A fungal protein increases plant inflorescence branching when both delivered by the fungus in maize and stably expressed in Arabidopsis, suggesting a conserved pathway for function.
Abstract
The biotrophic fungus Sporisorium reilianum causes head smut of maize (Zea mays) after systemic plant colonization. Symptoms include the formation of multiple female inflorescences at subapical nodes of the stalk because of loss of apical dominance. By deletion analysis of cluster 19-1, the largest genomic divergence cluster in S. reilianum, we identified a secreted fungal effector responsible for S. reilianum-induced loss of apical dominance, which we named SUPPRESSOR OF APICAL DOMINANCE1 (SAD1). SAD1 transcript levels were highly up-regulated during biotrophic fungal growth in all infected plant tissues. SAD1-green fluorescent protein fusion proteins expressed by recombinant S. reilianum localized to the extracellular hyphal space. Transgenic Arabidopsis (Arabidopsis thaliana)-expressing green fluorescent protein-SAD1 displayed an increased number of secondary rosette-leaf branches. This suggests that SAD1 manipulates inflorescence branching architecture in maize and Arabidopsis through a conserved pathway. Using a yeast (Saccharomyces cerevisiae) two-hybrid library of S. reilianum-infected maize tissues, we identified potential plant interaction partners that had a predicted function in ubiquitination, signaling, and nuclear processes. Presence of SAD1 led to an increase of the transcript levels of the auxin transporter PIN-FORMED1 in the root and a reduction of the branching regulator TEOSINTE BRANCHED1 in the stalk. This indicates a role of SAD1 in regulation of apical dominance by modulation of branching through increasing transcript levels of the auxin transporter PIN1 and derepression of bud outgrowth.
Plants are known to exhibit a broad range of different morphologies under different environmental conditions. In addition to changing height or leaf area, they are also able to alter their morphological appearance and develop leaves instead of floral organs or change their bud outgrowth pattern. Pathological changes in the developmental program are often caused by plant infection with biotrophic pathogens. Sporisorium reilianum, a biotrophic fungal pathogen of maize (Zea mays) and sorghum (Sorghum bicolor), interferes with regular development of inflorescences of its host plants and leads to phyllody (Matheussen et al., 1991) that is caused by changes in floral organ identity, floral meristem identity, and floral meristem determinacy (Matheussen et al., 1991; Ghareeb et al., 2011). In addition, the fungus triggers suppression of apical dominance, which leads to a higher number of female inflorescences (ears) of maize (Ghareeb et al., 2011) and increased tillering of sorghum (Matheussen et al., 1991). In fact, the inflorescence tillering symptom resembles the crazy top disease of maize that is induced by the downey mildew fungus Sclerophthora macrospora (Malaguti et al., 1977). Broom-like structures on its host plants are also induced by Moniliophthora perniciosa that induces hyperplasia and hyperproliferation of axillary shoots, resulting in witches’ broom disease of cacao (Theobroma cacao; Meinhardt et al., 2008). Similarly, infection of a whole range of dicotyledonous plants with biotrophic phytopathogenic bacteria of the taxon Candidatus Phytoplasma asteris leads to the reversion of floral organs into leaves or the emergence of a dense cluster of branches resembling broom as a consequence of lack of apical dominance (Hoshi et al., 2009).
Branching architecture is mainly regulated by auxin, strigolactone, and cytokinin, which affect axillary meristem initiation and outgrowth (McSteen, 2009; Gallavotti, 2013). Axillary meristem initiation is preceded by a depletion of auxin as shown for Arabidopsis (Arabidopsis thaliana) and tomato (Solanum lycopersicum; Wang et al., 2014). After meristem initiation, the emerging axillary meristem is characterized by a local accumulation of auxin, which is shown in maize (Gallavotti et al., 2008b). Basipetal auxin transport from the apex toward the root suppresses axillary meristem outgrowth, leading to apical dominance (Davies et al., 1966). Outgrowth of axillary buds is also inhibited by strigolactones synthesized in the shoots directly or long distance transported from the root to the shoot (Turnbull et al., 2002; Crawford et al., 2010). However, bud outgrowth is promoted by cytokinins produced in the nodal stem (Nordström et al., 2004; Tanaka et al., 2006). Cytokinins also modulate branching indirectly through regulation of meristem size (Shani et al., 2006). Although many details about the contribution of particular hormones to regulation of axillary meristem development have been elucidated, the exact mode of hormone action is still ambiguous (Cheng et al., 2013; Barazesh and McSteen, 2008).
Increasing knowledge about the mode of action of auxins, cytokinins, and strigolactones led to the proposal of two models, the auxin transport canalization model and the second messenger model, that explain apical dominance mechanistically (Domagalska and Leyser, 2011). In the auxin transport canalization model, polar auxin transport (PAT) plays the central role in apical dominance, regardless of the auxin concentration. During basipetal movement, auxin flux is canalized gradually into a tight thread of cells, the auxin canal, resulting in high PAT (Sachs, 1981). To be activated, the bud must also generate PAT and export auxin to the auxin canal in the stem (Domagalska and Leyser, 2011). Strigolactones also impact on bud activity, because they reduce PAT (Prusinkiewicz et al., 2009). In the second messenger model, auxin regulates cytokinins and strigolactones (second messengers) that migrate to the axillary bud and controls its activity (Domagalska and Leyser, 2011). Several studies have been conducted that support either of the two models (Domagalska and Leyser, 2011).
Axillary meristem initiation and outgrowth are also under genetic control of transcription factors and signaling components. In maize, determination of axillary meristems is influenced by DELAYED FLOWERING1 (DLF1), Zea FLORICAULA/LEAFY1 (ZFL1), and ZFL2 transcription factors (Coe et al., 1988; Sheridan, 1988; Bomblies and Doebley, 2006; Muszynski et al., 2006). The outgrowth of these axillary meristems is controlled by TEOSINTE BRANCHED1 (TB1), a suppressor of axillary branches (Hubbard et al., 2002). In addition, the Ser/Thr-protein kinase BARREN INFLORESCENCE2 (BIF2) is involved in auxin signaling, and disruption leads to a defect in inflorescence branch meristem initiation (McSteen and Hake, 2001; McSteen et al., 2007). The auxin efflux carrier PIN-FORMED1 (PIN1) also plays a regulating role in axillary meristem outgrowth (McSteen, 2009). Both PIN1 and BIF2 act on PAT and thereby, control bud outgrowth (Bennetzen and Hake, 2009).
In rare cases, mechanisms have been proposed to explain pathogen-mediated changes in plant branching architectures. Infection with S. reilianum decreased the GA content of sorghum, suggesting that low GA concentration is the reason for increased tillering of infected plants (Matheussen et al., 1991). A misregulation of GA biosynthesis genes was observed in S. reilianum-colonized maize inflorescences with early signs of floral reversion (Ghareeb et al., 2011). Additionally, misregulated genes were homologous to genes involved in auxin and cytokinin mobilization in other plants (Ghareeb et al., 2011). However, some examples exist where the architecture of the plant is altered by pathogen effector proteins that are secreted during plant colonization. SECRETED AY-WB PROTEIN54 was identified as a secreted protein from Candidatus Phytoplasma asteris that induces leafy flower development when expressed in Arabidopsis (MacLean et al., 2011). Likewise, the tengu-su inducer (TENGU) effector of Phytoplasma was shown to induce witches’ broom when expressed in Arabidopsis, and the TENGU-expressing transgenic plants showed down-regulation of auxin-responsive genes. This suggests that TENGU inhibits auxin-related pathways, which in turn, influence plant development (Hoshi et al., 2009).
The maize smut fungus Ustilago maydis is capable of auxin biosynthesis, and fungal auxin contributes to the total auxin content of colonized tumorous leaf tissues (Reineke et al., 2008). Interestingly, the fungal capacity of auxin formation did not affect the nature of the symptoms induced on its host plant maize (Reineke et al., 2008), suggesting that symptom formation may instead be influenced by fungal effector proteins secreted into the colonized tissues.
Analysis of the U. maydis genome sequence revealed the existence of gene clusters encoding secreted proteins with an effect on virulence (Kämper et al., 2006). After sequence elucidation of the S. reilianum genome, a genomic comparison identified divergence clusters encoding proteins weakly conserved between U. maydis and S. reilianum (Schirawski et al., 2010). The largest divergence cluster (19-1; Schirawski et al., 2010) corresponded to the U. maydis gene cluster 19A, and deletion resulted in the lack of U. maydis-specific tumor formation in seedling leaves (Kämper et al., 2006) but not in inflorescences (Skibbe et al., 2010).
In this study, we identified one gene (sr10077; SUPPRESSOR OF APICAL DOMINANCE1 [SAD1]) of cluster 19-1 of S. reilianum as responsible for the loss of apical dominance phenotype induced by S. reilianum wild-type strains. SAD1 is highly up-regulated upon fungal colonization of maize and potentially interacts with a large number of intracellular maize proteins. Lack of SAD1 in S. reilianum leads to decreased transcript levels of the polar auxin transporter PIN1 in roots and increased levels of the bud-outgrowth regulator TB1 in stalks of infected maize. When expressed heterologously as an SAD1-GFP fusion protein in Arabidopsis, SAD1 leads to an increase in secondary inflorescence branches. This suggests that SAD1 affects apical dominance by a conserved mechanism that includes regulation of gene transcription and modulation of auxin transport.
RESULTS
Cluster 19-1 Harbors an SAD
The divergence cluster 19-1 was identified as the largest region of genomic difference between S. reilianum and U. maydis (Schirawski et al., 2010). Because deletion of the gene cluster in U. maydis resulted in the loss of the U. maydis-specific symptom of tumor formation on seedling leaves (Kämper et al., 2006), we investigated whether the corresponding region in S. reilianum also encoded symptom specificity determinants.
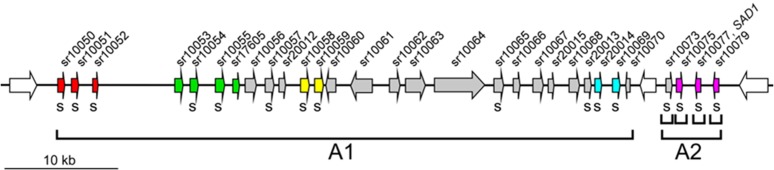
Cluster 19-1 of S. reilianum covers about 55 kb and is split in two parts by the presence of a highly conserved gene (sr10071; Fig. 1) encoding a predicted protein with strong sequence identity to tubulin β-chains of different fungi. Replacement of the first part (A1) with the phleomycin resistance cassette and replacement of the second part (A2) with the hygromycin resistance cassette resulted in strains with reduced virulence on the maize ‘Gaspe Flint’ (H. Ghareeb, Y. Zhao, and J. Schirawski, unpublished data). We used compatible mating type combinations of deletion strains for inoculation experiments of maize to analyze loss of the S. reilianum-specific symptom formation. Each inoculation experiment was done in three biological replicates with about 25 plants inoculated per experiment and strain combination.
Figure 1.
Overview of S. reilianum cluster 19-1 gene organization. Diagram showing the gene organization of cluster 19-1 of S. reilianum. The gene cluster comprises 30 genes (filled colored and gray arrows oriented in their direction of transcription with gene numbers indicated along the top). Homologous genes within this cluster are highlighted with identical coloring. The code below the genes indicates prediction for secretion (s indicates target P, reliability class 1-3; Emanuelsson et al., 2007). The delineation of gene subcluster deletions (A1 and A2) as well as the used individual gene deletions within part A2 are indicated by brackets below the scheme.
In all experiments, wild-type and deletion strains lacking either part A1 (ΔA1 strains) or part A2 (ΔA2 strains) were able to cause smutted ears and ears showing phyllody (H. Ghareeb, Y. Zhao, and J. Schirawski, unpublished data). In addition, we carefully dissected all ears by removal of the surrounding husk leaves (Supplemental Fig. S1). Outgrowth at subapical nodes of the shank was unwrapped and counted as additional ears if the structures could be macroscopically identified as ears (Supplemental Fig. S1D). Numbers of apical and subapical ears were added to calculate the number of ears per plant. Outgrowth of ears at subapical nodes is controlled by apical dominance. Suppression of apical dominance caused by S. reilianum occurred only with wild-type and ΔA1 strains but not with ΔA2 strains (Fig. 2A). Loss of apical dominance occurred predominantly at subapical nodes of a branch carrying an infected female inflorescence at the apex (Ghareeb et al., 2011; Supplemental Fig. S1B). At six weeks postinoculation (wpi), when the phenotype can be clearly observed, the wild type-inoculated plants showed a significantly increased (3.3 ± 0.14; P value < 0.05) number of ears per plant relative to mock-inoculated plants (2.6 ± 0.18). Relative to wild-type strains, infection with ΔA2 deletion strains led to a significant (P value < 0.01) reduction in the number of ears per plant (Fig. 2A). The produced number of ears did not change significantly relative to mock-infected plants (Fig. 2A). This indicates that the A2 part of S. reilianum cluster 19-1 encodes determinants for suppression of apical dominance.
