The protein kinase CIPK23 activates high-affinity K+ uptake in roots and is essential for growth in K+-limiting conditions.
Abstract
Plant growth and development requires efficient acquisition of essential elements. Potassium (K+) is an important macronutrient present in the soil solution at a wide range of concentrations. Regulation of the K+ uptake systems in the roots is essential to secure K+ supply. It has been shown in Arabidopsis (Arabidopsis thaliana) that when the external K+ concentration is very low (<10 µm), K+ nutrition depends exclusively on the high-affinity K+ transporter5 (HAK5). Low-K+-induced transcriptional activation of the gene encoding HAK5 has been previously reported. Here, we show the posttranscriptional regulation of HAK5 transport activity by phosphorylation. Expression in a heterologous system showed that the Ca2+ sensors calcineurin B-like (CBL1), CBL8, CBL9, and CBL10, together with CBL-interacting protein kinase23 (CIPK23), activated HAK5 in vivo. This activation produced an increase in the affinity and the Vmax of K+ transport. In vitro experiments show that the N terminus of HAK5 is phosphorylated by CIPK23. This supports the idea that phosphorylation of HAK5 induces a conformational change that increases its affinity for K+. Experiments of K+ (Rb+) uptake and growth measurements in low-K+ medium with Arabidopsis single mutants hak5, akt1, and cipk23, double mutants hak5 akt1, hak5 cipk23, and akt1 cipk23, and the triple mutant hak5 akt1 cipk23 confirmed the regulatory role of CIPK23 in planta.
Potassium (K+) is an essential macronutrient for plants, required for cell metabolism and extension growth (Marschner, 2012). It fulfills important functions related to turgor maintenance, activation of proteins, neutralization of negative charges, cytoplasmic pH homeostasis, maintenance of transmembrane voltage gradients, and phloem sugar loading (White and Karley, 2010). It is the most abundant inorganic cation in plant tissues, comprising up to 4% to 6% of plant dry weight (Leigh and Wyn Jones, 1984).
The K+ activity in the cell cytoplasm is maintained fairly constant around 100 mm (Walker et al., 1996). This is in sharp contrast with the highly variable K+ concentrations of the soil solutions that may range between 0.1 and 1 mm (White and Karley, 2010). Importantly, the K+ concentrations in the depletion zone around the roots may be even lower, which results in K+ gradients between the cytoplasm and the external solution of up to 10,000-fold.
To overcome such steeps K+ gradients and secure K+ supply under widely variable conditions, root cells are furnished with different K+ uptake systems. Classical studies in barley (Hordeum vulgare) identified low- and high-affinity components of K+ uptake (Epstein et al., 1963). Subsequent molecular studies have described in Arabidopsis (Arabidopsis thaliana) two systems as the major contributors to K+ uptake, the high-affinity K+ transporter HAK5 and the inward-rectifier K+ channel AKT1 (Alemán et al., 2011). The Arabidopsis model is probably extendable to other dicots and monocots, although in monocots, a third system of the high-affinity K+ transporter (HKT) family, HKT2;1, could also be involved in K+ uptake (Corratgé-Faillie et al., 2010). According to the Arabidopsis model, HAK5 is the only system operating at external K+ concentrations below 10 µm. Between 10 and 200 µm K+, HAK5 contributes to K+ acquisition assisted by AKT1 when the plasma membrane potential is negative enough to allow channel-mediated K+ uptake. At K+ concentrations higher than 500 µm, HAK5 is not relevant and AKT1 contribution becomes more important. In the absence of HAK5 and AKT1, other uncharacterized system(s) may provide a pathway for K+ uptake to support residual plant growth (Pyo et al., 2010; Alemán et al., 2011). Interestingly, a recent report suggests that AKT1 may also act as a K+ sensor in the root architecture response to nutrient supply, possibly by linking auxin transport to plasma membrane potential (Kellermeier et al., 2014).
Plant adaptation to different environmental conditions and nutrient supplies requires a precise regulation of the systems involved in nutrients uptake. Therefore, regulation of these transporters at the transcriptional and/or posttranscriptional level must be expected. With the exception of the wheat (Triticum aestivum) AKT1 homolog TaAKT1 (Buschmann et al., 2000), the transcription of the genes encoding AKT1 channels do not respond to the external supply of K+. AKT1 regulation seems to rely on posttranslational modifications, mainly phosphorylation/dephosphorylation mediated by the protein kinase complex CBL-interacting protein kinase23 (CIPK23)/calcineurin B-like proteins1-9 (CBL1-9) and the AKT1-interacting PP2C1 (AIP1) phosphatase (Li et al., 2006; Xu et al., 2006; Cheong et al., 2007; Lee et al., 2007). The reduction in the external K+ concentration could produce a specific Ca2+ signature in the cytosol that would be recorded by the Ca2+-binding CBL1-9 proteins, promoting CIPK23 recruitment to the plasma membrane to phosphorylate and activate AKT1. This activation process is reverted by the AIP1 phosphatase (Chérel et al., 2014). Other mechanisms of AKT1 regulation include interaction with other channel subunits such as K+ channel1 (KC1; Geiger et al., 2009), the syntaxin of plants121 (SYP121; Honsbein et al., 2009), or direct binding to CBL proteins such as CBL10 (Ren et al., 2013).
Regulation of HAK5 transporters has been exclusively described at the transcriptional level. Induction of HAK5 genes by low K+ begins with a hyperpolarization of the plasma membrane potential (Nieves-Cordones et al., 2008). Subsequent steps that lead to gene induction include increases in ethylene and reactive oxygen species (Shin and Schachtman, 2004; Jung et al., 2009; Kim et al., 2010). Several transcription factors and their target sequences in the HAK5 promoter have been identified (Kim et al., 2012; Hong et al., 2013).
Although no posttranscriptional regulation for HAK transporters has been shown, such a regulation needs to be evoked to explain results from different studies. Thus, while under K+-sufficient conditions, HAK5 is mainly detected in the endoplasmic reticulum, upon K+ deprivation, the protein is relocated to the plasma membrane. This suggests that low-K+-induced HAK5 trafficking between the endoplasmic reticulum and the plasma membrane is a mechanism of control of HAK5 activity (Qi et al., 2008). Other studies have shown that in hydroponically grown plants subjected to N, P, or S starvation by removing these nutrients from the growth solution for 7 d, the HAK5 gene was up-regulated, but no HAK5-mediated high-affinity K+ uptake was observed (Rubio et al., 2014). Only when, in addition to N, P, or S starvation, plants are subjected to K+ deprivation, HAK5-mediated high-affinity K+ uptake took place. This indicates that a low-K+ signal is required to produce the posttranscriptional activation of the transporter.
