± 0.33 | 80.33 ± 0.33 | 81.67 ± 0.88 | 9.74 ± 2.34 | 15.18 ± 2.45 | 21.80 ± 0.71 |
| OX0 | 6.34 ± 1.33 | 97.47 ± 2.06 | 10.67 ± 0.33 | 82.33 ± 1.20 | 84.00 ± 1.00 | 8.73 ± 0.87 | 9.52 ± 0.95 | 20.34 ± 0.62 |
| DR0 | 6.72 ± 1.27 | 95.23 ± 1.36 | 11.33 ± 1.33 | 81.67 ± 0.67 | 83.67 ± 0.67 | 7.96 ± 3.80 | 8.88 ± 4.28 | 17.89 ± 3.93 |
| OX1 | 3.84 ± 0.67** | 100.60 ± 2.17 | 10.33 ± 1.45 | 80.00 ± 1.53 | 81.67 ± 1.86 | 8.00 ± 1.42 | 8.78 ± 1.50 | 21.39 ± 0.30 |
| OX2 | 2.41 ± 0.92*** | 93.40 ± 2.84 | 12.33 ± 0.88 | 83.67 ± 0.88 | 86.00 ± 1.00 | 8.21 ± 1.12 | 8.93 ± 1.34 | 20.38 ± 0.72 |
| DR5-1 | 1.68 ± 0.68*** | 87.90 ± 2.51* | 7.80 ± 2.08 | 83.00 ± 1.15 | 85.67 ± 0.67 | 3.86 ± 1.07 | 4.21 ± 1.14 | 16.32 ± 0.48 |
| DR5-2 | 1.61 ± 0.29*** | 95.37 ± 1.84 | 6.67 ± 0.67* | 82.33 ± 0.67 | 85.00 ± 0.00 | 4.93 ± 0.40 | 5.48 ± 0.39 | 19.79 ± 1.15 |
| DR3-1 | 1.61 ± 0.31*** | 90.53 ± 1.79 | 9.67 ± 1.33 | 85.00 ± 0.00** | 87.00 ± 0.58** | 6.62 ± 1.37 | 7.33 ± 1.65 | 21.42 ± 0.73 |
| DR3-2 | 0.84 ± 0.18*** | 97.20 ± 1.73 | 9.00 ± 1.00 | 84.67 ± 0.33** | 86.33 ± 0.33* | 7.76 ± 0.73 | 8.41 ± 0.67 | 20.01 ± 0.68 |
OsMADS26 Is Required for Resistance against Blast Fungus and Bacterial Blight
Because OsMADS26 was found to be a stress-related gene in rice (Lee et al., 2008b, 2011), we further evaluated the response of the OsMADS26 transgenic lines to pathogen infection.
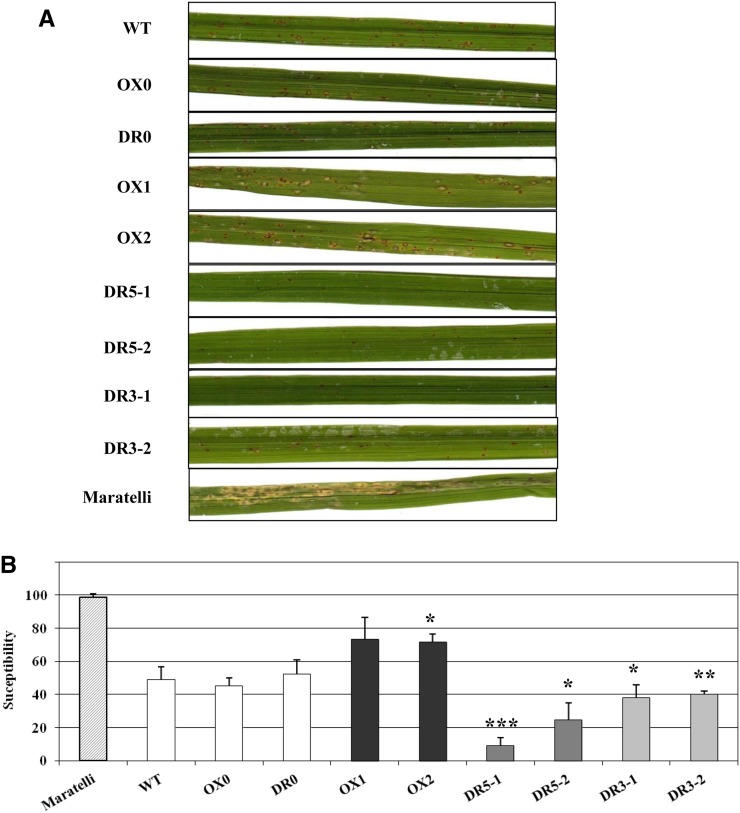
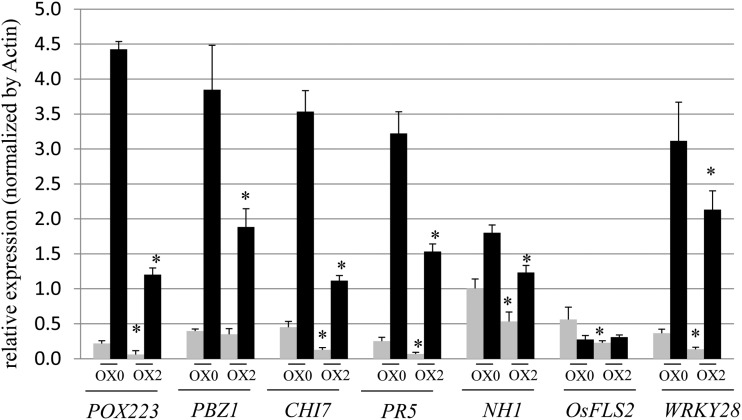
First, plantlets of the different OsMADS26 lines were inoculated with the moderately virulent fungal isolate GUY11 of M. oryzae (Delteil et al., 2012). This isolate triggers lesions in the leaf blade of cv Nipponbare consisting of an average of 50% grayish lesions surrounded by brown margins that are characteristic of successful invasion of the fungus (disease). The others are small and dark spots characteristic of unsuccessful invasion events (the wild type, OX0, and DR0 plants in Fig. 5A). Differences in the degree and development of disease symptoms caused by M. oryzae between transformed and untransformed plants were clearly visible at 7 d postinoculation (dpi; Fig. 5A). The two overexpressing lines (OX1 and OX2) presented more disease symptoms compared with the controls (the wild type and OX0). In contrast, all of the down-regulated lines displayed many small and dark spots characteristic of resistance and very few disease symptoms. These observations were further confirmed by calculating the percentage of susceptible lesion versus the total number of observed lesion on each infected leaf (Fig. 5B). Thus, this suggested that OsMADS26 negatively regulates blast resistance. In addition, the susceptibility to M. oryzae of OX0, OX2, and DR3-1 lines was challenged in a nethouse in Vietnam on 10-week-old plants inoculated with the VT15 Vietnamese isolate virulent on cv Nipponbare (Supplemental Fig. S3). In this experiment, the number of susceptible lesions was significantly higher in OX2 line and slightly lower in DR3-1 line than in the control (OX0), confirming the opposite phenotypes observed for overexpressing and down-regulated OsMADS26 lines. The expression of a set of selected major defense-related genes, PEROXIDASE22.3 (POX22.3; Vergne et al., 2007), CHITINASE7 (CHI7; Kaku et al., 2006), PATHOGENESIS-RELATED PROTEINS5 (PR5), NONEXPRESSOR OF PATHOGENESIS-RELATED1 HOMOLOG1 (NH1; Chern et al., 2005), (FLAGELLIN SENSING2), OsWRKY28, and PROBENAZOLE-INDUCIBLE1 (PBZ1; Delteil et al., 2012), was examined in OX2 lines 2 dpi with M. oryzae GY11 isolate or mock treatment (Fig. 6). This showed that, in mock-treated and inoculated plants, the expression of most of these genes (POX223, CHI7, PR5, NH1, FLS2, and WRKY28) was significantly reduced in the OX2 line compared with OX0 before and/or after infection. This result suggests that OsMADS26 acts as a negative regulator of defense gene expression.
Figure 5.
