An Arabidopsis kinase possesses a unique repeat motif and suppresses resistance to the bacterial pathogen Pseudomonas syringae.
Abstract
Receptor-like cytoplasmic kinases (RLCKs) are a subset of plant receptor-like kinases lacking both extracellular and transmembrane domains. Some of the 46 members in the Arabidopsis (Arabidopsis thaliana) RLCK subfamily VII have been linked to plant innate immunity; however, most remain uncharacterized. Thus, multiple subfamily VII members are expected to be involved in plant immune signaling. Here, we investigate the role of AvrPphB SUSCEPTIBLE1-LIKE13 (PBL13), a subfamily VII RLCK with unique domain architecture. Unlike other characterized RLCKs, PBL13 transfer DNA insertion lines exhibit enhanced disease resistance after inoculation with virulent Pseudomonas syringae. The pbl13-2 knockout also exhibits elevated basal-level expression of the PATHOGENESIS-RELATED GENE1 defense marker gene, enhanced reactive oxygen species (ROS) burst in response to perception of bacterial microbial patterns, and accelerated flagellin-induced activation of mitogen-activated protein kinases. Recombinant PBL13 is an active kinase, and its primary autophosphorylated sites map to a 15-amino acid repeat motif unique to PBL13. Complementation of pbl13-2 with PBL13-3xFLAG converts the enhanced resistance and elevated ROS phenotypes back to wild-type levels. In contrast, kinase-dead PBL13K111A-3xFLAG was unable to rescue pbl13-2 disease phenotypes. Consistent with the enhanced ROS burst in the pbl13-2 knockout, PBL13 is able to associate with the nicotinamide adenine dinucleotide phosphate, reduced oxidase RESPIRATORY BURST OXIDASE HOMOLOG PROTEIN D (RBOHD) by split-luciferase complementation assay, and this association is disrupted by flagellin treatment. We conclude that the PBL13 kinase negatively regulates plant innate immunity to pathogenic bacteria and can associate with RBOHD before pathogen perception. These data are consistent with the hypothesis that PBL13 acts to prevent inappropriate activation of defense responses in the absence of pathogen challenge.
Plants exist in environments abundantly populated with microbes. Many of these microbes form positive or negative associations with plants. To protect themselves from pathogenic microorganisms forming negative associations, plants actively defend themselves using an innate immune surveillance system consisting of receptors with extracellular domains as well as intracellular receptors (Dodds and Rathjen, 2010). Receptors possessing extracellular domains are typically Receptor-Like Kinases (RLKs) or Receptor-Like Proteins and recognize conserved microbial features termed pathogen-associated molecular patterns (PAMPs) or microbe-associated molecular patterns (MAMPs; Boller and Felix, 2009; Zipfel, 2014). Receptors for MAMPs are known as pattern recognition receptors (PRRs), and MAMP recognition culminates in microbe-associated molecular pattern-triggered immunity (MTI; Zipfel, 2014). MTI is also frequently referred to as PAMP-triggered immunity. Hallmarks of MTI include the production of reactive oxygen species (ROS), a cellular influx of calcium, mitogen-activated protein kinase (MAPK) activation, up-regulation of defense-related genes, and callose deposition (Zipfel, 2014). Intracellular immune receptors are generally nucleotide binding-leucine-rich repeat (NLR) proteins that detect the presence of pathogen effectors delivered into plant cells (Dodds and Rathjen, 2010; Elmore et al., 2011; Spoel and Dong, 2012). NLRs can directly detect effectors or monitor (guard) for effector-mediated perturbations of key host targets (Jones and Dangl, 2006). Effector recognition culminates in effector-triggered immunity (ETI). There is significant overlap in MTI and ETI signaling and transcriptional reprogramming, but their amplitudes are different (Tsuda and Katagiri, 2010; Thomma et al., 2011; Pritchard and Birch, 2014). ETI-based responses are considered more robust and frequently associated with a form of cell death, termed the hypersensitive response, localized to sites of pathogen infection (Elmore et al., 2011).
Over the years, a large number of plant immune receptors and downstream components has been discovered, but a detailed understanding of downstream signaling upon immune receptor activation remains elusive. Currently, it is known that multiple protein kinases, including MAPKs, calcium-dependent protein kinases, and RLKs, play critical roles in immune signaling (Tena et al., 2011). One class of important immune signaling kinases are receptor-like cytoplasmic kinases (RLCKs), a subcategory of the RLKs (Lin et al., 2013). RLCKs possess a single-kinase domain and can be anchored to the plasma membrane by acylation. Of all of the RLCK subfamilies, subfamily VII is the largest with 46 members, and several members have been shown to be involved in plant immunity. The best characterized RLCK required for immune responses is BOTRYITIS INDUCED KINASE1 (BIK1), a prominent player in perception of the bacterial MAMP flagellin (Lu et al., 2010; Zhang et al., 2010). BIK1 exists in complex with both the flagellin immune receptor FLAGELLIN-SENSITIVE2 (FLS2) and its coreceptor BRASSINOSTEROID INSENSITIVE1 ASSOCIATED RECEPTOR KINASE1 (BAK1) at resting state (Lu et al., 2010). Upon flagellin perception by FLS2, BAK1 is recruited to FLS2, and transphosphorylation events among the three kinase domains culminate in dissociation of BIK1 from the activated receptor complex (Chinchilla et al., 2007; Heese et al., 2007; Lu et al., 2010). BIK1 proceeds to phosphorylate the plasma membrane NADPH oxidase RESPIRATORY BURST OXIDASE HOMOLOG PROTEIN D (RBOHD), which is necessary for the extracellular ROS burst (Kadota et al., 2014; Li et al., 2014). Analysis of BIK1 paralogs revealed that AvrPphB SUSCEPTIBLE1 (PBS1) and AvrPphB SUSCEPTIBLE1-LIKE1 (PBL1) function additively with BIK1 to regulate the ROS burst (Zhang et al., 2010). RLCKs in subfamily VII have also been shown to be important for perception of chitin, a fungal MAMP (Zhang et al., 2010; Yamaguchi et al., 2013; Ao et al., 2014; Shinya et al., 2014). Rice (Oryza sativa) RLCK185, OsRLCK176, and Arabidopsis (Arabidopsis thaliana) PBL27 were also found to associate with the chitin coreceptor, indicating that RLCKs represent an important signaling module downstream of multiple PRR complexes (Yamaguchi et al., 2013; Ao et al., 2014; Shinya et al., 2014).
In addition to associating with PRR complexes, RLCKs have been found to associate with plant NLR immune receptors. PBS1 was one of the first subfamily VII RLCKs characterized, and it is monitored by the Arabidopsis NLR RESISTANCE TO PSEUDOMONAS SYRINGAE5 (RPS5; Ade et al., 2007). Cleavage of PBS1 by the Pseudomonas syringae Cys protease effector AvrPphB results in activation of RPS5 and thereby, ETI (Shao et al., 2003; DeYoung et al., 2012). Another Arabidopsis RLCK VII, RESISTANCE TO PSEUDOMONAS SYRINGAE PV. MACULICOLA1 (RPM1)-INDUCED PROTEIN KINASE (RIPK), interacts with the plasma membrane-associated protein RPM1 INTERACTING4 (RIN4; Liu et al., 2011). RIN4 is targeted by several P. syringae effectors and monitored by the NLR proteins RPM1 and RPS2 (Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003). The P. syringae effector AvrB induces hyperphosphorylation of RIN4 through RIPK and potentially, other RLCK VII members (Liu et al., 2011). In particular, phosphorylation of RIN4 at the conserved Thr-166 residue activates ETI through RPM1 (Chung et al., 2011; Liu et al., 2011). In tomato (Solanum lycopersicum), the RLCK PTO associates with the NLR PSEUDOMONAS RESISTANCE AND FENTHION SENSITIVITY (PRF) and mediates recognition of the P. syringae effectors AvrPto and AvrPtoB (Oh and Martin, 2011; Ntoukakis et al., 2013; Mathieu et al., 2014).
Pathogen effectors are critical virulence factors, inhibiting plant immune responses and promoting pathogen growth in the absence of NLR perception (Giraldo and Valent, 2013; Macho and Zipfel, 2015). Targeting of RLCKs by multiple bacterial effectors suggests that they may be common virulence targets. In addition to PBS1, AvrPphB also cleaves BIK1, PBL1, and six other RLCK VIIs (Zhang et al., 2010). The Xanthomonas campestris pv campestris effector AvrAC is a UMP transferase that uridylates BIK1, PBL1, and RIPK at highly conserved Ser and Thr residues within the kinase activation loop, thereby attenuating RLCK kinase activity (Feng et al., 2012). Single Arabidopsis bik1 and pbl1 knockout (KO) lines are compromised in MTI, and the bik1/pbl1 double KO exhibits weaker MTI responses (Zhang et al., 2010). Furthermore, transgenic plants expressing either AvrPphB or AvrAC are compromised in MTI, indicating that multiple RLCKs are critical for immune signaling (Zhang et al., 2010; Feng et al., 2012).
Interestingly, effectors may also recruit RLCKs to negatively regulate immunity by modulating activity of host immune regulators. In a susceptible genetic background lacking RPM1, AvrB-induced RLCK-mediated phosphorylation of RIN4 at Thr-166 inhibits MTI responses. This event is epistatic to phosphorylation of RIN4 Ser-141, which is critical for activation of MTI (Chung et al., 2014). Phosphorylation of RIN4 Ser-141 is diminished in bik1/pbl1 double mutants (Chung et al., 2014). Together, these data indicate that effectors can target RLCKs to alter kinase specificity or activity.