Figure 2.
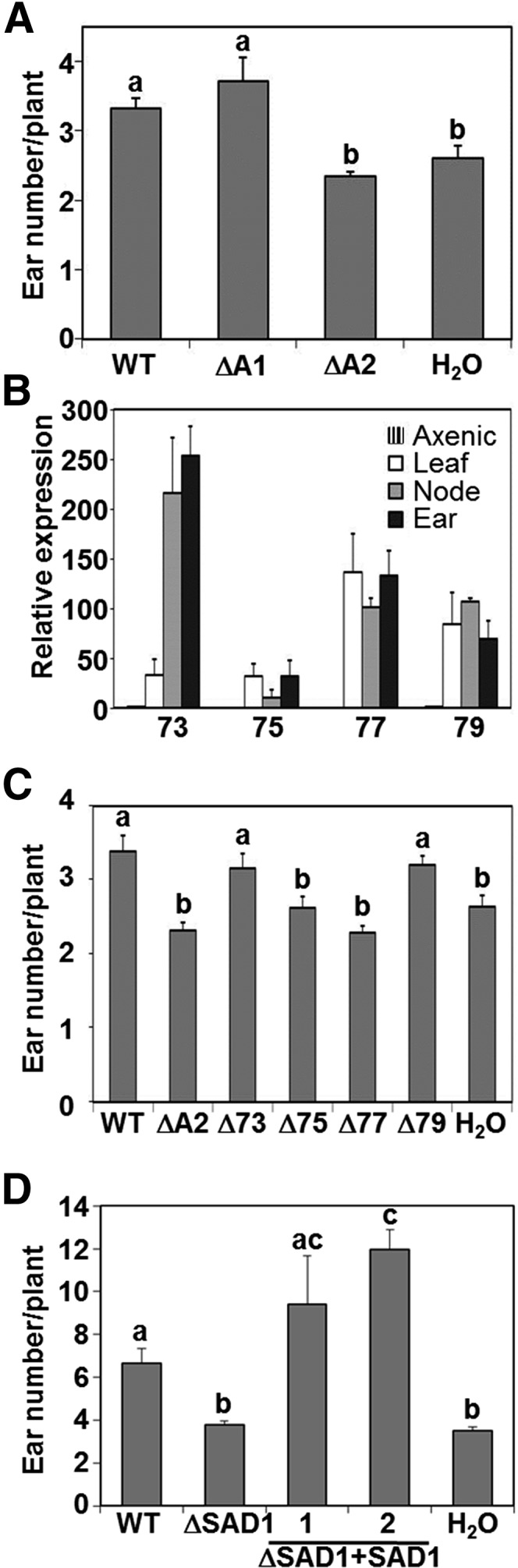
Part A2 of divergence cluster 19-1 of S. reilianum contains determinants for suppression of apical dominance. A and C, Influence of A1 and A2 deletion strains on development of female inflorescences. Plants were grown in a greenhouse, and ear numbers per plant were assessed at 6 wpi using water, compatible wild-type strains (WT), or strains lacking part A1 (ΔA1) or A2 (ΔA2) of cluster 19-1 (A) or strains lacking the single genes of part A2: sr10073 (Δ73), sr10075 (Δ 75), sr10077/SAD1 (Δ77), or sr10079 (Δ79; C). The identity of used strains is given in “Materials and Methods.” Values represent averages (± sem) of three independent experiments using 20 to 25 plants for each replicate and each strain combination. B, Real-time qRT-PCR analysis of gene transcript levels of the four genes of part A2 of divergence cluster 19-1 of S. reilianum at different stages. In liquid culture, transcripts of sr10075 (75) and sr10077 (SAD1; 77) could not be detected, whereas transcripts levels of sr10073 (73) and sr10079 (79) were low. RNA was extracted from leaves collected at 3 dpi, nodes at 2 wpi, and ears at wpi with compatible wild-type strains. Values are averages ± sem of three independent experiments using pools of 10 plants for each experiment relative to the S. reilianum ppi gene sr11196. D, Reintroduction of SAD1 (sr10077) into the S. reilianum ΔSAD1 deletion strains restores S. reilianum-induced suppression of apical dominance. Maize ‘Gaspe Flint’ seedlings were inoculated with water or compatible wild-type, Δsr10077 deletion (ΔSAD1), and Δsr10077+sr10077 complementation (ΔSAD1+SAD1) strains, and symptoms were evaluated at 8 wpi, when the plants show a higher number of inflorescences than at 6 wpi. Plants were grown in a phytochamber. Values are averages of three experiments of 25 plants ± sem. Letters above the data indicate statistically significant differences (P ≤ 0.05).
The part A2 contains four genes (sr10073, sr10075, sr10077, and sr10079) encoding putatively secreted proteins, of which the last three show weak (20% amino acid identity) homology to each other (Fig. 1). By real-time PCR analysis, we showed that all four genes were highly up-regulated upon fungal plant colonization in leaves at 3 d postinoculation (dpi), nodes at 2 wpi, and ears at 4 wpi (Fig. 2B). To identify which gene of the part A2 of cluster 19-1 was responsible for suppression of apical dominance by S. reilianum, we analyzed strains lacking one of the four genes of part A2 (H. Ghareeb, Y. Zhao, and J. Schirawski, unpublished data). Compatible mating type combinations of verified strains were used to inoculate maize seedlings in three independent experiments. At 6 wpi, we investigated the number of female inflorescences produced by plants inoculated with the four individual gene deletion mutants, lacking sr10073 (Δ73), sr10075 (Δ75), sr10077 (Δ77), or sr10079 (Δ79), compared mock- and the wild type-inoculated plants. Relative to mock-inoculated plants, plants inoculated with Δ73 or Δ79 strains showed the same significantly increased number of ears per plant as wild-type strains (Fig. 2C). In contrast, plants inoculated with strains lacking either sr10075 or sr10077 exhibited a lower number of ears per plant than the wild type-inoculated plants (Fig. 2C).
To ensure that loss of suppression of apical dominance is caused by gene deletion and not by a different accidental mutation in the genome, we confirmed contribution of sr10077 by reintroducing its open reading frame-including promoter and terminator regions in two compatible Δ77 deletion strains. Successful integration of sr10077 at a known ectopic locus (the MAIZE-INDUCED GENE1 [MIG1] locus; “Materials and Methods”) was verified by PCR and Southern blot. Southern-blot analysis showed that all obtained complementation strains contained multiple copies of sr10077. By quantitative reverse transcription (qRT)-PCR, we detected 11 to 19 copies of the introduced gene in each complementation strain (not shown). The complementation strains were tested for their ability to suppress apical dominance in maize. Because we noticed that the phenotype becomes more severe with time, we changed the evaluation time point from 6 to 8 wpi. Inoculation of maize with the complementation strains led to a significant increase (P < 0.001) in the number of ears per plant in comparison with the deletion strains and the mock control (Fig. 2D). In addition, maize inoculation with the complementation strains containing multiple copies of sr10077 led to an even higher number of ears per plant than inoculation with wild-type strains (Fig. 2D). These results indicate that suppression of apical dominance imposed by wild-type strains of S. reilianum is associated with sr10077. Therefore, we named the gene SAD1.
SAD1 Is Secreted from Fungal Hyphae in Planta
Analysis of the SAD1 amino acid sequence with the program SignalP 4.1 predicted the presence of a secretion signal peptide that is likely cleaved between amino acid positions 24 and 25 of SAD1 (probability = 0.833; Petersen et al., 2011; Supplemental Fig. S2). Protein secretion of pathogen effectors is a prerequisite for an interaction of the effector with plant proteins. To validate predicted SAD1 secretion, a construct expressing a C-terminal fusion of SAD1 with GFP, SAD1-GFP, under the control of SAD1 promoter was integrated at the MIG1 locus in two compatible S. reilianum strains lacking SAD1 (∆SAD1 deletion strains) to generate the complementation strains (∆SAD1+SAD1-GFP). Southern-blot analysis showed that the strains had integrated multiple copies of the SAD1-GFP construct at the MIG1 locus.
To test whether the SAD1-GFP fusion protein is secreted from plant tissue-colonizing fungal hyphae, we collected female inflorescences from plants inoculated with the ∆SAD1+SAD1-GFP complementation strains and control strains expressing GFP under control of the SAD1 promoter (PSAD1:GFP). Fluorescence microscopic analysis of manually sectioned samples revealed fluorescence around fungal hyphae expressing the SAD1-GFP fusion protein (Fig. 3A). To quantify the finding, we sorted visible hyphae as belonging to one of three types (Supplemental Fig. S3). Hyphae of type 1 had a weak GFP signal with a low signal-to-noise ratio that was associated to very slim hyphae (Supplemental Fig. S3A, left), hyphae of type 2 had a strong GFP signal around fungal hyphae and at hyphal tips (Supplemental Fig. S3A, center) that colocalized with the cell membrane staining dye FM 4-64 (Supplemental Fig. S4), and hyphae of type 3 had a strong signal within the boundaries of the fungal hyphae (Supplemental Fig. S3A, right). A GFP signal corresponding to very slim hyphae was present for nearly 50% of all hyphae detected for both strain combinations expressing either GFP or SAD1-GFP (Fig. 3B). A strong GFP signal around fungal hyphae and at hyphal tips could be observed for a small fraction of the strains expressing GFP (Fig. 3B, GFP) and about 50% of the hyphae of ∆SAD1 deletion strains expressing the SAD1-GFP fusion protein (Fig. 3B, SAD1-GFP). In contrast, an intracellular GFP signal was present in about 60% of hyphae expressing GFP, whereas no such GFP signals were observed with strains expressing the SAD1-GFP fusion protein (Fig. 3B). The differential localization pattern of GFP and SAD1-GFP fluorescence confirmed that the SAD1-GFP fusion protein does not localize to the fungal cytoplasm and is secreted by the fungal hyphae during plant colonization.
Figure 3.
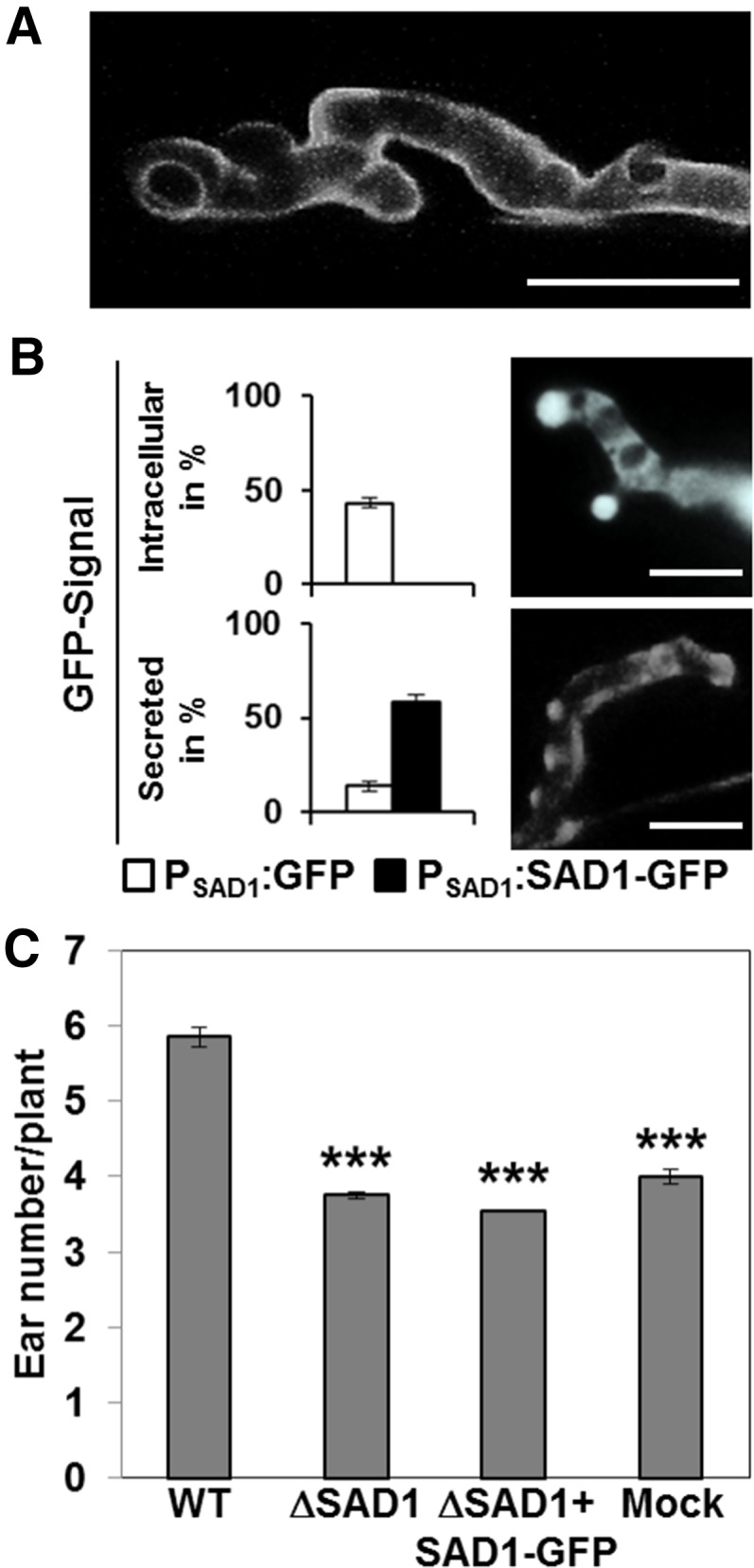
SAD1-GFP is secreted by S. reilianum during growth in planta. A, Fluorescence microscopy of ears infected with S. reilianum ∆SAD1+SAD1-GFP strains. GFP fluorescence surrounds the fungal hyphae. Picture is a Z stack of confocal images. B, Quantification and comparison of GFP signals from hyphae expressing intracellular GFP (upper right) or secreted SAD1-GFP (lower right). Upper left and lower left show quantification of hyphae showing intracellular or secreted GFP signals. Hyphae were counted in two experiments of five different plants. In total, 148 hyphae were counted for GFP, and 128 hyphae were counted for SAD1-GFP. Error bars indicate the sem. Bars = 10 µm. C, Ear number per plant infected with compatible ∆SAD1 deletion or ∆SAD1+SAD1-GFP complementation strains in comparison with the wild type (WT)- and water (mock)-inoculated plants at 7 wpi. The values are means of two independent experiments (n per experiment = 25 ± 2). ***, Statistically significant differences to the control (P ≤ 0.0001).