Interestingly, the role of the CIPK23/CBL1-9 complex in regulating K+ acquisition seems to be not restricted to the activation of the AKT1-mediated pathway. An additional unknown transporter was proposed as a target of that complex based on the lower K+ concentrations shown by cipk23 shoots compared with those of akt1 (Xu et al., 2006). Given that AKT1 and HAK5 are the two major systems mediating K+ uptake (Rubio et al., 2010), HAK5 emerges as a likely candidate for the above-mentioned unknown transporter. Here, we demonstrate that HAK5 is activated in yeast (Saccharomyces cerevisiae) by CIPK23 complexes comprising the regulatory partners CBL1, CBL8, CBL9, and CBL10. This activation is strictly dependent on the phosphorylation of the HAK5 transporter by CIPK23. Analyses of Arabidopsis mutant lines confirm that CIPK23 controls the high-affinity K+ uptake mediated by HAK5.
RESULTS
Coexpression of CIPK23 and CBL1 Activates HAK5
The activity of the Arabidopsis AKT1 channel is positively regulated by CIPK23-CBL1-9 protein complexes (Xu et al., 2006; Lee et al., 2007). Although AKT1 is the final target of the cascade and mutant plants lacking AKT1 should display the strongest phenotype, it was found that K+ content in the shoots of cipk23 mutant plants was even lower than in the shoots of akt1 mutants. This suggested that CIPK23 was also regulating an unknown K+ transport system involved in K+ uptake or its distribution within the plant (Xu et al., 2006).
We hypothesized that HAK5 protein could be this unknown K+ transport system modulated, in parallel with AKT1, by CIPK23. In addition, a previous report showed that the expression of HAK5 in a yeast mutant strain lacking the endogenous K+ uptake systems TRK1 and TRK2 produced a modest complementation of defective growth at low K+ (Rubio et al., 2000). This result could be explained if HAK5 was not fully functional in yeast and required activation by ancillary proteins, such as CIPK23 and CBL1.
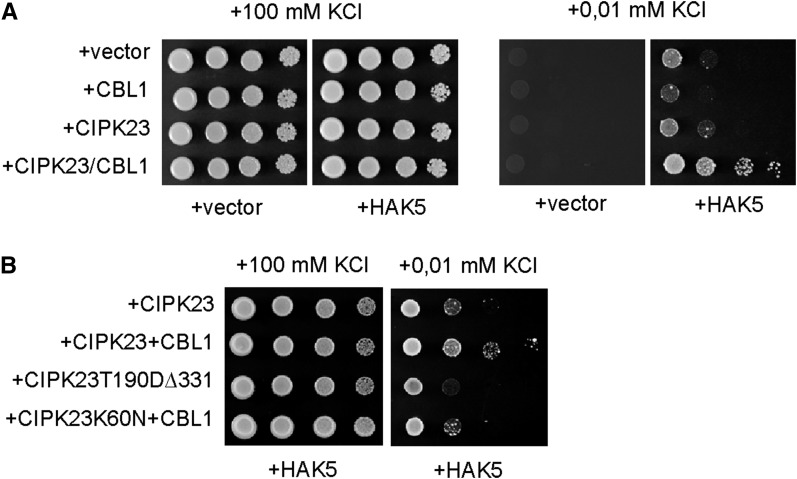
To test our hypotheses, we coexpressed CIPK23 and CBL1 in the absence or the presence of HAK5, in the yeast mutant strain 9.3 (trk1, trk2), defective in K+-uptake. Yeast growth of transformants was assayed in low-K+ medium. As shown in Figure 1A, the concurrent expression of the three proteins dramatically increased the capacity of transformed cells to grow in low K+, above levels imparted by HAK5 alone. Cells expressing the three proteins were able to grow in media containing as low as 10 µm K+. This enhanced growth was observed only when HAK5 was present, demonstrating that CBL1 and CIPK23 were not unmasking an endogenous yeast activity. Moreover, any other combination of the proteins failed to produce a substantial increase in growth. Omitting either CIPK23 or CBL1 prevented the HAK5-dependent enhancement of growth in low K+ (Fig. 1A). These results suggested that both the protein kinase CIPK23 and the Ca2+ sensor CBL1 were necessary and together were sufficient for in vivo activation of the high-affinity K+ transporter HAK5.
Figure 1.
Reconstitution in yeast of the HAK5 K+ uptake pathway. A, CIPK23 and CBL1 improved the growth in low K+ of yeast cells expressing the HAK5 ion transporter. Cells of strain 9.3 (trk1, trk2) were transformed with the empty vector (vector) or the indicated combination of cDNAs. B, The hyperactive CIPK23(T190D,∆331) and the kinase-dead mutant CIPK23(K60N) failed to activate HAK5. Yeast transformants were grown overnight in medium with 0.1 m KCl. Five microliters of serial decimal dilutions were spotted onto plates of AP medium supplemented with the indicated KCl concentration.
The CIPK23/CBL1 Complex Increases the Vmax and Reduces the Km Values for HAK5-Mediated Rb+ Uptake
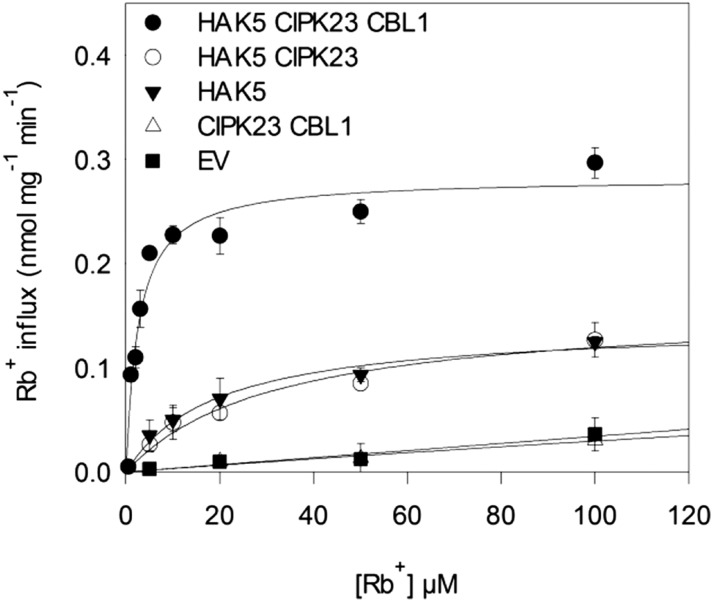
To further characterize the activation of HAK5 by the CIPK23/CBL1 complex, Rb+ uptake experiments were performed in yeast of genotype trk1 trk2 lacking the endogenous high-affinity K+ uptake. This allowed a precise kinetic characterization of HAK5-mediated K+ uptake. After growing the yeast cells overnight in K+-sufficient medium, a 7-h K+ starvation treatment was imposed. The initial rates of Rb+ uptake were determined at different external Rb+ concentrations in the range from 0 to 100 µm (Fig. 2), and the data were fitted to Michaelis-Menten kinetics. Control cells expressing the empty vector showed low rates of Rb+ uptake compatible with the low-affinity K+ uptake system remaining in the 9.3 strain (Fig. 2). Expression of CIPK23/CBL1 did not alter the rate of Rb+ uptake observed in yeast transformed with the empty vector, indicating that no endogenous K+ transport system is induced by the expression of CIPK23 and CBL1. When cells expressed HAK5, an increase in the Rb+ uptake rates that showed saturation kinetics was produced, in agreement with HAK5-meditated K+ uptake. The coexpression of HAK5 with CIPK23 did not produce any effect with respect to the expression of HAK5 alone. However, when the cells expressed HAK5 together with CIPK23 and CBL1, an outstanding increase in Rb+ uptake rates was produced. Importantly, the expression of the CIPK23/CBL1 complex produced a significant effect on the kinetic parameters of HAK5-mediated Rb+ uptake. The affinities for Rb+ with HAK5 alone or HAK5 together with CIKP23 were not significantly different, as shown by the Km values of 19.3 ± 4.5 and 31.9 ± 11.7 µm Rb+, respectively. However, the coexpression of HAK5 together with CIPK23 and CBL1 significantly reduced the Km value to 2.6 ± 0.6 µm. In addition, the maximal rate of uptake was also significantly increased in the HAK5/CIPK23/CBL1-expressing cells, reaching a value of 0.28 ± 0.6 nmol mg–1 min–1 compared with 0.14 ± 0.01 or 0.16 ± 0.02 nmol mg–1 min–1 for HAK5- or HAK5/CIPK23-expressing cells, respectively. These data suggest that the enhancement of growth at low K+ of yeast cells expressing HAK5, CIPK23, and CBL1 (Fig. 1) derived from a modification of the kinetic properties of the transporter induced by the regulatory complex CIPK23/CBL1 (Fig. 2).