Although recent work has provided important insight regarding the role of RLCKs in the positive regulation of plant innate immunity, many members of this kinase subfamily remain uncharacterized. For example, although BIK1 and PBL1 are dispensable for MAPK3/6 activation, transgenic plants expressing AvrAC are unable to fully inhibit MAPK3/6, suggesting that other RLCKs fulfill the role for MAPK3/6 activation (Feng et al., 2012). These additional RLCK members likely also play key roles in immune signaling. Furthermore, RLCKs that negatively regulate immune responses remain to be identified. Here, we report that transfer DNA (T-DNA) insertions in PBL13, an RLCK VII with unique domain architecture, result in enhanced disease resistance to P. syringae pv tomato DC3000 (Pto DC3000) in the model plant Arabidopsis. These insertion lines also exhibit elevated MTI responses, including MAPK activation, ROS burst, and defense gene expression. We found that PBL13’s regulation of plant immunity is dependent on its kinase activity. Furthermore, we show that PBL13 is an active kinase that, in vitro, is heavily phosphorylated at a unique C-terminal repeat region. Lastly, we show that PBL13 associates with RBOHD in planta, and flagellin treatment disrupts this association, providing a mechanistic insight into the role of PBL13 in plant innate immunity.
RESULTS
PBL13 Is an RLCK VII with Unique Domain Architecture
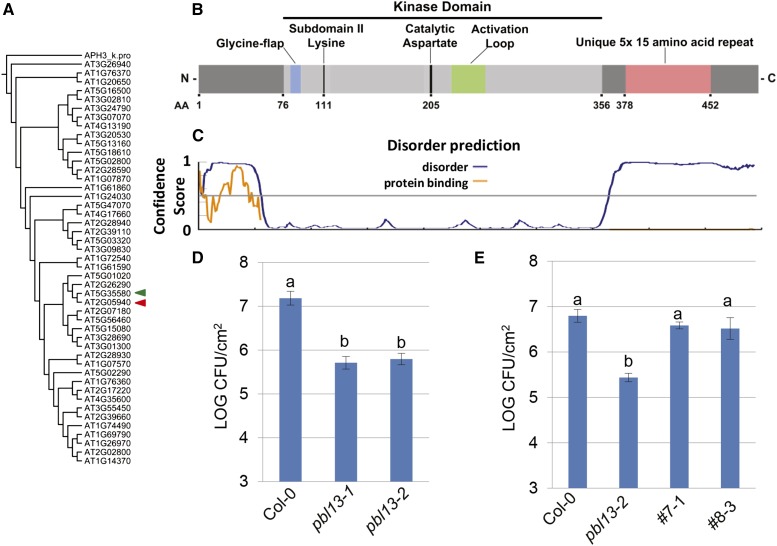
To identify unique RLCKs involved in innate immune regulation, existing microarray data sets were mined, identifying RLCKs from subfamily VII that are differentially regulated at the transcript level upon biotic stress (Supplemental Fig. S1). The investigated array data included experiments where plants were treated with virulent Pto DC3000, nonpathogenic bacteria (Pto DC3000 ∆hrcC, Pto DC3000 expressing avrRpm1, and P. syringae pv phaseolicoa), elicitors, and MAMPs. Twenty-four RLCKs were at least 1.5-fold up-regulated in one or more conditions. Seven RLCKs were found to be at least 1.5-fold down-regulated in one or more conditions. Intriguingly, one of the up-regulated RLCK VIIs, PBL13 (AT5G35580), had unique domain architecture. PBL13 is induced during treatment with Pto DC3000 (AvrRpm1) as well as with the elicitors Harpin Z and NECROSIS-INDUCING PHYTOPHTHORA PROTEIN1 and P. syringae pv phaseolicola (Supplemental Fig. S2). PBL13 possesses a 15-amino acid motif, K(P/T)RRE(V/T)K(E/D)TSLQNFD, at its C terminus that is repeated five times in tandem (Fig. 1B). BLAST analysis revealed that PBL13’s repeat motif has no homology to additional proteins of known function. Utilizing the DISOPRED Prediction Server, this repeat motif is predicted to be disordered in contrast to PBL13’s kinase domain (Fig. 1C; Ward et al., 2004; Buchan et al., 2013). The multiple metastable conformations that are adopted by disordered proteins are hypothesized to facilitate target recognition with high specificity and low affinity (Marín et al., 2013).
Figure 1.
The PBL13 RLCK possesses unique domain architecture and is a negative regulator of resistance to Pto DC3000. A, Phylogeny of RLCK subfamily VII kinase domains. RLCK VII kinase domains were analyzed using Phylogeny.fr and rooted with Staphylococcus spp. APH(3′)-IIIa (P0A3Y6). PBL13 (AT5G35580; green triangle) is most closely related to RIPK (AT2G05940; red triangle). B, Domain architecture of PBL13 drawn to scale. PBL13 possesses all canonical features of Ser/Thr kinases. PBL13 also possesses a unique 15-amino acid (AA) repeat located after the kinase domain that is repeated five times. C, Disorder prediction of PBL13 using the DISOPRED server, with the horizontal axis corresponding to the length of PBL13 in B. D, Bacterial titers in Col-0 and pbl13 T-DNA insertion lines 4 d postinoculation with Pto DC3000. Four-week-old Col-0, pbl13-1 truncation, and pbl13-2 KO plants were syringe infiltrated with 1 × 105 colony-forming units (CFUs) mL−1 Pto DC3000. Statistical differences were detected by Fisher’s lsd (α = 0.05). Error bars indicate se (n = 5). Results are representative of three independent experiments. E, Complementation of the pbl13-2 KO with 35S::gPBL13-3xFLAG. Col-0 and homozygous T3 lines 7-1 and 8-3 were inoculated as described above. Statistical differences were detected by Fisher’s lsd (α = 0.05). Error bars indicate se (n = 6). Results are representative of three independent experiments.
BLAST analyses to identify PBL13 orthologs in related Brassicas and other plants revealed that the emergence of PBL13’s C-terminal domain is relatively recent. No homolog possessing this repeat motif is present in tomato or white mustard (Sinapis alba), whereas a PBL13 ortholog exists in Arabidopsis lyrata with only one copy of the motif. To determine the relationship of PBL13 to other RLCK VIIs, we performed phylogenetic analyses using the kinase domains of subfamily VII rooted to Staphylococcus aureus 3’5’-aminoglycoside phosphotransferase [APH(3’)-IIIa]. PBL13 was found to be most closely related to RIPK (Fig. 1A). The kinase domains of PBL13 and RIPK share 85% identity, whereas the next closest related RLCK VII, PBL12, shares 70% identity (Fig. 1A).
pbl13 T-DNA Mutants Are More Resistant to Pto DC3000
To investigate whether PBL13 has a role in plant immune responses, available T-DNA insertion lines for PBL13 were obtained and screened for altered disease phenotypes upon syringe infiltration with Pto DC3000. After subsequent analyses of bacterial titers, both T-DNA insertion lines pbl13-1 and pbl13-2 exhibited at least 10-fold lower bacterial titers compared with wild-type Columbia-0 (Col-0) at 4 d postinoculation (Fig. 1D). pbl13-1 is a mutant expressing a transcript that is truncated shortly after the kinase-encoding region, whereas pbl13-2 is a true KO (Supplemental Fig. S3). The expression of truncated PBL13 in pbl13-1 is significantly higher than in Col-0, but interrogation of the T-DNA border sequence indicated that the insertion results in a stop codon shortly after the insertion (Supplemental Fig. S3C). To confirm that the pbl13 disease phenotype is caused by the associated T-DNA insertion, pbl13-2 KO was complemented with genomic PBL13-3xFLAG under control of the 35S promoter (35S::gPBL13-3xFLAG) construct. Two independent homozygous T3 lines were recovered, 7-1 and 8-3, in which PBL13-3xFLAG could be detected by western blotting (Supplemental Fig. S4). These lines were subsequently inoculated by syringe infiltration with Pto DC3000. PBL13-3xFLAG was able to complement pbl13-2 and restored wild-type bacterial growth (Fig. 1E).
PBL13 Is an Active Kinase with Activity That Is Required for Defense Suppression
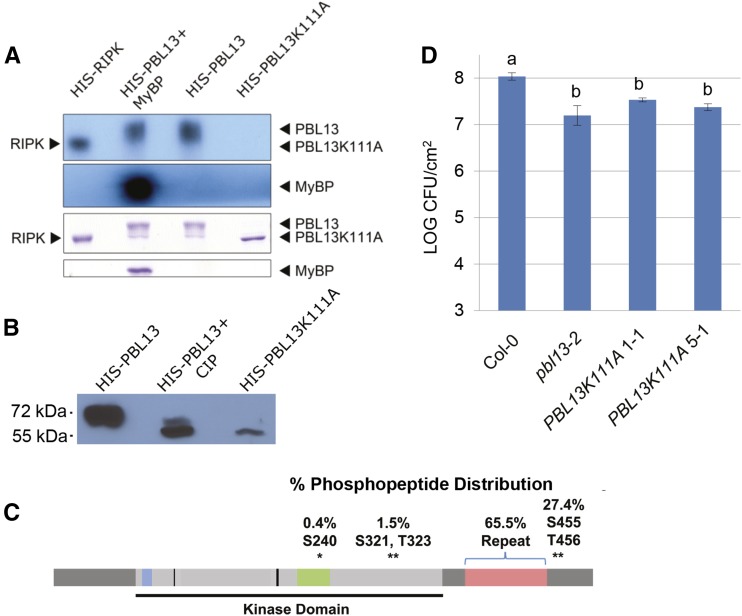
There are 11 conserved domains/residues commonly found in active protein kinases (Hanks et al., 1988). PBL13 has all of the major features of an active Ser/Thr protein kinase. For example, PBL13 possesses the Gly flap and associated invariant Lys as well as the conserved Asp residue in its active site (Fig. 1B; Hanks et al., 1988). To confirm PBL13 kinase activity, recombinant His6-PBL13 was purified from Escherichia coli and subjected to in vitro kinase activity assays. A kinase-dead variant of PBL13, PBL13K111A, was also purified. PBL13K111A possesses a mutation to Ala in the invariant Lys residue (K111) that is critical for orienting ATP for catalysis (Roskoski, 2012). Mutation of this Lys residue generally abolishes kinase activity (Hanks et al., 1988). Wild-type His6-PBL13 is capable of autophosphorylation and also, transphosphorylation of the common kinase substrate myelin basic protein (MyBP), whereas PBL13K111A does not exhibit detectable kinase activity (Fig. 2A). We also used the known RLCK RIPK, which was previously shown to be an active protein kinase, in our assays as a positive control (Fig. 2A; Liu et al., 2011).