OsMADS26 negatively regulates resistance against M. oryzae. OX (OX1 and OX2; black bars) and DR (DR5-1, DR5-2, DR3-1, and DR3-2; gray bars) OsMADS26 lines and corresponding control lines transformed with empty vectors or untransformed lines (OX0, DR0, and the wild type [WT]; white bars) and cv Maratelli, a highly susceptible cultivar, were tested. A, Symptom severity in leaves of transgenic and control plants inoculated with the GUY11 strain of M. oryzae. Photographs were taken 7 dpi. B, Percentage of susceptible versus total lesions observed in M. oryzae-infected leaves 7 dpi. Means and se were from 10 inoculated plants for each line. Results shown are from one of two independent experiments that produced similar results. A Student’s t test was done to establish whether one given transgenic line was different from its corresponding null segregant line. *, Significant difference with P < 0.05; **, significant difference with P < 0.01; ***, significant difference with P < 0.001.
Figure 6.
Expression of defense genes is down-regulated in OsMADS26 OX before and after infection by M. oryzae. Three-week-old rice seedlings of OsMADS26 overexpressing (OX2) line and control line (OX0) were challenged with the moderately virulent isolates of M. oryzae GY11 (black bars) or mock treated (gray bars). The RNA was extracted at postinoculation. The expression of each gene was normalized using the actin gene as control. The POX223, PBZ1, CHI7, and PR5 genes are coding for pathogenesis-related proteins used as classical markers of defense. The NH1, OsFLS2, and WRKY28 genes are coding for regulator proteins of defense in rice. The means and sd were calculated from three independent experiments. A Student’s t test was done to establish whether the relative expression level in the OX2 lines was different with the line used as control. *, P < 0.01.
Second, to evaluate whether constitutive deregulation of OsMADS26 affects the susceptibility to a bacterial pathogen, we challenged the overexpressing and down-regulated OsMADS26 lines with X. oryzae pv oryzae. Similar data were obtained for resistance to bacterial blight X. oryzae pv oryzae as with M. oryzae. In this case, the length of the necrotic and yellowing zone extending from the wounded extremity of the infected leaves was measured 14 dpi. The symptoms had a significantly higher severity for OX1 and OX2 lines compared with the control lines (Supplemental Fig. S4, A and B). Conversely, the symptoms developed by down-regulated lines (DR5-1, DR5-2, DR3-1, and DR3-2) were limited to a short necrosis just below the inoculation zone (Supplemental Fig. S4, A and B), suggesting that these lines were strongly resistant to X. oryzae pv oryzae and supporting a negative role of OsMADS26 on blight resistance.
Third, we tested whether the response to the Rice Yellow Mottle Virus (RYMV; Kouassi et al., 2005) could be affected by OsMADS26 OX or DR. We did not observe any difference in the development of symptoms or virus accumulation between the overexpressing lines, the down-regulated lines, and their respective controls (Supplemental Fig. S5), suggesting that misregulation of OsMADS26 expression had no impact on the resistance against RYMV.
OsMADS26 Inhibition Favors Plant Tolerance against Drought Stress
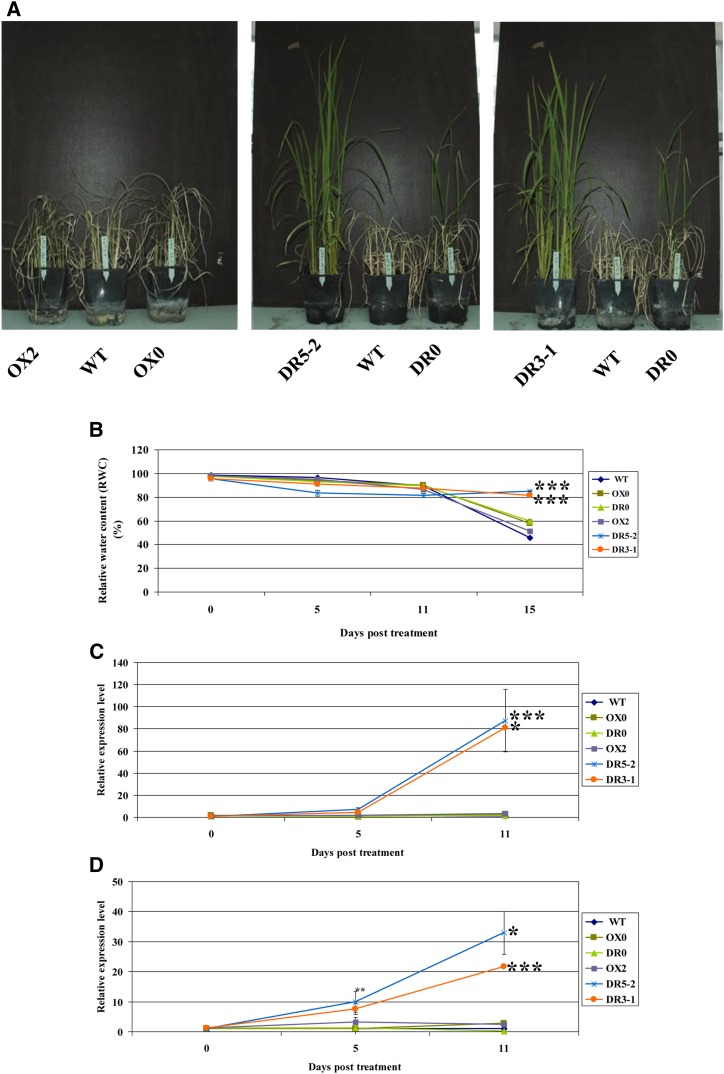
Because mannitol stress induces the expression of OsMADS26 (Fig. 1, B and C), we investigated the tolerance of overexpressing and down-regulated lines to the drought stress. After the drought stress, plants were rewatered for a period of 2 weeks to allow recovery. Although plants of all of the control and OsMADS26-overexpressing lines were mostly wilted and died, OsMADS26-down-regulated plants fully recovered from the water stress (Fig. 7A).
Figure 7.
OsMADS26 negatively regulates water stress tolerance. Six independent lines: overexpressing (OX2) or down-regulated (DR5-2 and DR3-1) OsMADS26 and corresponding control lines transformed with empty vectors (OX0 and DR0) or the wild type (WT) were used for this experiment. A, Drought stress was applied on 20-d-old plants growing in the greenhouse in pots by stopping watering during 18 d followed by 15 d of rewatering. The pictures were taken 15 d after rewatering. B, RWC of plants was measured on the last expanded leaf before and at 5, 11, and 15 d after watering stopping. Mean values and se were calculated from five individual plants for each line. C and D, qRT-PCR expression analysis of drought- and salt-responsive rice genes RAB21 (C) and SALT (D) in control and transgenic plants before and during drought stress. RNA were extracted from leaves of two plants of each line that had closest RWCs. We did not measure gene expression 15 d after the water deficit period, because the control and MADS26-overexpressing plants were already highly damaged. Means and se were from two individual plants for each line. A Student’s t test was done to establish whether the RWC or the gene expression level in transgenic lines was different from corresponding control line. *, Significant difference with P < 0.05; **, significant difference with P < 0.01; ***, significant difference with P < 0.001.
All of the lines exhibited at the beginning of the experiment a similar relative water content (RWC; nearly 95%) that decreased to approximately 85% after 11 d of water deficit (Fig. 7B). However, 15 d after water deprivation, the leaf RWCs of all of the control and OsMADS26-overexpressing lines dropped to a 47% to 62% range, whereas the two OsMADS26-down-regulated lines maintained significantly higher RWCs, falling within an 81% to 84% range. This suggests that the inhibition of OsMADS26 expression enhances the capacity of the rice plant to maintain its water content under water deficit.
The expression of two drought-responsive genes was analyzed: RESPONSIVE TO ABA21 (RAB21), a rice dehydrin and SALT STRESS-INDUCED PROTEIN (SALT; Claes et al., 1990; Oh et al., 2005). Their expression levels were similar in all lines before or 5 d after the water stress. After 11 d of water stress, however, their expression was significantly higher in the two OsMADS26-down-regulated lines compared with control and OsMADS26 OX lines (Fig. 7, C and D). This suggests that OsMADS26 may play a negative role in the regulation of some drought stress-responsive genes in response to water deficit.