Symptom evaluation at 8 wpi revealed that the ∆SAD1+SAD1-GFP complementation strains led to the same low number of ears per plant as mock- and ∆SAD1-inoculated plants (Fig. 3C), indicating that the SAD1-GFP fusion protein expressed in S. reilianum is not functional. To test the hypothesis that the protein is nonfunctional because of the increased size, we also tested smaller tags (MYC [AVIAN MYELOCYTOMATOSIS VIRUS ONCOGENE CELLULAR HOMOLOG] and HA [INFLUENZA HEMAGGLUTININ]). Infection of maize plants with S. reilianum strains ∆SAD1+MYC-SAD1 and ∆SAD1+SAD1-HA showed no increased number of ears per plant and thus, no complementation of SAD1 (Supplemental Fig. S5). The inoperativeness of the fusion protein precludes further functional analysis.
Heterologous Expression of SAD1 in Arabidopsis Changes Inflorescence Branching Architecture
To examine whether the SAD1 protein had an impact on plant development that is independent of the presence of S. reilianum, we generated homozygous transgenic Arabidopsis plants that expressed an intracellular GFP-SAD1 fusion protein (i.e. without the predicted secretion signal peptide [see below] under control of the 35S promoter [P35S:GFP-SAD1∆SP]). The reason for expressing an N-terminal GFP-SAD1∆SP fusion was based on the observation that expression of an SAD1-GFP fusion in S. reilianum leads to a nonfunctional SAD1 protein. Nonfunctionality could be caused by either GFP covering an active domain of SAD1 at the C terminus or the interference of the tag with uptake of the fusion protein into the plant cells.
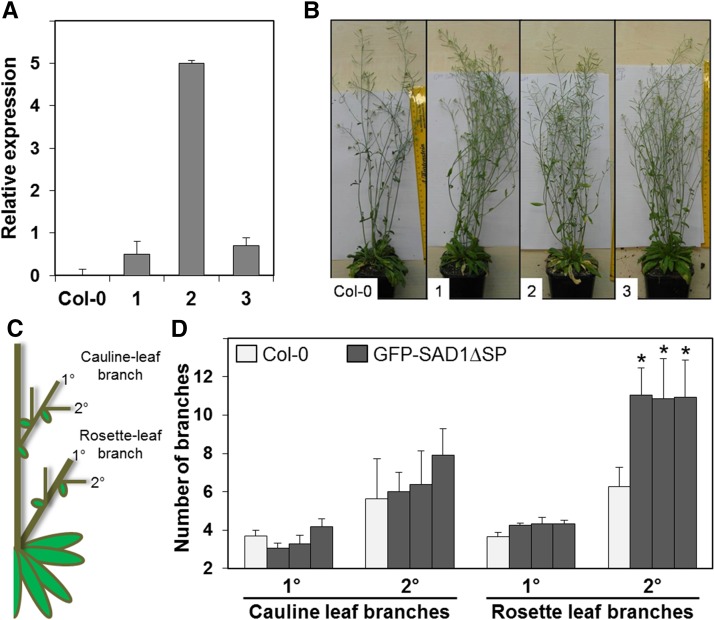
We verified production of SAD1 transcripts in the transgenic lines by real-time PCR (Fig. 4A). The plants grew without any obvious phenotype. However, at flowering time, their inflorescences were more branched than those of the progenitor plants (Fig. 4B). To quantify the changes that might occur in the branching pattern of the transgenic plants (Fig. 4C), the primary and secondary cauline-leaf branches and the primary and secondary rosette-leaf branches were counted at the end of the flowering period (8 weeks). Arabidopsis lines expressing intracellular GFP-SAD1∆SP had the same numbers of primary and secondary cauline-leaf branches and primary rosette-leaf branches but displayed a significant (P value < 0.05) increase in the numbers of secondary rosette-leaf branches (Fig. 4D). This shows that the intracellularly expressed GFP-SAD1∆SP fusion protein affects the inflorescence branching pattern of transgenic Arabidopsis.
Figure 4.
Expression of GFP-SAD1ΔSP in Arabidopsis leads to altered inflorescence branching. A, Analysis of GFP-SAD1ΔSP gene transcript levels in three independently generated transgenic Arabidopsis GFP-SAD1ΔSP expression lines (1–3) relative to transcript abundance of the AtACT1 gene as determined by real-time qRT-PCR. Error bars give sem of three independent experiments. B, Phenotype of transgenic Arabidopsis expressing GFP-SAD1ΔSP. Arabidopsis Col-0 and transgenic Arabidopsis lines (1–3) expressing GFP-SAD1ΔSP under control of the 35S-promoter 8 weeks after sowing. The transgenic lines display more highly branched inflorescences. C, Schematic model of an Arabidopsis plant with illustrated primary (1°) and secondary (2°) cauline- and rosette-leaf branches. D, Quantification of primary (1°) and secondary (2°) cauline-leaf and rosette-leaf branches of the wild type (Col-0) and the three independent GFP-SAD1ΔSP-expressing Arabidopsis lines used in B. Data are averages, with error bars indicating the sem of three experiments of 10 plants each. *, Statistically significant differences to the control (P < 0.05).
In Transgenic Arabidopsis, GFP-SAD1∆SP Localizes to the Cytoplasm and Accumulates in Nuclei in a Tissue-Specific Manner
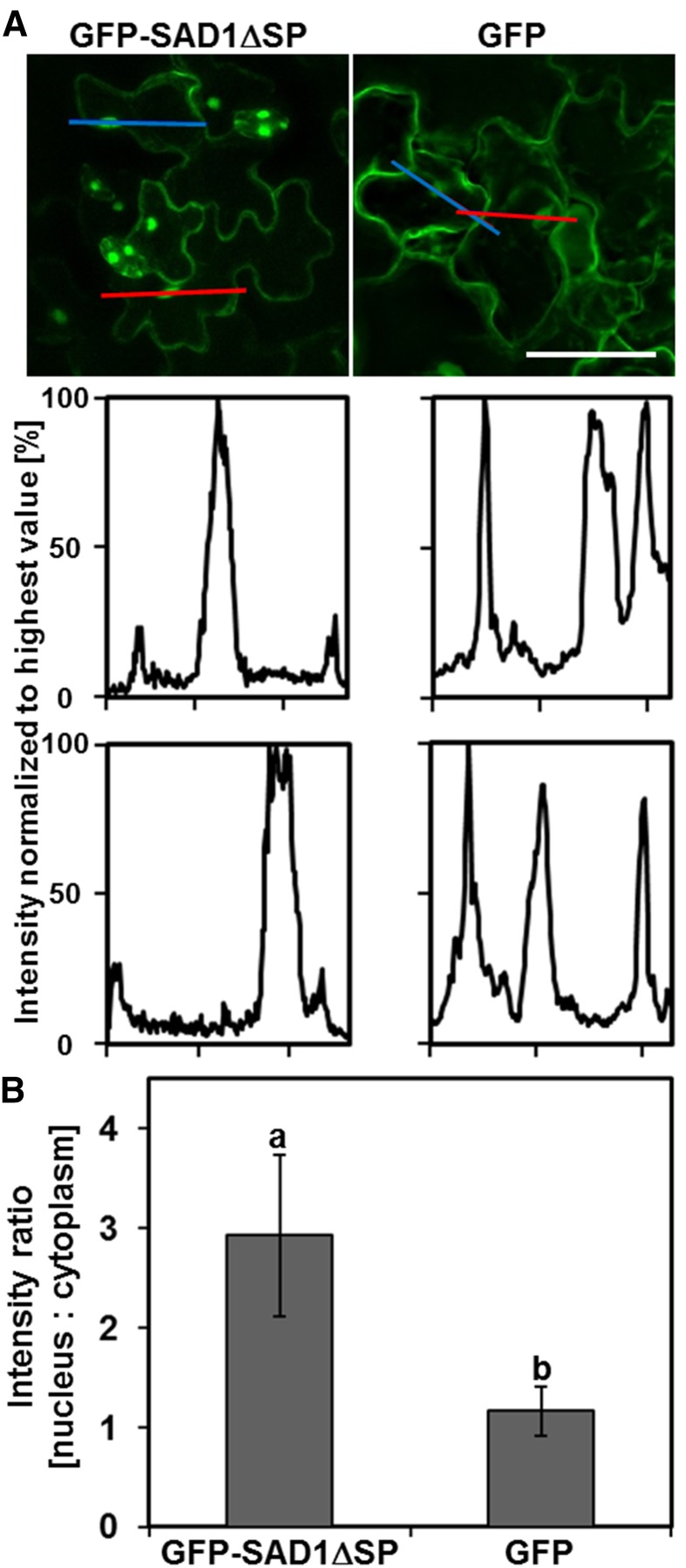
To know the subcellular localization of the functional GFP-SAD1∆SP fusion protein expressed in Arabidopsis, we compared localization of GFP-SAD1∆SP and GFP, both expressed under control of the cauliflower mosaic virus 35S promoter. GFP fluorescence was observed inside the cytoplasm and the nucleus for both constructs (Fig. 5A). Because the fluorescence microscopic pictures do not allow separation of cytoplasm and plasma membrane, there is the possibility that GFP-SAD1 additionally associates with the plasma membrane. Localization to the cell wall and the apoplast was excluded by plasmolysis experiments (Supplemental Fig. S6). In some leaf cells, the signal for GFP-SAD1∆SP was barely visible in the cytoplasm but strong in the nucleus (Fig. 5A). To check whether GFP-SAD1∆SP accumulates inside the nucleus, we quantified the fluorescence intensity of cytoplasm and nucleus for cells expressing GFP-SAD1∆SP and GFP (Fig. 5). Intensity profiles of single cells show three peaks: two associated with the cytoplasm and one associated with the nucleus of the cell (Fig. 5A). In plants expressing GFP, we observed three peaks with the same fluorescence intensity, confirming that the small GFP protein can freely diffuse in between the two cellular compartments. In cells expressing GFP-SAD1∆SP, we observed a low signal intensity in the cytoplasm relative to a strong signal intensity in the nucleus. The nucleus-to-cytoplasm intensity ratio for cells expressing GFP-SAD1∆SP was 3.2 ± 0.7 (sd; n = 35) compared with 1.4 ± 0.2 (sd; n = 40) for GFP-expressing cells (Fig. 5B; P < 0.001). This shows that fusion of SAD1∆SP to GFP leads to nuclear accumulation of the protein in plant cells, possibly by active import of the fusion protein into the nucleus or inhibiting export from the nucleus.
Figure 5.
Heterologously expressed GFP-SAD1ΔSP fusion protein accumulates in the nuclei of Arabidopsis cells. A, Fluorescence signals of plants expressing GFP-SAD1∆SP (top left) or GFP (top right). Red and blue colored lines indicate two examples for regions of fluorescence intensity measurements. Intensity measurements (middle and bottom) show GFP signal accumulation in the nucleus (middle peak) of plants expressing GFP-SAD1∆SP (bottom left) or GFP (bottom right) along the red (middle) or blue (bottom) region of interest indicated in top. B, Quantification of GFP intensity ratios between nucleus and cytoplasm of leaf cells from Arabidopsis plants expressing GFP-SAD1∆SP or GFP. Cells expressing the GFP-SAD1∆SP fusion protein show an average signal intensity ratio of nucleus:cytoplasm of about 3:1. Cells expressing GFP show a ratio of 1:1; n = 35 cells for GFP-SAD1∆SP, and n = 40 cells for GFP. Five different plants were analyzed for each construct. Error bars show sd. Letters above the data indicate statistically significant differences (P ≤ 0.001). Bars = 50 µm.