Figure 2.
PBL13 is an active kinase, and kinase activity is required for negative regulation of defense responses. A, Recombinant PBL13 is able to autophosphorylate and phosphorylate MyBP; 3 μg of recombinant His6-PBL13, His6-PBL13 and MyBP, and kinase-dead His6-PBL13K111A were incubated with γ-32P-ATP, separated by SDS-PAGE, and then visualized by autoradiography. His6-RIPK, an RLCK with known kinase activity, is included as a positive control. B, Recombinant His6-PBL13K111A, wild-type His6-PBL13, and wild-type His6-PBL13 incubated with calf alkaline phosphatase (CIP) were separated by SDS-PAGE and detected using anti-His immunoblotting. The kinase-inactive His6-PBL13K111A runs at the predicted Mr of 56, whereas wild-type His-6-PBL13 runs at 70. Incubation with CIP shifted His6-PBL13 back to its predicted Mr. C, Recombinant His6-PBL13 was subjected to an in vitro kinase assay with unlabeled ATP, and phosphorylated spectra were identified and mapped by MS. In total, 252 spectra were identified as phosphorylated. The percentages of phosphopeptide spectra associated with phosphorylated residues confidently identified by MS are depicted above PBL13. The bracket denotes residues found within PBL13’s C-terminal repeat region. *, Specific residues. D, The kinase-inactive mutant His-6-PBL13K111A cannot complement the pbl13-2 KO. Four-week-old Col-0, pbl13-2, and homozygous T3 pbl13-2 lines expressing 35S::gPBL13K111A-3XFLAG were syringe infiltrated with colony-forming units (CFUs) mL−1 Pto DC3000, and bacterial titers were analyzed 4 d postinoculation. Statistical differences were detected with Fisher’s lsd (α = 0.05). Error bars indicate se (n = 6). Results are representative of three independent experiments.
Recombinant His6-PBL13 protein has a predicted Mr of 56. However, when recombinant His6-PBL13 was separated by SDS-PAGE, it ran at a significantly larger Mr (approximately 70; Fig. 2, A and B). In contrast, kinase-dead His6-PBL13K111A ran at its predicted Mr of 56 (Fig. 2, A and B). Phosphorylated proteins are known to exhibit mobility shifts during SDS-PAGE analyses; BIK1 and PBL1 are examples of this phenomenon (Lu et al., 2010; Zhang et al., 2010). Because PBL13 is an active kinase and capable of autophosphorylation, we investigated whether autophosphorylation was responsible for the mobility shift. Recombinant His6-PBL13 was incubated with calf intestinal phosphatase and compared with untreated His6-PBL13 and His6-PBL13K111A. Subsequent western-blot analysis with anti-His revealed that phosphatase-treated His6-PBL13 migrates in a similar fashion to His6-PBL13K111A (Fig. 2B). These data indicate that recombinant His6-PBL13 phosphorylation is required for its higher Mr (Fig. 2B). Interestingly, dephosphorylated His6-PBL13 accumulates to lower levels by western-blot analysis, and His6-PBL13K111A protein production is not as robust as the wild type in E. coli (Fig. 2B). Possibly, unphosphorylated PBL13 is compromised in stability.
Having established that mutating Lys-111 of PBL13 to Ala abolishes kinase activity, we proceeded to test if genomic PBL13K111A could complement pbl13-2. Two independent homozygous T3 lines were generated from pbl13-2-expressing genomic PBL13K111A (35S::gPBL13K111A-3xFLAG) 1-1 and 5-1. One line expressing PBL13K111A (line 5-1) with similar stability as 35S::gPBL13-3xFLAG and a second line expressing PBL13K111A with detectable protein expression (line 1-1) were identified (Supplemental Fig. S4A). These lines were subjected to syringe infiltration with Pto DC3000, and pathogen proliferation was quantified by measuring bacterial titers 4 d postinoculation. Both lines retained enhanced disease resistance and exhibited levels of bacterial proliferation similar to pbl13-2 (Fig. 2D). These data show that kinase activity of PBL13 is necessary for its regulation of plant immunity.
PBL13 Autophosphorylation Primarily Localizes to Its C-Terminal Repeat Motif
PBL13 is heavily autophosphorylated in vitro. Previous studies focused on BIK1, OsRLCK176, and OsRLCK185 indicate that phosphorylation of a cluster of Ser and Thr residues residing within the kinase activation loop are necessary for RLCK activity (Lu et al., 2010; Zhang et al., 2010; Yamaguchi et al., 2013; Ao et al., 2014). These phosphorylated residues are highly conserved among RLCK VII members (Zhang et al., 2010; Yamaguchi et al., 2013; Ao et al., 2014). Mass spectrometry (MS) was used to ascertain which residues were phosphorylated in recombinant PBL13. His6-PBL13 and His6-PBL13K111A were purified from E. coli and subjected to tryptic in-gel digestion. The resulting samples were enriched for phosphopeptides and analyzed by MS. In total, 252 phosphopeptide spectra were identified by MS/MS from the His6-PBL13 sample, whereas none were detected from the His6-PBL13K111A sample. In flow-through samples depleted of phosphopeptides, 208 MS/MS spectra were detected from the His6-PBL13 sample (23% coverage), whereas 589 spectra were detected from the His6-PBL13K111A sample (45% coverage). Interestingly, 65% of the phosphopeptide spectra localized to PBL13’s unique repeat motif, whereas only 1 spectrum mapped to Ser-240 within the activation loop (Fig. 2C). Autophosphorylation within the activation loop has also been previously reported for other RLCKs (Zhang et al., 2010; Yamaguchi et al., 2013; Ao et al., 2014). Peptides with multiple phosphorylated residues may not efficiently elute from TiO2 beads, and it is likely that additional regions in PBL13’s kinase domain are also phosphorylated. The remaining phosphorylated spectra (27.4%) corresponded to either singly or doubly phosphorylated Ser-455 and Thr-456 (Fig. 2C; Table I). Ser-455 and Thr-456 are immediately C-terminal to the repeat motif and are not conserved across other RLCKs. To obtain confident phosphosite localizations, phosphopeptide spectra were manually validated and run through the LuciPHOr algorithm to further obtain the most confident localization within PBL13 (Table I; Fermin et al., 2013). Although we can confidently localize phosphosites within a given spectrum, in some cases, the nature of PBL13’s repeat motif prevents definitive assignment of phosphorylation to a particular residue within the repeat. However, given the overrepresentation of spectra mapping to the repeat motif, heavy phosphorylation of this motif likely contributes to the mobility shift seen in recombinant PBL13.
Table I. PBL13 phosphopeptides identified by LC-MS/MS analysis.
Recombinant His6-PBL13 and His6-PBL13K111A were subjected to in vitro kinase assays followed by trypsin digestion and TiO2 phosphopeptide enrichment before LC-MS/MS analyses. Phosphopeptide spectra were analyzed using the LuciPHOr algorithm for perform phosphosite localization. Reported phosphopeptides required a LuciPHOr global false localization rate score of less than 0.05 and were manually validated. No phosphorylated peptides were identified from His6-PBL13K111A samples; s, t, and y denote phosphorylated Ser, Thr, and Tyr, respectively, whereas m and e denote oxidized Met and dehydrated Glu, respectively.