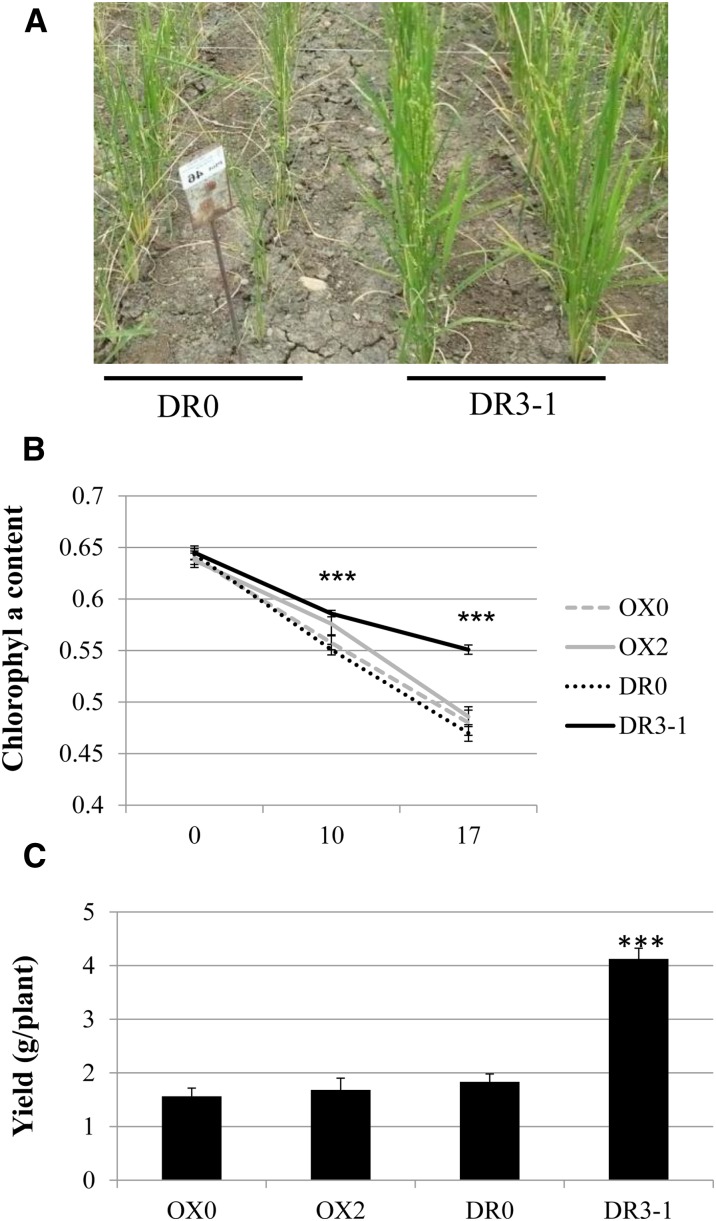
In addition, we challenged in the field the capacity of OX0, OX2, DR0, and DR3-1 lines to tolerate water deficit. The DR3-1 line presented a much better tolerance to water deficit conditions associated with a slower decrease of chlorophyll-a content and a better capacity to maintain yield under drought than the other lines (Fig. 8). Other measurements (leaf rolling, chlorophyll content, and biomass) confirmed that DR3-1 plants had an increased capacity to sustain drought stress (Supplemental Fig. S6). This confirmed that a constitutive DR of OsMADS26 increases the capacity of the plant to tolerate water deficit.
Figure 8.
OsMADS26 DR confers tolerance to water deficit under field conditions. Plants were grown in the field at the International Center for Tropical Agriculture, and a drought stress was applied (“Materials and Methods”). The shape of the plant 17 d after stress is shown (A), and the chlorophyll fluorescence (B) was measured at the indicated times after stress in three independent blocks on three plants. Yield was measured at the end of the experiment (C). The means and sd are shown, and a Student’s t test (n = 9) was used to evaluate statistical difference between the overexpressing OX2 and down-regulated DR3-1 transgenic lines with their respective controls OX0 and DR0. ***, P < 0.001.
Transcriptome Profiling of OsMADS26-Overexpressing and -Down-Regulated Lines
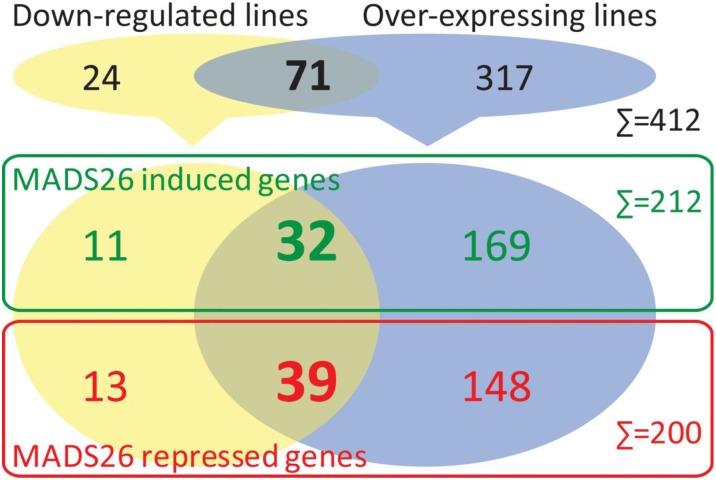
Preliminary evidence of altered expression of stress-related genes in OsMADS26-overexpressing and down-regulated lines led us to further identify the pathways potentially regulated by OsMADS26 through transcriptome profiling. Transcriptome profiles were established from two independent biological replicates per line. Genes significantly and reproducibly induced or repressed (fold change > 2 and P value ≤ 0.05) across lines and replicates compared with their values in the appropriate controls were selected for further analysis (for more information, see “Materials and Methods”). We finally selected genes at least one time inversely regulated in OX compared with DR lines or reproducibly overexpressed or repressed in OX or control lines. To compare our results with other available data, we converted the rice probes into Michigan State University's Rice Genome Annotation Project transcriptional units (Supplemental Table S1). This represented a total of 400 nonredundant genes. A total of 71 nonredundant genes presented an inverted regulation profile in OX and DR lines (Fig. 9; Supplemental Table S1). Overall, 212 genes were down-regulated in DR lines and/or up-regulated in OX lines. These genes should belong to pathways induced by OsMADS26. On the contrary, 200 genes were up-regulated in DR lines and/or down-regulated in OX lines. These genes should belong to pathways inhibited by OsMADS26.
Figure 9.
Genome-wide gene expression regulations in OsMADS26-overexpressing or -down-regulated lines. Number of genes significantly differentially expressed in the microarray experiment; 71 (32 and 39, respectively) genes presented an inverted regulation profile in OX and DR lines. Green and red colors depict genes induced or repressed, respectively, by OsMADS26 expression.
We then looked for overlaps between a set of >6,800 probes that were known to be transcriptionally regulated upon pathogen infection (Vergne et al., 2008) and the 400 genes that were significantly misregulated in DR and/or OX lines (Supplemental Table S1). We found that 53% of the 200 genes up-regulated in DR and/or down-regulated in OX lines are known to be transcriptionally regulated during pathogen challenge, whereas only 30% were expected by chance in a random selection of 2,000 genes (P < 0.001 as evaluated with a χ2 test; Vergne et al., 2008). In contrast, there was no such enrichment in the 212 genes up-regulated in DR lines and/or down-regulated in OX lines. Thus, OsMADS26 seems to down-regulate the transcription of a large number of genes known to be involved in disease resistance. Similarly, a large proportion (41%) of genes misregulated in OsMADS26 lines was found in a previously published drought data set (Minh-Thu et al., 2013). The extent of this overlap is proportional to the one observed with genes found to be deregulated in dexamethasone-inducible OsMADS26 lines (39%; Lee et al., 2008b). Our analysis thus resulted in a list of putative OsMADS26 target genes that may be involved in the regulation of biotic or abiotic stress resistance.
DISCUSSION
Alteration of OsMADS26 Expression Does Not Deeply Affect cv Nipponbare Plant Development
The OsMADS26-overexpressing lines presented a delayed development at the seedling stage, but their development in the greenhouse and field was almost similar to the development of control plants aside from a slight reduction in tiller number (Table I). This contrasts with the previous study by Lee et al. (2008b), which reported that OX of OsMADS26 driven by the same constitutive promoter triggered several dramatically abnormal developmental phenotypes, including anthocyanin accumulation or lethality. A tentative explanation might be in the use of different genetic backgrounds (cv Nipponbare versus cv Dongjin) for expressing OsMADS26. To our knowledge, there is at least one report where OX in different rice genetic backgrounds resulted in the opposite effects (Tao et al., 2009). Alternatively, it is possible that our transformation procedure (Sallaud et al., 2003) that differs from that used by Lee et al. (2008b) has counterselected plants presenting a severe reduction of their development or lethality because of very high levels of expression. Although we cannot explain the strong phenotypic differences between our overexpressing lines and the lines analyzed by Lee et al. (2008b), these differences may explain at least in part why we found little overlap between our experiments and their microarray experiments (16 genes in total; see below). Similarly, except for a delay in development observed at early stages, the overall development of the down-regulated lines was not strongly modified (Table I).