Figure 2.
Initial rates of Rb+ uptake in yeast cells expressing HAK5, CIPK23, and CBL1. Cells of trk1 trk2 genotype transformed with the empty vector (EV; black squares), CIPK23/CBL1 (white triangles), HAK5 (black triangles), HAK5/CIPK23 (white circles), or HAK5/CIPK23/CBL1 (black circles) were grown overnight in AP medium supplemented with 5 mm K+. After 6 h of K+ starvation, cells were suspended in assay buffer supplemented with different Rb+ concentrations. Initial rates of Rb+ uptake were determined and plotted against the corresponding Rb+ concentrations. Data were fitted to Michaelis-Menten equations and the Km and Vmax values calculated. Data represent the average of at least three repetitions, and error bars denote se.
The Kinase Activity and C-Terminal Regulatory Domain of CIPK23 Are Required for Activation of HAK5
CIPK/CBL complexes regulate their target proteins by phosphorylation or by establishing physical interactions (Pandey et al., 2014). Although these two regulatory mechanisms are not mutually exclusive, one of them is usually more predominant than the other for each specific case. In some instances, the activation does not require the enzymatic activity of the CIPK, and expression of full-length CIPK lacking kinase activity or partial CIPK peptides without the kinase domain are sufficient to regulate the target protein (Cheng et al., 2004; Held et al., 2011). In others, there is a strict requirement of phosphorylation by the CIPK kinase, and expression of the kinase moiety is sufficient to mimic the regulation of the complete CIPK protein (Li et al., 2006; Xu et al., 2006; Ho et al., 2009; Quintero et al., 2011). To gain insights into the regulatory mechanism of HAK5 by CIPK23/CBL1, we generated two different mutants of CIPK23. The allele CIPK23(T190D,Δ331) combines a T190D mutation in the kinase activation loop of CIPK23 with a C-terminal truncation at residue 311 that removes the C-terminal autoinhibitory domain present in this family of kinases (Chaves-Sanjuan et al., 2014). These two mutations yielded a hyperactive kinase that is independent of CBL binding (Chaves-Sanjuan et al., 2014). The second allele, CIPK23(K60N), contains a substitution of Lys-60 with Asn in the catalytic site required for phosphotransfer and produces an inactive kinase (Li et al., 2006). Complementation of yeast cells expressing CIPK23(T190D,Δ331) or CIPK23(K60N)/CBL1 together with HAK5 was assayed. None of the CIPK23 mutant proteins produced any significant improvement of the growth in low K+ of 9.3 yeast cells containing HAK5 compared with the expression of wild-type CIPK23 (Fig. 1B). This result showed that neither the kinase activity nor the physical interaction had a predominant role to activate HAK5, and both mechanisms were required simultaneously.
CIPK23 Phosphorylates HAK5
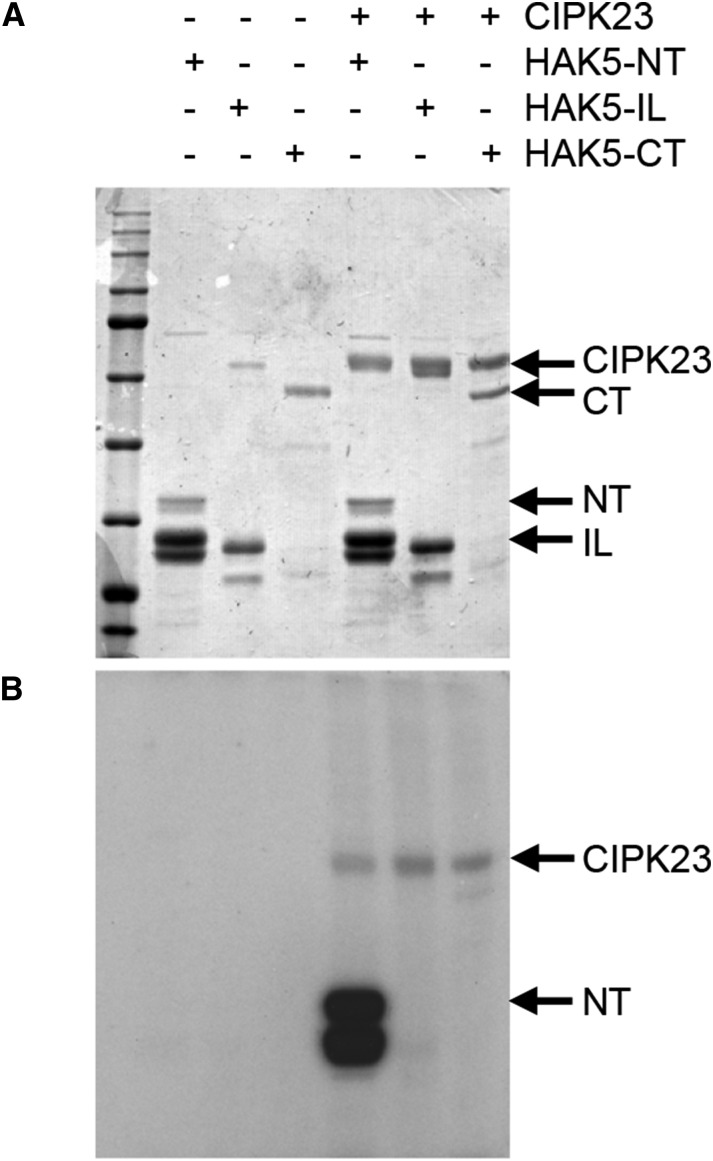
To demonstrate that CIPK23 phosphorylates HAK5, in vitro kinase assays were carried out. Attempts to express and purify a His-tagged version of the full-length HAK5 protein from yeast were unsuccessful. Analysis of the HAK5 polypeptide sequence with the TMHMM v2.0 prediction server (Krogh et al., 2001) revealed the existence of 12 putative transmembrane helices and three major hydrophilic regions, the N-terminal part from amino acids 1 to 95 (NT), an internal loop between TM2 and TM3 comprising residues 123 to 182 (IL), and the C-terminal tail from residues 530 to 784 (CT). Glutathione S-transferase (GST) fusions of these three cytosolic regions were expressed and purified from Escherichia coli cells. NT and IL fusion proteins gave rise to products of the expected length and smaller species, indicating degradation due to protein instability. Phosphorylation assays were performed with the different HAK5 peptides as substrates and the CIPK23 derivative CIPK23(T190D,Δ331) as the kinase. This mutant protein was used because it showed a higher protein kinase activity than the wild type and it was independent of CBL1 (Chaves-Sanjuan et al., 2014). A clear phosphorylation signal was observed only in the sample, where both the NT peptide and CIPK23(T190D,Δ331) were present in the reaction (Fig. 3). This result indicated that CIPK23 catalyzed a strong phosphorylation of HAK5 at the N-terminal region of the transporter.