| Residue | Peptide Sequence |
|---|---|
| S240 | (K)DGPQGDDTHVsTR(V) |
| S321 | (R)LEDQYsEtGAR(K) |
| T323 | (R)LEDQYSEtGARK(A) |
| T383 and S384 or T443 and S444 | (R)REVKEtsLQNFDKPR(R/N) |
| T383 or 428 or T443 | (R)REVKEtSLQNFDK(P/T) |
| T383 or T443 | (R)EVKEtSLQNFDKPR(R/N) |
| S384 or S429 or S444 | (R)EVKETsLQNFDK(P/T) |
| S384 or S444 | (R)eVKETsLQNFDKPR(R/N) |
| T395, T398 | (R)REtKVtSLQNFDK(T) |
| T395 | (R)REtKVTSLQNFDK(T) |
| T398 | (R)RETKVtSLQNFDKTR(R) |
| T406 | (K)VTSLQNFDKtRR(E) |
| T413 | (R)EVKDtSLQNFDK(T) |
| T421 | (R)EVKDTSLQNFDKtRR(E) |
| S455 AND T456 | (R)NVstTDNHQKFR(S) |
| S455 | (R)NVsTTDNHQK(F) |
| Y481 | (R)NGyNSPmRNEAGGERY- |
PBL13 Localizes to Microsomes and Does Not Relocalize upon flagellin22 Treatment
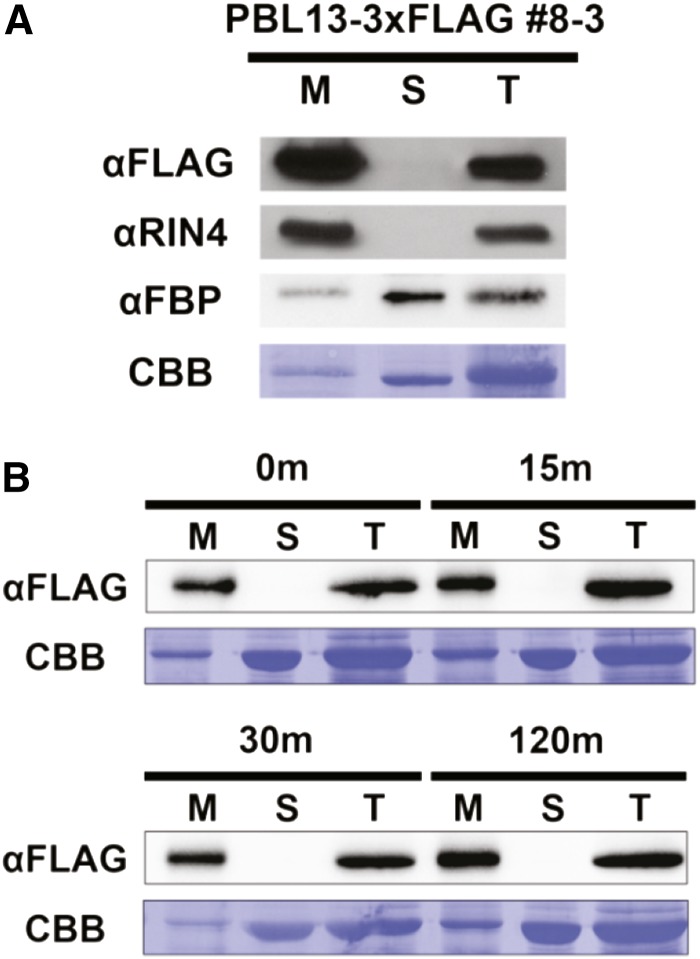
A number of RLCK VII members, including BIK1, RIPK, CAST AWAY, and PBS1, localize to the plasma membrane (Veronese et al., 2006; Burr et al., 2011; Liu et al., 2011; Takemoto et al., 2012). These RLCKs are anchored to the plasma membrane by N-terminal myristoylation or palmitoylation. Nearly all PBS1-like RLCK VIIs have been reported to have putative or validated myristoylation/palmitoylation motifs (Zhang et al., 2010). PBL13, like RIPK, is predicted to be palmitoylated at its N terminus. PBL13’s subcellular localization was initially investigated by microscopy through GFP fusions. Unfortunately, these fusions were unstable in planta, and free GFP was consistently detected. Thus, biochemical cell fractionation using the 35S::PBL13-3xFLAG line 8-3 was used to investigate general subcellular localization. Comparison of PBL13-3xFLAG localization using total leaf extract, soluble, and microsomal membrane fractions was performed by western-blot analysis. Antibodies recognizing CYTOSOLIC FRU-1,6-BISPHOSPHATASE and RIN4 were used as soluble and membrane markers, respectively (Fig. 3A). PBL13-3xFLAG localizes to membranes, and PBL13 was solely detected in microsomal fractions, supporting evidence that PBL13 is potentially palmitoylated and targeted to the plasma membrane (Fig. 3A). Because palmitoylation is a reversible modification, PBL13 relocalization during immune signaling was investigated. Leaves of 4-week-old 35S::PBL13-3xFLAG 8-3 lines were infiltrated with 1 μm of the 22 amino acid immunogenic flagellin epitope (flg22) and sampled at 0-, 15-, 30-, and 120-min time points. No change in PBL13 localization after flg22 treatment was observed, indicating that PBL13 does not relocalize to the cytosol upon flagellin perception and activation of MTI (Fig. 3B).
Figure 3.
PBL13 is membrane associated. A, Total, soluble, and microsomal cellular fractions were isolated from the 35S::gPBL13-3xFLAG complemented line 8-3. Anti-FLAG immunoblotting identifies PBL13 as associated with the microsomal membrane fraction. αRIN4 (AT3G25070) was used as a membrane marker, and αcytosolic fructose-1,6-bisphosphatase (FBP; AT1G43670) was used as a soluble marker. Coomassie Brilliant Blue (CBB) stain is provided to indicate protein loading. B, Total, soluble, and microsomal cellular fractions were isolated from the 35S::gPBL13-3xFLAG complemented line 8-3 after treatment with flg22 treatment. Five-week-old plants were syringe infiltrated with 1 µm flg22, and leaves were sampled at 0-, 15-, 30-, and 120-min time points and subjected to cellular fractionation. Three micrograms of protein from each fraction was analyzed by SDS-PAGE and western blotting. M, Membrane; S, soluble; T, total.
The pbl13-2 KO Exhibits More Rapid MAPK Activation and a More Robust ROS Burst
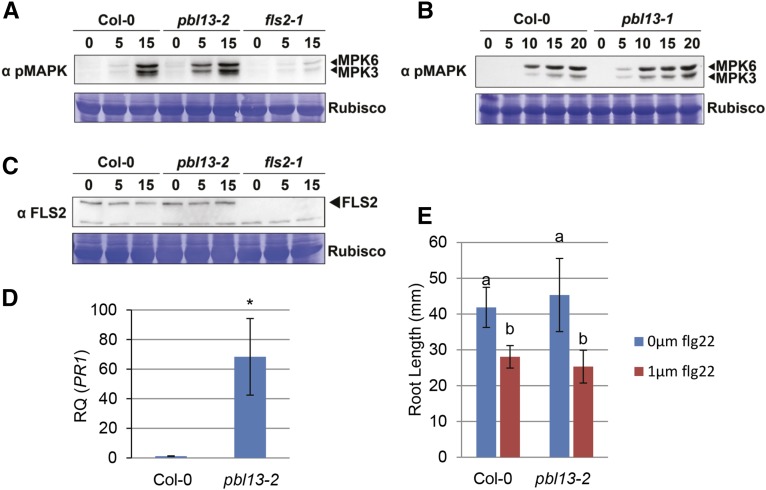
We hypothesized that basal defense gene expression may be enhanced in pbl13-2, because this KO line exhibits enhanced resistance to virulent Pto DC3000. It was previously found that bik1 KO plants, although defective in MTI signaling, were more resistant to virulent Pto DC3000 because of elevated basal levels of the defense hormone salicylic acid (Veronese et al., 2006). The bik1 KO exhibits dwarf stature, a phenotype associated with autoimmunity and high levels of salicylic acid (Zhang et al., 2010). In contrast, the pbl13-2 KO does not exhibit developmental defects; thus, changes in basal immune signaling may be subtle (Supplemental Fig. S3E). Quantitative reverse transcription PCR was used to detect differences in basal expression of the defense marker gene PATHOGENESIS-RELATED GENE1 (PR1). Four- to five-week-old Arabidopsis leaves were sampled for quantitative PCR (qPCR) analyses. The pbl13-2 KO exhibited significantly higher levels of PR1 expression compared with the Col-0 control (Fig. 4D).
Figure 4.
The pbl13-2 KO has accelerated MAPK3/6 activation and elevated basal PR1 expression. A and B, Activation of MAPK3/6 is accelerated in pbl13 mutants relative to Col-0 A, MAPK3/6 activation in pbl13-2. B, MAPK3/6 activation in pbl13-1. Four-week-old plants were sprayed with 1 μm flg22, and tissues were processed at indicated time points. Phosphorylated MAPK3 and MAPK6 were detected with α-p44/42 MAPK antibody. C, No difference in FLS2 levels is detected between 4-week-old Col-0 and pbl13-2 after treatment with 1 μm flg22. D, Real-time qPCR analyses of PR1 in Col-0 and pbl13-2. pbl13-2 exhibits higher levels of PR1 expression relative to the wild type (Col-0). Statistical differences were detected with a Student’s t test (α = 0.05; n = 3 biological replicates). Error bars indicate sd; *, P < 0.05. Expression was normalized to ELONGATION FACTOR 1α. RQ, Relative quantification. E, pbl13-2 does not exhibit enhanced flagellin-induced seedling root length inhibition. Seedlings were grown on one-half-strength concentration Murashige and Skoog medium in the presence and absence of 1 μm flg22. Root length was measured at 12 d. Statistical differences were detected by Fisher’s lsd (α = 0.01). Error bars indicate sd.