OsMADS26 Is a Negative Regulator of Both Biotic and Abiotic Stresses
Our data showed that OsMADS26-down-regulated lines displayed decreased susceptibility to two major pathogens of rice (Fig. 5; Supplemental Figs. S3 and S4) as well as an increased water deficit tolerance and a better recovery capacity after a drought stress (Figs. 7 and 8; Supplemental Fig. S2). The observation of consistent phenotypes in the OsMADS26-down-regulated lines obtained with two independent constructs targeting 5′ or 3′ UTR reduces the risk of misinterpretation related to transinterference with transcripts of other genes. Because the observed phenotypes are similar between the different down-regulated lines, we can assume that they are the consequence of a specific degradation of OsMADS26 mRNAs.
Up to 60% and 40% average disease symptom reductions were observed in down-regulated lines inoculated with X. oryzae pv oryzae and M. oryzae, respectively (Fig. 5; Supplemental Fig. S4). This corresponds to a high level of disease reduction compared with the range attained in transgenic lines obtained through misregulation of a set of defense-associated genes (Delteil et al., 2010). Consistently, an increased susceptibility of OsMADS26 OX lines to M. oryzae was also observed in the nethouse experiments, whereas the tested OsMADS26-down-regulated lines presented a reduction of susceptible lesions compared with the DR0 control (Supplemental Fig. S3). This shows that the negative regulation of OsMADS26 on the resistance mechanisms to M. oryzae can be observed at different developmental stages, with different virulent isolates and independent of the growth conditions. It is interesting to stress that there is a coincidence between the tissue localization of OsMADS26 transcripts and the cell barriers that pathogens have to cross in the plant (Fig. 2). For instance, OsMADS26 is expressed in the epidermis, a barrier that M. oryzae has to cross to perform its lifecycle. Transcripts of OsMADS26 also accumulated in cells around the vessels where X. oryzae pv oryzae develops. To our knowledge, this is the first report of the involvement of a MADS gene in disease resistance in plants. The resistance of rice against RYMV was not affected by OsMADS26 DR. Resistance against bacteria and fungi on the one hand and virus on the other hand involves different mechanisms, such as RNA silencing for the latter and pathways producing antimicrobial molecules for the former. Thus, OsMADS26 negatively participates in resistance to a wide range of rice pathogens but not to RYMV.
Other than this strong effect on biotic stress resistance, the OsMADS26-down-regulated lines showed an increased ability to maintain their RWCs under soil water deficit and recover from a severe drought stress as well as a better capacity to maintain yield in drought conditions in the field (Figs. 7 and 8; Supplemental Fig. S6). The preferential localization of OsMADS26 transcripts (Fig. 2) in peripheral tissues, such as epidermis and bulliform cells in leaves and exodermis in roots, supports a role for this transcription factor in the response mechanism to environmental clues. To our knowledge, OsNAC6 (for NAM [no apical meristem], ATAF [Arabidopsis transcription activation factor], CUC [cup-shaped cotyledon]) and OsNAC10 are the only transcription factors for which the deregulation had a joint benefit on both biotic and abiotic stress tolerances (Nakashima et al., 2007; Sun et al., 2013). OsNAC6-overexpressing rice plants showed an improved tolerance to dehydration and high-salt stresses as well as increased tolerance to blast disease. However, constitutive overexpressers also exhibit growth retardation and low reproductive yields in contrast to OsMADS26-down-regulated lines that presented only discrete developmental changes.
OsMADS26 Alters the Transcription of a Wide Range of Biotic and Abiotic Stress-Related Genes
We showed that the expression of a set of defense genes is lower in OX OsMADS26 lines than in the control before and after inoculation with a virulent isolate of M. oryzae (Fig. 6). This was confirmed by microarray analysis (Supplemental Table S1), where several other genes coding for pathogenesis-related proteins were down-regulated in OX OsMADS26 lines. Similarly, the expression of a set of drought resistance-related genes is higher in OsMADS26 DR lines after the application of a water deficit (Fig. 7). This suggests a direct or indirect involvement of OsMADS26 as a repressor of stress-responsive genes.
By using transcriptome analysis, we investigated whether the modified response to biotic and abiotic stresses was associated with a more global differential expression of stress-related genes before application of the stress itself. Using the Archipelago database referencing genes in rice involved in disease resistance (Vergne et al., 2008) or the drought-responsive genes data set (Minh-Thu et al., 2013), we could establish that a large proportion of the genes differentially regulated in down-regulated and overexpressing lines is known to be regulated by biotic (53%) or abiotic (41%) stresses. This was similar (49% and 39%, respectively) to what was found by Lee et al. (2008b) after dexamethasone-induced OX of OsMADS26. Thus, these transcriptome analyses show that OsMADS26 participates in the transcriptional regulation of defense-related genes. The low overlap with the data set obtained by Lee et al. (2008b) probably reflects the fact that we determined the genes regulated at steady-state levels after constitutive OX or DR of OsMADS26 expression, whereas Lee et al. (2008b) identified the genes deregulated upon a sudden increase of OsMADS26 transcription triggered by the dexamethasone induction treatment. Based on their transcriptome analysis, Lee et al. (2008b) stressed that OsMADS26 may be involved in the regulation of genes involved in jasmonate and ethylene stress hormone biosynthesis. Here, we found that rice LIPOXYGENASE8 (Os08g39840) is consistently up-regulated in DR lines and down-regulated in both OX OsMADS26 lines and dexamethasone-induced OsMADS26 lines (Lee et al., 2008b). This gene was reported to be regulated during the early stage of M. oryzae infection (Peng et al., 1994; Agrawal et al., 2004), by wounding (Marla and Singh, 2012), and during the senescence process (Kong et al., 2006). Two genes involved in ethylene biosynthesis, rice 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE3 (Os09g27750) and ACIREDUCTONE DIOXYGENASE1 (Os10g28350), are down-regulated in OX OsMADS26 lines. Rice 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE3 and ACIREDUCTONE DIOXYGENASE1 are strongly up-regulated by ethylene and contribute to maintain elevated ethylene rate in stressed plants (Rzewuski and Sauter, 2008). Similarly, the ethylene-responsive ETHYLENE RESPONSE FACTOR063 transcription factor (Os09g11480; Ma et al., 2013) was found to be down-regulated in OX OsMADS26 lines, suggesting that these lines are impaired for ethylene biosynthesis and response.
Other stress-related transcription factors were found to be differentially regulated in OX and/or DR OsMADS26 lines. OsNAC103 (Os07g48450), known to be up-regulated by water deficit treatment, salt stress, and jasmonate (Fang et al., 2008; Nuruzzaman et al., 2012) was found to be up- and down-regulated in DR and OX lines, respectively. OsNAC045 (Os11g03370) down-regulated in OX lines is up-regulated in response to salt or cold stress (Fang et al., 2008). OsWRKY24 (Os01g61080) represses abscisic acid and GA signaling in aleurone cells (Xie et al., 2005; Zhang et al., 2009) and is induced by chilling stress (Yun et al., 2010). It is up-regulated in DR lines and down-regulated in OX lines. OsWRKY53 (Os05g39720), down-regulated in OX lines, is induced by elicitors, jasmonate, and M. oryzae infection and during the Xa21-mediated resistance to X. oryzae pv oryzae. Its OX enhances rice resistance to M. oryzae (Chujo et al., 2007, 2014). Interestingly, we identified that RH1 (Os05g30500) is up-regulated in the OX line. RH1 is a Negative Regulator of Resistance homolog that can interact with and inhibit NH1/OsNONEXPRESSOR OF PATHOGENESIS-RELATED1, which is a master regulator of defense genes and systemic acquired resistance (Chern et al., 2012). The Wall-Associated Kinase25 (Os03g12470) was down-regulated in OX plants. This is consistent with the published function of this gene as a positive regulator of X. oryzae pv oryzae resistance (Seo et al., 2011). Finally, the rice ROOT MEANDER CURLING (Os04g56430) receptor-like kinase known to be highly induced by salt treatment (Serra et al., 2013) was up-regulated in DR plants and down-regulated in OX plants. Whether OX or DR OsMADS26 plants are more resistant to salt stress remains to be established.
Taken together, this shows that OsMADS26 contributes to the regulation of several stress-related transcriptional and regulatory pathways and that its OX or DR impacts on the expression of a wide range of biotic and abiotic defense-related genes, which is consistent with the observed phenotypes of DR and OX lines.
Is OsMADS26 a Hub for Stress Resistance Regulation in Plants?
Our data indicate that OsMADS26 probably mainly acts as a negative regulator of stress response. This has also been reported for OsMADS22 and OsMADS55, which act as negative regulators of the brassinosteroid response (Lee et al., 2008a). Whereas the DR of OsMADS26 transcription upon rice blast infection (Fig. 3), irrespective of the virulence of the isolate, can constitute a basal defense response, its up-regulation during osmotic stress (Fig. 1) is more difficult to interpret. We propose that this up-regulation of OsMADS26 could be part of a negative feedback loop that would dampen abiotic stress response.