Figure 3.
CIPK23 phosphorylates HAK5 in vitro. GST fusions encompassing the HAK5 N-terminal region (NT), the internal loop (IL), or the cytosolic tail (CT) were subjected to phosphorylation by CIPK23 and resolved by SDS/PAGE. A, Coomassie Brilliant Blue staining of gel showing proteins in the assay. B, Autoradiograph showing incorporation of 32P into the N-terminal region of HAK5.
Different Members of the Arabidopsis CBL Family Form a Complex with CIPK23 That Activates HAK5
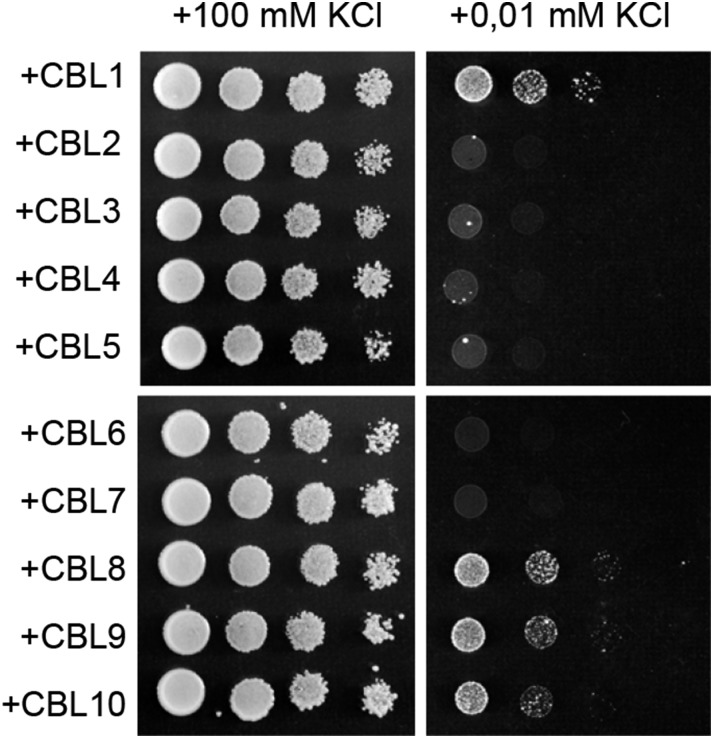
The protein kinase CIPK23 can interact independently with the Ca2+ sensors CBL1 and CBL9, and both protein complexes increase AKT1 channel activity when expressed in Xenopus laevis oocytes (Li et al., 2006; Xu et al., 2006). Protein interaction assays have shown that CIPK23 can associate with other members of the CBL protein family. However, there is a discrepancy regarding the implication of these alternative complexes in AKT1 regulation. According to Xu et al. (2006), only the CIPK23/CBL1 and CIPK23/CBL9 pairs are able to up-regulate AKT1 function. By contrast, Lee et al. (2007) found that CIPK23/CBL2 and CIPK23/CBL3 complexes are also competent to active AKT1 in Xenopus spp. oocytes. To test the specificity of the CBL proteins in the regulation of the HAK5 transporter, all the members of the Arabidopsis CBL family were expressed along with CIPK23 and HAK5 in the yeast 9.3 strain. Growth in low-K+ medium showed that, in addition to CBL1, proteins CBL8, CBL9, and CBL10 were able to complement the growth defect of the yeast cells (Fig. 4). Plasma membrane localization of these four CBL proteins has been reported (Quan et al., 2007; Batistic et al., 2010) which is consistent with their suggested role of promoting CIPK23 recruitment to the plasma membrane to regulate HAK5 function.
Figure 4.
Several members of the Arabidopsis CBL family of Ca2+ sensors interact with CIPK23 and activate HAK5. Yeast cells expressing HAK5 and CIPK23 were further transformed to coexpress the indicated CBL protein. The growth test was performed as indicated in Figure 1.