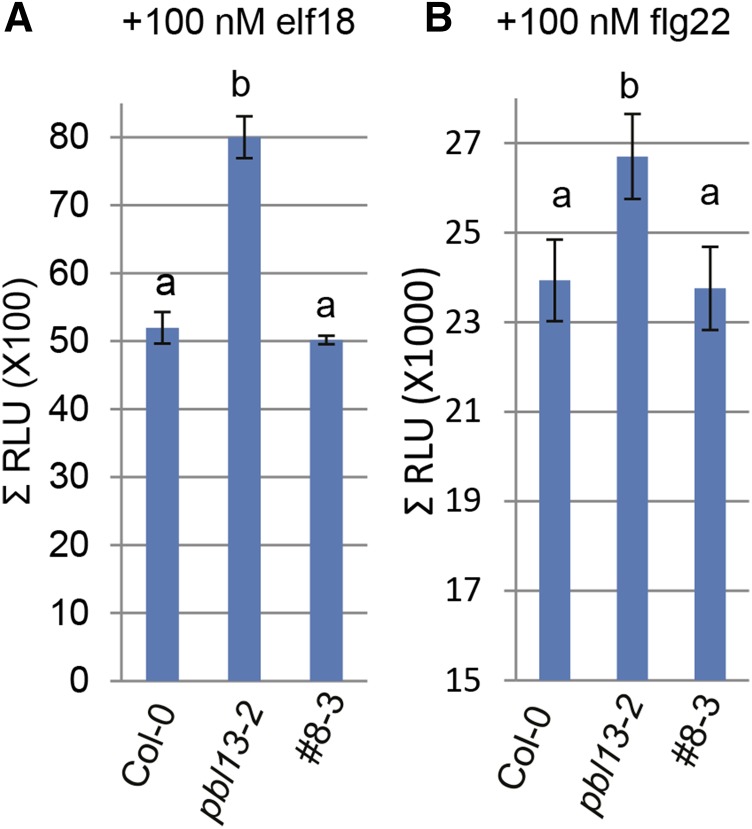
In addition to basal defense gene expression, we also analyzed the pbl13-2 KO for alterations in MAMP-induced MAPK activation, ROS production, FLS2 accumulation, and seedling root length inhibition: all hallmarks of MTI activation (Asai et al., 2002; Chinchilla et al., 2007). Treatment of both pbl13-1 truncation mutant and pbl13-2 KO with flg22, the immunogenic epitope of bacterial flagellin, resulted in accelerated activation of MAPK3 and MAPK6 at 5 min (Fig. 4, A and B). Additionally, pbl13-2 also exhibited a significantly enhanced ROS burst relative to wild-type Col-0 upon treatment with both flg22 and the 18-amino acid immunogenic epitope of bacterial elongation factor-tu (elf18; Fig. 5). The PBL13-3xFLAG complementation line 8-3 was rescued for this phenotype and exhibited wild-type levels of ROS production (Fig. 5). However, pbl13-2 did not exhibit significant differences in flg22-induced FLS2 accumulation or inhibition of seedling root growth (Fig. 4, C and E). Taken together, these data suggest that PBL13 functions to negatively regulate basal defense gene expression, MAMP-triggered MAPK activation, and the extracellular ROS burst.
Figure 5.
The pbl13-2 KO exhibits enhanced elf18- and flg22-triggered ROS production. elf18- (A) and flg22-triggered (B) production of ROS in pbl13-2 is enhanced relative to Col-0. Two- to 3-week-old plants were treated with 100 nm flg22, and extracellular ROS was quantified using a luminol-based assay. The graph depicts total relative light units (RLUs) detected over a 30-min period after treating with flg22 or elf18. Expression of PBL13-3xFLAG in pbl13-2 (lines 7-1 and 8-3) complements the mutant phenotype. Error bars indicate se (n ≥ 21). Statistical differences were detected by Fisher’s lsd (α = 0.05).
PBL13 Associates with RBOHD in Nicotiana benthamiana
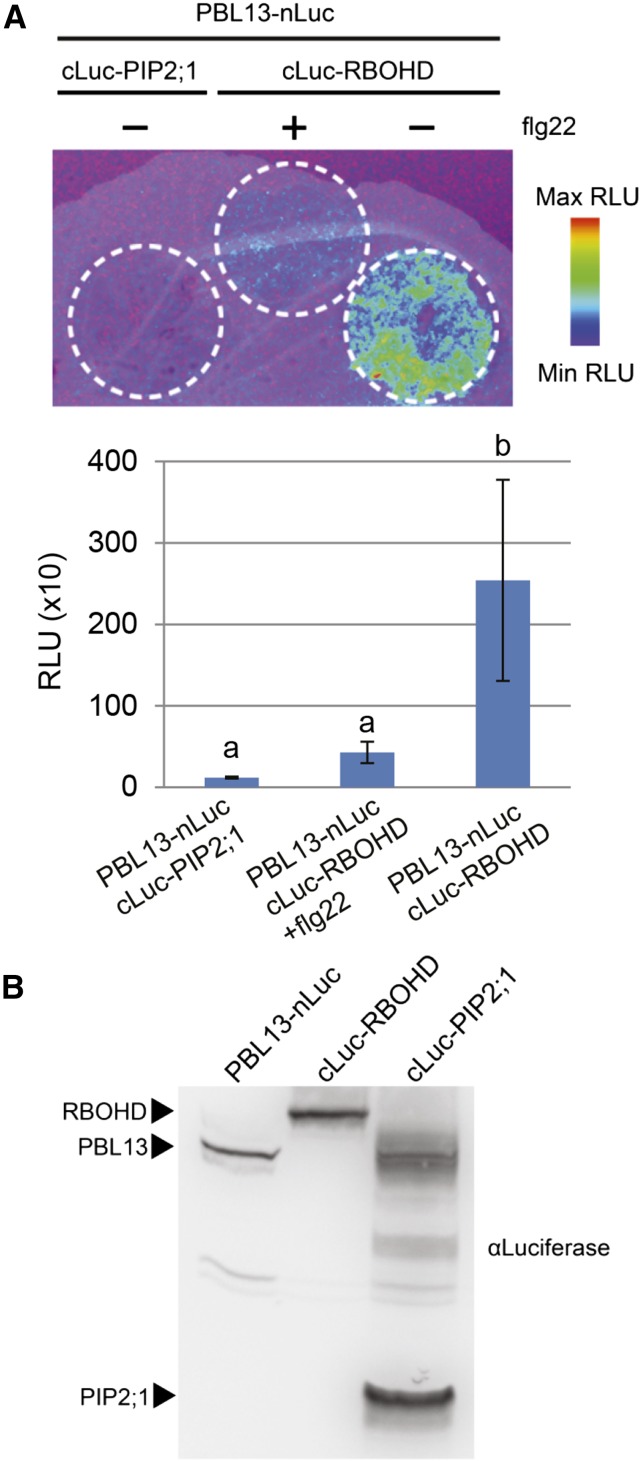
RLCKs regulating MTI have been found to associate with PRR complexes and positively regulate immune responses in Arabidopsis and rice (Yamaguchi et al., 2013; Ao et al., 2014; Kadota et al., 2014; Li et al., 2014; Shinya et al., 2014). In light of the enhanced early MTI responses observed in the pbl13-2 KO, we reasoned that PBL13 may associate with PRR complexes to negatively regulate immune responses. Previously, FLS2 and BIK1 had been shown to associate with Arabidopsis RBOHD through split-luciferase complementation imaging (Li et al., 2014). Using this system, we tested PBL13’s ability to associate with Arabidopsis FLS2, BIK1, BAK1, and RBOHD in N. benthamiana. No luminescent signal could be detected when leaf spots coexpressing PBL13-cLuc with FLS2-nLuc, BIK1-nLuc, or BAK1-nLuc were infiltrated with 1 mm luciferin (Supplemental Figs. S5 and S6). As a negative control, we used the integral plasma membrane aquaporin PLASMA MEMBRANE INTRINSIC PROTEIN2-1 (PIP;2-1). No luminescence was detected in samples coexpressing PIP2;1 fused to the c-terminus of luciferase (cLuc-PIP2;1) and PBL13 fused to the n-terminus of luciferase (PBL13-nLuc). We then proceeded to test PBL13’s ability to associate with RBOHD. Luminescence was detected in samples coexpressing PBL13-nLuc and cLuc-RBOHD, suggesting a specific interaction between PBL13 and RBOHD. We also found that supplementing the luciferin solution with 100 nm flg22 suppressed PBL13-RBOHD-dependent luminescence (Fig. 6). From these data, we conclude that PBL13 associates with RBOHD and that this complex is disrupted by flg22-induced immune signaling.
Figure 6.
PBL13 associates with RBOHD and dissociates upon flg22 treatment. A, PBL13-nLuc associates with cLuc-RBOHD but not cLuc-PIP2;1 in N. benthamiana. flg22 treatment results in dissociation of the PBL13-RBOHD complex. Constructs were transiently expressed in N. benthamiana, and luminescence was detected after treatment with luminol solution ±100 nm flg22. Upper, Luminescence visualized using a CCD camera. Lower, Luminescence was quantified using a luminometer. Error bars indicate sd (n = 3). Statistical differences were detected by Fisher’s lsd (α = 0.05). RLU, Relative light unit. B, Split-luciferase fusion proteins are detected after transient expression in N. benthamiana.
DISCUSSION
Protein kinases are important components of plant immunity, and they regulate plant responses to pathogens at multiple levels. Kinase-mediated immune regulation ranges from initial perception of MAMPs by transmembrane PRRs to mediating downstream immune signaling by RLCKs, calcium-dependent protein kinases, and MAPKs (Tena et al., 2011). The majority of characterized RLCK VII members function as positive regulators of MTI (Lu et al., 2010; Zhang et al., 2010; Yamaguchi et al., 2013; Ao et al., 2014; Shinya et al., 2014). In this study, we focused on the genetic and biochemical characterization of PBL13, an RLCK from subfamily VII possessing a unique C-terminal repeat motif. Interestingly, pbl13 T-DNA mutant lines exhibited enhanced resistance to Pto DC3000, although PBL13 transcription is induced in response to pathogen perception. Subsequent experiments showed that PBL13 functions as a negative regulator of MTI, and this is dependent on PBL13’s kinase activity. The induction, amplitude, and duration of plant immune responses are tightly controlled (Belkhadir et al., 2014). This is exemplified in attenuation of FLS2 signaling postflagellin perception because of ubiquitination by PLANT U-BOX12 (PUB12) and PUB13, two U-box E3 ubiquitin ligases (Lu et al., 2011). Induction of plant immune responses is an energetic process, and its activation negatively impacts plant growth and development (Lozano-Durán et al., 2013; Belkhadir et al., 2014). For example, treatment with flg22 strongly inhibits seedling growth. Therefore, we hypothesize that PBL13 acts to negatively regulate immune signaling in absence of pathogen challenge to prevent untimely activation of defenses. PBL13 may then also function to dampen immune signaling downstream of pathogen perception, allowing for fine control of defense responses.