Nevertheless, it cannot be excluded that OsMADS26 might have both activating and inhibiting activity on stress response genes depending on posttranslational modifications or interaction with other regulatory proteins. Indeed, MADS-box proteins are combinatorial transcription factors, and their regulatory specificity is affected by the interaction with other DNA binding or accessory factors (Messenguy and Dubois, 2003). In this context, OsMADS26 could be a hub that integrates different signals, contributes to a short-term activation of defense mechanisms, and becomes, afterward, partly responsible for their cancellation. In this respect, it will be interesting to identify the proteins that can interact in vivo with OsMADS26.
CONCLUSION
Our data show that OsMADS26 is a negative regulator of different stresses of major agronomical importance in rice. They also represent the description of a new range of functions for MADS genes in plants and open the door toward the achievement of drought-tolerant and disease-resistant plants. To reach this goal, it will be very interesting to identify in rice tilling population plants with OsMADS26 null alleles and test their resistance against stresses. These alleles could be introduced in future breeding programs.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Dehulled and surface-sterilized seeds of rice (Oryza sativa) ‘Nipponbare’ were incubated in sterile distilled water in a growth chamber (16 h of light per day; 500 µE m−2 s−1; 28°C/25°C day-night cycle) for 2 d at 25°C. Imbibed seeds were transferred in square petri dishes (245 × 245 mm; CORNING; seven seeds per dish) containing 250 mL of one-half-strength Murashige and Skoog (DUCHEFA) medium (MS) solidified with 8 g L−1 agarose type II (SIGMA). These dishes were transferred and placed vertically in a growth chamber at 28°C under 16 h of light. Roots and shoots of 7-d-old seedlings were collected and used for in situ hybridization and RNA isolation for qRT-PCR or transcriptome analyses. Salt and osmotic stresses were applied by supplementing the culture medium with 150 mm NaCl (DUCHEFA) or 100 mm mannitol (DUCHEFA), respectively.
Plants were grown in 3-L pots filled with EGO 140 Soil Substrate (TREF; www.Trefgroup.com) in a containment greenhouse (16-h-light/8-h-dark cycles at 28°C–30°C). For plant phenotyping, the plants belonging to the different lines were randomly distributed in the greenhouse. Twenty DAG, plant height and tiller number were measured once a week until the early flowering stage. The latter stage was defined as the date when the first spike emerges from the flag leaf sheath on a plant. The flowering date corresponds to the date when spikes are observed on 50% of the tillers of a plant. After harvesting, the dry weight of the aerial part of the plant part was determined after drying the plant tissues at 70°C for 96 h. Panicles of each plant were also individually weighted after a drying treatment at 37°C for 3 d. The 1,000-seed weight was evaluated using seeds borne by the master tiller panicle. This experiment was repeated twice using three plants per line.
Specific culture conditions used for evaluation of pathogen and drought tolerance are detailed in the corresponding sections.
Plasmid Construction for Plant Transformation
The isolation of OsMADS26 (Os08g02070) cDNA from rice ‘Nipponbare’ was achieved by qRT-PCR. Total RNA was extracted from 100 mg of leaf tissue of 7-d-old seedlings grounded in liquid nitrogen using 1 mL of TRIzol (INVITROGEN) following the recommendation of the supplier. A PCR amplification was performed with a couple of specific primers designed in the 5′ and 3′ UTR of OsMADS26 (Supplemental Fig. S7). The amplified cDNA was cloned using the pGEM-T Easy Cloning Kit (Promega). From the cDNA, further PCR reactions were done using specific primers to amplify a 215-bp fragment located in the 5′ UTR of OsMADS26 named gene sequence tag1 (GST1) and a 321-bp fragment comprising the end of the last exon and the major part of the 3′ UTR region named GST2 (Supplemental Fig. S7). PCR cycling conditions were 94°C for 4 min (1 cycle) and 94°C for 1 min, an annealing step at various temperatures depending on the melting temperature of the primers used (typically melting temperature of −5°C) for 1.5 min, and 72°C for 1 min (35 cycles) with a 5-min final extension step at 72°C. PCR was performed in a final volume of 25 µL with 0.25 units of Taq polymerase in MgCl2-free buffer (PROMEGA), 2 mm MgCl2, 200 nm each dNTP, appropriate oligonucleotides (1 µm), and cDNA (2 µL) or pGEM-T-PC8 plasmid (10 ng). The BP-tailed OsMADS26-amplified cDNA was cloned with the Gateway BP recombinase (INVITROGEN) in a modified pCAMBIA 1300 binary vector for OX named PC5300.OE, where the Ccdb gene surrounded by the BP recombination sites was cloned between the constitutive promoter of ubiquitin gene from maize (Zea mays) and the terminator of the nopaline syntase gene from Agrobacterium tumefaciens (J.C. Breitler, unpublished data). After cloning, the presence of the OsMADS26 cDNA in frame was ascertained by sequencing. The plasmid named PC5300.OE-PC8 was transferred into A. tumefaciens strain EHA105. For RNAi, the BP-tailed amplified GST1 or GST2 was cloned by BP recombination in the pDON207 Entry Plasmid (INVITROGEN) and transferred with the LR Recombinase (INVITROGEN) in the small interfering RNA binary plasmid pANDA (Miki and Shimamoto, 2004). The insertion of the GSTs in pANDA was controlled by sequencing. The resulting plasmids, named pANDA-DR5 and pANDA-DR3, were mobilized into A. tumefaciens strain EHA105 for plant transformation.
Transgenic plants were obtained by coculture of seed embryo-derived callus with A. tumefaciens strain EHA105 carrying the adequate binary plasmids following the procedure detailed in Sallaud et al. (2003). Single-locus and homozygous T2 lines were selected on the basis of the segregation of the antibiotic resistance gene carried by the T-DNA and Southern-blot analysis.
The expression of OsMADS26 in selected transgenic lines was analyzed by qRT-PCR using specific primers (Supplemental Table S1).
Real-Time qRT-PCR Analysis
Total RNA was extracted from 100 mg of grounded leaf tissues with 1 mL of TRIzol (INVITROGEN) following the recommendation of the supplier; 2 µg of RNA was treated by RQ1 DNAse (PROMEGA) to remove residual genomic DNA. The first strand cDNA synthesis was performed in 20 µL of final volume using the kit Superscripts III (INVITROGEN) following the manufacturer’s instructions.
For qRT-PCR analysis, specific forward and reverse primers were designed to amplify a fragment of 200 to 400 bp in the 3′ UTR of each studied gene using the Vector NTI (version 10.1) software with default parameters. Primer sequences are given in Supplemental Table S2. qRT-PCR was performed with a LighCycler 480 (ROCHE) using the SYBR Green Master Mix (ROCHE). The reaction was carried out in 96-well optical reaction plates (ROCHE). The reaction mix contained 7.5 µL of SYBR Green QPCR Master Mix (ROCHE), 250 nm each primer (forward and reverse), and 3 µL of 10-fold diluted cDNA template. All reactions were heated to 95°C for 5 min followed by 45 cycles of 95°C for 10 s and 60°C for 30 s. Melt curve analysis and gel electrophoresis of the PCR products were used to confirm the absence of nonspecific amplification products. The primer efficiencies observed for the couples of primers used ranged between 1.86 and 2.05. Transcripts from Expressed Protein (EXP; Os06g11070) or actin (Os03g50890) genes were also detected and used as an endogenous control to normalize expression of the other genes. EXP or actin was chosen as the reference gene, because their expression seemed to be the most stable in different tissues and physiological conditions (Caldana et al., 2007). We verified that, in all of our experiments, the threshold cycle (Ct) value of the EXP and actin genes remained stable, irrespective of the treatment applied to the plants, and ranges between 26 and 28. Relative expression level was calculated by subtracting the Ct values for EXP or actin from those of the target gene (to give ΔCt) and then ΔΔCt and calculating 2−ΔΔCt (Giulietti et al., 2001). Reactions were performed on technical triplicates from duplicated biological experiments.
