PTEN proteins regulate rhizoid and gametophore development in Physcomitrella patens.
Abstract
Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) is a lipid phosphatase implicated in cellular proliferation and survival. In animal cells, loss of PTEN leads to increased levels of phosphatidylinositol (3,4,5)-trisphosphate, stimulation of glucose and lipid metabolism, cellular growth, and morphological changes (related to adaptation and survival). Intriguingly, in plants, phosphatidylinositol (3,4,5)-trisphosphate has not been detected, and the enzymes that synthesize it were never reported. In this study we performed a genetic, biochemical, and functional characterization of the moss Physcomitrella patens PTEN gene family. P. patens has four PTENs, which are ubiquitously expressed during the entire moss life cycle. Using a knock-in approach, we show that all four genes are expressed in growing tissues, namely caulonemal and rhizoid cells. At the subcellular level, PpPTEN-green fluorescent protein fusions localized to the cytosol and the nucleus. Analysis of single and double knockouts revealed no significant phenotypes at different developmental stages, indicative of functional redundancy. However, compared with wild-type triple and quadruple pten knockouts, caulonemal cells grew faster, switched from the juvenile protonemal stage to adult gametophores earlier, and produced more rhizoids. Furthermore, analysis of lipid content and quantitative real-time polymerase chain reaction data performed in quadruple mutants revealed altered phosphoinositide levels [increase in phosphatidylinositol (3,5)-bisphosphate and decrease in phosphatidylinositol 3-phosphate] and up-regulation of marker genes from the synthesis phase of the cell cycle (e.g. P. patens proliferating cell nuclear antigen, ribonucleotide reductase, and minichromosome maintenance) and of the retinoblastoma-related protein gene P. patens retinoblastoma-related protein1. Together, these results suggest that PpPTEN is a suppressor of cell growth and morphogenic development in plants.
Human Phosphatase and Tensin Homolog (PTEN; located on chromosome 10q23, EC 3.1.3.67) was first identified as a protein frequently disrupted in multiple sporadic tumor types (Li et al., 1997; Steck et al., 1997). Its role as a tumor suppressor gene belonging to the most frequently mutated genes in human cancer is now firmly established (Chalhoub and Baker, 2009). PTEN is a dual phosphatase that can act both on polypeptide and phosphoinositide (PPI) substrates, with a high preference toward PPIs, specifically the signaling molecule phosphatidylinositol (3,4,5)-trisphosphate [PtdIns(3,4,5)P3], which it dephosphorylates to phosphatidylinositol (4,5)-bisphosphate [PtdIns(4,5)P2] (Maehama and Dixon, 1998; Myers et al., 1998). In animal cells, PTEN acts in a haploinsufficient manner and is a negative regulator of the phosphatidylinositol 3-kinase (PI3K)-serine/threonine-protein kinase (AKT) signaling pathway. Mutation of PTEN in mice leads to the accumulation of PtdIns(3,4,5)P3 (Goebbels et al., 2010) and activation of a subset of proteins that contain pleckstrin homology domains, including AKT family members and phosphatidylinositol-dependent kinase1 (PDK1), which play crucial roles in cell survival, cell proliferation, angiogenesis, and anabolic metabolism (Song et al., 2012). In addition, PTEN has been implicated in a wide array of other functions. Nuclear PTEN regulates cellular senescence by enhancing the activity of the anaphase-promoting complex/cyclosome (Song et al., 2011), and by up-regulating the transcription of the eukaryote homolog of bacterial RecA (Rad51), a key protein involved in double-strand break repair, PTEN helps control chromosomal integrity (Shen et al., 2007). In the cytoplasm of mammalian cells, PTEN is involved in the PI3K/AKT/target of rapamycin (mTOR) pathway, which integrates nutrient status and stress responses with cell growth (Slomovitz and Coleman, 2012). PTEN also regulates actin remodelling and is involved in controlling the size of DNA-damaged cells (Kim et al., 2011).
Structurally, animal PTENs consist of four functional domains: a short N-terminal PtdIns(4,5)P2-binding domain, a phosphatase domain, a C2 domain, and a C-terminal tail containing a PEST sequence (Pro, Glu, Ser, and Thr), which was shown to have regulatory features (Chalhoub and Baker, 2009). The majority of the missense mutations found in human tumors occur in the phosphatase domain affecting PTEN catalytic activity, highlighting the importance of its phosphatase activity in tumor suppression (Eng, 2003). Due to PTEN’s pivotal role, the regulation of its gene (which in humans is a single copy gene) includes epigenetic silencing, transcriptional repression, microRNA regulation, and posttranslational modifications such as phosphorylation, acetylation, oxidation, and ubiquitination (Song et al., 2012). While early studies suggested an exclusively cytosolic localization for PTEN protein (Furnari et al., 1997; Li et al., 1997), subsequent work has shown PTEN to associate with several organelles, such as the nucleus, mitochondria, and endoplasmic reticulum (Trotman et al., 2007; Bononi et al., 2013; Liang et al., 2014).
PTEN function has been studied in diverse model organisms. In Mus musculus and in Drosophila melanogaster, PTEN homozygous knockouts are embryonic lethal, but in Caenorhabditis elegans, they are not. However, in all three organisms, reduced levels of PTEN are linked to the stimulation of the PI3K/AKT pathway driving cell survival, cell proliferation, and angiogenesis (Huang et al., 1999; Rouault et al., 1999; Dupont et al., 2002). In Dictyostelium discoideum, PTEN is involved in the regulation of chemotaxis and motility to cAMP gradients, a process for which proper local gradients of PtdIns(3,4,5)P3 in the plasma membrane are essential (Funamoto et al., 2002; Iijima and Devreotes, 2002).
In contrast to animal cells, PtdIns(3,4,5)P3 has never been detected in plant cells. The only reports of PtdIns(3,4,5)P3 production in plants are in vitro activity assays using heterologous-expressed Arabidopsis phosphatidylinositol phosphate5-kinase1 (AtPIP5K1) and PpPIPK1 kinases (Elge et al., 2001; Saavedra et al., 2009). However, the phosphatidylinositol (3,4)-bisphosphate [PtdIns(3,4)P2] substrate for this reaction is likely not present in plants, and sequences coding for phosphatidylinositol 3-phosphate (PtdIns3P) kinase Class I, the enzyme responsible for the synthesis of PtdIns(3,4,5)P3, have not been identified in sequenced plant genomes (Boss and Im, 2012). The current data thus suggest that, in plants, PTEN may act on substrates other than PtdIns(3,4,5)P3 and/or exerts its function through another signaling cascade. In the plant lineage, two distinct phylogenetic groups of PTEN genes have been described (Grunt et al., 2008; Pribat et al., 2012). These two groups are based on different amino acid composition in the catalytic phosphatase domain. The Arabidopsis (Arabidopsis thaliana) genome contains three PTEN genes, AtPTEN1, in the type I group, and AtPTEN2a and AtPTEN2b in the type II group. AtPTEN1 is specifically expressed in pollen and was shown to dephosphorylate PtdIns(3,4,5)P3 in vitro, but no other PPI species were tested as substrates (Gupta et al., 2002). Silencing AtPTEN1 levels by RNA interference caused pollen cell death after mitosis, showing its essential role for pollen viability (Gupta et al., 2002). Using a transient overexpression approach in tobacco (Nicotiana tabacum) pollen tubes, AtPTEN1 was suggested to regulate autophagy by disrupting the dynamics of PtdIns3P (Zhang et al., 2011). The other Arabidopsis isoforms, AtPTEN2a and AtPTEN2b, have been biochemically characterized. AtPTEN2a was found to be the most active with strong preferences for 3-phosphorylated PPI substrates, specifically PtdIns3P, PtdIns(3,4)P2, and PtdIns(3,5)P2 but not PtdIns(3,4,5)P3. AtPTEN2b was reported to be almost inactive toward PPIs substrates (Pribat et al., 2012). Thus, the function of the PTEN-type class II group remains elusive.
To address this question, we have studied PTEN function in the moss Physcomitrella patens, a multicellular plant with a much simpler developmental pattern than most flowering plants. The gametophytic generation consists of protonemata (filamentous tissue that germinates from the spore) and gametophores (shoots that develop off protonemata). Protonemata are composed of two cells types: chloronemata and caulonemata, which differ in chloroplast number and morphology, cell length, tip growth rate, and orientation of the cell plates to the long axis of the filament (Vidali and Bezanilla, 2012). The gametophore is composed of a photosynthetic nonvascularized stem, which carries the phyllids, the reproductive organs, and the filamentous rhizoids, and on top of the gametophore, the sporophytic generation develops (Schaefer and Zrÿd, 2001). P. patens has four PTEN genes, all belonging to the class II group. Phylogenetically and based on sequence similarity, these genes can be divided into two subgroups: PpPTENA and PpPTENB in one group and PpPTENC and PpPTEND in the other. Previously, it was shown that PpPTENA but not PpPTEND could functionally replace the N-terminal domain of a class II formin (For2A), rescuing the compromised growth phenotype obtained after silencing all For2As (van Gisbergen et al., 2012). This study showed that PpPTENA but not PpPTEND binds PtdIns(3,5)P2 in vitro and that the N-terminal PTEN domain functions to localize formin to sites on the membrane rich in PtdIns(3,5)P2 (van Gisbergen et al., 2012).
Here, we performed a functional characterization of the PpPTEN family using single, double, triple, and quadruple pten knockout mutants as well as overexpression lines. Our data shows that while PpPTEN genes are expressed throughout the moss life cycle, their expression is enriched in caulonemata and rhizoids. Our results further suggest that PpPTENs function to regulate the transition from the two-dimensional filamentous protonemal stage to the three-dimensional adult gametophore, possibly by regulating transition to the synthesis (S) phase of the cell cycle.
RESULTS
The P. patens Genome Contains Four PTEN-Like Class II Type Genes
Using Arabidopsis PTEN protein sequences as queries, we identified four PTEN orthologs in the P. patens genome (v1.6, http://phytozome.jgi.doe.gov/pz/portal.html). An extensive phylogenetic analysis previously performed on proteins bearing PTEN domains from different organisms (Grunt et al., 2008) identified two different clades for plant PTENs: AtPTEN1-type and AtPTEN2/AtPTEN3-type, later renamed as type I and type II PTENs (Pribat et al., 2012). The PTEN type I clade incorporates sequences from angiosperms, gymnosperms, lycophytes, and from Chlamydomonas reinhardtii, whereas the PTEN type II clade comprises sequences from angiosperms, lycophytes, and bryophytes. The four PpPTEN sequences belong to the type II group, which clustered into two subgroups, with PpPTENA and PpPTENB in one and PpPTENC and PpPTEND in the other (Supplemental Fig. S1; Grunt et al., 2008); the proximity of these subgroups is in agreement with a recent duplication of the P. patens genome (Rensing et al., 2008).