The pbl13-2 KO exhibits a robust disease phenotype and elevated basal PR1 expression. We were also able to detect accelerated MAPK activation and an enhanced extracellular ROS burst in pbl13-2. Furthermore, complementation of pbl13-2 with PBL13-3xFLAG was able to restore a wild-type ROS burst. In plant immunity, the extracellular ROS burst is primarily controlled by the NADPH oxidase RBOHD, which has been shown to be present in complex with PRRs (Zhang et al., 2007; Kadota et al., 2014; Li et al., 2014). Recent studies have shown that the RLCK BIK1 positively regulates this process by dynamically associating with and phosphorylating RBOHD upon flagellin perception (Kadota et al., 2014; Li et al., 2014). Phosphorylation of distinct residues within a protein can have antagonistic effects, promoting either activation or deactivation. For example, phosphorylation of the penultimate Thr of the plasma membrane AUTOINHIBITED H+-ATPASE2 results in activation, whereas phosphorylation of Ser-941 inhibits activity (Elmore and Coaker, 2011). Additionally, phosphorylation of RIN4 Ser-141 promotes MTI responses, whereas phosphorylation of Thr-166 has an antagonistic effect (Chung et al., 2014). Thus, it is possible that multiple competing RLCKs, including PBL13, are involved in differential regulation of RBOHD through phosphorylation of distinct residues. In light of our finding that PBL13 associates with RBOHD in N. benthamiana, we hypothesize that, in the absence of pathogen stimuli, PBL13 may associate with RBOHD and inhibit ROS production. Treatment with flg22 disrupts this complex, potentially enabling BIK1, PBL1, and calcium-dependent protein kinases to positively regulate RBOHD activity (Kobayashi et al., 2007; Dubiella et al., 2013; Gao et al., 2013; Kadota et al., 2014; Li et al., 2014). The requirement of these sequential events for RBOHD activation could ensure elegant control of the timing and amplitude of the extracellular ROS burst.
Phylogenetic analyses of the RLCK VII subfamily revealed that PBL13 is most related to RIPK, a kinase involved in RPM1-mediated resistance (Chung et al., 2011; Liu et al., 2011). We previously reported that the ripk KO also exhibits enhanced disease resistance when challenged with virulent Pto DC3000 (Liu et al., 2011). However, ripk exhibits enhanced disease resistance primarily after surface inoculation, indicating that these lines are compromised in initial pathogen entry (Liu et al., 2011). There was no significant difference in Pto DC3000 titers between ripk and wild-type Arabidopsis after syringe infiltration (Liu et al., 2011). In contrast, the pbl13-2 KO exhibits a strong enhanced disease resistance phenotype upon syringe infiltration with Pto DC3000, with bacterial titers reduced at least 10-fold at 4 d postinoculation. These data suggest that PBL13 and RIPK may primarily have distinct modes of action. Although PBL13 is closely related to RIPK throughout its kinase domain, PBL13 also possesses a unique C-terminal repeat region, where a stretch of 15 amino acids is repeated five times in tandem. Furthermore, the fidelity of the PBL13 repeat region is extremely high, with only three polymorphic amino acids within an individual repeat. From an evolutionary standpoint, the emergence of this repeat is relatively recent, because this motif is present in Arabidopsis and A. lyrata but absent in tomato and white mustard.
Our experiments showed that PBL13 is an active kinase. Recombinant PBL13 could autophosphorylate as well as transphosphorylate MyBP. Extensive PBL13-mediated autophosphorylation was detected by a significant, reversible mobility shift of PBL13 in vitro. Interestingly, the mobility shift of PBL13 is only detectable in vitro, suggesting that PBL13’s kinase activity and phosphorylation of this motif are carefully controlled in planta. Identification of phosphorylated residues within PBL13 using MS verified that PBL13 is a Ser/Thr protein kinase, because all but one phosphorylated residue were found to be a Ser or Thr residue. Phosphorylation mapping of PBL13 also revealed that Ser-240, which is present within the activation loop and conserved among the RLCK VIIs, is autophosphorylated (Zhang et al., 2010). This residue corresponds to Ser-236 within BIK1, one of a triplet of Ser/Thr residues residing in BIK1’s activation loop that is phosphorylated upon PAMP perception (Zhang et al., 2010). Lines expressing BIK1S236A exhibit reduced phosphorylation after flagellin treatment and inhibited PAMP-triggered transcriptional reprogramming (Zhang et al., 2010). In addition to Ser-240, we detected an overabundance of phosphorylated peptides mapping to PBL13’s C-terminal tandem repeat region, showing that this motif is heavily autophosphorylated. The predicted disordered nature of PBL13’s repeat region may account for its hyperphosphorylation, because sites of posttranslational modifications have been found to correlate with intrinsic disordered regions of proteins (Gao and Xu, 2012). Furthermore, posttranslational modification of intrinsically disordered protein regions can regulate target binding specificity (Dyson and Wright, 2005).
CONCLUSION
We find that PBL13 functions as a negative regulator of plant immunity, and this is dependent on its kinase activity. pbl13-2 KOs exhibit enhanced basal levels of PR1 expression and produce increased flagellin-induced ROS production. PBL13 associates with RBOHD in planta, and this association is disrupted by treatment with bacterial flagellin. Furthermore, PBL13 is autophosphorylated on its C terminus as well as in its activation domain in vitro. The identification of PBL13 substrates as well as the elucidation of the phosphorylation dynamics of PBL13 targets and the kinase itself will significantly enhance our understanding of how this RLCK modulates plant innate immune responses.
MATERIALS AND METHODS
Microarray Analyses
To investigate RLCK VII expression patterns, raw AtGenExpress Biotic Stress data files were normalized using the Robust Multichip Average method with the biocLite R package (Bioconductor; Bolstad et al., 2003). Replicates of the absolute signal intensities were then averaged by geometric mean. Ratios of treatment to control values were log2 transformed, and data were visualized using MultiExperiment Viewer 4.0 (TM4 Microarray Software Suite). PBL13 transcript expression data were extracted from the Arabidopsis Electronic Fluorescent Pictograph Browser (Winter et al., 2007).
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) T-DNA insertion lines pbl13-1 (GABI_586B09) and pbl13-2 (SALK_203577) were obtained from the Nottingham Arabidopsis Stock Centre and the Arabidopsis Biological Resource Center, respectively. PCR using gene- and T-DNA-specific primers (Supplemental Table S1) was performed to identify individuals homozygous for T-DNA insertions. Gene-specific primers were then used for semiquantitative reverse transcription PCR to determine KO or truncation status. For disease and MTI marker assays, Arabidopsis (Arabidopsis thaliana) seeds were stratified for 2 d at 4°C, sown on soil, and then grown in controlled environmental chambers set to 23°C and 70% relative humidity with a 10-h-light/14-h-dark photoperiod (100 μmol m−2 s−1).
Transgenic Lines
To generate 35S::PBL13-3xFLAG and 35S::PBL13K111A-3xFLAG constructs, a modified pEARLEY103 vector was used. In brief, the pEARLEY103 GATEWAY cassette GFP fragment was removed using XhoI/SpeI and replaced with the XhoI/SpeI GATEWAY 3xFLAG fragment cut from pCR2.1/des/3xFLAG (Li et al., 2013), producing pEARLEY/des/3xFLAG. Genomic PBL13 was then PCR amplified from Col-0 genomic DNA without a stop codon, TOPO cloned into pENTR/D-TOPO (Invitrogen), and recombined into pEARLEY/des/3xFLAG by LR reaction (Invitrogen). The resulting 35S::PBL13-3xFLAG construct was then mutated by PCR-based site-directed mutagenesis to produce 35S::PBL13K111A-3xFLAG. Binary vectors were then transformed into Agrobacterium tumefaciens strain GV3101, and complementation lines were generated by floral dip (Clough and Bent, 1998). Transformants were selected for one-half-strength Murashige and Skoog medium supplemented with 20 μg mL−1 BASTA (Sigma Aldrich). Expression of PBL13-3xFLAG was then confirmed by western blotting in T3 homozygous lines. Anti-FLAG horseradish peroxidase (HRP) Conjugate (Sigma) was used at a 1:2,000 concentration in conjunction with SuperSignal West Pico Chemiluminescent Substrate (Pierce) for detection.
Kinase Motif and Protein Disorder Prediction
Conserved protein Ser/Thr kinase motifs were found by searching the PBL13 amino acid sequence against the PROSITE Pattern and Motif Database (Sigrist et al., 2002). Intrinsic disorder was predicted using the DISOPRED component of the PSIPRED Prediction Server, with the false discovery rate set to 5% (Ward et al., 2004; Buchan et al., 2013).
Disease Assays
Pto DC3000 was grown at 28°C for 2 d on nutrient yeast-glycerol agar (100 μg mL−1 rifampicin and 25 μg mL−1 kanamycin). Four- to five-week-old Arabidopsis leaves were syringe infiltrated with 1 × 105 colony-forming units mL−1 bacteria as previously described (Liu et al., 2011). Analyses of bacterial titers were performed 4 d postinoculation. Statistical differences were detected with Fisher’s lsd (α = 0.05). All experiments were repeated at least three times with a minimum of five biological replicates.