The CIPK23/CBL1 Complex Also Activates Homologs HAK5 from Various Plant Species
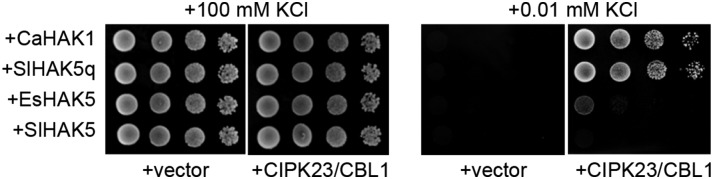
Homologs of the Arabidopsis HAK5 have been identified in other plant species such as tomato (Solanum lycopersicum; SlHAK5; Nieves-Cordones et al., 2007), pepper (Capsicum annuum; CaHAK1; Martínez-Cordero et al., 2004), and the Arabidopsis close relative Eutrema salsuginea (EsHAK5; Alemán et al., 2009). Previous studies have shown CaHAK1- and EsHAK5-mediated functional complementation in yeast, allowing cell growth at 0.1 mm K+ (Martínez-Cordero et al., 2004; Alemán et al., 2009). Although SlHAK5 did not promote yeast growth at low K+, a chimeric transporter, named SlHAK5q, with only 15 amino acids of CaHAK1 in the N terminus of the recombinant protein, led to yeast growth in the presence of 0.1 mm K+ (Nieves-Cordones et al., 2008). Here, growth of yeast cells expressing the mentioned HAK transporters alone or together with the CIPK23/CBL1 complex was analyzed (Fig. 5). In yeast peptone dextrose media supplemented with 100 mm K+, all yeast transformants grew well. In minimal arginine phosphate-based (AP) media with 10 µm K+, none of the cells expressing the CaHAK1, SlHAK5, SlHAK5q, or EsHAK5 alone grew. However, cells coexpressing the CIPK23/CBL1 complex together with CaHAK1 or SlHAK5q showed a remarkably robust growth. Although at lower rates, cells expressing the EsHAK5 also grew. Finally, SlHAK5 was not able to mediate cell growth, even when coexpressed with the CIPK23/CBL1 complex. These results suggest that HAK5 activation by CIPK23/CBL1 is evolutionarily conserved, although the phosphorylation site and/or target sequence recognition may vary among distant plant species.
Figure 5.
Growth of yeast expressing the Arabidopsis CIPK23/CBL1 kinase complex and the HAK5 homologs of pepper, tomato, and E. salsuginea was tested in low-K+ medium. The yeast strain 9.3 (trk1, trk2) was transformed with the empty vector or containing the CIPK23 and CBL1 cDNAs. A second plasmid containing the cDNAs of pepper (CaHAK1), tomato (SlHAK5), or E. salsuginea (EsHAK5) or a quimeric tomato HAK5 that contained the 15 first amino acids of CaHAK1 (SlHAK5q) was included. The growth test was performed as indicated in Figure 1. Indicated are the K+ concentrations of the media used for yeast growth in each case.
Arabidopsis Mutants Lacking CIPK23 Are Deficient in HAK5-Mediated High-Affinity K+ Uptake
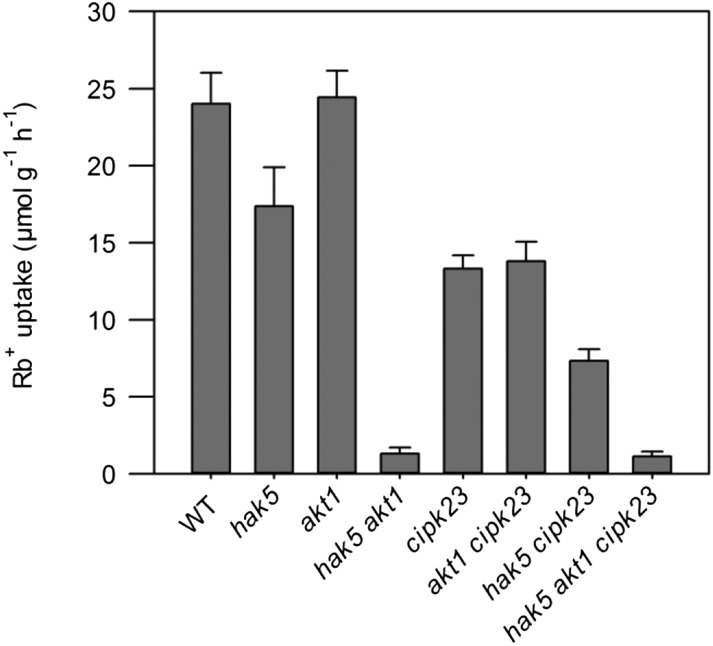
The results from yeast experiments showed the activation of the high-affinity K+ transporter HAK5 by the CIPK23/CBL1 complex in a heterologous system. In addition, in vitro assays demonstrated that the N terminus of HAK5 was phosphorylated by CIPK23. To directly demonstrate HAK5 activation in planta, transfer DNA insertional mutants in HAK5, AKT1, and CIPK23 were used to characterize high-affinity K+ uptake. In addition to the hak5, akt1, and cipk23 single mutants (Rubio et al., 2008, 2010; Pyo et al., 2010), double mutants and the triple mutant lines were obtained and similarly assayed. Rb+ depletion from a 50 µm hydroponic solution was determined in plants starved of K+ for 7 d. Under these conditions, the initial rates of Rb+ uptake mediated by HAK5 and AKT1 could be determined as described previously (Rubio et al., 2008). Uptake assays showed that at 50 µm, the lack of HAK5 reduced Rb+ uptake to 72% compared with the wild type, which implies that under these conditions HAK5 mediated at least one-third of total uptake (Fig. 6). While the absence of AKT1 alone had no effect, Rb+ uptake in the hak5 akt1 double mutant was almost negligible (5% of the wild type). These data are in agreement with the notion that these two systems are the major contributors to K+ uptake and that HAK5 and AKT1 function in a reciprocal compensatory manner at concentrations in the 10 to 50 µm range (Rubio et al., 2008, 2010). In other words, HAK5 activity fully compensated for the absence of AKT1, whereas the converse counterbalance was only partial. The cipk23 mutant line showed 55% of Rb+ uptake rate relative to the wild type, indicating that CIPK23 was governing 45% of total uptake. Despite the compensatory effects between HAK5 and AKT1 systems, the contribution of CIPK23 regulation to each Rb+ uptake system could be estimated from the comparisons of uptake rates in the single hak5 mutant versus hak5 cipk23 double mutant (reporting AKT1 activity) and of akt1 versus akt1 cipk23 (corresponding to HAK5 activity). These differential rates of Rb+ uptake indicated that CIPK23 governed approximately 43% of the transport mediated by HAK5 and 57% of the uptake that could be assigned to AKT1. Finally, the triple mutant hak5 akt1 cipk23 showed a residual Rb+ uptake (5% of the wild type), similar to the double hak5 akt1 mutant, implying that no additional Rb+ (K+) transport processes controlled by CIPK23 are active at 50 µm Rb+. Collectively, these data demonstrate the critical role of CIPK23 in the activation of HAK5 and AKT1 transporters upon K+ limitation.
Figure 6.
Initial rates of K+ (Rb+) uptake in the Arabidopsis wild type (WT) and knockout lines in genes HAK5, AKT1, and CIPK23. Seeds of Arabidopsis, the single mutants hak5, akt1, and cipk23, the double mutants hak5 akt1, akt1 cipk23, and hak5 cipk23, and the triple mutant hak5 akt1 cipk23 were germinated and grown for 30 d in K+-sufficient nutrient solution containing 1.4 mm K+. After 7 d of K+ starvation, plants were transferred to solutions with 50 µm Rb+ for 7 h, during which time, samples of external solution were taken. Rb+ concentrations of these samples were determined and used to calculate the rate of Rb+ absorption from the amount of Rb+ depleted in the external solution per unit of root dry weight and time. Shown are average values of at least six repetitions, and error bars denote se.