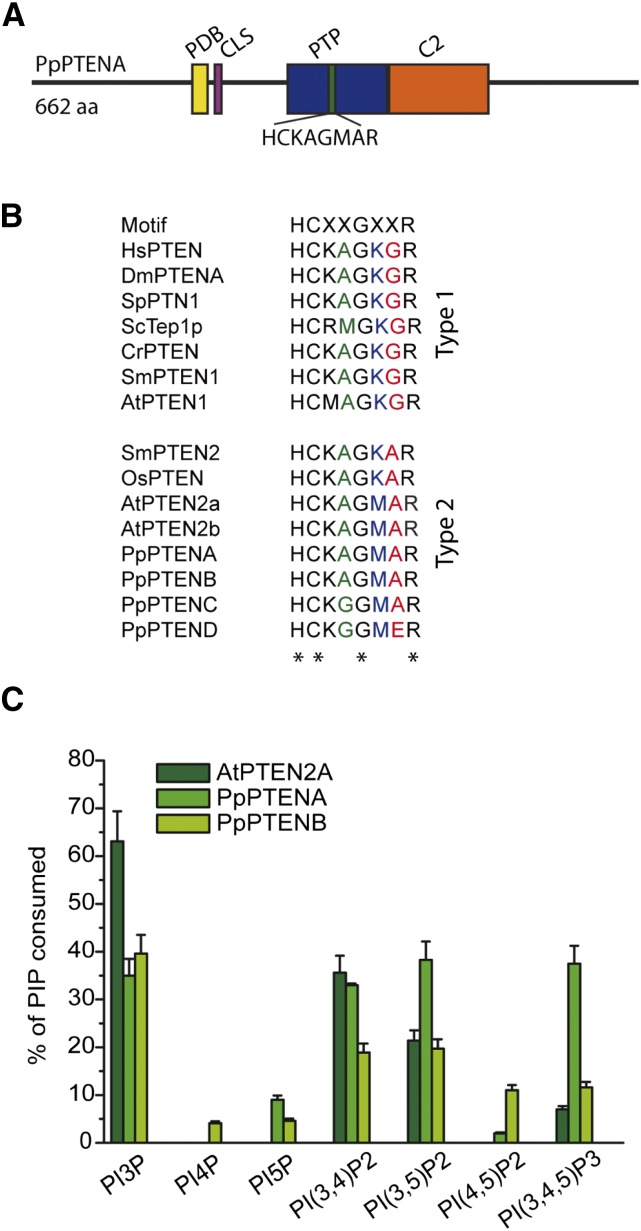
Protein similarity is 81.6%, between PpPTENA and PpPTENB, and 41.4%, between the second subgroup PpPTENC and PpPTEND. Sequence analysis using the National Center for Biotechnology Information Conserved Domain Search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) identified, in all four PpPTENs, a conserved phosphatase catalytic domain and a C2 domain (Fig. 1A), in accordance with the description of PTEN. PTEN proteins exhibit, in their catalytic domain, the phosphatase signature motif HCXXGXXR (where X is any amino acid) that is found in all protein Tyr phosphatases (Yin and Shen, 2008; Chalhoub and Baker, 2009; Song et al., 2012) and forms the phosphate-binding loop responsible for the hydrolysis of PPIs. In human PTEN, the sequence corresponding to this phosphatase signature motif is HCKAGKGR (comprising residues 123–130). This sequence has residues that are essential for catalytic activity; the mutation C124S inactivates both protein and lipid phosphatase activity (Maier et al., 1999), and G129A abrogates the lipid but not the protein phosphatase activity (Liaw et al., 1997). While in type I PTENs, this sequence is highly conserved, the type II group has two substitutions, K128 to Met and G129 to Ala (Fig. 1B). Importantly, these substitutions were shown to be responsible for changes in PPI substrate preferences for AtPTEN2a (Pribat et al., 2012). Noteworthy, PpPTENC and PpPTEND have another substitution of A126 to Gly, and in PpPTEND, G129 is replaced by a Glu residue (Fig. 1B). A short phosphatidylinositol (4,5)-bisphosphate-binding domain (PDB) domain and a putative cytosolic signal (Denning et al., 2007) were also identified at the N terminus of PpPTENA and PpPTENB, sequences that do not share a high degree of conservation in PpPTENC and PpPTEND (Supplemental Fig. S2). Neither PEST sequences nor PDZ (for Postsynaptic-density protein of 95 kD, discs large, zona occludens1) domain (interaction motif) were identified in PpPTENs. However, the C termini of PpPTENs contain a high number of Thr and Ser residues, which could reflect a mode of regulation of these proteins that is similar to the PEST sequences of their animal counterparts.
Figure 1.
Modular and biochemical characterization of P. patens class II PTENs. A, Modular structure of PpPTEN proteins. PtdIns(4,5)P2-binding domain (PBD), cytosolic signal (CLS), phosphatase domain (PTP), and C2 domain (C2). B, PTEN phosphatase signature motif HCXXGXXR (phosphate-binding loop) responsible for the hydrolysis of PPIs. In HsPTEN the sequence corresponding to this phosphatase signature motif is HCKAGKGR (comprising residues 123–130). C, Biochemical characterization of PPI phosphatase activity of recombinant GST-AtPTEN2a and GST-PpPTENs and GST proteins. The activity was measured by the amount of free inorganic phosphate released from the substrate using a malachite green assay. Reactions were performed in triplicates, and results are represented by bars as the mean ± sd. PI4P, Phosphatidylinositol-4-phosphate; PI5P, phosphatidylinositol-5-phosphate; PI(3,4)P2, phosphatidylinositol-(3,4)-bisphosphate; PI(3,5)P2, phosphatidylinositol-(3,5)-bisphosphate; PI(4,5)P2, phosphatidylinositol-(4,5)-bisphosphate; PI(3,4,5)P3, phosphatidylinositol-(3,4,5)-trisphosphate; ScTep1p, tensin-like phosphatase; WT, wild type.
PpPTENA and PpPTENB Have Lipid Phosphatase Activity
It is well known that PTENs are dual phosphatases that can act on both polypeptide and PPI substrates, with a high preference toward the three-position of the inositol ring of PPIs. To test the specificity of PpPTENs against PPI substrates, their open reading frames were expressed in Escherichia coli as fusion proteins with a glutathione S-transferase (GST)-tag, and their phosphatase activity was tested in vitro toward the seven PPI isomers (Fig. 1C). GST alone was used as a negative control, and the previously characterized AtPTEN2a (Pribat et al., 2012) was included in the assay as a positive control due to its close phylogenetic relationship with the PpPTENs. Recombinant GST-PpPTENs were successfully expressed, with the exception of GST-PpPTENC, despite multiple attempts. Therefore, no functional statements can be made for PpPTENC. The other isoforms, expressed as fusions to N-terminal GST-tags, were purified, and size and identity of the recombinant proteins were confirmed by western blot using anti-GST antibodies (Supplemental Fig. S3). We found that PpPTENA efficiently (and to similar extent) dephosphorylated PtdIns3P, PtdIns(3,4)P2, PtdIns(3,5)P2, and PtdIns(3,4,5)P3, whereas PpPTENB preferentially dephosphorylated PtdIns3P with decreasing activity toward PtdIns(3,4)P2 and PtdIns(3,5)P2 (Fig. 1C). Interestingly, PpPTEND had no phosphatase activity against any of the seven PPI isomers tested. This could be due to a specific substitution observed in PpPTEND (G129E), which, in humans, was shown to abrogate PTEN lipid phosphatase activity while maintaining protein phosphatase activity (Myers et al., 1997).
Among the monophosphorylated PPI species, both PpPTENA and PpPTENB only dephosphorylated the phosphate in the D3 position. Similarly, PpPTENA and PpPTENB preferred the bisphosphorylated PPI in the D3 position [PtdIns(3,4)P2 and PtdIns(3,5)P2] compared with PtdIns(4,5)P2 as substrate. These data indicate that PpPTENs have a strong preference for dephosphorylating the D3 position. Given that AtPTEN2a, PpPTENA, and PpPTENB share identical phosphate-binding loop sequences, it was surprising that PpPTENA dephosphorylated PtdIns(3,4,5)P3 in vitro, suggesting that substrate selection likely extends beyond the phosphate-binding loop.
PpPTEN Genes Are Expressed throughout the Moss Life Cycle
The simple life cycle of P. patens, with a clear cellular differentiation, makes this moss ideal to study PTEN expression from a developmental point of view. The expression pattern of PpPTEN genes was analyzed in the gametophytic generation, and expression of the four genes was detected in both chloronemal and caulonemal cells and in gametophores (Supplemental Fig. S4). Our results confirmed the PpPTEN expression data deposited in the Genevestigator database, which additionally includes expression of the whole PTEN family in the sporophyte.
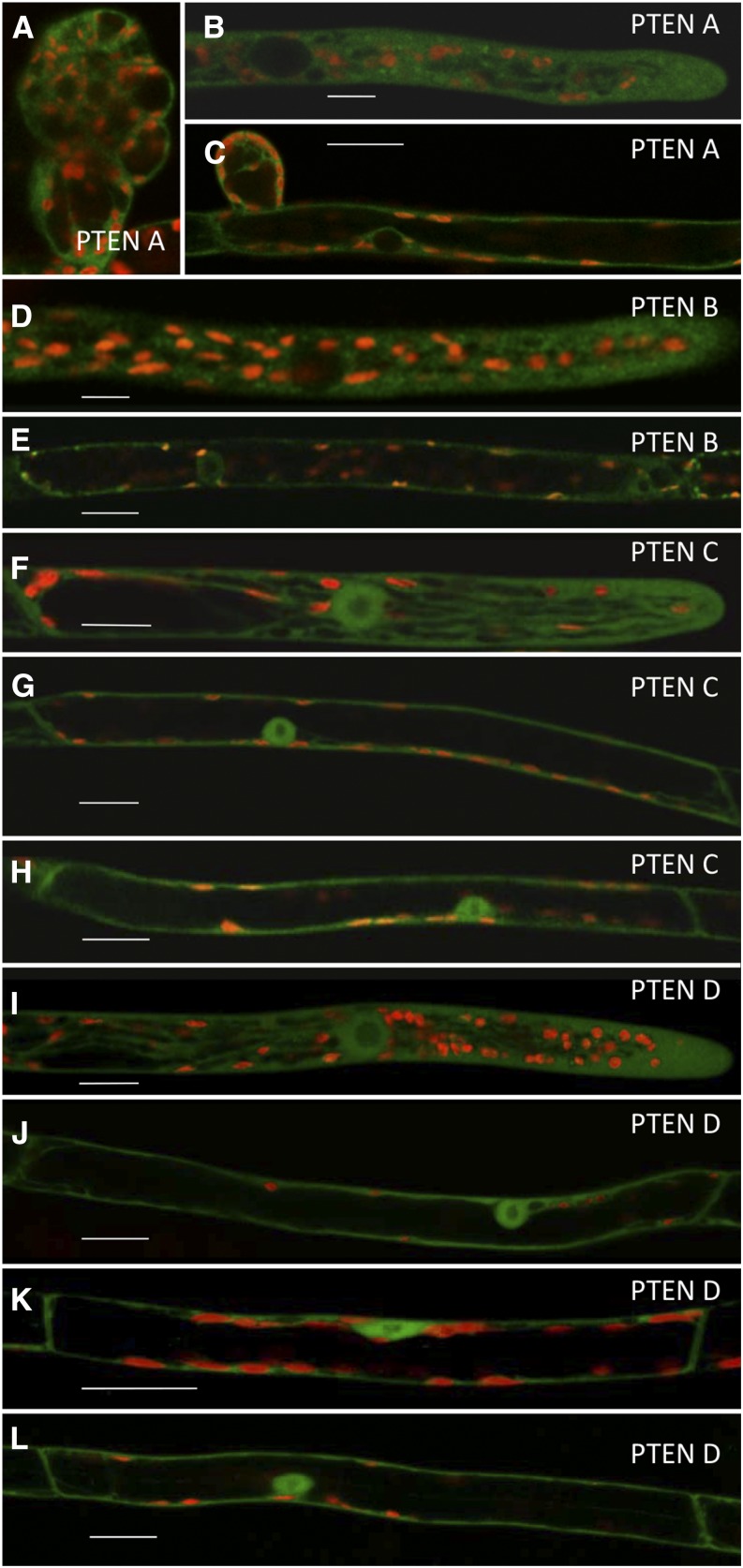
To investigate PpPTEN’s cellular and subcellular localization, we integrated sequences encoding monomeric enhanced GFP (mEGFP) in frame and downstream of each PpPTEN gene. Using this strategy, expression of the PpPTEN fusion proteins is under the control of their endogenous promoters, thereby minimizing possible overexpression artifacts. Stable PpPTENpro:PpPTEN-mEGFP lines were obtained for the four PpPTEN genes, all of which were verified by PCR analysis (Supplemental Fig. S5B). At a tissue level and except for differences in fluorescence intensities within the different lines, the same cellular localization pattern was observed for all PpPTEN genes. In agreement with a role in cellular growth and proliferation, PpPTEN-GFP fusions exhibited higher fluorescence in the apical cells of chloronemata and caulonemata and new developing buds, whereas in developed gametophores, the signal was mainly observed in the emerging rhizoids cells (Fig. 2; Supplemental Fig. S5, D and E). In the case of PpPTENApro:PpPTENA-mEGFP, the fluorescent signal was often too weak for adequate imaging. We therefore generated also a PpPTENpro:PpPTEN-3XmEGFP to complement our observations. Using this construct for ectopic expression, fluorescence was visible in developing buds, as well as in actively growing caulonemal and rhizoid cells (Fig. 2, A–C).