Phylogenetic Analysis
The amino acid sequences for the kinase domains of RLCK subfamily VII and Staphylococcus spp. aminoglycoside phosphotransferase APH(3′)-IIIa (Uniprot identification code P0A3Y6) were obtained from Lehti-Shiu et al., 2009. Sequences were analyzed using the Phylogeny.fr web tool (Dereeper et al., 2008). Alignments were made using MUSCLE with default settings, and a phylogenetic tree was built using the maximum likelihood-based PhyML software (Edgar, 2004; Guindon et al., 2010). The resulting tree was then visualized in FigTree (http://tree.bio.ed.ac.uk/software/figtree) and rooted to APH(3′)-IIIa.
Real-Time qPCR
Four- to five-week-old Col-0 and pbl13-2 lines were used for qPCR analysis. One leaf per plant was used for total RNA extraction with three biological replications; 1 μg of total RNA was used for complementary DNA synthesis using M-MLV Reverse Transcriptase (Promega) coupled with oligo(dT) primers. The CFX96 Touch RT-PCR Detection System (BioRad) was used for quantification. Primer pairs for PR1 (AT2G14610) are provided in Supplemental Table S1. A primer pair that targets the Arabidopsis ELONGATION FACTOR 1α (AT5G60390) was used as an endogenous control (Supplemental Table S1). Statistical differences were detected by Student’s t test (α = 0.05). Results are representative of at least two independent experiments.
Flagellin-Induced Seedling Root Length and MAMP-Triggered ROS Burst Assay
For flagellin-induced seedling root inhibition, Arabidopsis seedlings were grown on one-half-strength concentration Murashige and Skoog medium in the presence and absence of 1 μm of flg22. Root length was then measured at 12 d. Statistical differences were detected with Fisher’s lsd (α = 0.01) with a minimum n = 30. Experiments were repeated twice with similar results.
To assay MAMP-induced ROS burst, the third, fourth, and fifth true leaves of 2- to 3-week-old Arabidopsis plants were sampled with a 1 cork borer (4.76 mm; Carolina) and floated overnight on sterile water in a white 96-well plate (Corning Costar). Immediately before measuring ROS burst, water was removed, and samples were allowed to dry for 5 min. ROS burst was triggered by adding a solution containing 100 nm elf18 or flg22 (85% purity; Genscript), 17 mg mL−1 luminol (Sigma), and 10 mg mL−1 horseradish peroxidase (Sigma). Luminescence was measured using the TriStar LB 941 Multimode Microplate Reader (Berthold). Statistical differences were detected with Fisher’s lsd (α = 0.05). All experiments were repeated at least three times with similar results, with at least 21 samples per replication.
Activated MAPK Assay and FLS2 Accumulation
Four-week-old Arabidopsis plants were sprayed with 10 μm flg22 in water with 0.025% (v/v) Silwett l-77. Leaf samples were then taken at 0-, 5-, and 15-min time points for pbl13-2, whereas additional samples at 10- and 20-min time points were taken for pbl13-1. Samples were ground in liquid nitrogen and resuspended in 50 mm HEPES (pH 7.5), 50 mm NaCl, 10 mm EDTA, 0.2% Triton X-100, 1× Protease Inhibitor Cocktail (Sigma), and 1× Halt Phosphatase Inhibitor Cocktail (Thermo). After centrifugation, Laemmli sample buffer was added to the supernatant to a concentration of 1× and then boiled for 5 min. Activated MAPK3 and MAPK6 were then detected by western blotting using anti-p44/42 MAPK antibody (Cell Signaling Technology). FLS2 was detected by western blotting using anti-FLS2 (Heese et al., 2007) as primary antibody overnight and goat anti-rabbit-HRP (Bio-Rad) as secondary antibody.
Expression and Purification of Recombinant Proteins
To produce HIS6-PBL13 and HIS6-RIPK, PBL13 and RIPK were cloned into pET-28a(+) (Novagen) as BamHI/XhoI and NdeI/BamHI fragments, respectively. HIS6-PBL13K111A was generated by PCR-based site-directed mutagenesis. Expression vectors were transformed into Escherichia coli BL21(DE3) for production of recombinant protein. Protein expression was induced by addition of 0.5 mm isopropylthio-β-galactoside to a 250-mL culture at optical density at 600 nm = 0.4 followed by incubation for 4 h at 28°C. Cells were centrifuged and resuspended in lysis buffer containing 20 mm Tris-HCl (pH 8), 500 mm NaCl, and 10 μg mL−1 lysozyme. Cleared lysate was then incubated with Ni-NTA Agarose (Qiagen) for 1 h at 4°C. Beads were then washed four times with 20 mm Tris-HCl (pH 8) and 500 mm NaCl (buffer A). Stepwise elution of protein was then performed with 80 and 250 mm imidazole in buffer A.
In Vitro Kinase Assay and Mobility Shift Analysis
Kinase reactions were carried out in 20 mm Tris-HCl (pH 7.5), 10 mm MgCl2, 1 mm CaCl2, 1 mm dithiothreitol, and 100 μm ATP; 3 μg of each recombinant protein was used per reaction. Radioactive kinase assays were carried out with the addition of 10 μCi of [32P]γATP (Perkin Elmer). Reactions were incubated for 30 min at 30°C and stopped with the addition of 7.5 μL of 5× Laemmli sample buffer. Radioactive samples were separated on 12% Precise Tris-HEPES SDS-PAGE Gels (Thermo Scientific) and visualized by autoradiography. For analysis of phosphorylation-related mobility shift, 1 μg of recombinant protein was treated with calf intestinal phosphatase (New England BioLabs) and then incubated at 37°C for 30 min. The reaction was stopped with addition of 3× Laemmli sample buffer and compared with untreated samples by western blot using anti-His primary antibody (Invitrogen) and anti-mouse-HRP secondary (BioRad). Blots were visualized after incubation with SuperSignal West Pico Chemiluminescent Substrate (Pierce).
MS
Cold kinase reactions were separated on 12% Precise Tris-HEPES SDS-PAGE Gels (Thermo) and then stained with Novex Colloidal Blue (Invitrogen). Bands were excised, and in-gel tryptic digestion was performed as described in Elmore et al., 2012. Selective enrichment for phosphopeptides was then performed on samples using the TiO2 Phosphopeptide Enrichment and Clean-Up Kit (Thermo Scientific) according to manufacturer’s specifications. Phosphopeptide-enriched fractions were submitted to the Genomics Center Proteomics Core at the University of California, Davis, for liquid chromatography (LC)-MS/MS analysis.
LC separation was performed on a Proxeon Easy-nLC II HPLC (Thermo Scientific) with a Proxeon Nanospray Source. The digested peptides were reconstituted in 2% acetonitrile and 0.1% trifluoroacetic acid, and 10 µL of each sample was loaded onto a 100-µm × 25-mm Magic C18 100-Å, 5-Unit Reverse-Phase Trap, where they were desalted online before being separated on a 75-µm × 150-mm Magic C18 200-Å, 3-Unit Reverse-Phase Column. Peptides were eluted using a gradient of 0.1% formic acid (buffer A) and 100% acetonitrile (buffer B) with a flow rate of 300 nL min−1. A 60-min gradient of 5% to 35% buffer B was run over 45 min, a 35% to 80% gradient of buffer B was run over 5 min, an 80% gradient of buffer B was run over 1 min, an 80% to 5% gradient of buffer B was run over 1 min; and finally, it was held at 5% buffer B for 8 min.
Mass spectra were collected on an Orbitrap Q Exactive Mass Spectrometer (Thermo Fisher Scientific) in data-dependent mode with one MS precursor scan followed by 15 MS/MS scans. A dynamic exclusion of 5 s was used. MS spectra were acquired with a resolution of 70,000 and a target of 1 × 106 ions or a maximum injection time of 20 ms. MS/MS spectra were acquired with a resolution of 17,500 and a target of 5 × 104 ions or a maximum injection time of 250 ms. Peptide fragmentation was performed using higher energy collision dissociation with a normalized collision energy value of 27. Unassigned charge states as well as +1 and ions greater than +5 were excluded from MS/MS fragmentation.
Phosphopeptide Identification and Phosphosite Localization
Tandem mass spectra were analyzed as previously described in Elmore et al., 2012. Spectra corresponding to phosphopeptides with a Peptide Prophet Score exceeding 95% were detected in Scaffold 4 (Proteome Software). Phosphosite localization was performed using the LuciPHOr algorithm (Fermin et al., 2013). Phosphopeptide localizations are reported only if the corresponding phosphopeptide was confirmed by manual validation and received a LuciPHOr global false localization score less than 0.05.