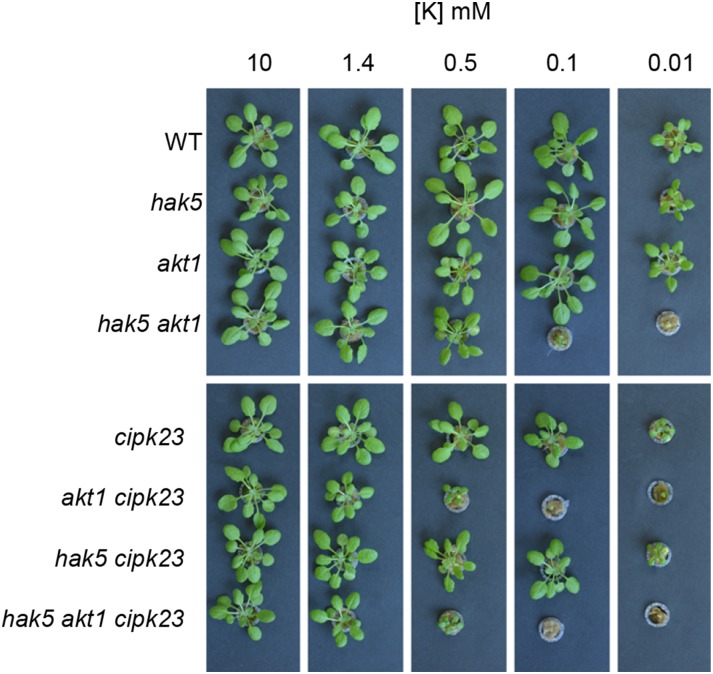
The above Rb+ uptake experiments determined the initial rates of Rb+ uptake as affected by the different mutations. However, the role of the CIPK23 kinase in the regulation of the transporters for providing long-term K+ uptake supporting plant growth may be more complex. To address this point, long-term growth experiments with plants grown hydroponically were conducted. Seeds of the different lines were directly germinated in nutrient solutions with different K+ concentrations for 30 d. In the presence of 10 mm K+, all plant lines showed a similar size (Fig. 7). However, at 1.4 mm K+, slight differences in plant size were observed that resulted evident, as the external K+ concentration was reduced further. In the presence of 0.5 mm K+, the hak5 line showed the biggest size of all mutant lines. The lack of AKT1 or CIPK23 alone did not produce an important effect, but when both CIPK23 and AKT1 were absent, either in the cipk23 akt1 or the hak5 akt1 cipk23 lines, a remarkable reduction in plant growth was observed. Notably, these two mutant lines grew much slower than the hak5 akt1 line at 0.5 mm K+, suggesting the existence of additional target(s) of CIPK23. In the presence of 10 µm K+, the hak5 line grew slower than the wild type and akt1 lines. Growth of cipk23 and hak5 cipk23 lines was greatly reduced and that of hak5 akt1, akt1 cipk23, and hak5 akt1 cipk23 was completely arrested. The effects of the different mutations on plant growth were quantified by determining shoots and roots fresh weights, which are shown in Supplemental Figure S1. In addition, when sufficient plant material was available for reliable analyses, the K+ concentrations were determined (Supplemental Fig. S2). It could be observed that whereas at 10 mm external K+ all lines produced shoots with similar K+ concentrations, at 0.5 mm K+, important differences were observed. Plants of the akt1 cipk23 and hak5 akt1 cipk23 lines showed strong reductions in shoot K+ concentrations (Supplemental Fig. S2), in agreement with their smallest size (Fig. 7; Supplemental Fig. S1). Together, these results show that CIKP23 governs the growth of Arabidopsis under K+ limitation by coordinately regulating the two major K+ uptake systems operating in roots, HAK5 and AKT1, and suggest the existence of additional target(s) of CIPK23 supporting growth in the low-millimolar range.
Figure 7.
Plant growth of Arabidopsis (wild type [WT]) and knockout lines in HAK5, AKT1, and CIPK23 genes in a range of K+ concentrations. Seeds of the same lines used in Figure 6 were directly germinated in nutrient solution containing the indicated K+ concentrations. The picture shows plant size after 30 d of growth.
DISCUSSION
Recent studies have demonstrated that CIPK23/CBL1-9 complexes activate the inward-rectifying K+ channel AKT1. Here, we report that the same CIPK/CBL modules also up-regulates the K+ transporter HAK5, responsible for the high-affinity K+ uptake in the roots. This result highlights the crucial role of the protein kinase CIPK23 and the calcium sensors CBL1 and CBL9 in regulating the two major systems involved in K+ uptake in roots. In addition to the Arabidopsis HAK5 transporter, the HAK5 homologs from E. salsuginea, pepper, and tomato, are also activated by the CIPK23/CBL1 complex (Fig. 5). Importantly, activation of the rice (Oryza sativa) OsAKT1 (Li et al., 2014) or the grapevine (Vitis vinifera) VvK1.1 channels (Cuéllar et al., 2010) by CIPK23/CBL1 has also been described. Thus, the important regulatory role on HAK5- and AKT1-mediated K+ uptake shown here for the Arabidopsis CIPK23/CBL complex can be extended to other plant species and, most importantly, to crops. The CIPK23/CBL modules emerge as central regulator of process as essential for plant growth and development as K+ nutrition. Recently, a report showed activation of the Dionaea muscipula AKT1 and HAK5 by CIPK23/CBL9 as a mechanism for regulating K+ acquisition in the specialized gland cells of this plant species (Scherzer et al., 2015). This suggests that, in addition to its central role in root K+ uptake, the CIPK23/CBL1-9 complex may play roles in specific K+ uptake capacities of certain plant species.
The CIPK23 kinase is also involved in the regulation of NO3– uptake. Regulation of the NO3– transceptor CHL1 is achieved by interaction with the CIPK23 kinase. CHL1 can function as a high- and a low-affinity NO3– transporter, and the phosphorylation of CHL1 at T101 by CIPK23 keeps the primary NO3– response at low levels (Ho et al., 2009; Krouk et al., 2010). Therefore, the results presented here and in previous reports point to the CIPK23 kinase as a central regulator of the systems involved in the acquisition of two of the most important nutrients for plants, i.e., K+ and NO3–. Regulation of uptake of these two nutrients by the same kinase supports the emerging hypothesis of a crosstalk between the signaling pathways modulating the acquisition of different nutrients (Wang et al., 2002; Ho and Tsay, 2010; Hu et al., 2015). Moreover, in addition to sharing regulators, direct physical interactions between NO3– and K+ transporters could exist (Ho and Frommer, 2014). These findings could constitute the molecular bases for the long-standing observation of an interaction between NO3– and K+ uptake and distribution in plants (Lin et al., 2008; Marschner, 2012).