Figure 2.
Subcellular imaging of stable GFP-PpPTEN expression (under their endogenous promoter) in different developmental stages of the moss gametophytic phase. Red indicates chlorophyll autofluorescence. A to C, Confocal scanning laser microscopy (CSLM) imaging of PTENA in developing bud (A), rhizoid apical cell (B), and caulonemal subapical cell (C). D to E, CSLM imaging of PTENB in rhizoid apical (D) and subapical cell (E). F to H, CSLM imaging of PTENC in rhizoid apical (F) and subapical cell (G) and in caulonemal subapical cell (H). I to L, CSLM imaging of PTEND in rhizoid apical (I) and subapical cell (J), chloronema subapical cell (K), and caulonemal subapical cell (L). Bar = 20 μm.
At a subcellular level, and in agreement with animal PTEN localization, a primarily cytosolic signal was observed for all PpPTEN genes (Fig. 2, A–L). The isoforms PpPTENC and PpPTEND were also clearly observed in the nucleus of chloronemal, caulonemal, and rhizoid cells (Fig. 2, F–L). Weak PpPTENB-mEGFP fluorescence was found in the nucleus (Fig. 2E), in agreement with its putative cytosolic signal functioning also as a noncanonical signal for nuclear trafficking (Denning et al., 2007). Similar observations were made for PpPTENA-mEGFP in few caulonemal subapical cells, but PpPTENA-3XmEGFP fluorescence was found only in the cytosol, which is likely a consequence of the fusion to three tandem GFP molecules, thereby preventing normal protein transit through the nuclear pores. Although a nuclear localization for PTENA and PTENB is not to be excluded, these observations suggest that the two subgroups of PpPTENs may have evolved different functions and localizations, with PpPTENA and PpPTENB playing their role mostly in the cytosol and PpPTENC and PpPTEND acting both at nuclei and cytosol. Additionally, we also observed fluorescent punctate structures in the cytoplasm of cells expressing both PpPTENA-mEGFP and PpPTENB-mEGFP (Fig. 2, B and E), which could be related to the localization of specific PPIs intracellular pools.
Triple and Quadruple pten Knockout Mutants’ Transition from Protonemata to Gametophores Occurs Earlier Than the Wild Type
To characterize the functional role of PpPTEN genes, we generated pten knockout lines by homologous recombination using targeted gene disruption. Putative knockout lines resistant to the corresponding antibiotic cassettes were selected and further analyzed by PCR and real-time (RT)-PCR to confirm the disruption of the gene of interest (Supplemental Figs. S6 and S7; Supplemental Table S1). When more than one PpPTEN was targeted, the addition of each gene was performed progressively.
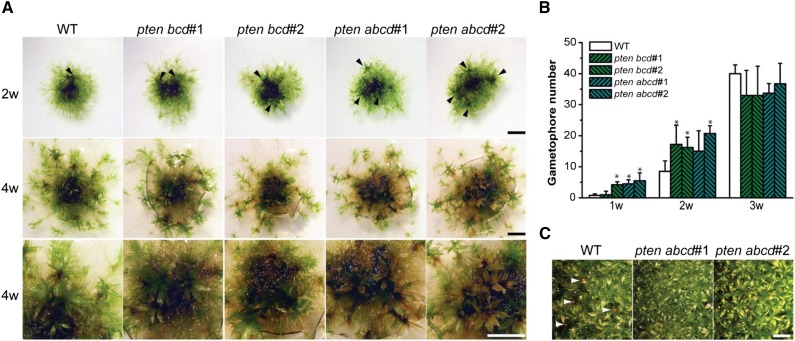
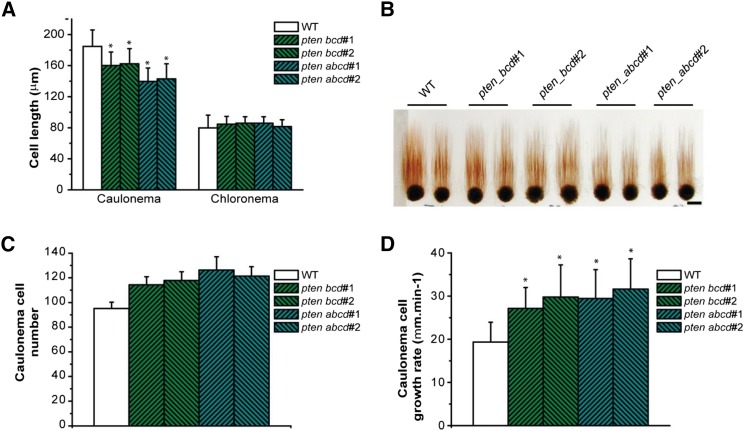
Analysis of single ptena, ptenb, ptenc, and ptend knockout lines, under normal growth conditions, exhibited no significant differences compared with the wild type (data not shown). Because the four PpPTEN genes are expressed in all tissues, functional redundancy is the likely explanation for these results. In agreement with this hypothesis, double knockout lines, either generated from members of same subgroup (e.g. ptencd) or different subgroup (e.g. ptenbd), exhibited nonsignificant differences to the wild type when grown under normal conditions. Thus, we generated triple (ptenbcd) and quadruple (ptenabcd) knockout lines (Supplemental Figs. S6 and S7; Supplemental Table S1). Two lines for each knockout were isolated from independent transformations and used for detailed analyses. We found that triple and quadruple pten knockout lines developed gametophores earlier than the wild type (Fig. 3A). After 2 weeks of growth on minimal media, knockout protonemata of approximately 1-mm diameter developed almost twice the number of gametophores when compared with the wild type (Fig. 3B). This observation suggests that, at earlier stages of moss development, the disruption of pten leads to a faster transition from protonemata to gametophores. However, later in development, wild-type colonies had more gametophores and were larger (Supplemental Fig. S8). After 4 to 5 weeks of growth in minimal media, quadruple pten knockouts exhibited signs of cell death (e.g. brownish color), which was not observed in the wild type (Fig. 3A). Furthermore, even though sporophytes could be developed in quadruple knockout lines, only one to two could be detected in approximately 1 cm2 of gametophores grown in jiffy pellets (Fig. 3C).
Figure 3.
Triple and quadruple pten knockout mutants exhibit earlier transition from the juvenile protonemata to the gametophore stage. A, One-millimeter pieces of protonemata tissue of wild-type (WT) and pten knockout mutants grown on minimal media for 2 weeks (2w, top row; gametophores are indicated by arrowheads) and 4 weeks (4w, middle and bottom rows; bottom row shows details of gametophores). B, Number of gametophores of wild-type and pten knockout mutants grown on minimal media for 1 (1w), 2 (2w), and 3 (3w) weeks. After 4 weeks, intensive growth and gametophore proximity prevented accurate measurements. Bars represent the mean ± sd (n = 5 colonies; three independent experiments were performed). Asterisks indicate values that are significantly different from the wild type (P < 0.05, Student’s t test). C, Sporophyte induction (arrowheads) in the wild type and in quadruple pten knockouts (severely affected). Bars = 0.2 cm.
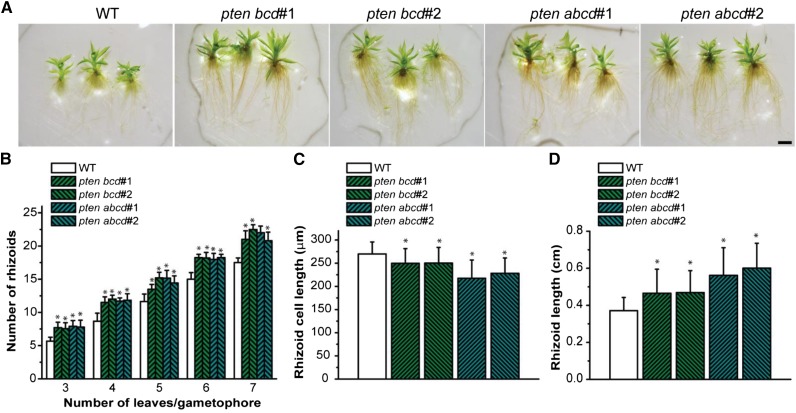
Notably, triple and quadruple pten knockout lines had enhanced rhizoid production on their gametophores (Fig. 4, A and B), which were longer and darker brown but composed of shorter cells (Fig. 4, C and D), when compared with the wild type. These observations suggest that pten mutant cells enter into cell division sooner (higher division rate) upon reaching a lower critical size threshold compared with the wild type.
Figure 4.
Triple and quadruple pten knockout mutants exhibit higher rhizoid number and longer rhizoids with shorter cells. A, Twenty-day-old gametophytes of the wild type (WT) and pten knockout mutants growing in minimal media. B, Number of rhizoids versus development stage (n° leaves) of gametophores of the wild type and pten knockout mutants. Values represent mean ± sd (n ≥ 10). Asterisks indicate values that are significantly different from the wild type (P < 0.05, Student’s t test). C, Rhizoid length of the wild type and pten knockout mutants. Values represent mean ± sd (n ≥ 10). Asterisks indicate values that are significantly different from the wild type (P < 0.05, Student’s t test). D, Rhizoid cell length of the wild type and pten knockout mutants. Values represent mean ± sd (n ≥ 30). Asterisks indicate values that are significantly different from the wild type (P < 0.001, Student’s t test). Bar = 0.1 cm.