In Situ Hybridization
For OsMADS26 probe preparation, we used the same primers designed for OsMADS26 qRT-PCR amplification (Supplemental Table S1). A 18S ribosome coding sequence was used as positive hybridization control and PCR amplified from cDNA using the primer couple Rib-Up (5′-CCGACCCTGATCTTCTGTGAAGGG-3′) and Rib-Down (5′-CAAGTCAGACGAACGATTTGCACG-3′). Primers containing the above specific sequences but extended at their 5′ ends with the T7 RNA polymerase promoter sequence (5′-GCGAAATTAATACGACTCACTATAGGGAGA-3′) were also designed and named OsMADS26-T7-Up, OsMADS26-T7-Down, RibT7-Up, and RibT7-Down. Finally, one primer corresponding to the T7 end was also designed and named E-T7 (5′-GCGAAATTAATACGACTCAC-3′). To generate sense and antisense probes, specific cDNAs were amplified by PCR with one primer Up and one primer T7-Down or with one primer Down and one primer T7-Up, respectively. These cDNAs were used to generate sense or antisense digoxigenin-labeled RNA probes by in vitro transcription using the T7 primer (T7 MAXIScript Kit; AMBION). Plant samples were fixed in 4% (v/v) paraformaldehyde in phosphate buffer (0.2 m, pH 7.5); inclusion, section preparation, and hybridization were done as previously described (Jabnoune et al., 2009). Sections were observed with a DM6000 (LEICA) Microscope under white light. Photographs were taken with a Retiga 2000R Camera (QIMAGING), and images were processed through Volocity 4.0.1 (IMPROVISION). In situ hybridization experiments have been conducted on the Plate-Forme d’Histocytologie et d’Imagerie Cellulaire Végétale (http://phiv.cirad.fr/) using microscopes of the Montpellier Rio Imaging platform (www.mri.cnrs.fr).
Microarray Hybridization and Analysis
For microarray hybridization experiments, total RNA was extracted from 100 mg of frozen leaves and roots after removal of the remaining seeds from 7-d-old seedlings using a RNeasy Plant Mini Kit (QIAGEN) according to the manufacturer’s instructions. Residual genomic DNA was removed with the RNAse-Free DNase Set (QIAGEN) during RNA purification. Two independent biological experiments were used for each studied plant line.
Microarray hybridization and data processing were carried out with Affymetrix Custom Service (AFFYMETRIX) by following the standard protocol for Affymetrix DNA chip as previously described (Coudert et al., 2011). The complete transcriptome data are accessible through Gene Expression Omnibus Series accession number GSE52640 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE52640). Expression values were normalized with the robust multiarray average method (Irizarry et al., 2003). Differential analysis and extraction of Affymetrix microarray analysis suite 5 call flags were done with linear models and empirical Bayes and Student's t tests relative to a threshold methods within affy and limma R packages (www.r-project.org; Gautier et al., 2004; Smyth, 2004; Smyth et al., 2005; McCarthy and Smyth, 2009). Raw P values were adjusted with the Benjamini-Hochberg method to control the false discovery rate (Benjamini and Hochberg, 1995). Empirical Bayes method with the Benjamini-Hochberg correction was kept for further analysis, because it allowed us to confirm the DRs and up-regulations of OsMADS26 in the two replicates in the DR and OX lines, respectively. Orygenes DataBase (http://orygenesdb.cirad.fr/; Droc et al., 2006) was used to retrieve gene annotation corresponding to selected Affymetrix probes. Microarray control probe sets and probe sets without annotation were discarded for further analysis. Only probe sets with present detection call were kept for subsequent analysis. The two biological repetitions for each type of DR or OX transgenic lines were compared with the corresponding controls. A gene was considered significantly regulated if it presented a fold change ≥ 2 and a Benjamini-Hochberg-corrected P ≤ 0.05 in at least two of the four different contrasts. Genes showing inconsistent regulations, such as (1) inverse regulation in two biological repeats of the same type of DR or OX line or (2) similar regulation in the two different types of DR and OX line, were discarded. A set of up-regulated genes from DNA chip analysis was confirmed by qRT-PCR analysis as previously described using specific primers (Supplemental Table S1).
Disease Resistance Assays
The GUY11 (Centre de Coopération Internationale en Recherche Agronomique pour le Développement Collection) or VT15 (Laboratoire Mixte International Rice Functional Genomics and Plant Biotechnology Collection) isolates of Magnaporthe oryzae were used for inoculation. GUY11 and VT15 isolates are compatible with rice ‘Nipponbare’ and generate moderate susceptibility symptoms. For gene expression studies (Fig. 3), we used the fully virulent FR13 isolate and the avirulent isolate CL3.6.7 (Delteil et al., 2012). In laboratory, inoculations were performed on four- to five-leaf stage plantlets as described in Berruyer et al., 2003; rice japonica ‘Maratelli’ was used as a susceptible control in the experiments in addition to the studied transgenic lines. The data presented are representative of data obtained from three independent replicated experiments. For gene expression studies (Fig. 3), we used the fully virulent FR13 isolate and the avirulent isolate CL3.6.7 (Delteil et al., 2012). Leaves were collected before and after inoculation in liquid nitrogen and used for RNA extraction and qRT-PCR analysis to measure the expression level of different defense genes using specific primers (Supplemental Table S2).
For nethouse experiments in Vietnam, plants were grown in pots (28 L) filled with organic soil (10 kg by pots) and supplemented with nitrogen (2 g by pots) 3 and 9 weeks after planting. After germination in water, plants were planted (five plants by pots and one pot by line) following a randomized design, where OX, DR, and control lines were interspersed with cv Maratelli and cv Sariceltick susceptible lines. Plants were grown in a nethouse and natural conditions and irrigated permanently to saturation. After 6 weeks of growth, plants were sprayed twice a week during 6 weeks using a fresh M. oryzae VT15 isolate spore solution (500,000 spores mL−1 and 1% [w/v] gelatin). Symptoms were observed 15 weeks after sowing. Leaves were collected and scanned, and the number of susceptible lesions was numbered according to Berruyer et al., 2003.
Resistance assays against Xanthomonas oryzae pv oryzae were carried out on 8-week-old rice plants. The X. oryzae pv oryzae strain PXO99A (Salzberg et al., 2008) was inoculated using the leaf-clipping method as previously described (Kauffman et al., 1973). The data presented are representative of two independent experiments. Before inoculation and after symptom development, infected leaves were collected in liquid nitrogen and used for RNA extraction and qRT-PCR analysis to measure the expression level of different defense genes using specific primers (Supplemental Table S2).
For resistance assay against RYMV, 10 plants per line were inoculated by finger rubbing the leaves in presence of Carborundum (600 mesh) with purified RYMV particles at a concentration of 100 μg mL−1 as previously described (Quilis et al., 2008). Virus accumulation in tissues was measured by ELISA analysis using an antibody against the RYMV coat protein (N’Guessan et al., 2000). Presented data are representative of two independent replicated experiments.
Resistance Assay to Water Deficit
Plants were germinated directly in soil and grown in the greenhouse. Each pot was filled with EGO 140 soil Substrate (TREF; www.Trefgroup.com), planted with five seedlings, and watered with the same volume of water. After 1 month, plants were subjected to 18 d of withholding water followed by 15 d of rewatering. Drought tolerance was evaluated by determining the percentage of plants that survived or continued to grow after the period of recovery. This experiment was performed using 20 plants per line and repeated three times.
During the water stress period, the RWCs of plants were monitored using a 7-cm-long segment of the last expanded leaf in a random set of five plants per line according to Barr and Weatherley, 1962. The other leaves were also harvested, frozen in liquid nitrogen, and stored at −80°C for RNA extraction and qRT-PCR analysis of stress-related genes expression using two plants per line exhibiting closest RWCs. qRT-PCR analysis was conducted as described earlier with specific primers of genes identified as drought and high salinity stress markers in rice: RAB21, a rice dehydrin (AK109096), and SALT (AF001395) genes (Claes et al., 1990; Oh et al., 2005). The primer sequences used are given in Supplemental Table S1.
Upland field experiments were carried out under a confined rain-out shelter field facility at the International Center for Tropical Agriculture. This field trial was laid out in a randomly complete block design with three replicates. Drought stress was imposed from panicle initiation (56 d after direct seeding) and continued approximately 3 weeks or until severe leaf rolling and wilting appeared in nontransgenic controls. Then, the plants were rewatered until physiological maturity. The intensity of drought was monitored through volumetric soil water. Leaf rolling scores were recorded on a one to nine International Rice Research Institute (IRRI) scale standardized for rice. The following agronomic traits were scored according to the criteria established in the Standard Evaluation System for Rice (IRRI, 2002): plant height (centimeters), single-plant dry biomass (grams), and single-plant yield were recorded. The degree of relative chlorophyll content in the fully expanded flag leaf was determined using an SPAD-502 Chlorophyll Meter (Minolta Co.) under stress at different stages of crop development. Chlorophyll-a fluorescence parameters were also measured using a Fuorpen FP100 Chlorophyll Fluorometer. Variable PSII fluorescence in the dark-adapted state (Fv)/maximum PSII fluorescence in the dark-adapted state (Fm) represented the maximal photochemical efficiency. Leaves were kept in the dark for 20 min before measurement. Fv/Fm was calculated with the following formula: Fv/Fm = (Fm − initial [minimum] PSII fluorescence in the dark-adapted state)/Fm.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number OsMADS26 (Os08g02070).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Stability of OsMADS26 overexpression or underexpression.