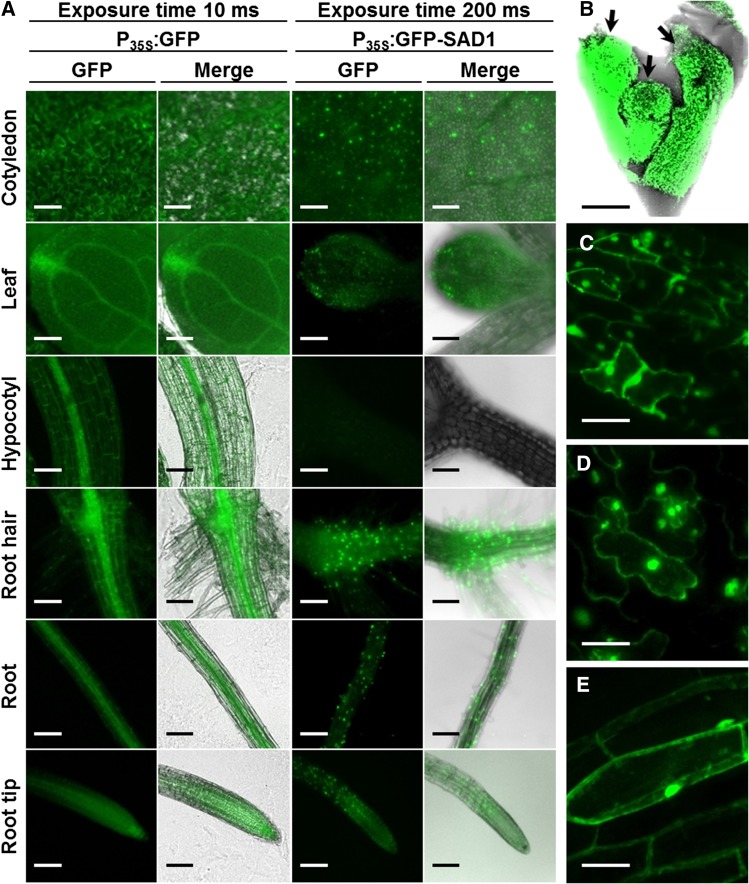
Interestingly, intensity of signals of GFP-SAD1∆SP-expressing plants differed dramatically from plants expressing GFP alone. In 10- to 12-d-old transgenic Arabidopsis seedlings that had just produced the first pair of true leaves, GFP-expressing plants produced a prominent signal throughout the entire plant, even at low exposure times of 10 ms. Using the same exposure time revealed a very weak GFP signal for plants of all three transgenic lines expressing GFP-SAD1∆SP (Supplemental Fig. S7). A strong fluorescence signal from GFP-SAD1∆SP could only be observed in guard cells of the cotyledons. When the exposure time was increased to 200 ms, GFP-SAD1∆SP fluorescence became visible in leaves and roots but not in hypocotyls (Fig. 6A). In leaves, GFP-SAD1∆SP fluorescence was stronger in guard cells than in pavement cells, whereas the signal intensity was equally strong in different leaf cells expressing GFP alone. Additionally, the signal intensity of vascular bundles in leaves and roots was weaker for GFP-SAD1∆SP than for GFP alone (Fig. 6A). In older plants (6 weeks), fluorescence of the GFP-SAD1∆SP fusion protein could readily be detected in floral buds (Fig. 6B) and floral organs (Fig. 6C) as well as in leaves (Fig. 6D) and roots (Fig. 6E), and the protein localized to both the cytoplasm and the nucleus. Interestingly, fluorescence varied from well detectable to not detectable in individual inflorescences. The distribution of the GFP-SAD1∆SP-derived fluorescence in different plant tissues that should express the protein in every cell suggests that, in Arabidopsis, expression or stability of the fusion protein is translationally or posttranslationally controlled.
Figure 6.
Microscopic overview of GFP-SAD1ΔSP distribution in transgenic Arabidopsis plants. A, Twelve-day-old Arabidopsis plants expressing GFP (left) or GFP-SAD1∆SP (right). Pictures were taken with an exposure time of 10 (left) or 200 ms (right). Plants expressing GFP show a fluorescence signal in every part of the plant. GFP-SAD1∆SP was not detected in hypocotyls. B, Fluorescence microscopic picture of three floral buds (arrows) of Arabidopsis expressing GFP-SAD1∆SP. C to E, GFP fluorescence of GFP-SAD1ΔSP expressed in petal cells of the flower (C), leaves (D), and root cells (E) of 6-week-old plants. Fluorescence is observed in the nucleus and the cytoplasm. Bars = 100 µm (A and B) and 25 µm (C–E).
SAD1 Can Activate Transcription of Reporter Genes in Yeast
To investigate whether SAD1 has the potential to activate gene transcription, we expressed SAD1 lacking its signal peptide as a fusion to the galactose metabolism protein4 (GAL4) DNA-binding domain (BD; BD-SAD1∆SP) in the strain Y2HGold of the Matchmaker Gold Yeast Two-Hybrid System expressing the GAL4 activation domain (AD). In cell extracts of this strain, presence of a 41-kD fusion protein corresponding to BD-SAD1∆SP could clearly be detected (Supplemental Fig. S8). In contrast to control strains that only expressed AD and the GAL4 BD and in which no reporter genes were activated, two of the four reporter genes were activated in the strains expressing AD- and BD-SAD1∆SP. Colonies grew on medium containing Aureobasidin A resulting from activation of AUREOBASIDIN A RESISTANCE GENE (AUR1-C) and led to the formation of a blue color on plates containing 5-bromo-4-chloro-3-indolyl alpha-d-galactopyranoside resulting from activation of MELIBIASE1 (MEL1; Supplemental Fig. S9A). In contrast, cells were unable to grow on media lacking adenine or His, showing that AMINOIMIDAZOLE RIBONUCLEOTIDE CARBOXYLASE2 (ADE2) and IMIDAZOLEGLYRECOL-PHOSPHATE DEHYDRATASE3 (HIS3) were not activated (Supplemental Fig. S9B). Because expression of HIS3 and ADE2 is driven by the G1 and G2 promoter elements, respectively, whereas the expression of both AUR1-C and MEL1 is driven by the M1 promoter element, this indicates that BD-SAD1∆SP is able to autoactivate the transcription of the M1 promoter but not that of the G1 and G2 promoters, showing that SAD1 has the potential to activate gene expression in yeast (Saccharomyces cerevisiae). Control experiments where SAD1 lacking its signal peptide was fused to the GAL4 AD (AD-SAD1ΔSP) showed that AD-SAD1ΔSP is not able to autoactivate MEL1 expression (Supplemental Fig. S10). This likely indicates that SAD1 can influence nuclear processes, including activation of gene transcription, not by direct binding to DNA but by interaction with nuclear proteins.
SAD1 Modulates Transcription of Plant Genes Involved in Auxin Transport and Branching Control
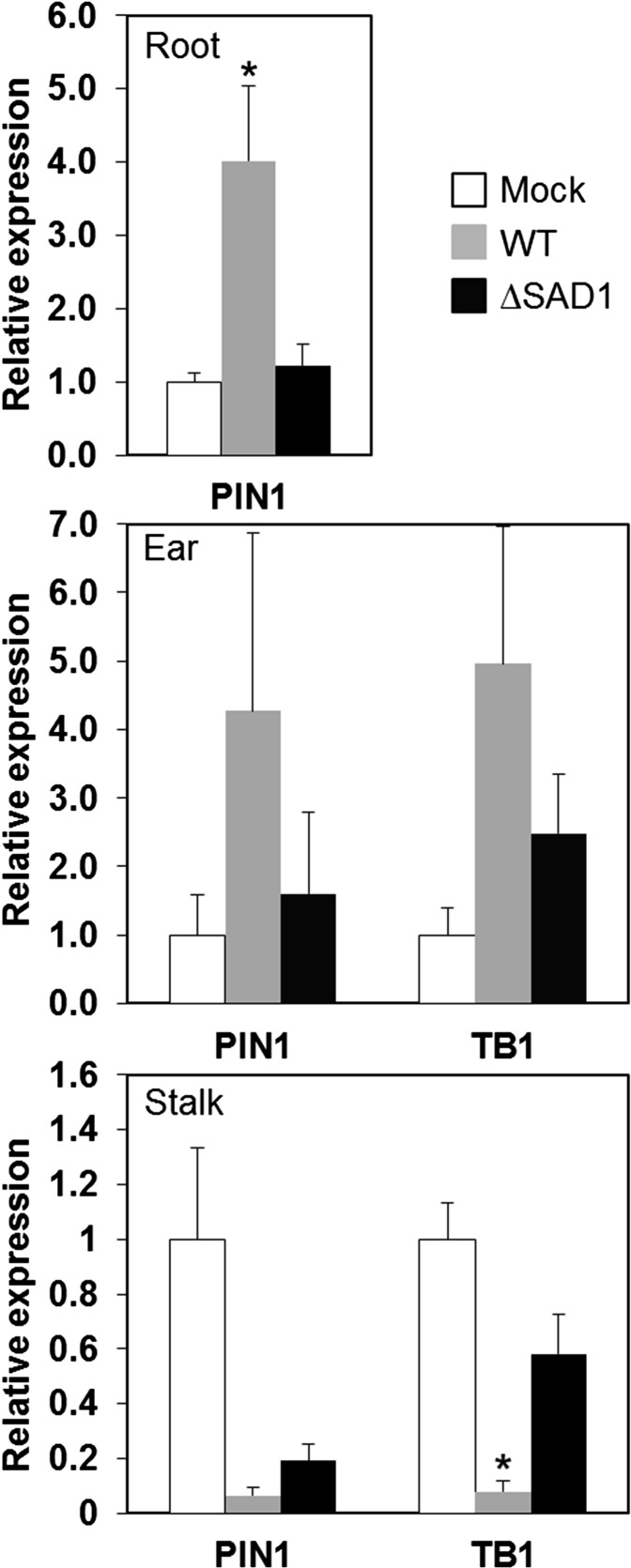
To find out whether SAD1 regulates nuclear processes in maize by modulating plant gene expression, we analyzed the transcript level pattern of TB1, a dosage-dependent inhibitor of axillary bud outgrowth in maize, and PIN1 encoding an auxin efflux transporter (Hubbard et al., 2002; Gallavotti et al., 2008b). Transcript levels were determined by qRT-PCR in different tissues of maize plants inoculated with water (mock), S. reilianum wild-type, or ∆SAD1 deletion strains. Tissues were sampled at 7 wpi, a time point at which changes in the branching architectures of the wild type-infected maize plants are prominent. Transcript abundance was quantified in three different tissues (roots, stalks, and ears) relative to mock-inoculated plants. For quantification of PIN1 transcripts, a pair of primers was used that amplified part of the first exons of ZmPIN1a, ZmPIN1b, and ZmPIN1c (Supplemental Fig. S11; Carraro et al., 2006; Forestan et al., 2010). In roots of the wild type-inoculated maize plants, PIN1 transcript levels were significantly increased (P value < 0.05), which was not the case for plants inoculated with strains lacking SAD1 (Fig. 7, top). In ears, no significant difference could be detected in abundance of PIN1 or TB1 transcripts in the wild type- or ∆SAD1-inoculated plants (Fig. 7, middle). In stalks, where an altered plant gene expression should have the most imminent effect on plant morphology, the level of PIN1 transcripts was significantly reduced (P value < 0.05) in both the wild type- and ΔSAD1-inoculated plants, indicating am SAD1-independent effect. In contrast, the transcript level of TB1 was significantly (P value < 0.05) decreased in the stalks of the wild type- but not ΔSAD1- or mock-inoculated plants (Fig. 7, bottom). These results show that presence of SAD1 in S. reilianum influences expression of the maize auxin transporter in roots and the main repressor of bud outgrowth in stalks of ears.
Figure 7.
Real-time PCR analysis of PIN1 and TB1 transcript abundance. Gene expression was measured in roots (top), ears (middle), and stalks (bottom) of maize inoculated with water (mock) wild-type (WT) and ∆SAD1 (ΔSAD1) deletion strains of S. reilianum. Values given are expression values relative to mock-inoculated plants and means of three biological replicates with pools of three plants per replicate and tissue ± sem. *, Statistical difference to both mock and ΔSAD1 inoculations within the same tissue (P value < 0.05).