Caulonemal Cell Growth Is Accelerated in pten Knockout Lines
Because triple and quadruple pten knockouts developed gametophores earlier than the wild type, we carefully analyzed caulonemal cells, as this is the cell type that differentiates and produces buds (Cove, 2005). Caulonemal cells are easily distinguished from chloronemal cells, which are the first cells to emerge from the spore. Compared with chloronemal cells, caulonemal cells grow faster and have fewer chloroplasts. Caulonemal cells also have oblique cross walls between the cells of the filament (Vidali and Bezanilla, 2012). To differentiate between chloronemal and caulonemal cells, we stained the cell walls with calcofluor. Triple and quadruple pten knockout lines had average chloronemal cell length values similar to the wild type (Fig. 5A). However, under the same growth conditions, the average caulonemal cell length in triple and quadruple pten knockouts was, respectively, 15% and 25% lower than that of wild-type cells (Fig. 5A), a pattern similar to our rhizoid length observations. We subsequently used the ability of caulonemal (but not chloronemal cells) to grow in the dark to follow them in more detail. We observed that, even though single caulonemal cells of triple and quadruple pten knockouts were shorter, the whole filament was approximately the same length, suggesting that caulonemal cells in pten knockouts divide more frequently than in the wild type (Fig. 5, B and C). This observation was confirmed by time lapse imaging of apical tip-growing caulonemal cells (n = 15), which allowed us to verify that wild-type caulonemal cells grew at a mean rate of 19.34 μm h–1 (values of the magnitude previously described [Menand et al., 2007]), while triple and quadruple pten knockout caulonemal cell growth rates were 27.12 and 29.45 μm h–1 (n = 15), respectively (Fig. 5D). These data show that in the absence of PTEN, cells divide more frequently at a shorter cell length, suggesting that PTEN function is implicated in controlling entrance into the cell cycle.
Figure 5.
Triple and quadruple pten knockout mutants exhibit shorter caulonemal cells but a faster growth rate. A, Chloronema and caulonemal cell length grown for 7 d from fragmented moss in minimal media and normal growth conditions. Values represent mean ± sd (n ≥ 30). Asterisks indicate values that are significantly different from the wild type (WT; P < 0.001, Student’s t test). B, One-millimeter pieces of protonemata tissue of the wild type and pten knockout mutants grown in the dark for 4 weeks. C, Caulonema cell number grown in the dark for 4 weeks estimated by the ratio of caulonema filament length to mean cell length. Values represent mean ± sd (n ≥ 30). Asterisks indicate values that are significantly different from the wild type (P < 0.001, Student’s t test). D, Caulonemal cell growth rate in minimal media under normal growth conditions. Values represent mean ± sd (n ≥ 15). Asterisks indicate values that are significantly different from the wild type (P < 0.001, Student’s t test). Bar = 0.5 cm.
It is important to note that protonemal morphology was not affected in triple and quadruple pten knockout. This was verified by generating and following their regeneration over 15 d for the wild type and triple and quadruple pten knockouts (Supplemental Fig. S9). The branching pattern was uniform, with the third subapical cell typically containing a single branch. Albeit, we observed a reduction in both, the distance between the first branch and the tip of the filament and the distance between the first and second branch in pten knockouts, as a consequence of shorter caulonemal cells.
The Levels of Key PpPTEN PPI Substrates Are Altered in pten Knockouts
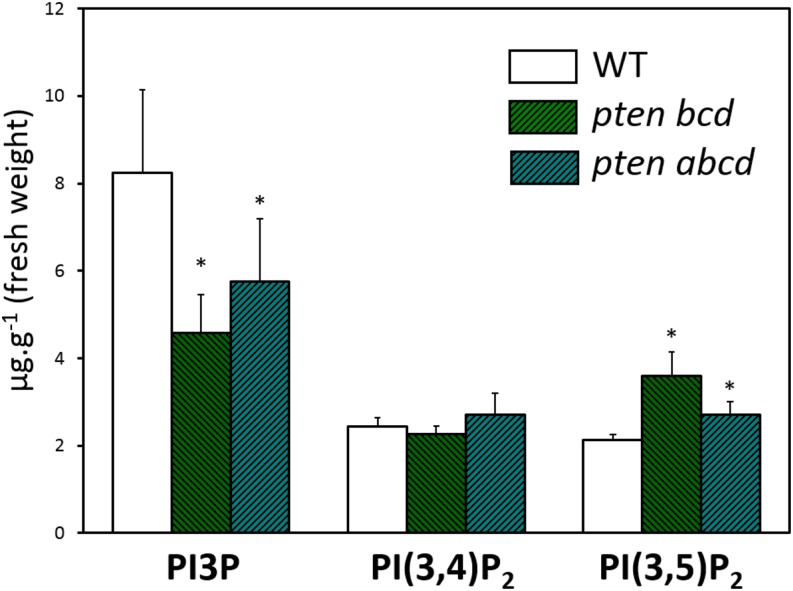
As shown above, in vitro, PpPTENA and PpPTENB use PtdIns3P, PtdIns(3,4)P2, and PtdIns(3,5)P2 as substrates. We therefore investigated whether endogenous levels of PPIs were altered in vivo as a consequence of pten gene disruption. PPIs were quantified in the wild type and in pten triple and quadruple knockout lines using 10-d-old moss protonemata (Fig. 6). We found that in knockout lines, the levels of PtdIns3P were lower and PtdIns(3,4)P2 was unchanged, whereas PtdIns(3,5)P2 levels were higher than in wild-type controls. The overall levels of these three phosphorylated PPIs are very low in all cell types, so putative changes must be carefully interpreted considering cellular variability and the limitations of the methods used. The changes we detected in the three PPI forms in triple and quadruple knockout lines suggest a tight control in the levels of synthesis versus degradation rates of these PPIs. It is thus possible that the phenotypes described for the knockout lines result directly from the absence of the pten transcripts and/or the protein per se (e.g. interaction with targets) and not only from their phosphatase activity.
Figure 6.
Lipid analysis in triple and quadruple pten knockout mutants. The levels of PtdIns3P, PtdIns(3,4)P2, and PtdIns(3,5)P2 from protonemata tissue of wild-type (WT) controls and the pten lines grown under normal growth conditions were extracted and separated by thin-layer chromatography. PPIs were determined by quantifying the amount of associated fatty acids. Data represent the means ± se of two independent experiments. Asterisks indicate values that are significantly different from the wild type (P < 0.05, Student’s t test).
Effect of PpPTEN Overexpression in a Wild-Type Background Mirror Those of PpPTEN Knockouts
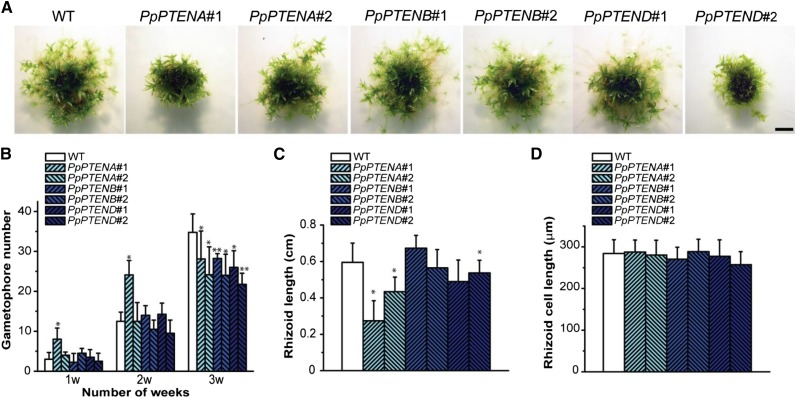
To investigate the effect of altered PpPTEN activity, we expressed each gene individually at high levels using the maize (Zea mays) ubiquitin promoter and nopaline synthase terminator (Anterola et al., 2009). Gene targeting in a wild-type background was used to integrate the pUbi-PpPTEN expression cassette into the PP108 locus, which has been shown not to affect moss growth and development when disrupted (Schaefer and Zrÿd, 1997). Putative overexpression lines resistant to the hygromycin cassette were selected and further analyzed by PCR to confirm the disruption of the PP108 locus and by RT-PCR to check PpPTEN transcript levels in each transformed line (Supplemental Fig. S10; Supplemental Table S1). Two different lines for each oe-PpPTEN were selected, and their development was followed for 4 weeks (Fig. 7A). The transgenic lines exhibited a phenotype with an intensity that correlated roughly with the levels of PpPTEN expression. As observed in Figure 7A, oe-PpPTENA line number 1 exhibited a stronger phenotype than line number 2, correlating with the levels of transcript (Supplemental Fig. S10C). The two oe-PpPTENB lines with similar levels of transcript displayed minor differences to the wild type, whereas oe-PpPTEND line number 2 exhibited a stronger phenotype and higher levels of transcript. The observation that overexpressing PTEND, the form lacking measurable phosphatase activity, also produced a phenotype makes it tempting to speculate that more relevant than a change in PPI levels is the presence of the PTEN proteins. For oe-PpPTENC, only one line was successfully selected, and this exhibited a phenotype identical to PpPTEND line number 2 (Supplemental Fig. S10D).
Figure 7.
Overexpression of PpPTEN in a wild-type (WT) background. A, One-millimeter pieces of protonemata tissue of the wild type (WT) and PpPTENA, PpPTENB, and PpPTEND overexpression lines grown on minimal media for 4 weeks. B, Number of gametophores of the wild type and pten overexpressor lines at 1, 2, and 3 weeks of growth on minimal media. Values represent mean ± sd. Asterisks indicate values that are significantly different from the wild type (*P < 0.05 and **P < 0.01, Students t test). C, Rhizoid length of the wild type and pten overexpressor lines. Values represent mean ± sd (n ≥ 18). Asterisks indicate values that are significantly different from the wild type (P < 0.05, Student’s t test). D, Rhizoid cell length of the wild type and pten overexpressor lines. Values represent mean ± sd (n ≥ 30). Bar = 0.2 cm.
The developmental pattern and number of gametophores in the oe-PpPTEN lines (grown in minimal media) also resembled the knockout lines, an initial faster development of gametophores followed by a reduction in their number when compared with the wild-type protonemata (Fig. 7B versus Fig. 3B). Concerning protonemata cell length of oe-PpPTEN lines, again a pattern similar to knockout lines was observed with no significant difference to the wild type (data not shown). However, whole rhizoid length was shorter in lines with high levels of transcripts (Fig. 7C) while maintaining rhizoid cell length similar to the wild type (Fig. 7D).