Supplemental Figure S2. Phenotype of OsMADS26 overexpressing and down-regulated lines.
Supplemental Figure S3. Blast resistance evaluation of OsMADS26 lines in field conditions.
Supplemental Figure S4. Blight resistance evaluation of OsMADS26 lines.
Supplemental Figure S5. Rice Yellow Mottle Virus resistance evaluation of OsMADS26 lines.
Supplemental Figure S6. Drought resistance evaluation of OsMADS26 lines in field condition.
Supplemental Figure S7. OsMADS26 cDNA, GST1, and GST2 position and primer sequences used for PCR amplification.
Supplemental Table S1. Microarray values and analysis.
Supplemental Table S2. Primers used for RT-qPCR.
Supplementary Material
Acknowledgments
We thank Véronique Pantesco (Microarray Core Facility of the Institute of Research of Biotherapy, Centre Hospitalier de Recherche Universitaire-Institut National de la Santé et de la Recherche Médicale-Université Montpellier 1; Montpellier, France) for processing the Affymetrix microarrays.
Glossary
- cDNA
complementary DNA
- Ct
threshold cycle
- DAG
days after germination
- dpi
days postinoculation
- Fm
maximum PSII fluorescence in the dark-adapted state
- Fv
variable PSII fluorescence in the dark-adapted state
- hpi
hours postinoculation
- MS
Murashige and Skoog medium
- qRT
quantitative reverse transcription
- RNAi
RNA interference
- RWC
relative water content
- RYMV
Rice Yellow Mottle Virus
- T-DNA
transfer DNA
- UTR
untranslated region
Footnotes
This work was supported by the Hoa Sen Lotus French-Vietnamese Collaboration Program (no. 18346RB), the Agropolis Fondation and Fondazione Cariplo (Rice Connections grant no. 1201–001), the French Agence Nationale de la Recherche (Program Investissement d’Avenir no. ANR–10–LABX–0001–01), the French Embassy in Vietnam (Evariste Galois Program for PhD fellowship to G.N.K.), the Centre de Coopération Internationale en Recherche Agronomique pour le Développement (PhD fellowship to G.N.K.), and the Indian Government (Boycast Postdoctoral Fellowship to P.K.P.).
Articles can be viewed without a subscription.
References
- Agrawal GK, Tamogami S, Han O, Iwahashi H, Rakwal R (2004) Rice octadecanoid pathway. Biochem Biophys Res Commun 317: 1–15 [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Liljegren SJ, Pelaz S, Gold SE, Burgeff C, Ditta GS, Vergara-Silva F, Yanofsky MF (2000) MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J 24: 457–466 [DOI] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S (2007) MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr HD, Weatherley PE (1962) A re-examination of the relative turgidity technique for estimating water deficit in leaves. Aust J Biol Sci 15: 413–428 [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300 [Google Scholar]
- Berruyer R, Adreit H, Milazzo J, Gaillard S, Berger A, Dioh W, Lebrun MH, Tharreau D (2003) Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor Appl Genet 107: 1139–1147 [DOI] [PubMed] [Google Scholar]
- Caldana C, Scheible WR, Mueller-Roeber B, Ruzicic S (2007) A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez Montes RA, Coello G, González-Aguilera KL, Marsch-Martínez N, de Folter S, Alvarez-Buylla ER (2014) ARACNe-based inference, using curated microarray data, of Arabidopsis thaliana root transcriptional regulatory networks. BMC Plant Biol 14: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MK, Hsu WH, Lee PF, Thiruvengadam M, Chen HI, Yang CH (2011) The MADS box gene, FOREVER YOUNG FLOWER, acts as a repressor controlling floral organ senescence and abscission in Arabidopsis. Plant J 68: 168–185 [DOI] [PubMed] [Google Scholar]
- Chern M, Bai W, Sze-To WH, Canlas PE, Bartley LE, Ronald PC (2012) A rice transient assay system identifies a novel domain in NRR required for interaction with NH1/OsNPR1 and inhibition of NH1-mediated transcriptional activation. Plant Methods 8: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M, Fitzgerald HA, Canlas PE, Navarre DA, Ronald PC (2005) Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol Plant Microbe Interact 18: 511–520 [DOI] [PubMed] [Google Scholar]
- Chujo T, Miyamoto K, Ogawa S, Masuda Y, Shimizu T, Kishi-Kaboshi M, Takahashi A, Nishizawa Y, Minami E, Nojiri H, et al. (2014) Overexpression of phosphomimic mutated OsWRKY53 leads to enhanced blast resistance in rice. PLoS One 9: e98737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Takai R, Akimoto-Tomiyama C, Ando S, Minami E, Nagamura Y, Kaku H, Shibuya N, Yasuda M, Nakashita H, et al. (2007) Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim Biophys Acta 1769: 497–505 [DOI] [PubMed] [Google Scholar]
- Claes B, Dekeyser R, Villarroel R, Van den Bulcke M, Bauw G, Van Montagu M, Caplan A (1990) Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell 2: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo MJS, Luth D, Blankenship KM, Anderson OD, Blechl AE (1993) Activity of a maize ubiquitin promoter in transgenic rice. Plant Mol Biol 23: 567–581 [DOI] [PubMed] [Google Scholar]
- Coudert Y, Bès M, Le TV, Pré M, Guiderdoni E, Gantet P (2011) Transcript profiling of crown rootless1 mutant stem base reveals new elements associated with crown root development in rice. BMC Genomics 12: 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delteil A, Blein M, Faivre-Rampant O, Guellim A, Estevan J, Hirsch J, Bevitori R, Michel C, Morel JB (2012) Building a mutant resource for the study of disease resistance in rice reveals the pivotal role of several genes involved in defence. Mol Plant Pathol 13: 72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delteil A, Zhang J, Lessard P, Morel JB (2010) Potential candidate genes for improving rice disease resistance. Rice (N Y) 3: 56–71 [Google Scholar]
- Droc G, Ruiz M, Larmande P, Pereira A, Piffanelli P, Morel JB, Dievart A, Courtois B, Guiderdoni E, Périn C (2006) OryGenesDB: a database for rice reverse genetics. Nucleic Acids Res 34: D736–D740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang SC, Fernandez DE (2002) Effect of regulated overexpression of the MADS domain factor AGL15 on flower senescence and fruit maturation. Plant Physiol 130: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, You J, Xie K, Xie W, Xiong L (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genomics 280: 547–563 [DOI] [PubMed] [Google Scholar]
- Fernandez DE, Heck GR, Perry SE, Patterson SE, Bleecker AB, Fang SC (2000) The embryo MADS domain factor AGL15 acts postembryonically. Inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 12: 183–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) affy: analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C (2001) An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 25: 386–401 [DOI] [PubMed] [Google Scholar]
- Guo S, Xu Y, Liu H, Mao Z, Zhang C, Ma Y, Zhang Q, Meng Z, Chong K (2013) The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun 4: 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Liang W, Yin C, Cui X, Zong J, Wang X, Hu J, Zhang D (2011) Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23: 515–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- International Rice Research Institute (IRRI) (2002) Standard Evaluation System for Rice, International Rice Research Institute, Los Baños, Philippines [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A (2009) TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Jabnoune M, Espeout S, Mieulet D, Fizames C, Verdeil JL, Conejero G, Rodriguez-Navarro A, Sentenac H, Guiderdoni E, Abdelly C, et al. (2009) Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol 150: 1955–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman H, Reddy APK, Hsieh SPY, Merca SD (1973) An improved technique for evaluating resistance to rice varieties of Xanthomonas oryzae. Plant Dis Rep 57: 537–541 [Google Scholar]
- Kong Z, Li M, Yang W, Xu W, Xue Y (2006) A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol 141: 1376–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouassi NK, N’Guessa P, Albar L, Fauquet CM, Brugidou C (2005) Distribution and characterization of Rice yellow mottle virus: a threat to African farmers. Plant Dis 89: 124–133 [DOI] [PubMed] [Google Scholar]
- Lee I, Seo YS, Coltrane D, Hwang S, Oh T, Marcotte EM, Ronald PC (2011) Genetic dissection of the biotic stress response using a genome-scale gene network for rice. Proc Natl Acad Sci USA 108: 18548–18553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Choi SC, An G (2008a) Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. Plant J 54: 93–105 [DOI] [PubMed] [Google Scholar]