Interaction Partners of SAD1 Are Involved in Development, Transcription, Ubiquitination, and Signaling
To get a better insight into the role of SAD1 in planta, we aimed to identify the protein's interaction partners. We generated a normalized yeast two-hybrid library with complementary DNA (cDNA) generated from axenically grown S. reilianum and different S. reilianum-colonized maize tissues that result in C-terminal fusions to the GAL4 AD domain. Transformation of the library into the yeast strain Y187 resulted in 1.3 × 109 independent clones. The library-containing strains were mated to the pGBKT7-SAD1-containing Y2HGold strain of the Matchmaker Gold Yeast Two-Hybrid System expressing BD-SAD1∆SP, which resulted in 5 × 105 independent diploid colonies auxotrophic for Trp and Leu and thus, contains bait and prey plasmids. Of these, 434 diploids grew on medium also lacking adenine and His (Supplemental Fig. S12), indicating that they contained plasmids encoding proteins that interacted with SAD1. The plasmid insert sequences of 384 clones were determined, and the sequences were compared among each other. This resulted in 179 nonredundant sequences encoding potential SAD1 interaction partners. We verified 139 interactions by reintroduction of representative plasmids of each group into Y187 followed by mating with the Y2HGold strain containing pBKT7-SAD1. Of the 139 potential interaction partners, 74 were recovered more than once. Of these, only one (found three times) was of fungal origin, excluding a possible general stickiness of the bait protein. By comparison with nucleotide databases (BLASTN and NCBI; Altschul et al., 1990), we were able to assign a putative function to 45 of the identified proteins. Interestingly, more than one-half of these annotated interaction partners have a putative function in development, transcription, ubiquitination, or signaling (Table I). These results suggest that SAD1 can interact with multiple intracellular maize proteins with a putative cytoplasmic or nuclear localization.
Table I. List of SAD1 interaction partners putatively involved in development, transcription, ubiquitination, or signaling.
| Name Interaction Partner | Frequencya | Accession No. | Annotation |
|---|---|---|---|
| Development | |||
| SAD1-IP4 | 5 | NM_001111851.1 | Maize AGAMOUS1 |
| SAD1-IP1 | 4 | AF467541 | Putative aldehydedehydrogenase MIS1 |
| SAD1-IP2 | 3 | AY883559 | Teosinte glume architecture1 |
| SAD1-IP3 | 2 | NM_001177122 | Fimbriata-like protein |
| Nuclear processes | |||
| SAD1-IP17 | 5 | NM_001175076 | Nucleolar complex protein 2 homolog |
| SAD1-IP5 | 4 | NM_001112378 | Helicase RH2 protein-like |
| SAD1-IP6 | 4 | NM_001158297 | Polypyrimidine tract-binding protein |
| SAD1-IP7 | 3 | NM_001155221 | Arg/Ser-rich splicing factor10 |
| SAD1-IP8 | 3 | EU955917 | RNA-binding protein-like |
| SAD1-IP9 | 3 | EU962463 | Zinc finger protein-like1 mRNA |
| SAD1-IP10 | 2 | NM_001154418 | Zinc finger, C3HC4 type family protein |
| SAD1-IP11 | 2 | NM_001175194 | Transcription factor PIF3 |
| SAD1-IP12 | 2 | EU964022 | DNA-directed RNA polymerase II subunit J |
| Ubiquitination | |||
| SAD1-IP22 | 7 | EU965267 | Similar to SAM domain of Anks family protein |
| SAD1-IP23 | 4 | NM_001147846 | Probable E3 ubiquitin-protein ligase |
| SAD1-IP24 | 2 | NM_001155504 | Probable E3 ubiquitin-protein ligase |
| SAD1-IP25 | 2 | U29161 | MubG7 ubiquitinfusionproteingene |
| SAD1-IP26 | 2 | BT066423 | Ubiquitin-associated protein |
| SAD1-IP27 | 2 | NM_001152740 | E3 ubiquitin-protein ligase RGLG2 |
| SAD1-IP28 | 2 | EU940814.1 | E3 ubiquitin-protein ligase KEG |
| SAD1-IP29 | 2 | BT067186 | S-phase kinase-associated protein 1-interacting partner14 |
| Signaling | |||
| SAD1-IP33 | 6 | NM_001153317 | Probable phosphatase 2C |
| SAD1-IP34 | 2 | NM_001111427 | Pyruvatedehydrogenase (lipoamide) kinase1 |
| SAD1-IP35 | 2 | EU954821 | Casein kinase II subunit β-4 |
| SAD1-IP36 | 2 | NM_001158779 | Kinase APK1B |
The frequency of recovered sequences.
DISCUSSION
Comparison of the S. reilianum and U. maydis smut fungal genomes revealed the existence of divergence regions, often correlating to previously identified regions encoding proteins with a predicted secretion signal and no known enzymatic function (Kämper et al., 2006; Schirawski et al., 2010). Deletion of the largest virulence effector-encoding region in U. maydis resulted in strains that were unable to induce tumors on seedling leaves (Kämper et al., 2006) and had thus lost one prominent U. maydis-specific symptom. This prompted us to investigate whether the corresponding region in S. reilianum encoded determinants for S. reilianum-specific symptom formation. In comparison with U. maydis, S. reilianum has an extended endophytic phase, in which it lives and spreads inside the plant without causing obvious damage (Schirawski et al., 2010). Symptoms include the replacement of individual flowers or complete inflorescences by spore-filled sori, the presence of floral reversion leading to phyllody, and the occurrence of multiple female inflorescences at subapical nodes, which is caused by suppression of apical dominance (Ghareeb et al., 2011). Gene deletion strains lacking the left and larger (ΔA1) or the right and smaller part (ΔA2) of cluster 19-1 were still able to induce spores and phyllody in inflorescences of maize (H. Ghareeb, Y. Zhao, and J. Schirawski, unpublished data). However, the ΔA2 deletion strains did not lead to suppression of apical dominance. Individual gene deletion of each of the four genes present in the 19A2 region (H. Ghareeb, Y. Zhao, and J. Schirawski, unpublished data) followed by symptom analysis showed that absence of SAD1 led to absence of the loss of apical dominance phenotype in infected maize plants. Reintroduction of SAD1 in multiple copies in the ΔSAD1 deletion strains resulted in a severe increase in the number of female inflorescences per plant, suggesting that SAD1 functions as an SAD in maize. Thus, SAD1 is a fungal effector that affects the branching architecture of maize and represents a molecular link between pathogen proteins and symptom formation.
To affect development of host tissues, SAD1 should be expressed and secreted during colonization of maize by S. reilianum. Both requirements are met, because SAD1 is highly expressed in leaves, nodes, and ears of colonized plants (Fig. 2B). In addition, the functionality of the bioinformatically predicted secretion signal peptide (Supplemental Fig. S2) was indicated by localization around fungal hyphae of an SAD1-GFP fusion protein expressed in S. reilianum and colocalization with the cell membrane in maize inflorescences (Fig. 3; Supplemental Figs. S3 and S4). Thus, SAD1 fulfills the requirements to influence plant development by directly interacting with components of the plant cell.
Yeast two-hybrid analysis revealed intracellular plant proteins as potential interaction partners. Although some interaction partners likely occur in the plant cell cytoplasm (i.e. those involved in signaling or ubiquitination; Table I), others are likely located in the plant cell nucleus (i.e. those involved in nuclear processes; Table I). This strongly suggests that SAD1 functions inside plant cells and might have a decisive role inside plant cell nuclei. Experiments involving heterologous expression of SAD1 fusion proteins in Arabidopsis support this suggestion. A GFP-SAD1∆SP fusion protein expressed without secretion signal peptide could be detected in the cytoplasm and accumulated in plant cell nuclei when stably expressed in Arabidopsis (Figs. 4 and 5). In addition, expression in yeast of SAD1 as a fusion to the GAL4 BD led to transcriptional activation of reporter genes (Supplemental Fig. S9), and interaction of SAD1 with the DNA-directed RNA polymerase II subunit J of maize in a yeast two-hybrid screen indicated that the protein may act as a transcriptional activator. Because SAD1 was not able to substitute for the GAL4 BD (Supplemental Fig. S11), promoter activation might be through protein-protein interaction. Because we did not use a split-ubiquitin assay for protein-protein interaction detection, we cannot rule out that SAD1 interacts with membrane-bound plant proteins inside or even outside the plant cell and functions by activating downstream signaling pathways.
Heterologous expression of SAD1 as a GFP-SAD1∆SP fusion in stable transgenic Arabidopsis resulted in plants that formed an increased number of secondary rosette leaf branches (Fig. 4C). This is remarkable, because it shows that SAD1 alone without the help of any other fungal proteins is able to modify inflorescence branching architecture in Arabidopsis. In addition, this indicates that the fusion protein is functional when localized to plant intracellular compartments. The increased secondary branching that occurs in both Arabidopsis-expressing SAD1 and S. reilianum-infected maize indicates a function of SAD1 through a pathway conserved in the monocot maize and the dicot plant Arabidopsis.
Function of SAD1 may depend on its transfer to the plant cell nucleus, because the concentration of the GFP-SAD1∆SP fusion protein was much higher in nuclei than in the cytoplasm (Fig. 5). Reasons for the nuclear accumulation could be increased cytoplasmic degradation, retention of the protein in nuclei, or specific translocation from the cytoplasm to the nucleus in plant cells. Further experimentation is needed to find out which scenario is valid for SAD1. Intracellular localization of SAD1 might be dependent on posttranslational modification, such as phosphorylation. Examples include the PROLINE-RICH TYROSINE KINASE2 of neurons that localizes to the nucleus of rat pheochromocytoma cells upon phosphorylation (Faure et al., 2013) and the EXTRACELLULAR SIGNAL-REGULATED KINASE1, where phosphorylation by a casein kinase2 promotes nuclear import (Plotnikov et al., 2011). Bioinformatic analysis predicted several phosphorylation sites within SAD1, two phosphorylation sites for protein kinase C, and one for casein kinase 2 (Supplemental Fig. S2). In addition, three putative Ser/Thr-protein kinases, one putative casein kinase II, and one putative phosphatase of maize were found to interact with SAD1 in the yeast two-hybrid assay (Table I). At this point, it remains speculative whether phosphorylation is involved in regulation of activity, stability, or localization of SAD1.
Although it is unclear if and how the SAD1 protein secreted from fungal hyphae enters plant cells, the facts that we find intracellular plant proteins as interaction partners and that an inflorescence branching phenotype is observed when the protein is expressed in Arabidopsis strongly hint at an intracellular function of SAD1 in maize. Once inside the plant cell, SAD1 accumulates in the nucleus and could affect apical dominance by altering transcription of genes involved in biosynthesis, signaling, or transport of auxin, cytokinin, strigolactones, or GAs. Measurement of auxin concentrations in emerging inflorescences at 4 wpi, a time point at which the appearance of multiple ears per branch is initiated (F. Drechsler and J. Schirawski, unpublished data), showed a 30% increase in auxin concentration of infected inflorescences (Ghareeb et al., 2011). Because auxin transport rather than absolute auxin concentration is the main contributor to apical dominance, we investigated transcript levels of the auxin efflux transporter PIN1. In ears, PIN1 transcripts were slightly but not significantly increased by the presence of SAD1. PIN1 transcript levels in the stalk were down-regulated, regardless of the presence of SAD1 (Fig. 7). This shows that reduction of PIN1 transcripts in the aerial parts of the plant is SAD1 independent. In contrast, presence of SAD1 during infection with S. reilianum leads to increased PIN1 transcript abundance in roots (Fig. 7). Increased PIN1 transcript abundance in the root may increase auxin sink strength in the stalk and thus, enable axillary buds to establish a PAT stream and continue bud outgrowth as predicted by the auxin canalization model (Domagalska and Leyser, 2011). To determine whether SAD1 has an effect on PAT, PIN1 protein distribution in roots of infected plants needs to be verified.
In addition to PIN1, we measured transcript levels of the bud outgrowth inhibitor TB1. TB1 has been identified as a maize domestication gene that contributes to the increased apical dominance in maize in comparison with its highly branched ancestor, teosinte (Studer et al., 2011). TB1 suppresses axillary bud outgrowth in maize, and TB1 deletion mutants show excessive side branching (Doebley et al., 1997). In our experiments, TB1 transcripts were reduced in stalks of the wild type-inoculated plants in comparison with mock- or ∆SAD1-inoculated plants (Fig. 7). Down-regulation of TB1 in the presence of SAD1 explains the observed increase in subapical inflorescence bud outgrowth in infected maize. In Arabidopsis, a homolog of TB1 was identified as BRANCHED1 (BRC1) that is a central regulator of branching (Aguilar-Martínez et al., 2007). Expression of BRC1 was shown to be enhanced by overproduction of auxin in Arabidopsis 35S:YUCCA plants, leading to a lower amount of branches compared with wild-type plants (Finlayson, 2007). In contrast, it was shown in maize that mutation of the auxin biosynthetic genes SPARSE INFLORESCENCE1 (SPI1) encoding a flavin monooxygenase or VANISHING TASSEL2 encoding a Trp aminotransferase leads to reduced auxin levels (Phillips et al., 2011), and fewer tillers are produced in spi1 and spi1/tb1 mutant plants (Gallavotti et al., 2008a). This indicates that inactivation of auxin biosynthesis genes leads to opposite effects in Arabidopsis and maize (i.e. increased branching in Arabidopsis and decreased tillering in maize; Gallavotti, 2013), whereas in both maize and Arabidopsis, branching is dependent on TB1/BRC1 (Finlayson, 2007; Doebley et al., 1997). Because expression of SAD1 results in an increase in bud outgrowth in both maize and Arabidopsis, SAD1 likely plays a role in bud outgrowth that is independent of auxin.