S-Phase Cell Cycle Genes Are Up-Regulated in the Quadruple pten Knockout Mutants
The combination of data obtained from the analysis of pten knockouts and overexpressor lines led us to hypothesize that PpPTEN genes may act as growth suppressors in P. patens. Their absence (or misregulation) would lead to faster growth, stimulated cell division, and early developmental transitions but also faster aging and cell death. To test this hypothesis, we performed quantitative real-time (qRT)-PCR to study the expression of three genes typically used as S-phase markers of the cell cycle: proliferating cell nuclear antigen (PpPCNA), minichromosome maintenance (PpMCM), and ribonucleotide reductase (PpRNR). The genes coding for ubiquitin-conjugating enzyme E2 (Ub-E2) and adenine phosphoribosyltransferase (Ade-PRT), previously shown to display minimal expression level variations under different stages of the gametophytic phase of P. patens (Le Bail et al., 2013), were used as reference genes. Analysis was conducted on three biological replicates from the 12-d-old wild type and quadruple knockout ground protonemata grown in minimal media, which, at this stage, is rich in caulonemata.
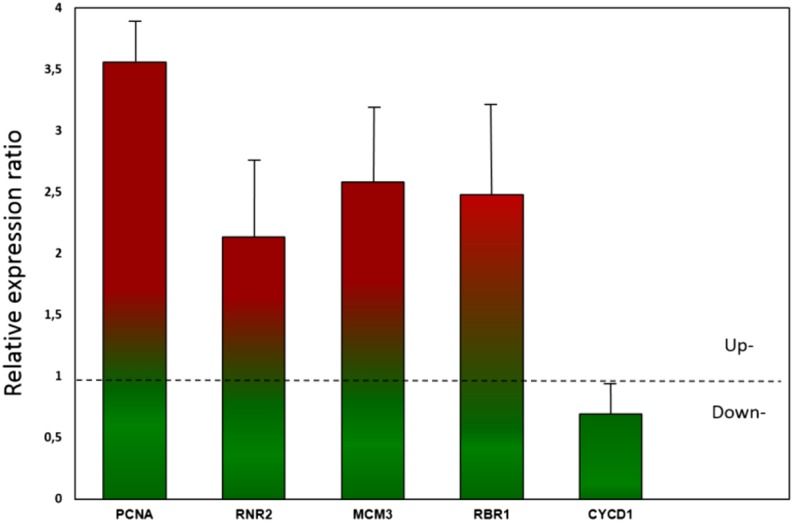
We found that the three genes PpPCNA, PpMCM, and PpRNR were up-regulated in pten quadruple knockouts compared with the wild type, supporting the hypothesis that a depletion of PpPTEN leads to a stimulation of cell proliferation (Fig. 8). In agreement, we also found in quadruple knockouts an up-regulation of the retinoblastoma-related protein1 gene (PpRBR1) and slight down-regulation of Cyclin D1 gene (a protein involved in the phosphorylation of retinoblastoma).
Figure 8.
Quadruple pten knockout mutants show up-regulation of cell-cycle genes. Relative gene expression ratio of qRT-PCR analysis in pten abcd quadruple knockout tissue versus the wild type. The ratio of the normalized target gene in the sample was calculated according to the method by Pfaffl (2001), where values greater than 1 and values less than 1 (dotted line) represent up-regulation and down-regulation, respectively. Data represent the mean ratio ± se of three independent experiments. CYCD1, Cyclin D1.
DISCUSSION
Previously, an important role for PPIs was reported in regulating growth of P. patens (Saavedra et al., 2011). Here, we focused on the role of P. patens PTEN genes during cellular growth and moss development. P. patens has four PTEN genes all belonging to the type II group, and the evolutionary position of mosses in the plant lineage, together with the high efficiency for gene targeting, makes this model organism ideal to study PTEN function.
PpPTENs Are Functionally Redundant Phosphatases Ubiquitously Expressed in Moss Tissues
PpPTEN modular structure is overall well conserved compared with PTEN sequences from other organisms. The four PpPTENs exhibited conserved phosphatase and C2 domains along with a short PDB domain and a cytosolic signal (CLS), with several differences between the two subgroups (I and II) and with the animal counterpart. Both PpPTENA and PpPTENB contain a clear PDB motif and cytosolic signal similar to that of the animal counterpart (Supplemental Fig. S1). However, these sequences do not share a high degree of conservation in PpPTENC and PpPTEND. Although PpPTENs exhibit conserved residues at the positions 124 and 130 (human PTEN numbering), differences are also observed within the phosphate-binding loop sequence of the two subgroups. Both PpPTENC and PpPTEND exhibit a substitution of A126G, which probably has no effect on catalytic activity, but an additional substitution specific for PpPTEND, G129E, replaced a neutral amino acid with one having a strong negative charge, which could support the absence of lipid phosphatase activity reported here. Altering human PTEN with this change resulted in lack of lipid phosphatase activity without disrupting protein phosphatase activity (Myers et al., 1997).
The in vitro characterization of recombinant PTEN isoforms from P. patens (Fig. 1) suggests that the moss PTENs, like other reported plant PTENs, have a more promiscuous lipid phosphatase activity than the mammalian isoforms (Pribat et al., 2012). In agreement with other reports (Zhang et al., 2011; Pribat et al., 2012), we found that PtdIns3P, PtdIns(3,4)P2, and PtdIns(3,5)P2 are all dephosphorylated by PTENs (A and B). Technical difficulties (associated with the bacterial expression of GST-PpPTENC) prevented us from assaying PTENC activity, but considering its conserved structure, similar phosphatase activity is predicted. It should be noted that PpPIPK1 and PpPIPK2 (Saavedra et al., 2009) have also been shown to have a broader substrate range when compared with mammalian (Anderson et al., 1999) or Arabidopsis counterparts (Heilmann and Heilmann, 2015). The limited specificity of PPI formation or breakdown might represent an important distinction between the PPI systems of plants and mammals (Saavedra et al., 2011).
All PpPTEN genes were found to be ubiquitously expressed in actively growing tissues (e.g. chloronemal and caulonemal cells and in young growing cells of adult gametophores), suggesting a role in the control of cellular growth. This pattern resembles Arabidopsis PTEN type II expression, which is found in young tissues but not in mature leaves or siliques, and contrasts to AtPTEN1 type I, which is restricted to reproductive tissues (Gupta et al., 2002; Pribat et al., 2012). At the subcellular level, all four PpPTENs localized to the cytosol, with PpPTENC and PpPTEND showing nuclear localization as well. A nuclear localization for PpPTENA and PpPTENB is not to be excluded. Faint nuclear signals were also observed for these two isoforms (Fig. 2D), but the low fluorescence signals obtained with PpPTEN-mEGFP (A and B) made imaging difficult. Imaging of tandem PpPTEN-3XmEGFP constructs did not reveal nuclear localization, but their large size likely prevents normal protein trafficking. In mammalian cells, PTEN is found both in cytosol and nucleus despite the absence of a classical nuclear localization signal. The mechanisms of PTEN trafficking are unknown, with several hypotheses currently under debate, from a simple diffusion process (Liu et al., 2005) to active shuttling regulated by a putative nuclear localization signal that mediates PTEN nuclear entry through the Major Vault Protein (Chung et al., 2005). As previously mentioned, both PDB and CLS domains contribute to PTEN localization in mammalian cells (Walker et al., 2004), with specific mutations in their sequence (e.g. F21A) causing preferential nuclear localization (Denning et al., 2007). Therefore, the differences observed in the PpPTENs’ subcellular localization are likely a consequence of the evolutionary changes in their PDB and CLS domains. PpPTENA-mEGFP and PpPTENB-mEGFP were also found to accumulate in discrete punctate structures, which could reflect association with specific PPI pools. For example, Zhang et al. (2011) reported that Arabidopsis PTEN localized to phosphatidylinositol-3-phosphate (PI3P)-positive vesicles. These can also represent preferential sites for signaling crosstalk with other proteins, namely For2As (van Gisbergen et al., 2012; Cvrčková et al., 2015).
PpPTEN Deletion Perturbs Cellular Growth, Causing Faster Development and Aging
Due to the high degree of similarity and the overall tissue expression pattern, functional redundancy was expected for PpPTEN genes. A significant phenotype caused by disruption of PpPTEN genes was only observed in triple or quadruple knockouts. Double knockout lines, either generated from members of same subgroup (e.g. ptencd) or different subgroups (e.g. ptenbd), exhibited nonsignificant differences to the wild type when grown under normal conditions, suggesting that members of different PpPTEN subgroups act in the same pathway.
Triple and quadruple pten knockout plants were characterized by shorter caulonemal cells with a faster growth (when compared with the wild type), an early switch from the juvenile protonemal stage to the development of adult gametophores, and enhanced rhizoid production. Concomitantly, earlier signs of cell death were also observed. These observations are consistent with a functional role of PpPTENs in the regulation of cell cycle events as reported in animal cells (Song et al., 2012).
Cell cycle and cell differentiation are closely linked in P. patens (Schween et al., 2003). We also observed disturbed growth and altered developmental pattern upon PpPTENs overexpression, suggesting that their cellular activity must be balanced at a critical level and that deviations in either direction compromise homeostasis. A similar observation was reported in P. patens RHD SIX-LIKE (RSL) genes in which caulonemal differentiation was caused either by a double Pprsl3 Pprsl4 mutation or the overexpression of PpRSL4 (Pires et al., 2013). This pattern is also characteristic for factors that control localized processes, such as polar tip growth, as previously observed by us. In Arabidopsis pollen tubes and root hairs, knocking down or overexpressing phosphatidylinositol-4-monophosphate 5-kinases resulted in abnormal morphologies and reduced growth (Kusano et al., 2008; Ischebeck et al., 2008; Sousa et al., 2008; Stenzel et al., 2008).
PpPTENs Exert Their Role through Changes in PPI Levels and Regulation of S-Phase Cell Cycle Genes
It remains unclear how PPIs might exert regulatory effects on cell division. The observed overexpression phenotypes, similar for the four PpPTENs, suggest that their effect is not solely due to dephosphorylation of PPI substrates. As reported here (Fig. 1C), PpPTENs exhibited different affinities for PPI isomers, including no lipid phosphatase activity for PpPTEND. Analogous observation was made for the moss Ser/Thr protein kinase PDK1, which was reported to lack a phospholipid-binding domain (Dittrich and Devarenne, 2012), suggesting that in nonvascular land plants, lipid regulation evolved differently.
Unlike some observations in animal cells (Goebbels et al., 2010), a clear increase in a PPI isomer was not observed in our moss pten knockout lines. Only PtdIns(3,5)P2 showed increased levels in the pten knockouts (Fig. 6), whereas other PPIs were not significantly altered or showed even reduced levels. The promiscuity of plant PTENs versus different PPI substrates in vitro makes it thus difficult to link the observed growth effects to changes in any particular PPI in vivo. Interpretation is further limited by a lack of knowledge of how some of the lipids shown to be substrates for PpPTENs are metabolically connected. Compensatory effects by other enzymes in response to misregulated PPI levels are likely, as have previously been proposed for Arabidopsis mutants with defects in the expression of PPI phosphatases of the suppressor of actin family (Nováková et al., 2014). Rapid metabolite turnover and system compensation are characteristics of resilient organisms like Physcomitrella spp. Nevertheless, it is tempting to speculate that the reduced PtdIns3P levels measured in triple and quadruple knockouts result from a misregulation of PI3K. In animal cells, PTEN is a major antagonist of PI3K activity and of the PI3K/AKT/mTOR, an intracellular signaling pathway important in regulating the cell cycle (Song et al., 2012). Plants do not contain AKT kinase genes, but the mTOR/ribosomal S6 kinase is well conserved, and it has recently been shown to negatively regulate cell division as part of a signaling pathway connected to the RBR1-adenovirus E2 promoter (E2F) transcriptional machinery (Henriques et al., 2010).