- Lee S, Woo YM, Ryu SI, Shin YD, Kim WT, Park KY, Lee IJ, An G (2008b) Further characterization of a rice AGL12 group MADS-box gene, OsMADS26. Plant Physiol 147: 156–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF (2000) SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Ma B, He SJ, Duan KX, Yin CC, Chen H, Yang C, Xiong Q, Song QX, Lu X, Chen HW, et al. (2013) Identification of rice ethylene-response mutants and characterization of MHZ7/OsEIN2 in distinct ethylene response and yield trait regulation. Mol Plant 6: 1830–1848 [DOI] [PubMed] [Google Scholar]
- Mao L, Begum D, Chuang HW, Budiman MA, Szymkowiak EJ, Irish EE, Wing RA (2000) JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 406: 910–913 [DOI] [PubMed] [Google Scholar]
- Marla SS, Singh VK (2012) LOX genes in blast fungus (Magnaporthe grisea) resistance in rice. Funct Integr Genomics 12: 265–275 [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Smyth GK (2009) Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics 25: 765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabi R, Ding S, Xu JR (2008) MADS-box transcription factor mig1 is required for infectious growth in Magnaporthe grisea. Eukaryot Cell 7: 791–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F, Dubois E (2003) Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316: 1–21 [DOI] [PubMed] [Google Scholar]
- Miki D, Shimamoto K (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Minh-Thu PT, Hwang DJ, Jeon JS, Nahm BH, Kim YK (2013) Transcriptome analysis of leaf and root of rice seedling to acute dehydration. Rice (N Y) 6: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel G, Breton C, Thiersault M, Burlat V, Jay-Allemand C, Gantet P (2007) Transcription factor Agamous-like 12 from Arabidopsis promotes tissue-like organization and alkaloid biosynthesis in Catharanthus roseus suspension cells. Metab Eng 9: 125–132 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51: 617–630 [DOI] [PubMed] [Google Scholar]
- N’Guessan PN, Pinel A, Caruana ML, Frutos R, Sy A, Guesquière A, Fargette D (2000) Evidence of the presence of two erotypes of rice yellow mottle sobemovirus in Côte d’Ivore. Eur J Plant Pathol 106: 167–178 [Google Scholar]
- Nuruzzaman M, Sharoni AM, Satoh K, Moumeni A, Venuprasad R, Serraj R, Kumar A, Leung H, Attia K, Kikuchi S (2012) Comprehensive gene expression analysis of the NAC gene family under normal growth conditions, hormone treatment, and drought stress conditions in rice using near-isogenic lines (NILs) generated from crossing Aday Selection (drought tolerant) and IR64. Mol Genet Genomics 287: 389–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M, Kim YK, Nahm BH, Kim JK (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138: 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al. (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelucchi N, Fornara F, Favalli C, Masiero S, Lago C, Colombo L, Kater MM (2002) Comparative analysis of rice MADS-box genes expressed during flower development. Sex Plant Reprod 15: 113–122 [Google Scholar]
- Peng YL, Shirano Y, Ohta H, Hibino T, Tanaka K, Shibata D (1994) A novel lipoxygenase from rice. Primary structure and specific expression upon incompatible infection with rice blast fungus. J Biol Chem 269: 3755–3761 [PubMed] [Google Scholar]
- Puig J, Meynard D, Khong GN, Pauluzzi G, Guiderdoni E, Gantet P (2013) Analysis of the expression of the AGL17-like clade of MADS-box transcription factors in rice. Gene Expr Patterns 13: 160–170 [DOI] [PubMed] [Google Scholar]
- Quilis J, Peñas G, Messeguer J, Brugidou C, San Segundo B (2008) The Arabidopsis AtNPR1 inversely modulates defense responses against fungal, bacterial, or viral pathogens while conferring hypersensitivity to abiotic stresses in transgenic rice. Mol Plant Microbe Interact 21: 1215–1231 [DOI] [PubMed] [Google Scholar]
- Ribot C, Hirsch J, Balzergue S, Tharreau D, Nottéghem JL, Lebrun MH, Morel JB (2008) Susceptibility of rice to the blast fungus, Magnaporthe grisea. J Plant Physiol 165: 114–124 [DOI] [PubMed] [Google Scholar]
- Rzewuski G, Sauter M (2008) Ethylene biosynthesis and signaling in rice. Plant Sci 175: 32–42 [Google Scholar]
- Sallaud C, Meynard D, van Boxtel J, Gay C, Bès M, Brizard JP, Larmande P, Ortega D, Raynal M, Portefaix M, et al. (2003) Highly efficient production and characterization of T-DNA plants for rice (Oryza sativa L.) functional genomics. Theor Appl Genet 106: 1396–1408 [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, Tsuge S, Furutani A, Ochiai H, Delcher AL, Kelley D, et al. (2008) Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YS, Chern M, Bartley LE, Han M, Jung KH, Lee I, Walia H, Richter T, Xu X, Cao P, et al. (2011) Towards establishment of a rice stress response interactome. PLoS Genet 7: e1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra TS, Figueiredo DD, Cordeiro AM, Almeida DM, Lourenço T, Abreu IA, Sebastián A, Fernandes L, Contreras-Moreira B, Oliveira MM, et al. (2013) OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors. Plant Mol Biol 82: 439–455 [DOI] [PubMed] [Google Scholar]
- Shinozuka Y, Kojima S, Shomura A, Ichimura H, Yano M, Yamamoto K, Sasaki T (1999) Isolation and characterization of rice MADS box gene homologues and their RFLP mapping. DNA Res 6: 123–129 [DOI] [PubMed] [Google Scholar]
- Shore P, Sharrocks AD (1995) The MADS-box family of transcription factors. Eur J Biochem 229: 1–13 [DOI] [PubMed] [Google Scholar]
- Smaczniak C, Immink RG, Muiño JM, Blanvillain R, Busscher M, Busscher-Lange J, Dinh QD, Liu S, Westphal AH, Boeren S, et al. (2012) Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci USA 109: 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: 3. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Michaud J, Scott HS (2005) Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21: 2067–2075 [DOI] [PubMed] [Google Scholar]
- Sun L, Zhang H, Li D, Huang L, Hong Y, Ding XS, Nelson RS, Zhou X, Song F (2013) Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol Biol 81: 41–56 [DOI] [PubMed] [Google Scholar]
- Tao Z, Liu H, Qiu D, Zhou Y, Li X, Xu C, Wang S (2009) A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol 151: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia-López R, García-Ponce B, Dubrovsky JG, Garay-Arroyo A, Pérez-Ruíz RV, Kim SH, Acevedo F, Pelaz S, Alvarez-Buylla ER (2008) An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol 146: 1182–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Münster T, Winter KU, Saedler H (2000) A short history of MADS-box genes in plants. Plant Mol Biol 42: 115–149 [PubMed] [Google Scholar]
- Vergne E, Ballini E, Droc G, Tharreau D, Nottéghem JL, Morel JB (2008) ARCHIPELAGO: a dedicated resource for exploiting past, present, and future genomic data on disease resistance regulation in rice. Mol Plant Microbe Interact 21: 869–878 [DOI] [PubMed] [Google Scholar]
- Vergne E, Ballini E, Marques S, Sidi Mammar B, Droc G, Gaillard S, Bourot S, DeRose R, Tharreau D, Nottéghem JL, et al. (2007) Early and specific gene expression triggered by rice resistance gene Pi33 in response to infection by ACE1 avirulent blast fungus. New Phytol 174: 159–171 [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJM, McQuinn R, Chung M, Poole M, Rose J, Seymour G, Grandillo S, Giovannoni J, et al. (2009) Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, Shen QJ (2005) Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 137: 176–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wu F, Lin X, Du X, Chong K, Gramzow L, Schilling S, Becker A, Theißen G, Meng Z (2012) Live and let die: the B sister MADS-box gene OsMADS29 controls the degeneration of cells in maternal tissues during seed development of rice (Oryza sativa). PLoS One 7: e51435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun KY, Park MR, Mohanty B, Herath V, Xu F, Mauleon R, Wijaya E, Bajic VB, Bruskiewich R, de Los Reyes BG (2010) Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant Biol 10: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZL, Shin M, Zou X, Huang J, Ho TH, Shen QJ (2009) A negative regulator encoded by a rice WRKY gene represses both abscisic acid and gibberellins signaling in aleurone cells. Plant Mol Biol 70: 139–151 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.