Other factors could also contribute to SAD1-dependent bud outgrowth, such as the timing of the local accumulation of auxin, strigolactone, and cytokinin (Müller and Leyser, 2011). Reduction of strigolactone or increase in cytokinin concentrations would activate bud outgrowth (Cheng et al., 2013) and lead to the observed increase in the number of ears per plant. In Arabidopsis, it was shown that strigolactone decreases PIN1 levels by 2- to 6-fold, which is sufficient to alter apical dominance (Shinohara et al., 2013). Recently, effector proteins of a plant pathogenic phytoplasma bacterium were identified that influence different phases of meristem development in tomato (Wei et al., 2013). Additionally to the tested transcription factor TB1, there are multiple other transcription factors that are involved in inflorescence architecture. One very interesting example is DLF1 (Muszynski et al., 2006), a basic Leu zipper protein. Maize B73 with a DLF1 deletion exhibits the same branching phenotype in ears as we observed for cv Gaspe Flint infected with S. reilianum. We could not detect DLF1 in the yeast two-hybrid screen, and it is unknown whether SAD1 has an effect on transcription of DLF1. It will be challenging to unravel the exact target and mechanism of how the SAD1 effector influences plant branching architecture.
CONCLUSION
Apical dominance in maize is broken by infection with S. reilianum. We have identified a fungal protein responsible for this disease symptom that we named SAD1. SAD1 was found to be secreted from fungal hyphae. SAD1 transcripts were not detectable when the fungus was growing as a saprotroph but were heavily abundant as soon as the fungus entered the maize plant and spread through leaves, nodes, and inflorescences. Several factors indicate that SAD1 mediates suppression of apical dominance. First, S. reilianum strains lacking SAD1 no longer lead to suppression of apical dominance, although virulence is unaffected. Second, reintroduction of SAD1 in the S. reilianum ΔSAD1 deletion strain in multiple copies restored and even increased the suppression of apical dominance phenotype. Third, stable heterologous expression of the SAD1 protein as a fusion to GFP and lacking its secretion signal peptide in Arabidopsis resulted in increased inflorescence branching.
The mechanism of how SAD1 affects apical dominance in maize and Arabidopsis is less clear. It seems likely that the factor functions within plant cells. It is secreted from fungal hyphae, it has an effect when expressed within Arabidopsis cells, it localizes to the cytoplasm and the nucleus in Arabidopsis, and it can interact with maize cytoplasmic and nuclear proteins. If and how the factor—once secreted from fungal hyphae—ends up in maize cells is an open question as well as what exactly its effect is once it has entered. However, we could detect a positive influence of the presence of SAD1 on the PIN1 transcript level in roots of infected maize plants, which suggests increased auxin flow to the root as a mechanism supporting bud outgrowth. In addition, we observed SAD1-dependent down-regulation of the branching inhibitor TB1, which is well in line with the observed phenotype of increased branching induced by SAD1. Circumstantial evidence points to a role of SAD1 in bud outgrowth that is independent of auxin. Although SAD1 has the same effect in maize and Arabidopsis, disruption of auxin biosynthesis has an opposite effect on branching in maize and Arabidopsis. Elucidating the exact mechanism of how this interesting protein mediates suppression of apical dominance is one of the challenges ahead.
MATERIALS AND METHODS
Plant Lines, Growth Conditions, and Inoculation Experiments
Maize (Zea mays) ‘Gaspe Flint,’ an early-flowering dwarf maize line, was cultivated and used for inoculation with Sporisorium reilianum as previously described (Ghareeb et al., 2011). Sterilized Arabidopsis (Arabidopsis thaliana) Columbia-0 (Col-0) seeds were kept at 4°C for 1 to 3 d before sowing. Plants were grown in soil (Fruhstofer; Type T25) under long-day conditions at 22°C, 160 µmol, and 65% humidity in a phytochamber (Johnson Controls). Sterilized Arabidopsis seeds were kept for 2 d before plating. Plants were grown on one-half-strength Murashige and Skoog medium including vitamins (Duchefa-biochemie) supplemented with 1% (w/v) Suc. The Col-0 lines expressing GFP-SAD1∆SP were (1) 970.1, (2) 975.2, and (3) 979.4.
S. reilianum strains were prepared as stocks for storage in 25% (v/v) glycerol at −80°C, strains were freshly streaked on potato dextrose plates, and single colonies were used for inoculation of precultures in 10 g L−1 yeast (Saccharomyces cerevisiae) extract, 10 g L−1 peptone, and 10 g L−1 Suc (YEPS-light medium). For plant inoculation experiments, precultures of the compatible S. reilianum sp. zeae (SRZ) wild-type strains SRZ1_5-2 and SRZ2_5-1 (Schirawski et al., 2005; Zuther et al., 2012) and their deletion derivatives were used to inoculate potato dextrose broth (Difco) and grown at 28°C under constant shaking to an optical density at 600 nm (OD600) of 0.5 to 0.8. Cell pellets were resuspended in water to reach an OD600 of 2.0. Cultures of compatible strains carrying the same deletion were mixed in a 1:1 ratio, and the mixture was used to inoculate 7-d-old maize seedlings by syringe-assisted leaf whorl apposition (Gillissen et al., 1992).
Generation of Plasmids, Knockout, Complementation, and Localization Constructs
For generating the expression vectors for p35S:GFP-SAD1∆SP to be expressed in Arabidopsis, the primers forA and revA were used to amplify a 0.5-kb fragment containing the SAD1 open reading frame lacking the signal peptide (SAD1∆SP). The fragment was fused to GFP by cloning into a binary vector containing GFP using NotI and EcoRI. The fusion construct was moved as an AscI and EcoRI fragment to the binary plant expression vector pAMPAT-MCS (accession no. AY436765). For yeast two-hybrid analysis, SAD1∆SP was amplified with oHG210 and oHG211. The 0.5-kb PCR amplicon was digested with EcoRI and BamHI and cloned into the bait vector pGBKT7 (Clontech).
For complementation studies, a strategy to integrate the genes of interest at the mig1 locus (intergenic region between the genes sr14222 and sr14223) was established. For generation of the SAD1 complementation construct, right and left flanks of the MIG1 locus and the SAD1 gene, including promoter and terminator regions, were amplified using primers listed in Table II. The MIG1 flanks, SAD1, and the carboxin resistance cassette from the plasmid pMF1-c (Brachmann et al., 2004) were digested with SfiI and ligated. The ligation product was excised from gel and used to amplify the complementation construct using oligonucleotides listed in Table II.
Table II. List of oligonucleotides used in this study.
| Name | Sequence | Use to Amplify |
|---|---|---|
| oHG149 | TCGCAACGAGGAAACGAGAC | Left flank of mig1 locus |
| oHG183 | CAACCTCGAGCGGCCTACCCAATCTTCAACG | Left flank of mig1 locus |
| oHG151 | ATCTAGGCCTGAGTGGCCCTGACGGTAACGGCCGAAAC | Right flank of mig1 locus |
| oHG152 | CGCTTTTGCCACTGCTTCC | Right flank of mig1 locus |
| oHG153 | CCGTGGTATCTGAAGCAATC | Complementation and localization constructs |
| oHG154 | CCGCCTTGTTGTTCATGG | Complementation and localization constructs |
| oHG194 | TGCGCCGCTCAGCATGACATAC | Integrated constructs at mig1 locus |
| oHG195 | ACGTCGCTGACTGGAGGCTTTG | Integrated constructs at mig1 locus |
| oHG202 | ATCACGGCCTCTAAGGCCAATACGCAAACCGCCTCTC | egfp-nos terminator |
| oHG205 | GATCACGGAGTGGCAGGTGCAGTGAGCAAGGGCGAGGAG | egfp-nos terminator |
| oHG186 | GTGGAGGAGCCCTACATACC | sr10073 in qRT-PCR |
| oHG187 | CAGCGGGCTTATCAATGTGG | sr10073 in qRT-PCR |
| oHG188 | ATGCGCCTTCTACTCCAACG | sr10075 in qRT-PCR |
| oHG189 | CCGCTCTTTGCAACTCTTCG) | sr10075 in qRT-PCR |
| oHG190 | CATGAGAATGCCATGCTTCC | sr10077 in qRT-PCR |
| oHG191 | TTCATGGTGCATCACGATCC | sr10077 in qRT-PCR |
| oHG192 | ATTGGAGCCCATGCCTCACC | sr10079 in qRT-PCR |
| oHG193 | TGGCGTACACGGCGTATTCG | sr10079 in qRT-PCR |
| oHG143 | CCGCCAGAATCATGTCCAAC | ppi in qRT-PCR |
| oHG144 | CATGAACTGCGGGATGACAC | ppi in qRT-PCR |
| oBH73 | CCGCCAGAATCATGTCCAAC | ACT1 in qRT-PCR |
| oBH74 | CATGAACTGCGGGATGACAC | ACT1 in qRT-PCR |
| oHG252 | ACTTCATCTCCACCAACAACCC | PIN1 in qRT-PCR |
| oHG253 | AGAAGTCGCCGTACATGCCC | PIN1 in qRT-PCR |
| oHG264 | AGCCACCACTCATCGTTGTC | TB1 in qRT-PCR |
| oHG265 | CGTATCCTCCGTTGCCAAAG | TB1 in qRT-PCR |
| oHG210 | ATCTAGAATTCAGTGGCTCTCAGAGCGGCGG | SAD1ΔSP for yeast two hybrid |
| oHG211 | ATCTAGGATCCCTATACTGATAAATGGAGAGCAGG | SAD1ΔSP for yeast two hybrid |
| forA | AGCGGCCGCTAGTGGCTCTCAGAGCGG | SAD1∆SP for Arabidopsis expression |
| revA | TGAATTCCTATACTGATAAATGGAGAG | SAD1∆SP for Arabidopsis expression |
Transformation of S. reilianum and Generation of Recombinant Strains
The complementation and localization constructs were used to transform S. reilianum by protoplast transformation. Protoplast preparation and transformation of S. reilianum were performed according to the protocols used for Ustilago maydis (Schulz et al., 1990; Gillissen et al., 1992) with some modifications. A single fungal colony was used to inoculate 2 mL of liquid YEPS-light medium and incubated at 28°C for 8 to 12 h. This culture was used to inoculate 100 mL of YEPS-light medium, which was incubated at 28°C with constant shaking at 200 rpm. Cells were grown to an OD600 of 0.6 to 0.8 and subsequently pelleted by centrifugation at 3,500g (Beckmann Biofuge) for 5 min. The supernatant was discarded, and the pellet was resuspended in 50 mL of 20 mm Na-citrate, pH 5.8, and 1 m sorbitol sterile filtered (SCS buffer). Cells were centrifuged at 3,500g for 10 min, and the supernatant was discarded. Protoplasts were produced by resuspending the cells in 2 mL of Novozyme (2.5 mg mL−1) solution (Novo Nordisc) and incubating at room temperature for 5 to 10 min until approximately 50% of the cells produced protoplasts. The formation of protoplasts was confirmed by microscopy. Protoplasting was stopped by adding 20 mL of SCS buffer and centrifuging the solution at 2,300g for 15 min. The supernatant was discarded, and the pellet was washed twice with SCS buffer and once with 10 mm Tris-HCl, pH 7.5, 100 mm CaCl2, and 1 m sorbitol sterile filtered (STC buffer), respectively, by carefully resuspending the pellet in 20 mL of either buffer and centrifuging at 2,300g for 15 min. Finally, protoplasts were resuspended in 500 μL of ice-cold STC buffer, dispensed into 70-μL aliquots, and either used directly for transformation or stored at −80°C.