Our findings that at least three of the four PpPTENs (B, C, and D) localize also to the nucleus, together with the biochemical data discussed above, are in agreement with a functional role for PTENs in cell cycle extending beyond their lipid phosphatase activity (Lindsay et al., 2006). For example, nuclear PTEN was reported to positively regulate DNA repair independently of its phosphatase activity (Shen et al., 2007). Such hypothesis is supported by quantitative PCR data showing up-regulation of S-phase cell cycle genes. Both animals and plants, use the same retinoblastoma (RB)/E2F/dimerization partner (DP) pathway to control the G1-to-S transition (Inzé and De Veylder, 2006). In plants, RBR is phosphorylated in a cell cycle-specific manner by several Cyclin/cyclin-dependent kinase complexes, namely cyclin D (Boniotti and Gutierrez, 2001; Nakagami et al., 2002; Menges et al., 2006). The activated cyclin complexes phosphorylate RB and inhibit RB binding to E2F/DP transcription factor, leading to activation in transcription of genes needed for DNA replication during the S-phase of the cell cycle. In Arabidopsis, RBR was also shown to be a positive regulator of developmental switches promoting the transition from embryonic to autotrophic plant development (Gutzat et al., 2011). The shorter caulonemal cell size observed in pten knockout plants and its higher growth rate were thus possibly a consequence of an accelerated entry into the S-phase of the cell cycle, manifested by an up-regulation of PpPCNA, PpRNR2, PpMCM3, and RBR1 genes and a down-regulation of cyclin D1 gene. This is in agreement with the finding that the chloronemal tissue is arrested in G2, while caulonemal cells are mainly in G1 (Schween et al., 2003). Chloronema-to-caulonema transition upon changes in gene expression have been reported (Jang and Dolan, 2011). We cannot exclude the possibility of altered cell identities in pten knockout plants. However, our observation that chloronemal cell growth is not significantly affected in triple and quadruple knockouts does suggest that PTENs’ main effect is on cell cycle (transition G1/S) and not on cell differentiation.
Taken together, the data presented in this report support the hypothesis that PpPTEN genes are involved in the critical regulation of cell size and cell cycle transitions (Jorgensen and Tyers, 2004), playing a fundamental role in organism development and aging. It further raises new questions about the evolution of plant lipid regulation and how protein function is achieved beyond catalytic activity.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Gransden wild-type strain of Physcomitrella patens (Ashton and Cove, 1977) was used in this study. The wild type and mutant strains were grown axenically at 24°C under a 16-h-light/8-h-dark regime with a photon flux of 55 μmol m–2 s–1. P. patens protonemal tissue was propagated routinely at 7-d intervals on cellophane disks (AA packaging) overlaying petri dishes (90-mm diameter) that contained PP-NH4 medium (PP-NO3 [0.8 g L–1 CaNO3⋅4H2O, 0.25 g L–1 MgSO4⋅7H2O, 0.0125 g L–1 FeSO4⋅7H2O, 0.055 mg L–1 CuSO4⋅5H2O, 0.055 mg L–1 ZnSO4⋅7H2O, 0.614 mg L–1 H3BO3, 0.389 mg L–1 MnCl2⋅4H2O, 0.055 mg L–1 CoCl2⋅6H2O, 0.028 mg L–1 KI, 0.025 mg L–1 Na2MoO4⋅2H2O, and 0.25 mg L–1 KH2PO4 buffer, pH 7.0, with KOH] supplemented with 0.5 g L–1 [di]ammonium tartrate), and 7 g L–1 agar (Duchefa). Phenotypic analyses of the wild type and mutant strains were performed on PP-NO3 medium.
For gametophores and rhizoid analysis of plants, protonemal pieces of approximately 1 mm were placed on PP-NO3 media and grown for 3 to 4 weeks. Chloronemal and caulonemal cell length measurements were performed on three-first subapical cells of 6-d-old protonemata stained with 10 μg mL–1 fluorescence brightener 28 (Sigma-Aldrich).
For cell growth rate measurements, protonemal tissue was grown on petri dishes (30-mm diameter) on PP-NO3 media from 5 to 6 d. Images of apical caulonemal cells were acquired every 30 min during a 2.3-h period with a PCO Sensicam-QE camera (Labocontrole) attached to an Olympus IX-50 microscope (Labocontrole).
For the dark growth experiments, square petri dishes with 7-d-old moss protonemata pieces were grown for 10 d under continuous light on solid PP-NO3 supplemented with 5 mm (di)ammonium tartrate and 2% (w/w) Suc and then grown in darkness in vertical position for 4 weeks.
For sporophyte induction, fresh protonemata were grown in Jiffy7 pots covered by water below 1 cm at 24°C under a 16-h-light/8-h-dark regime. After 6 weeks, cultures were transferred to 19°C under short-day conditions (8-h-light per day) for sporophyte induction.
Data analysis was performed using IMAGEJ software (http://rsbweb.nih.gov/ij/).
Phylogenetic Analysis
The P. patens ssp. patens genome (v1.6, http://www.phytozome.net/) was screened for PTEN homologs using TBLASTN with query sequences from Arabidopsis (Arabidopsis thaliana). PTEN sequences were also searched for the green algae Chlamydomonas reinhardtii and additional 10 representative land plants (P. patens, Selaginella moellendorffii, Pinus taeda, Oryza sativa, Sorghum bicolor, Vitis vinifera, Arabidopsis, Glycine max, and Populus trichocarpa) and from Schizosaccharomyces pombe, Saccharomyces cerevisiae, and Homo sapiens genomes deposited at Phytozome v9.1 and at the National Center for Biotechnology Information. Amino acid sequences were aligned with ClustalW, and the evolutionary history was inferred using the maximum likelihood with the MEGA6 package (Tamura et al., 2013). The bootstrap consensus tree was inferred from 500 replicates. The data matrix was analyzed with heuristic search and the default search options.
RT-PCR
For cloning coding sequences (CDs), gene expression analysis by RT-PCR, or mutant genotyping, total RNA from moss tissue was isolated using the RNeasy Plant Mini kit (Qiagen), according to the manufacturer’s recommendations. One microgram of total RNA was treated with two units of DNaseI (RQ1, Promega) and then used as template for reverse transcription with Superscript III reverse transcriptase (Invitrogen) or RevertAid H Minus Reverse Transcriptase (Thermo Fisher Scientific) and primed with an oligo(dT) primer following the manufacturer’s protocol. A one-fortieth volume of the complementary DNA was subsequently used as template for PCR. Primers used for amplification are listed in Supplemental Table S1.
qRT-PCR
Total RNA was purified using the RNeasy Plant Mini kit (Qiagen) from 12-d-old ground protonemata grown in minimal media (ppNO3). One microgram of total RNA was treated with two units of DNase I (RQ1, Promega) and then used as template for reverse transcription with RevertAid H Minus Reverse Transcriptase (Thermo Fisher Scientific) and primed with an oligo(dT) primer. qRT-PCR was performed using a PikoReal 96-Well System (PikiReal Real-Time PCR System, Thermo Scientific) with Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fisher Scientific). The sequences of primers for qRT-PCR are described in Supplemental Table S1. The genes encoding Ub-E2 and Ade-PRT were used as controls for normalization of mRNA content between samples, according to previously published data (Le Bail et al., 2013). For each gene, nine reactions were performed including three biological replicates and three technical replicates. Melting curves were obtained to ensure that only a single product was amplified. The relative quantification of the target gene was obtained using Pfaffl method (Pfaffl, 2001) in which the relative expression ratio (R) is determined using the Efficiency (E) and the threshold cycle (Ct) deviation of an unknown sample versus a control compared with a reference gene according to the following equation: R = [(Etarget) ΔCt (control–test)]/[(Eref)ΔCt (control–sample)]. For statistical analysis, a Student’s t test was applied (values from the wild type were considered as expected with P < 0.05 and 1 degree of freedom).
Gene Isolation and Protein Expression
Full-length CDs of AtPTEN2a, PpPTENB, and PpPTEND were amplified with specific primers (Supplemental Table S1) and cloned into pENTR/D-TOPO vector (Invitrogen) following the manufacturer’s protocol. PpPTENA and PpPTENC CDs cloned in pENTR/D-TOPO vector were obtained from Magdalena Benzanilla (van Gisbergen et al., 2012). PTEN CDs were then transferred to the destination expression vector pDEST15 (Invitrogen) using LR clonase (Invitrogen). GST-PTEN open reading frames were verified by sequencing.
Escherichia coli Turner cells, harboring a plasmid for the expression of a recombinant fusion protein of GST with PTEN were grown in Luria Broth medium at 37°C and 180 rpm until an optical density at 600 nm of 0.6 to 0.8 was reached. At this point, isopropyl β-d-thiogalactoside was added to a final concentration of 0.2 mm. Cells were incubated overnight at 19°C and 200 rev min–1 and then collected by centrifugation and frozen at –80°C. For GST fusion protein purification, frozen cells were thawed on ice and resuspended in Tris-buffered saline extraction buffer (10 mm Tris-HCl, pH 8.0, 140 mm NaCl containing 1% [w/v] Triton X-100, 1 mm EDTA, 1 mm dithiothreitol, and 1 mm phenylmethylsulfonyl fluoride). The crude extract was sonicated for 120 s (0.5-s intervals) and centrifuged for 15 min at 10,000g at 4°C. The supernatant was incubated with 0.1 mL of glutathione ± Sepharose resin (Amersham Pharmacia Biotech) and pre-equilibrated with extraction buffer for 1 h at 4°C. The resin was then washed with extraction buffer twice, and then proteins bound to the resin were eluted with 10 mm glutathione (Sigma-Aldrich), 50 mm Tris-HCl, pH 8.0, and 5% (v/v) glycerol and stored at –80°C.
Lipid Phosphatase Activity Using Malachite Green Assays
PTEN hydrolyzing activity toward the different PPIs was determined in vitro by the malachite green-based assay that measures the released inorganic phosphate (Odriozola et al., 2007). Briefly, 1 μg of recombinant protein was incubated with 100 μm of one of the seven di-C8 PPI lipids [PI3P, phosphatidylinositol-4-phosphate, phosphatidylinositol-5-phosphate, phosphatidylinositol (3,4)-bisphosphate, phosphatidylinositol (3,5)-bisphosphate, phosphatidylinositol (4,5)-bisphosphate, or phosphatidylinositol (3,4,5)-trisphosphate (Echelon Biosciences)] for 60 min at 37°C in a final volume of 25 μL. The reaction buffer was 25 mm Tris-HCl, pH 7.4, 140 mm NaCl, 2.7 mm KCl, and 10 mm dithiothreitol. The reaction was stopped with 100 μL of malachite green reagent (Echelon Biosciences). The amount of phosphate released was determined by reading the A630 in an ELx800 Absorbance Microplate Reader (BioTek) and converted to a molar amount of product using a standard curve. The amount of protein used and the duration of the assay were within the linear range of the reaction. Free inorganic phosphate was measured in controls (buffer, substrate solutions, and recombinant proteins alone) and subtracted for calculating PTEN activity. GST was used as negative control. For the reaction with PI3P, 38% (v/v) of the substrate was consumed at the end of the reaction. The activity assay was performed in triplicates and are presented as the mean ± se of two independent experiments.