Potassium uptake from diluted solutions is greatly enhanced when plants are faced with K+ deprivation. So far, this enhancement was explained by the activation of AKT1 by CIPK23/CBL1-9 and by the transcriptional activation of the gene encoding HAK5. Here, we describe a posttranslational regulation for HAK5 that is crucial for plant survival at K+ concentrations lower than 10 µm (Fig. 7). Activation of the transport activity of HAK5 by the CIPK23/CBL1 complex requires both the Ser/Thr kinase catalytic domain and the C-terminal domain of CIPK23 (Fig. 1B). In other transport systems regulated by CIPK/CBL, there is a functional predominance of one of these two domains. For instance, the K+ channel AKT1 is controlled by the CIPK23/CBL1 complex, but substantial activation is attained when expressing only the kinase domain of CIPK23 (Li et al., 2006; Lee et al., 2007). By contrast, partial activation of the K+ channel AKT2, which is regulated by CIPK6/CBL4, is obtained by expressing the CIPK6 inhibitory domain alone (Held et al., 2011). Available evidence indicates that the relative dominance of the CIPK kinase activity is associated with a direct modulation of the transport activity of the target protein, whereas the dominance of kinase-target interaction involving the C-terminal part of CIPK correlates with the regulation of the correct localization of the transporter. AKT1 reaches the plasma membrane in the absence of CIPK23/CBL1, and full channel activity requires phosphorylation by this complex. In the case of AKT2, translocation of the channel from the endoplasmic reticulum to the plasma membrane requires the presence of CIPK6/CBL4. The activation mechanism of HAK5 by CIPK23/CBL1, which produces a substantial change of the catalytic properties Km and Vmax (Fig. 2), is consistent with a conformational change triggered by the action of the CIPK23 kinase domain. Analysis of Rb+ uptake in yeast expressing HAK5 alone or with the CIPK23/CBL1 complex revealed that activated HAK5 showed a remarkable affinity increase for its substrate (Fig. 2). The Km value decreased from about 20 µm when only HAK5 was expressed to 2.6 µm K+ when the transporter was coexpressed with CIPK23/CBL1. The affinity for K+ obtained in yeast, 2.6 µm K+, is higher than the one observed in Arabidopsis wild-type plants, between 21 and 24 µm (Shin and Schachtman, 2004; Gierth et al., 2005; Pyo et al., 2010). However, it should be taken into account that in Arabidopsis wild-type plants, both AKT1 and HAK5, are contributing with different affinities to K+ absorption at low K+. In addition, the specific growth conditions of each experiment and the degree of K+ starvation are known to have a great impact on the affinity of K+ uptake shown by the plants (Martínez-Cordero et al., 2004). Although with some caution because of compensatory effects, the use of mutant plant lines, as those used here, may help to determine the individual properties of these two K+ transport system in planta (Rubio et al., 2008). Interestingly, a Km value as low as 0.8 µm K+ has been reported for the akt1-1 line in which the only system mediating K+ uptake is HAK5 (Gierth et al., 2005). This value of 0.8 µm K+ is closer to the affinity obtained for the CIPK23/CBL1-activated HAK5 in yeast of 2.6 µm.
In addition to phosphorylation-induced conformational changes of HAK5 by the CIPK23 kinase, the absolute requirement of the C-terminal autoinhibitory region of CIPK23 for HAK5 activation suggests that localization of the transporter may also be regulated by the kinase. Assuming that this mechanism of regulation also takes place in yeast, it would explain why the expression of the CIPK23/CBL1 complex increases the Vmax of Rb+ uptake mediated by HAK5 in the fungus (Fig. 2). The notion of HAK5 regulation by means of its targeting is supported by different studies. A report showed that, in K+-sufficient plants, HAK5 protein was mainly detected in the endoplasmic reticulum, while K+ starvation produced an enrichment of HAK5 protein in the plasma membrane (Qi et al., 2008). In addition, the KT/HAK/KUP family of K+ transporters belongs to the amino acid-polyamine-organocation (APC) superfamily of secondary carriers (Vastermark et al., 2014). Regulation of APC transporter activity in eukaryotes maintains strong similarities with those found here for HAK5. Phosphorylation is critical for transport activity of APC transporters, and the phosphoaminoacids are located in the N-terminal cytosolic region. Moreover, transit of the transporter from the target membrane to endosomes also plays a fundamental regulatory role (Uemura et al., 2005; Markadieu and Delpire, 2014; Rudnick et al., 2014). Thus, it can be concluded that the regulation of HAK5 by CIPK23 may involve both phosphorylation and targeting of the transporter, an attractive hypothesis that deserves further investigation.
Here, we show that CIPK23 is a central regulator of K+ uptake in plants and that when the kinase is absent, the initial rates of Rb+ uptake of K+-starved plants (Fig. 6), plant growth, and internal K+ concentrations at low K+ are reduced (Fig. 7; Supplemental Figs. S1 and S2). Activation of the K+ transport systems is evident by analyzing the initial rates of Rb+ uptake and the growth of the different mutant lines in a range of K+ concentrations (Figs. 6 and 7). Up-regulation of AKT1 activity by CIPK23 is deduced from the decreased Rb+ uptake rate of the hak5 cipk23 line compared with the hak5 line (43%). The activation of HAK5 by CIPK23 is inferred from the reduced Rb+ uptake rate in the akt1 cipk23 line compared with the akt1 line (57%). This is further supported by the effect of the mutations on plant growth (Fig. 7; Supplemental Fig. S1) that shows a reduced growth of the hak5 cipk23 line compared with the hak5 line and of the akt1 cipk23 line compared with the akt1 line. It is worth noting that growth of the akt1 cipk23 line was much more reduced than that of the hak5 cipk23 line (Fig. 7; Supplemental Fig. S1), whereas the former line shows a higher initial rate of Rb+ uptake than the latter one (Fig. 6). These results may be explained by the different experimental approaches used to measure these two parameters. The initial rate of Rb+ uptake was obtained in a short period of time with plants grown on K+-sufficient solutions and starved of K+ before starting the experiment. By contrast, plant growth was determined by growing the plants on a range of K+ concentrations from seed germination. This latter approach highlighted long-term K+ nutrition and its many effects on plant growth. In fact, when the internal K+ concentrations of shoots are determined (Supplemental Fig. S2), it was observed that at 0.5 mm K+ the shoots of the akt1 cipk23 line contained much less K+ than those of the hak5 cipk23 line.