For transformation of the protoplasts, a 70-μL aliquot of protoplasts was mixed with approximately 5 μg of DNA and 1.5 μL of heparin sodium sulfate (50 mg mL−1) and kept on ice for 10 min. Protoplasts were carefully mixed with 500 μL of cold STC buffer containing 40% (v/v) polyethylene glycol 4,000 solution and incubated on ice for a further 15 min. The entire mixture was plated onto regeneration medium (10 g L−1 tryptone, 10 g L−1 yeast extract, 10 g L−1 Suc, 182.2 g L−1 sorbitol, and 20 g L−1 agar) supplemented with the appropriate antibiotic (1 µg mL−1 phleomycin, 5 µg mL−1 carboxin, or 150 µg mL−1 hygromycin). Plates were incubated at 28°C for 4 to 6 d or until distinct colonies appeared. Single colonies were picked using sterile toothpicks and streaked onto PD or regeneration medium plates supplemented with the appropriate antibiotic to obtain single colonies. Putative transformants were selected and verified by PCR using primers listed in Table II and Southern blot using the deletion construct as probe.
For each mutant, one to three different strains in each SRZ1_5-2 and SRZ2_5-1 background were used to inoculate maize plants. Strains used in this study for ∆19A1 mutants were (1) JS747 and JS751 and (2) JS748 and JS752, strains used for ∆19A2 mutants were (1) HG125 and HG127 and (2) HG126 and HG128, strains used for ∆sr10073 mutants were HG109 and HG113, strains used for ∆sr10075 mutants were HG80 and HG84, strains used for ∆sr10077 mutants were HG95 and HG99, strains used for ∆sr10079 mutants were HG89 and HG92 (H. Ghareeb, Y. Zhao, and J. Schirawski, unpublished data), strains used for ∆SAD1+SAD1 mutants were (1) HG163 and HG167 and (2) HG164 and HG168, strains used for PSAD1:GFP-containing mutants were HG289 and HG292, and strains used for ∆SAD1+SAD1-GFP mutants were (1) HG183 and HG186 and (2) HG185 and HG187)
For generation of S. reilianum strains expressing cytoplasmic GFP, protoplasts of the compatible S. reilianum wild-type strains SRZ2_5-1 and SRZ1_5-2 were transformed with pHSP70-SG (Spellig et al., 1996).
Arabidopsis Transformation
Seeds of Arabidopsis ecotype Col-0 were grown on soil in growth chambers (York) at 22°C to 25°C under a long-day regime of a 16-h-light/8-h-dark cycle at approximately 60% humidity. Recombinant DNA constructs were introduced into Arabidopsis plants by Agrobacterium tumefaciens-mediated transformation using A. tumefaciens strain EHA105 (Hood et al., 1993) and the floral-dip method (Clough and Bent, 1998). Independent transformants were selected according to resistance against aerosolic glufosinate ammonium (Bayer).
RNA Isolation and qRT-PCR Analysis
Transcript levels of cluster 19A2 genes (as indicated in Fig. 2B) were measured by isolating RNA from sporidia grown in liquid culture to an OD600 of 0.5 as well as isolating RNA from infected plant samples. Plant samples were collected from a 2-cm piece below the injection hole of the third leaf at 3 dpi (10 d after sowing [das]), developing nodes of the main stem at 15 dpi (21 das), and ears, including stalks, with a length of 1 to 3 cm at 7 wpi (56 das) from mock- or S. reilianum-inoculated plants. The samples were collected from at least 20 plants for each experiment and sampling time point, and the experiment was done three independent times.
For qRT-PCR analysis of the ZmPIN1 and TB1 genes (indicated in Fig. 7), three plant tissues were analyzed: roots, ears of a size of 1 to 3 cm, excluding stalks, and stalks after removal of the ears or axillary buds. For each biological replicate, samples were collected from five different S. reilianum- or water-inoculated plants at 7 wpi (56 das). RNA was extracted with Trizol (Invitrogen), DNase treatment was performed using RNase-Free DNase Set (Qiagen), and RNA was purified with RNeasy Plant Mini Kit (Qiagen). Gene transcript levels were determined relative to the peptidylprolylisomerase (ppi; sr11196) transcript level from S. reilianum for analysis of cluster 19A2 genes and relative to the ACTIN1 (ACT1) gene from maize for analysis of maize gene transcription. Because the family members of ZmPIN1 share high conservation of amino acid sequences and exon/intron structure (Carraro et al., 2006; Forestan et al., 2010, 2012), we designed oligonucleotide primers that targeted exon 1 of the three family members ZmPIN1a, ZmPIN1b, and ZmPIN1c (Supplemental Fig. S11). Primers are listed in Table II.
Statistical Data Analysis
For statistical analysis, all data were tested for normal distribution. The statistical differences between different treatments were analyzed by one-way ANOVA and Tukey’s test using the R software.
Yeast cDNA Library Construction
RNA for normalized cDNA library construction was obtained from an axenically grown mixture of the two compatible S. reilianum wild-type strains SRZ1_5-2 and SRZ2_5-1 and three different maize tissues infected with S. reilianum: inoculated leaves (2 cm below injection hole) at 3 dpi, infected nodes at 15 dpi, and infected ears at 4 wpi (smaller than 2 cm). The tissues were collected from three independent experiments, which each included 20 plants. The quality and purity of RNA from each individual experiment and tissue were checked by Bioanalyzer 2100 (Agilent) and NanoDrop ND-1000 (Thermo Fisher Scientific). Equal RNA amounts from the pooled RNA of each tissue were mixed to make the RNA sample for cDNA library construction. A normalized cDNA library was constructed by Bio S&T. Briefly, 100 µg of total RNA was used for mRNA extraction and cDNA synthesis followed by cDNA normalization using a modified SMART cDNA synthesis method (Clontech). The cDNA was amplified, purified, digested with SfiI for directional cloning into the pGADT7 (modified to include an SfiI site at the multiple cloning site), ligated, and transformed into Escherichia coli. Cells were plated into Luria-Bertani solid medium plates with ampicillin and incubated at 37°C overnight. Colonies were collected from plates with 10% (v/v) glycerol and the help of glass beads. The E. coli cells were kept at −80°C in 1-mL aliquots. A portion of the E. coli-containing cDNA library was grown overnight in Luria-Bertani liquid medium with ampicillin. The pGADT7 containing the cDNA library was isolated and used for transformation of yeast to construct the yeast two-hybrid library.
Yeast Two-Hybrid Library Generation
The yeast two-hybrid library was constructed according to Make Your Own Mate and Plate Yeast Two-Hybrid Library System and Yeastmaker Yeast Transformation System 2 (Clontech) with some modifications. A 2- to 3-mm single colony of the yeast strain Y187 was inoculated in 3 mL of yeast peptone dextrose adenine (YPDA) liquid medium and incubated at 30°C with 250 rpm constant shaking for 8 to 12 h. From this culture, 5 µL were used to inoculate 50 mL of YPDA liquid medium and incubated as previously indicated for 16 to 20 h until OD600 = 0.15 to 0.3 is reached. The culture was centrifuged at 700g for 5 min. The cell pellet was resuspended in 100 mL of YPDA liquid medium and left to grow at 30°C with 250 rpm constant shaking for 3 to 5 h until OD600 = 0.4 to 0.5 is reached. The culture was centrifuged at 700g for 5 min. The cell pellet was resuspended in 30 mL of water and centrifuged once again. The cell pellet was resuspended in 1.2 mL of Tris-EDTA buffer. The cells were used immediately for transformation of cDNA library.
For transformation, the 600-µL cell suspension was mixed with 15 µg of cDNA library, 20 µL of denatured DNA carrier (10 mg mL−1; Clontech), and 2.5 mL of polyethylene glycol-LiAc solution. The transformation mixture was incubated at 30°C for 45 min with gentle mixing every 15 min. Thereafter, 160 µL of dimethyl sulfoxide was added to the mixture and then incubated in a water bath at 42°C for 20 min with mixing every 10 min. The cells were centrifuged at 700g for 5 min, resuspended in 3 mL of YPD Plus Medium (Clontech), and incubated at 30°C with 250 rpm shaking for 90 min. The transformed culture was centrifuged once again. This transformation was performed 10 times individually, and the cell pellets were pooled and resuspended in 30 mL of 0.9% (w/v) NaCl. The cell suspensions (150 µL per plate) were plated on 200 SD/-Leu solid medium. Plates were incubated at 30°C for 4 d and then chilled at 4°C for 1 d. The colonies in each plate were collected using 5 mL of freezing medium (YPDA with 25% [v/v] glycerol and 50 µg mL−1 kanamycin) and a glass rod. Cell suspensions were collected in one container, well mixed, and distributed in 1-mL aliquots for direct use or 50-mL aliquots for long-term storage. Aliquots were stored at −80°C until used. Cell density of the library was calculated using a hemocytometer and titering 10−4, 10−5, 10−6, and 10−7 dilutions of the library on SD/-Leu solid plates.
Autoactivation Test and Yeast Two-Hybrid Screening
The yeast two-hybrid screening was performed according to Matchmaker Gold Yeast Two-Hybrid System (Clontech). To prepare for the yeast two-hybrid screening with SAD1, the SAD1 gene without the signal peptide was cloned into pGBKT7 (pGBKT7–SAD1) so that it fuses with the GAL4 BD. The plasmid was transformed into the yeast strain Y2HGold. As controls, the empty plasmid pGBKT7 and pGBKT7-53 (containing p53 coding sequence) as well as pGADT7 (empty prey plasmid), pGADT7-T (containing T antigen coding sequence), and pGADT7-Lam (containing the Lam coding sequence) were transformed into the Y2HGold and Y187 strains, respectively. The Y2HGold and Y187 strains can mate.
To test whether SAD1 has an autoactivation effect on the selectable markers of the yeast strain, the strain Y2HGold containing pGBKT7-SAD1 was mated with the strain Y187 containing pGADT7, and as control, the strain Y2HGold containing pGBKT7-53 was mated with the strain Y187 containing either pGADT7-T (resulting in positive interaction) or pGADT7-Lam (resulting in negative interaction). The mating was performed by mixing one 2- to 3-mm single colony from each strain in 2× YPDA liquid medium. The mixture was incubated at 30°C with 50 rpm shaking for 20 to 24 h. The mating events were selected on SD/-Leu/-Trp double dropout (DDO). The autoactivation of the selectable markers was analyzed by growing the zygotes on DDO with 125 to 300 ng of Aureobasidin A and 5-bromo-4-chloro-3-indolyl alpha-d-galactopyranoside or DDO additionally lacking adenine and His (quadruple dropout [QDO]) solid medium.
To screen for the interaction partners of SAD1, a single colony of the Y2HGold strain containing pGBKT7-SAD1 was inoculated in 70 mL of SD/-Trp liquid medium and incubated at 30°C with 250 rpm shaking until an OD600 of 0.8 was reached. The culture was centrifuged at 1,000g for 5 min; then the cell pellet was resuspended in 4 mL of SD/-Trp liquid medium and combined with 200 µL of Y187 containing the cDNA library (see above) in a sterile 2-L flask. To allow mating, 45 mL of 2× YPDA liquid medium supplemented with 50 μg mL−1 kanamycin was added, and the culture was incubated at 30°C with 50 rpm shaking for 24 h. The mated culture was centrifuged at 1,000g for 10 min and then resuspended in 10 mL of one-half-strength YPDA (with 50 μg mL−1 kanamycin). Each 200-µL aliquot of mating suspension was spread on high stringency medium (QDO). Additionally, dilution serials 10−3, 10−5, and 10−6 were prepared, and 100 µL of each dilution were spread on SD/-Leu, SD/-Trp, and DDO plates to estimate the mating efficiency and calculate the total number of screened clones. The plates were incubated at 30°C for 5 d.
The growing colonies were repurified twice on QDO with 300 ng mL−1 Aureobasidin. Colonies that survived were used to isolate plasmids, which were individually used to transform E. coli, and then plasmids were isolated from E. coli and sequenced. The sequences were grouped, and a representative plasmid (pGADT7-PREY) for each group was retransformed in the Y187 strain to generate Y187+pGADT7-PREY. The Y187+pGADT7-PREY strains were grown in 150 µL of liquid SD/-Leu medium in 96-well microtiter plates at 30°C and shaking at 280 rpm for 2 d. Meanwhile, the Y2HGold strains containing either pGBKT7-SAD1 or pGBKT7 were grown in 20 mL of SD/-Trp medium at 30°C and 250 rpm until an OD600 of 5 to 6 was reached. To verify the interaction, 75 µL of Y187+pGADT7-PREY was mixed with 75 µL of Y2HGold strains containing either pGBKT7-SAD1 or pGBKT7 in 96-well microtiter plates and incubated overnight at 30°C with shaking at 50 rpm. The cell mixtures were printed on DDO and QDO plates using a 96-prong replicator and incubated at 30°C for 3 to 4 d.