Lipid Analysis
Material was ground in liquid nitrogen. Phospholipids were extracted from 250 mg of the ground material and analyzed exactly as described previously (König et al., 2008). For statistical analysis, a Student’s t test was applied (values from the wild type were considered as expected with P < 0.05).
Generation of Targeting Constructs
Sequences for targeting constructs were all amplified from genomic DNA isolated from moss protonemata. The PpPTEN gene knock-in constructs were performed using Multisite Gateway four-fragment recombination (Invitrogen). In first position, a fragment of 1,000 bp just before the stop codon was amplified using primers containing attB1 and attB5r sites (Supplemental Table S1) and cloned into pDONR 221 P1-P5r using BP clonase. At second position, EGFP cloned into pDONR 221 P5-P4 was used. At third position, pDONR 221 P4r-P3r containing a hygromycin resistance cassette was used, and at fourth position, a fragment of 1,000 bp belonging to the 3′ untranslated region of PpPTENs was amplified using primers containing attB3 and attB2 sites (Supplemental Table S1) and cloned into pDONR 221 P3-P2 using BP clonase. Sequences of specific primers used for cloning the 5′ fragment and 3′ fragment for the knock-in constructs are listed in Supplemental Table S1. EGFP and hygromycin sequences cloned in pEntry vectors were obtained from Magdalena Benzanilla.
As target sequences for homologous recombination for PpPTEN gene disruption, two genomic DNA fragments of 1,000 bp from the 5′ and 3′ regions of PpPTEN genes were cloned with specific primers (Supplemental Table S1). The PpPTENA knockout construct was performed the using Multisite Gateway three-fragment recombination (Invitrogen). For this purpose, 5′ PpPTENA and 3′ PpPTENA genomic DNA fragments were cloned into pDONR 221 P1-P4 and pDONR 221 P3-P2 using BP clonase (Invitrogen). These entry vectors and Nos_loxGentlox-R3R4 vector were cloned into the pGEM. PpPTENB, PpPTENC, and PpPTEND knockout constructs were obtained by cloning with restriction enzymes using fragments amplified with primers listed in Supplemental Table S1 cloned into p35SZeo, pBHRF, and pBNRF, respectively. All generated clones were verified by sequencing.
For the overexpression analysis PpPTENA to PpPTEND, CDs cloned in pENTR/D-TOPO were transferred to the destination vector uj3-pTUbiGate (Anterola et al., 2009) using LR clonase II (Invitrogen).
Sequences of all targeting vectors used for transformation were verified by sequencing. In all cases, putative transformant lines growing in the corresponding antibiotic selective media from knock-in, knockout, and overexpression experiments were verified by PCR and RT-PCR with primers sequences listed in Supplemental Table S1.
Transformation of P. patens
Isolation of protoplasts, polyethylene glycol-mediated transformation, regeneration, and antibiotic selection were performed as described previously (Schaefer and Zrÿd, 1997). For polyethylene glycol-mediated protoplast transformation, 6-d-old protonemata were treated with 0.5% Driselase in 8.5% (w/v) mannitol for 30 min, passed through a 100-mm sieve, incubated for 15 min at room temperature, and passed through a 50-mm sieve. The protoplasts of the final flowthrough were washed twice in 8.5% mannitol before further use. Protoplasts were transformed at a concentration of 1.6 × 106 protoplasts mL–1. Each transformation consisted of 0.3 mL of protoplast suspension and 15 to 20 μg of linear DNA. To eliminate any episomal resistant colonies, two rounds of selection were undertaken using the appropriate antibiotic. To select for antibiotic-resistant cells, G418, Zeocin, Hygromycin, and Gentamycin all purchased from Duchefa were added at 50, 25, 25, and 200 mg L–1 to the medium, respectively.
Fluorescence Imaging
Moss protonemata was inoculated over cellophane of a PP-NO3 growth medium plate and grown for 7 to 20 d. At these two time points, small protonemata pieces or gametophores were transferred to a slide, and microscopic fluorescence images were acquired with a Leica SP-E confocal laser scanning microscope. Thin time course optical sections (approximately 3 µm thick) were acquired using a ×20 Plan Apo dry objective (numerical aperture = 0.75) and less than 20% laser intensity and operating in the mode 1,024 × 1,024 and 400 Hz (approximately one-third s per frame). GFP was imaged using the 488-nm excitation line, and the chlorophyll signal was imaged with the 532-nm excitation line. Images were then processed using the LAS AFL 2.6.0 (Leica Microsystems) and Image-Pro Plus 5.0 software (Media Cybernetics, Leiden, The Netherlands).
GFP fluorescence images from gametophores were acquired using a Zeiss SteREO Lumar.V12 and an Imaging Source DFK23u274 camera.
Sequence data from this article can be found at the Phytozome database: PpPTENA (Phpat.019G062100), PpPTENB (Phpat.022G039200), PpPTENC (Phpat.021G028600), and PpPTEND (Phpat.022G052200).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Maximum likelihood tree showing that the P. patens PTEN proteins fall into the type II class.
Supplemental Figure S2. PDB and CLS motifs of PpPTENs.
Supplemental Figure S3. Immunoblot of recombinant GST-PTENs.
Supplemental Figure S4. Expression analysis of PpPTENs.
Supplemental Figure S5. Genotyping and western-blot analysis of PpPTENs transformants.
Supplemental Figure S6. Constructs used for homologous recombination.
Supplemental Figure S7. PCR genotyping analysis of wild-type, triple, and quadruple pten knockouts.
Supplemental Figure S8. Total colony area values in square centimeters from wild-type, triple, and quadruple pten knockouts growing in minimal media for 5 weeks.
Supplemental Figure S9. Effects of pten deletion on moss structural morphology.
Supplemental Figure S10. Overexpressing PpPTEN mutants by disruption of the Pp108 locus.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank Drs. Andreia Figueiredo, Filipa Monteiro, and Ana Margarida Fortes (University of Lisbon) for the technical assistance in qRT-PCR, Dr. Mattias Thelander (Swedish University of Agricultural Sciences) for the pUBW302 vector, Dr. Mitsuyasu Hasebe (National Institute Basic Biology, Japan) for the p35S-Zeo vector, Dr. Pierre-François Perroud (Washington University, St. Louis) for the uj3-pTUbiGate vector, Dr. Fabien Nogue (Institut National de la Recherche Agronomique, Institut Jean-Pierre Bourgin) for the pBNRF and pBHRF vectors, and Dr. Mareike Heilmann (Martin-Luther-University Halle-Wittenberg) for helpful discussion on cell cycle regulation.
Glossary
- PtdIns(3,4,5)P3
phosphatidylinositol (3,4,5)-trisphosphate
- PtdIns(3,5)P2
phosphatidylinositol (3,5)-bisphosphate
- PtdIns(4,5)P2
phosphatidylinositol (4,5)-bisphosphate
- qRT
quantitative real-time
- PPI
phosphoinositide
- PtdIns3P
phosphatidylinositol 3-phosphate
- For2A
class II formin
- GST
glutathione S-transferase
- RT
real-time
- PI3P
phosphatidylinositol-3-phosphate
- CD
coding sequence
Footnotes
This work was supported by the Fundação Ciência e Tecnologia (Ministry of Science, Technology, and Higher Education/Programa de Investimentos e Despesas de Desenvolvimento da Administração Central, Portugal; Postdoctoral Fellowship/63619/2009 to L.S., PhD grant no. PD/BD/52493/2014 to R.C., and research funds PEst–OE/BIA/UI4046/2014 and UID/MULTI/04046/2013 to R.M.), the Fundação Ciência e Tecnologia/Deutscher Akademischer Austauschdienst program (grant no. daad130757760421333), and the National Science Foundation (MCB–1330171 to M.B.).
L.S. conceived and carried out the experiments, designed and carried out the data analysis, and cowrote the article; R.C. carried out the experiments and the data analysis; T.H. performed the lipid analysis; I.H. and M.B. cowrote and revised the article; R.M. conceived the experiments, carried out the data analysis, and cowrote and revised the article.
Articles can be viewed without a subscription.