Cellular Fractionation
Two hundred milligrams of leaf tissue from 4- to 5-week-old Arabidopsis plants, either untreated or syringe infiltrated with 1 µm flg22 for time course experiments, was frozen in liquid nitrogen and ground to a fine powder. Soluble and microsomal fractions were then obtained using the Minute Plasma Membrane Isolation Kit (Invent Biotechnologies). Total protein extract was obtained by homogenizing leaf samples in 1× Laemmli sample buffer. Protein concentrations were measured using the 660-nm Protein Assay (Pierce), and 3 μg of protein for each fraction was analyzed by western blotting with anti-cytosolic Fru-1,6-bisphosphatase (1:5,000; Agrisera), anti-FLAG-HRP (1:2,000), and affinity-purified anti-RIN4 (1:3,000; Liu et al., 2009). Blots were visualized after incubation with SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Split-Luciferase Complementation Assay
To generate PBL13-nLuc, PBL13-cLuc, and cLuc-PIP2;1 fusion proteins, genomic PBL13 and PIP2;1 were recombined into pCAMBIA/des/cLuc and pCAMBIA/des/nLuc (Chen et al., 2008). The resulting constructs along with FLS2-nLuc, BIK1-nLuc, BAK1-nLuc, and cLuc-RBOHD were then transformed into A. tumefaciens strain GV3101. Western blotting using antiluciferase (Sigma) was first performed to validate expression of fusion proteins in Nicotiana benthamiana. Split-luciferase fusion proteins were then coexpressed in N. benthamiana. FLS2-nLuc, BIK1-nLuc, BAK1-nLuc, cLuc-RBOHD, and cLuc-PIP2;1 were agroinfiltrated 48 h before imaging, whereas PBL13-luciferase fusions were agroinfiltrated 24 h before imaging to maximize fusion protein expression levels. Coagroinfiltrated spots were treated with 1 mm luciferin or 1 mm luciferin supplemented with 100 nm flg22 and immediately imaged for 60 s with a Carestation 4000R ImagePro. Three leaf discs were then excised from each coagroinfiltrated spot, and luminescence was quantified using a TriStar LB 941 Multimode Microplate Reader (Berthold). Statistical differences were detected using Fisher’s lsd (α = 0.05).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RLCK VII microarray meta-analysis.
Supplemental Figure S2. PBL13 expression during biotic stress.
Supplemental Figure S3. pbl13 T-DNA line characterization.
Supplemental Figure S4. Transgenic pbl13 immunoblots,
Supplemental Figure S5. PBL13 and FLS2, BAK1, BIK1 luciferase complementation imaging.
Supplemental Figure S6. Luciferase-fusion protein immunoblot.
Supplemental Table S1. Primer sequences.
Supplementary Material
Acknowledgments
We thank Brian Staskawicz and Jian-Min Zhou for providing split-luciferase constructs; Antje Heese and Cyril Zipfel for the anti-FLS2 antibody; Jun Liu, Theresa Dao, Jesus Banderas, and Pranaya Venkatapuram for contributions to this project; Judy Callis and Rick Bostock for comments regarding this article; and the University of California, Davis, Genome Center Proteomics Core Facility for assistance and MS service.
Glossary
- Col-0
Columbia-0
- ETI
effector-triggered immunity
- KO
knockout
- LC
liquid chromatography
- MAMP
microbe-associated molecular pattern
- MS
mass spectrometry
- MTI
microbe-associated molecular pattern-triggered immunity
- PAMP
pathogen-associated molecular pattern
- PRR
pattern recognition receptor
- qPCR
quantitative PCR
- ROS
reactive oxygen species
- T-DNA
transfer DNA
- MAPK
mitogen-activated protein kinase
Footnotes
This work was supported by the National Institutes of Health (grant no. R01GM92772 to G.C.), The Netherlands Organisation for Scientific Research (Rubicon Grant to T.W.H.L.), and the National Science Foundation (grant no. MCB–1054298 to G.C.).
Articles can be viewed without a subscription.
References
- Ade J, DeYoung BJ, Golstein C, Innes RW (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA 104: 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao Y, Li Z, Feng D, Xiong F, Liu J, Li JF, Wang M, Wang J, Liu B, Wang HB (2014) OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J 80: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y, Yang L, Hetzel J, Dangl JL, Chory J (2014) The growth-defense pivot: crisis management in plants mediated by LRR-RK surface receptors. Trends Biochem Sci 39: 447–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT (2013) Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res 41: W349–W357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr CA, Leslie ME, Orlowski SK, Chen I, Wright CE, Daniels MJ, Liljegren SJ (2011) CAST AWAY, a membrane-associated receptor-like kinase, inhibits organ abscission in Arabidopsis. Plant Physiol 156: 1837–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JDG, Felix G, Boller T (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Chung EH, da Cunha L, Wu AJ, Gao Z, Cherkis K, Afzal AJ, Mackey D, Dangl JL (2011) Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9: 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung EH, El-Kasmi F, He Y, Loehr A, Dangl JL (2014) A plant phosphoswitch platform repeatedly targeted by type III effector proteins regulates the output of both tiers of plant immune receptors. Cell Host Microbe 16: 484–494 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Qi D, Kim SH, Burke TP, Innes RW (2012) Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Cell Microbiol 14: 1071–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T (2013) Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci USA 110: 8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6: 197–208 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JM, Coaker G (2011) The role of the plasma membrane H+-ATPase in plant-microbe interactions. Mol Plant 4: 416–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JM, Lin ZJD, Coaker G (2011) Plant NB-LRR signaling: upstreams and downstreams. Curr Opin Plant Biol 14: 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JM, Liu J, Smith B, Phinney B, Coaker G (2012) Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Mol Cell Proteomics 11: M111.014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng F, Yang F, Rong W, Wu X, Zhang J, Chen S, He C, Zhou JM (2012) A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485: 114–118 [DOI] [PubMed] [Google Scholar]
- Fermin D, Walmsley SJ, Gingras AC, Choi H, Nesvizhskii AI (2013) LuciPHOr: algorithm for phosphorylation site localization with false localization rate estimation using modified target-decoy approach. Mol Cell Proteomics 12: 3409–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Xu D (2012) Correlation between posttranslational modification and intrinsic disorder in protein. Pac Symp Biocomput 2012: 94–103 [PMC free article] [PubMed] [Google Scholar]
- Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, McCormack M, Sheen J, et al. (2013) Bifurcation of Arabidopsis NLR immune signaling via Ca²⁺-dependent protein kinases. PLoS Pathog 9: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo MC, Valent B (2013) Filamentous plant pathogen effectors in action. Nat Rev Microbiol 11: 800–814 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241: 42–52 [DOI] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AME, He K, Li J, Schroeder JI, Peck SC, Rathjen JP (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, et al. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell 54: 43–55 [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Ohura I, Kawakita K, Yokota N, Fujiwara M, Shimamoto K, Doke N, Yoshioka H (2007) Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19: 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti-Shiu MD, Zou C, Hanada K, Shiu SH (2009) Evolutionary history and stress regulation of Plant Receptor-Like Kinase/Pelle genes. Plant Physiol 150: 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, et al. (2014) The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338 [DOI] [PubMed] [Google Scholar]
- Li W, Chiang YH, Coaker G (2013) The HopQ1 effector’s nucleoside hydrolase-like domain is required for bacterial virulence in Arabidopsis and tomato, but not host recognition in tobacco. PLoS One 8: e59684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Ma X, Shan L, He P (2013) Big roles of small kinases: the complex functions of receptor-like cytoplasmic kinases in plant immunity and development. J Integr Plant Biol 55: 1188–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G (2009) RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol 7: e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Lin ZJD, Coaker G (2011) A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R, Macho AP, Boutrot F, Segonzac C, Somssich IE, Zipfel C (2013) The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. eLife 2: e00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107: 496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zipfel C (2015) Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr Opin Microbiol 23: 14–22 [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF III, Wiig A, Dangl JL (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Marín M, Uversky VN, Ott T (2013) Intrinsic disorder in pathogen effectors: protein flexibility as an evolutionary hallmark in a molecular arms race. Plant Cell 25: 3153–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Schwizer S, Martin GB (2014) Pto kinase binds two domains of AvrPtoB and its proximity to the effector E3 ligase determines if it evades degradation and activates plant immunity. PLoS Pathog 10: e1004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntoukakis V, Balmuth AL, Mucyn TS, Gutierrez JR, Jones AME, Rathjen JP (2013) The tomato Prf complex is a molecular trap for bacterial effectors based on Pto transphosphorylation. PLoS Pathog 9: e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh CS, Martin GB (2011) Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci 16: 132–140 [DOI] [PubMed] [Google Scholar]
- Pritchard L, Birch PRJ (2014) The zigzag model of plant-microbe interactions: is it time to move on? Mol Plant Pathol 15: 865–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R., Jr (2012) MEK1/2 dual-specificity protein kinases: structure and regulation. Biochem Biophys Res Commun 417: 5–10 [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233 [DOI] [PubMed] [Google Scholar]
- Shinya T, Yamaguchi K, Desaki Y, Yamada K, Narisawa T, Kobayashi Y, Maeda K, Suzuki M, Tanimoto T, Takeda J, et al. (2014) Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J 79: 56–66 [DOI] [PubMed] [Google Scholar]
- Sigrist CJA, Cerutti L, Hulo N, Gattiker A, Falquet L, Pagni M, Bairoch A, Bucher P (2002) PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform 3: 265–274 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12: 89–100 [DOI] [PubMed] [Google Scholar]
- Takemoto D, Rafiqi M, Hurley U, Lawrence GJ, Bernoux M, Hardham AR, Ellis JG, Dodds PN, Jones DA (2012) N-terminal motifs in some plant disease resistance proteins function in membrane attachment and contribute to disease resistance. Mol Plant Microbe Interact 25: 379–392 [DOI] [PubMed] [Google Scholar]
- Tena G, Boudsocq M, Sheen J (2011) Protein kinase signaling networks in plant innate immunity. Curr Opin Plant Biol 14: 519–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BP, Nürnberger T, Joosten MH (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Katagiri F (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol 13: 459–465 [DOI] [PubMed] [Google Scholar]
- Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, Mengiste T (2006) The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18: 257–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 337: 635–645 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Yamada K, Ishikawa K, Yoshimura S, Hayashi N, Uchihashi K, Ishihama N, Kishi-Kaboshi M, Takahashi A, Tsuge S, et al. (2013) A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 13: 347–357 [DOI] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, et al. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185 [DOI] [PubMed] [Google Scholar]
- Zipfel C. (2014) Plant pattern-recognition receptors. Trends Immunol 35: 345–351 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.