Staining and Confocal Microscopy
Samples for microscopy were freshly collected and prepared by mounting approximately 1-cm pieces of infected leaves or thin sections of infected ears on a glass slide and immediately fixing the material on the slide with the help of a drop of water and a coverslip. The samples were immediately observed using a TCS-SP5 Confocal Microscope (Leica), a Zeiss Axioplan II Microscope or a Axio Observer.Z1 Microscope (Zeiss), or an AF 6000LX Fluorescence Microscope (Leica). For fluorescence microscopy of GFP, the fluorescein isothiocyanate filter (excitation at 450–490 nm and emission at 515–565 nm) was used. Image processing was performed using the imaging software MetaMorph v6.2 (Universal Imaging), AxioVision v4.3 (Zeiss), or Leica Application Suite Advanced Fluorescence v4.0 (Leica, Germany). For confocal microscopy of SAD1-GFP fusion proteins in planta, the samples were laser excited at 488 nm, and the emission signal was detected at 500 to 530 nm. For staining plant and fungal cell membranes, FM4-64 (Molecular Probes; Invitrogen) was used. The infected ears were manually thin sectioned, dipped in FM4-64 staining solution (17 µm FM4-64 in water), and incubated in the dark and on ice for 30 min. The FM4-64 was excited at 561 nm, and signal was detected at 640 to 750 nm. Images were processed using LAS AF v1.8 and v 4.0 (Leica). Deconvolution of Z stacks was performed using the LAS AF v4.0 (Leica) using 10 iterations with a refractive index of 1.33 for the HCX PL APO 40×/1.10 water objective and the blind mode.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Loss of apical dominance due to infection by S. reilianum.
Supplemental Figure S2. SAD1 amino acid sequence and domain analysis.
Supplemental Figure S3. S. reilianum strains expressing SAD1-GFP show extracellular GFP fluorescence.
Supplemental Figure S4. Colocalization of SAD1-GFP fluorescence with the membrane-staining dye FM4-64.
Supplemental Figure S5. Number of ears per plant of maize plants inoculated with S. reilianum strains expressing SAD1 tagged with MYC or HA.
Supplemental Figure S6. GFP-SAD1 does not localize to the cell wall in transgenic Arabidopsis cells expressing GFP-SAD1 and mCHERRY.
Supplemental Figure S7. Microscopic overview of GFP-SAD1ΔSP distribution in transgenic Arabidopsis plants.
Supplemental Figure S8. Immunodetection of BD-SAD1ΔSP and BD in Y2H-Gold strains containing the plasmids pGBKT7-BD-SAD1ΔSP and pGBKT7-BD, respectively, with antibody specific for c-MYC.
Supplemental Figure S9. Transcriptional activation of reporter genes in yeast by SAD1.
Supplemental Figure S10. The fusion protein AD-SAD1 is not able to autoactivate the MEL1 reporter gene in the yeast two-hybrid strain Y187.
Supplemental Figure S11. Nucleotide sequence alignment of qRT-PCR amplicons generated from exon1 of ZmPIN1a, ZmPIN1b, and ZmPIN1c.
Supplemental Figure S12. Identification of SAD1 interaction partners.
Supplementary Material
Acknowledgments
We thank Regine Kahmann and Thomas Brefort (Max Planck Institute for Terrestrial Microbiology Marburg) for stimulating discussions; Jennifer Krüger (Georg August Universität Göttingen) for technical assistance and help with the identification of homozygous Arabidopsis lines expressing GFP-SAD1ΔSP; Elmar Meyer (Max Planck Institute for Terrestrial Microbiology Marburg) for help with the generation of cytoplasmic GFP expressing S. reilianum strains; Yufei Wang (Georg August Universität Göttingen) for the generation of the PSAD1:GFP construct; Ivo Feussner (Georg August Universität Göttingen) and Regine Kahmann (Max Planck Institute for Terrestrial Microbiology Marburg) for greenhouse space; and Volker Lipka (Georg August Universität Göttingen) for use of the confocal microscope.
Glossary
- Col-0
Columbia-0
- das
days after sowing
- DDO
double dropout
- dpi
days postinoculation
- PAT
polar auxin transport
- QDO
quadruple dropout
- qRT
quantitative reverse transcription
- SCS buffer
20 mm Na-citrate, pH 5.8, and 1 m sorbitol sterile filtered
- STC buffer
10 mm Tris-HCl, pH 7.5, 100 mm CaCl2, and 1 m sorbitol sterile filtered
- wpi
weeks postinoculation
- YEPS-light medium
10 g L−1 yeast extract, 10 g L−1 peptone, and 10 g L−1 Suc
- cDNA
complementary DNA
Footnotes
This work was supported by the Max Planck Society, the RWTH Aachen University, the International Max Planck Research School for Environmental, Cellular and Molecular Microbiology (H.G.), the Göttingen Graduate School for Neurosciences, Biophysics, and Molecular Biosciences (H.G.), and the Georg August University through the German Initiative of Excellence (grant no. DFG ZUK45/1 to J.S.).
Articles can be viewed without a subscription.
References
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Barazesh S, McSteen P (2008) Hormonal control of grass inflorescence development. Trends Plant Sci 13: 656–662 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Hake SC (2009) Handbook of Maize: Its Biology. Springer, New York [Google Scholar]
- Bomblies K, Doebley JF (2006) Pleiotropic effects of the duplicate maize FLORICAULA/LEAFY genes zfl1 and zfl2 on traits under selection during maize domestication. Genetics 172: 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann A, König J, Julius C, Feldbrügge M (2004) A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol Genet Genomics 272: 216–226 [DOI] [PubMed] [Google Scholar]
- Carraro N, Forestan C, Canova S, Traas J, Varotto S (2006) ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol 142: 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Ruyter-Spira C, Bouwmeester H (2013) The interaction between strigolactones and other plant hormones in the regulation of plant development. Front Plant Sci 4: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coe EH, Neuffer MG, Hoisington DA (1988) The Genetics of Corn. American Society of Agronomy, Madison, WI [Google Scholar]
- Crawford S, Shinohara N, Sieberer T, Williamson L, George G, Hepworth J, Müller D, Domagalska MA, Leyser O (2010) Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137: 2905–2913 [DOI] [PubMed] [Google Scholar]
- Davies CR, Seth AK, Wareing PF (1966) Auxin and kinetin interaction in apical dominance. Science 151: 468–469 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Domagalska MA, Leyser O (2011) Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol 12: 211–221 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- Faure C, Ramos M, Girault JA (2013) Pyk2 cytonuclear localization: mechanisms and regulation by serine dephosphorylation. Cell Mol Life Sci 70: 137–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlayson SA. (2007) Arabidopsis Teosinte Branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot Teosinte Branched1. Plant Cell Physiol 48: 667–677 [DOI] [PubMed] [Google Scholar]
- Forestan C, Farinati S, Varotto S (2012) The maize PIN gene family of auxin transporters. Front Plant Sci 3: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestan C, Meda S, Varotto S (2010) ZmPIN1-mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development. Plant Physiol 152: 1373–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A. (2013) The role of auxin in shaping shoot architecture. J Exp Bot 64: 2593–2608 [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P (2008a) Sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci USA 105: 15196–15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Yang Y, Schmidt RJ, Jackson D (2008b) The relationship between auxin transport and maize branching. Plant Physiol 147: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghareeb H, Becker A, Iven T, Feussner I, Schirawski J (2011) Sporisorium reilianum infection changes inflorescence and branching architectures of maize. Plant Physiol 156: 2037–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillissen B, Bergemann J, Sandmann C, Schroeer B, Bölker M, Kahmann R (1992) A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68: 647–657 [DOI] [PubMed] [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2: 208–218 [Google Scholar]
- Hoshi A, Oshima K, Kakizawa S, Ishii Y, Ozeki J, Hashimoto M, Komatsu K, Kagiwada S, Yamaji Y, Namba S (2009) A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc Natl Acad Sci USA 106: 6416–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard L, McSteen P, Doebley J, Hake S (2002) Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162: 1927–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämper J, Kahmann R, Bölker M, Ma LJ, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Müller O, et al. (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444: 97–101 [DOI] [PubMed] [Google Scholar]
- MacLean AM, Sugio A, Makarova OV, Findlay KC, Grieve VM, Tóth R, Nicolaisen M, Hogenhout SA (2011) Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol 157: 831–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaguti G, Fernandez B, Nass H (1977) Downy mildew or crazy top of maize in Venezuela. Agronomia Tropical 27: 103–129 [Google Scholar]
- Matheussen AM, Morgan PW, Frederiksen RA (1991) Implication of gibberellins in head smut (Sporisorium reilianum) of Sorghum bicolor. Plant Physiol 96: 537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P. (2009) Hormonal regulation of branching in grasses. Plant Physiol 149: 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P, Hake S (2001) Barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 128: 2881–2891 [DOI] [PubMed] [Google Scholar]
- McSteen P, Malcomber S, Skirpan A, Lunde C, Wu X, Kellogg E, Hake S (2007) barren inflorescence2 Encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol 144: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt LW, Rincones J, Bailey BA, Aime MC, Griffith GW, Zhang D, Pereira GAG (2008) Moniliophthora perniciosa, the causal agent of witches’ broom disease of cacao: what’s new from this old foe? Mol Plant Pathol 9: 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muszynski MG, Dam T, Li B, Shirbroun DM, Hou Z, Bruggemann E, Archibald R, Ananiev EV, Danilevskaya ON (2006) delayed flowering1 Encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol 142: 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D, Leyser O (2011) Auxin, cytokinin and the control of shoot branching. Ann Bot (Lond) 107: 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström A, Tarkowski P, Tarkowska D, Norbaek R, Astot C, Dolezal K, Sandberg G (2004) Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proc Natl Acad Sci USA 101: 8039–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P (2011) vanishing tassel2 Encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23: 550–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov A, Chuderland D, Karamansha Y, Livnah O, Seger R (2011) Nuclear extracellular signal-regulated kinase 1 and 2 translocation is mediated by casein kinase 2 and accelerated by autophosphorylation. Mol Cell Biol 31: 3515–3530 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Prusinkiewicz P, Crawford S, Smith RS, Ljung K, Bennett T, Ongaro V, Leyser O (2009) Control of bud activation by an auxin transport switch. Proc Natl Acad Sci USA 106: 17431–17436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke G, Heinze B, Schirawski J, Buettner H, Kahmann R, Basse CW (2008) Indole-3-acetic acid (IAA) biosynthesis in the smut fungus Ustilago maydis and its relevance for increased IAA levels in infected tissue and host tumour formation. Mol Plant Pathol 9: 339–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs T. (1981) The control of the patterned differentiation of vascular tissues. Adv Bot Res 9: 151–262 [Google Scholar]
- Schirawski J, Heinze B, Wagenknecht M, Kahmann R (2005) Mating type loci of Sporisorium reilianum: novel pattern with three a and multiple b specificities. Eukaryot Cell 4: 1317–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirawski J, Mannhaupt G, Münch K, Brefort T, Schipper K, Doehlemann G, Di Stasio M, Rössel N, Mendoza-Mendoza A, Pester D, et al. (2010) Pathogenicity determinants in smut fungi revealed by genome comparison. Science 330: 1546–1548 [DOI] [PubMed] [Google Scholar]
- Schulz B, Banuett F, Dahl M, Schlesinger R, Schäfer W, Martin T, Herskowitz I, Kahmann R (1990) The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60: 295–306 [DOI] [PubMed] [Google Scholar]
- Shani E, Yanai O, Ori N (2006) The role of hormones in shoot apical meristem function. Curr Opin Plant Biol 9: 484–489 [DOI] [PubMed] [Google Scholar]
- Sheridan WF. (1988) Maize developmental genetics: genes of morphogenesis. Annu Rev Genet 22: 353–385 [DOI] [PubMed] [Google Scholar]
- Shinohara N, Taylor C, Leyser O (2013) Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol 11: e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbe DS, Doehlemann G, Fernandes J, Walbot V (2010) Maize tumors caused by Ustilago maydis require organ-specific genes in host and pathogen. Science 328: 89–92 [DOI] [PubMed] [Google Scholar]
- Spellig T, Bottin A, Kahmann R (1996) Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol Gen Genet 252: 503–509 [DOI] [PubMed] [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J (2011) Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet 43: 1160–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Takei K, Kojima M, Sakakibara H, Mori H (2006) Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J 45: 1028–1036 [DOI] [PubMed] [Google Scholar]
- Turnbull CG, Booker JP, Leyser HM (2002) Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J 32: 255–262 [DOI] [PubMed] [Google Scholar]
- Wang Q, Kohlen W, Rossmann S, Vernoux T, Theres K (2014) Auxin depletion from the leaf axil conditions competence for axillary meristem formation in Arabidopsis and tomato. Plant Cell 26: 2068–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Davis RE, Nuss DL, Zhao Y (2013) Phytoplasmal infection derails genetically preprogrammed meristem fate and alters plant architecture. Proc Natl Acad Sci USA 110: 19149–19154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuther K, Kahnt J, Utermark J, Imkampe J, Uhse S, Schirawski J (2012) Host specificity of Sporisorium reilianum is tightly linked to generation of the phytoalexin luteolinidin by Sorghum bicolor. Mol Plant Microbe Interact 25: 1230–1237 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.