References
- Anderson RA, Boronenkov IV, Doughman SD, Kunz J, Loijens JC (1999) Phosphatidylinositol phosphate kinases, a multifaceted family of signaling enzymes. J Biol Chem 274: 9907–9910 [DOI] [PubMed] [Google Scholar]
- Anterola A, Shanle E, Perroud PF, Quatrano R (2009) Production of taxa-4(5),11(12)-diene by transgenic Physcomitrella patens. Transgenic Res 18: 655–660 [DOI] [PubMed] [Google Scholar]
- Ashton NW, Cove DJ (1977) Isolation and preliminary characterization of auxotrophic and analog resistant mutants of moss Physcomitrella patens. Mol Gen Genet 154: 87–95 [Google Scholar]
- Boniotti MB, Gutierrez C (2001) A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J 28: 341–350 [DOI] [PubMed] [Google Scholar]
- Bononi A, Bonora M, Marchi S, Missiroli S, Poletti F, Giorgi C, Pandolfi PP, Pinton P (2013) Identification of PTEN at the ER and MAMs and its regulation of Ca2+ signaling and apoptosis in a protein phosphatase-dependent manner. Cell Death Differ 20: 1631–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss WF, Im YJ (2012) Phosphoinositide signaling. Annu Rev Plant Biol 63: 409–429 [DOI] [PubMed] [Google Scholar]
- Chalhoub N, Baker SJ (2009) PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol 4: 127–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Ginn-Pease ME, Eng C (2005) Phosphatase and tensin homologue deleted on chromosome 10 (PTEN) has nuclear localization signal-like sequences for nuclear import mediated by major vault protein. Cancer Res 65: 4108–4116 [DOI] [PubMed] [Google Scholar]
- Cove D. (2005) The moss Physcomitrella patens. Annu Rev Genet 39: 339–358 [DOI] [PubMed] [Google Scholar]
- Cvrčková F, Oulehlová D, Žárský V (2015) Formins: linking cytoskeleton and endomembranes in plant cells. Int J Mol Sci 16: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning G, Jean-Joseph B, Prince C, Durden DL, Vogt PK (2007) A short N-terminal sequence of PTEN controls cytoplasmic localization and is required for suppression of cell growth. Oncogene 26: 3930–3940 [DOI] [PubMed] [Google Scholar]
- Dittrich AC, Devarenne TP (2012) Characterization of a PDK1 homologue from the moss Physcomitrella patens. Plant Physiol 158: 1018–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont J, Renou JP, Shani M, Hennighausen L, LeRoith D (2002) PTEN overexpression suppresses proliferation and differentiation and enhances apoptosis of the mouse mammary epithelium. J Clin Invest 110: 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elge S, Brearley C, Xia HJ, Kehr J, Xue HW, Mueller-Roeber B (2001) An Arabidopsis inositol phospholipid kinase strongly expressed in procambial cells: synthesis of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in insect cells by 5-phosphorylation of precursors. Plant J 26: 561–571 [DOI] [PubMed] [Google Scholar]
- Eng C. (2003) PTEN: one gene, many syndromes. Hum Mutat 22: 183–198 [DOI] [PubMed] [Google Scholar]
- Funamoto S, Meili R, Lee S, Parry L, Firtel RA (2002) Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell 109: 611–623 [DOI] [PubMed] [Google Scholar]
- Furnari FB, Lin H, Huang HS, Cavenee WK (1997) Growth suppression of glioma cells by PTEN requires a functional phosphatase catalytic domain. Proc Natl Acad Sci USA 94: 12479–12484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebbels S, Oltrogge JH, Kemper R, Heilmann I, Bormuth I, Wolfer S, Wichert SP, Möbius W, Liu X, Lappe-Siefke C, et al. (2010) Elevated phosphatidylinositol 3,4,5-trisphosphate in glia triggers cell-autonomous membrane wrapping and myelination. J Neurosci 30: 8953–8964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunt M, Zárský V, Cvrcková F (2008) Roots of angiosperm formins: the evolutionary history of plant FH2 domain-containing proteins. BMC Evol Biol 8: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Ting JT, Sokolov LN, Johnson SA, Luan S (2002) A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell 14: 2495–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzat R, Borghi L, Fütterer J, Bischof S, Laizet Y, Hennig L, Feil R, Lunn J, Gruissem W (2011) RETINOBLASTOMA-RELATED PROTEIN controls the transition to autotrophic plant development. Development 138: 2977–2986 [DOI] [PubMed] [Google Scholar]
- Heilmann M, Heilmann I (2015) Plant phosphoinositides-complex networks controlling growth and adaptation. Biochim Biophys Acta 1851: 759–769 [DOI] [PubMed] [Google Scholar]
- Henriques R, Magyar Z, Monardes A, Khan S, Zalejski C, Orellana J, Szabados L, de la Torre C, Koncz C, Bögre L (2010) Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. EMBO J 29: 2979–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Potter CJ, Tao W, Li DM, Brogiolo W, Hafen E, Sun H, Xu T (1999) PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development 126: 5365–5372 [DOI] [PubMed] [Google Scholar]
- Iijima M, Devreotes P (2002) Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell 109: 599–610 [DOI] [PubMed] [Google Scholar]
- Inzé D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Ischebeck T, Stenzel I, Heilmann I (2008) Type B phosphatidylinositol-4-phosphate 5-kinases mediate Arabidopsis and Nicotiana tabacum pollen tube growth by regulating apical pectin secretion. Plant Cell 20: 3312–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang G, Dolan L (2011) Auxin promotes the transition from chloronema to caulonema in moss protonema by positively regulating PpRSL1and PpRSL2 in Physcomitrella patens. New Phytol 192: 319–327 [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Tyers M (2004) How cells coordinate growth and division. Curr Biol 14: R1014–R1027 [DOI] [PubMed] [Google Scholar]
- Kim JS, Xu X, Li H, Solomon D, Lane WS, Jin T, Waldman T (2011) Mechanistic analysis of a DNA damage-induced, PTEN-dependent size checkpoint in human cells. Mol Cell Biol 31: 2756–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König S, Hoffmann M, Mosblech A, Heilmann I (2008) Determination of content and fatty acid composition of unlabeled phosphoinositide species by thin-layer chromatography and gas chromatography. Anal Biochem 378: 197–201 [DOI] [PubMed] [Google Scholar]
- Kusano H, Testerink C, Vermeer JEM, Tsuge T, Shimada H, Oka A, Munnik T, Aoyama T (2008) The Arabidopsis Phosphatidylinositol Phosphate 5-Kinase PIP5K3 is a key regulator of root hair tip growth. Plant Cell 20: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bail A, Scholz S, Kost B (2013) Evaluation of reference genes for RT-qPCR analyses of structure-specific and hormone regulated gene expression in Physcomitrella patens gametophytes. PLoS One 8: e70998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. (1997) PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275: 1943–1947 [DOI] [PubMed] [Google Scholar]
- Liang H, He S, Yang J, Jia X, Wang P, Chen X, Zhang Z, Zou X, McNutt MA, Shen WH, et al. (2014) PTENα, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab 19: 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, et al. (1997) Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet 16: 64–67 [DOI] [PubMed] [Google Scholar]
- Lindsay Y, McCoull D, Davidson L, Leslie NR, Fairservice A, Gray A, Lucocq J, Downes CP (2006) Localization of agonist-sensitive PtdIns(3,4,5)P3 reveals a nuclear pool that is insensitive to PTEN expression. J Cell Sci 119: 5160–5168 [DOI] [PubMed] [Google Scholar]
- Liu F, Wagner S, Campbell RB, Nickerson JA, Schiffer CA, Ross AH (2005) PTEN enters the nucleus by diffusion. J Cell Biochem 96: 221–234 [DOI] [PubMed] [Google Scholar]
- Maehama T, Dixon JE (1998) The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 273: 13375–13378 [DOI] [PubMed] [Google Scholar]
- Maier D, Jones G, Li X, Schönthal AH, Gratzl O, Van Meir EG, Merlo A (1999) The PTEN lipid phosphatase domain is not required to inhibit invasion of glioma cells. Cancer Res 59: 5479–5482 [PubMed] [Google Scholar]
- Menand B, Calder G, Dolan L (2007) Both chloronemal and caulonemal cells expand by tip growth in the moss Physcomitrella patens. J Exp Bot 58: 1843–1849 [DOI] [PubMed] [Google Scholar]
- Menges M, Samland AK, Planchais S, Murray JA (2006) The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell 18: 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK (1998) The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci USA 95: 13513–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MP, Stolarov JP, Eng C, Li J, Wang SI, Wigler MH, Parsons R, Tonks NK (1997) P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc Natl Acad Sci USA 94: 9052–9057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A (2002) Phosphorylation of retinoblastoma-related protein by the cyclin D/cyclin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 14: 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nováková P, Hirsch S, Feraru E, Tejos R, van Wijk R, Viaene T, Heilmann M, Lerche J, De Rycke R, Feraru MI, et al. (2014) SAC phosphoinositide phosphatases at the tonoplast mediate vacuolar function in Arabidopsis. Proc Natl Acad Sci USA 111: 2818–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odriozola L, Singh G, Hoang T, Chan AM (2007) Regulation of PTEN activity by its carboxyl-terminal autoinhibitory domain. J Biol Chem 282: 23306–23315 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires ND, Yi K, Breuninger H, Catarino B, Menand B, Dolan L (2013) Recruitment and remodeling of an ancient gene regulatory network during land plant evolution. Proc Natl Acad Sci USA 110: 9571–9576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribat A, Sormani R, Rousseau-Gueutin M, Julkowska MM, Testerink C, Joubès J, Castroviejo M, Laguerre M, Meyer C, Germain V, et al. (2012) A novel class of PTEN protein in Arabidopsis displays unusual phosphoinositide phosphatase activity and efficiently binds phosphatidic acid. Biochem J 441: 161–171 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Rouault JP, Kuwabara PE, Sinilnikova OM, Duret L, Thierry-Mieg D, Billaud M (1999) Regulation of dauer larva development in Caenorhabditis elegans by daf-18, a homologue of the tumour suppressor PTEN. Curr Biol 9: 329–332 [DOI] [PubMed] [Google Scholar]
- Saavedra L, Balbi V, Dove SK, Hiwatashi Y, Mikami K, Sommarin M (2009) Characterization of phosphatidylinositol phosphate kinases from the moss Physcomitrella patens: PpPIPK1 and PpPIPK2. Plant Cell Physiol 50: 595–609 [DOI] [PubMed] [Google Scholar]
- Saavedra L, Balbi V, Lerche J, Mikami K, Heilmann I, Sommarin M (2011) PIPKs are essential for rhizoid elongation and caulonemal cell development in the moss Physcomitrella patens. Plant J 67: 635–647 [DOI] [PubMed] [Google Scholar]
- Schaefer DG, Zrÿd JP (1997) Efficient gene targeting in the moss Physcomitrella patens. Plant J 11: 1195–1206 [DOI] [PubMed] [Google Scholar]
- Schaefer DG, Zrÿd JP (2001) The moss Physcomitrella patens, now and then. Plant Physiol 127: 1430–1438 [PMC free article] [PubMed] [Google Scholar]
- Schween G, Gorr G, Hohe A, Reski R (2003) Unique tissue-specific cell cycle in Physcomitrella. Plant Biol 5: 50–58 [Google Scholar]
- Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, Yin Y (2007) Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 128: 157–170 [DOI] [PubMed] [Google Scholar]
- Slomovitz BM, Coleman RL (2012) The PI3K/AKT/mTOR pathway as a therapeutic target in endometrial cancer. Clin Cancer Res 18: 5856–5864 [DOI] [PubMed] [Google Scholar]
- Song MS, Carracedo A, Salmena L, Song SJ, Egia A, Malumbres M, Pandolfi PP (2011) Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell 144: 187–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Salmena L, Pandolfi PP (2012) The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol 13: 283–296 [DOI] [PubMed] [Google Scholar]
- Sousa E, Kost B, Malhó R (2008) Arabidopsis phosphatidylinositol-4-monophosphate 5-kinase 4 regulates pollen tube growth and polarity by modulating membrane recycling. Plant Cell 20: 3050–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, et al. (1997) Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet 15: 356–362 [DOI] [PubMed] [Google Scholar]
- Stenzel I, Ischebeck T, König S, Hołubowska A, Sporysz M, Hause B, Heilmann I (2008) The type B phosphatidylinositol-4-phosphate 5-kinase 3 is essential for root hair formation in Arabidopsis thaliana. Plant Cell 20: 124–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon-Cardo C, Erdjument-Bromage H, et al. (2007) Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128: 141–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gisbergen PA, Li M, Wu SZ, Bezanilla M (2012) Class II formin targeting to the cell cortex by binding PI(3,5)P(2) is essential for polarized growth. J Cell Biol 198: 235–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L, Bezanilla M (2012) Physcomitrella patens: a model for tip cell growth and differentiation. Curr Opin Plant Biol 15: 625–631 [DOI] [PubMed] [Google Scholar]
- Walker SM, Leslie NR, Perera NM, Batty IH, Downes CP (2004) The tumour-suppressor function of PTEN requires an N-terminal lipid-binding motif. Biochem J 379: 301–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Shen WH (2008) PTEN: a new guardian of the genome. Oncogene 27: 5443–5453 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li S, Zhou LZ, Fox E, Pao J, Sun W, Zhou C, McCormick S (2011) Overexpression of Arabidopsis thaliana PTEN caused accumulation of autophagic bodies in pollen tubes by disrupting phosphatidylinositol 3-phosphate dynamics. Plant J 68: 1081–1092 [DOI] [PubMed] [Google Scholar]