A careful analysis of the phenotypes shown by the mutant lines suggests additional effects of CIPK23 on K+ uptake that surpass the regulation of HAK5 and AKT1. In the presence of 0.5 mm K+, the triple mutant hak5 akt1 cipk23 grew slower than the double mutant hak5 akt1. This result opens the possibility that the system that is mediating K+ uptake in the absence of HAK5 and AKT1, and that allows plant growth when the external K+ concentration is sufficiently high (Caballero et al., 2012), is also regulated by CIPK23. The identity of this additional system/s is unknown, but a pharmacological characterization suggests that it may be a Ca2+-sensitive system that is regulated by cyclic nucleotides (Caballero et al., 2012). Two families of transporters, cyclic nucleotide gated channel and glutamate receptor, have been suggested as candidates to contain members contributing to K+ uptake in the hak5 akt1 lines. Activation of these types of transporters by CIPK kinases has not been described, but it may be possible because CIPK kinases show a broad range of targets (Steinhorst and Kudla, 2013). Another possibility is that a K+ transporter of the HAK family other than HAK5 is mediating the observed K+ uptake in the absence of HAK5 and AKT1. In that case and, after showing here activation of HAK5 by CIPK23, the regulation of such a HAK transporter would not be so surprising.
An open question is why two K+ transporters, i.e., HAK5 and AKT1, working at different ranges of external K+ are regulated by the same CIPK. The answer may be found in the activation mechanisms described for the CIPK kinases. The structural and biochemical data available show that the activity of CIPK kinases requires the coordinated release of the activation loop from the active site and of the CBL binding motif (NAF) from the nucleotide-binding site (Chaves-Sanjuan et al., 2014). Phosphorylation of the activation loop by upstream kinases and Ca2+-dependent CBL binding to the NAF motif promote the conformational changes that release the catalytic domain. Full CIPK activity is achieved when both regulatory processes take place, but partial CIPK activity can be attained when only one is acting. As a consequence, CIPK/CBL complexes with different states of activation might exist, depending on factors that affect the action of upstream kinases or CBL binding. Activation of AKT1 is observed in the presence of the kinase domain of CIPK23, suggesting that partially activated CIPK23/CBL complexes could up-regulate the channel (Lee et al., 2007). By contrast, the results presented here show that HAK5 activation is only possible when full-length CIPK23 and CBL1 are present (Fig. 1B). This indicates that the fully active CIPK23/CBL1 complex is required to activate the transporter. In conclusion, we speculate that mild K+ deprivation produces a partially activated CIPK23, which is competent for activating AKT1, and that severe K+ deprivation leads to full activation of CIPK23 and the subsequent activation of HAK5.
Identification of CIPK23 as a nutrient regulatory hub, involved in K+ and NO3– nutrition, has a potential interest for biotechnology. High crop yields depend on the application of fertilizers. Improving plant nutrient uptake could decrease the use of these compounds, which has two beneficial effects: It reduces the final cost of agrochemical inputs and minimizes the detrimental effects for the environment due to nutrient leaching. Because CIPK23 controls the uptake of K+ and NO3–, two of the main components of fertilizers, and probably the uptake of other nutrients, engineering CIPK23 to produce activated isoforms may conceivably produce a concomitant increase in the uptake efficiency of several important nutrients. Current research efforts are more centered in the modification of regulatory elements of nutrient acquisition that govern upstream reactions affecting entire pathways, rather than on the end points of the response as transporters (Bohnert et al., 2006). Studies as the one presented here may provide relevant tools for improving the efficiency of mineral nutrition of crops.
MATERIALS AND METHODS
Yeast Strain and Media
The yeast (Saccharomyces cerevisiae) strain 9.3 (MATa, ena1Δ::HIS3::ena4Δ, leu2, ura3, trp1, ade2, trk1Δ, trk2::pCK64) has been described previously (Bañuelos et al., 1995). Yeast cells were routinely grown in yeast peptone dextrose medium (1% [w/v] yeast extract, 2% [w/v] peptone, and 2% [w/v] Glc) or synthetic dextrose minimal medium supplemented with 50 mm KCl. Growth tests in low-K+ were performed in the alkali cation-free medium AP (Rodríguez-Navarro and Ramos, 1984) solidified with 2% (w/v) noble agar (Difco) and supplemented with KCl at the concentrations indicated.
Plasmid Constructs
A fragment of 3.8 kb containing the AtHAK5 complementary DNA (cDNA) under the control of the yeast PMA1 promoter was excised with PvuII from the vector pDR195 (Rubio et al., 2000) and subcloned in the yeast shuttle vector pRS425 digested also with PvuII (Christianson et al., 1992). The CIPK23 cDNA was amplified by PCR using the high-fidelity Accuzyme DNA polymerase (Bioline) using the primers CIPK23forw and CIPK23rev. The PCR product was digested with SmaI and XhoI and ligated into the yeast expression vector p414GPD (Mumberg et al., 1995). The CIPK23(K60N) mutant allele was obtained by overlap extension PCR using primers CIPK23K60Nfor and CIPK23K60Nrev. CIPK23(K60N) and CIPK23(T190D,∆331) (Chaves-Sanjuan et al., 2014) mutant cDNAs were ligated into p414GPD using SmaI and XhoI restriction enzymes. Primer sequences are provided in Supplemental Table S1.
The CBL1 cDNA was isolated from plasmid pGEX-2TK (Guo et al., 2002) as a 0.64-kb BamHI/EcoRI fragment and subcloned in the yeast expression vector pYPGE15 (Brunelli and Pall, 1993) digested with the same restriction enzymes.
CBL2, CBL3, CBL5, CBL6, CBL8, and CBL9 were subcloned as BamHI/EcoRI fragments in pYPGE15. CBL4(SOS3) and CBL10 (SCaBP8) cDNAs in pYPGE15 are already described (Guo et al., 2004; Quan et al., 2007).
Rb+ Uptake Experiments in Yeast
For Rb+ uptake rates determination, yeast cells of the 9.3 strain transformed to express various combinations of cDNAs encoding HAK5, CIPK23, and CBL1 were grown overnight at 28°C in Arg phosphate minimal medium containing 3 mm K+. When cultures reached an optical density at 550 nm between 0.2 and 0.3, cells were centrifuged, washed twice with distilled water, resuspended in AP medium without K+, and incubated for 6 h at 28°C with shaking. Then, cells were centrifuged and resuspended in uptake buffer that consisted of 10 mm MES brought to pH 6.0 with Ca(OH)2 supplemented with 2% Glc in a shaker bath at 28°C. At zero time, Rb+ was added to reach different external concentrations in different flasks. Samples of cells were collected by filtration on nitrocellulose filters (0.8-µm pore) at different time points, and their internal cationic contents were acid extracted by incubation in 0.1 m HCl. Internal Rb+ concentration in cells was determined by atomic emission spectrophotometry with a Perkin Elmer Analyst 400 spectrophotometer and referred to the corresponding dry weight of the cells. The initial rates of Rb+ uptake were calculated and plotted for each external concentration of Rb+. The data were fitted to Michaelis-Menten equations, and the Km and Vmax values were calculated by nonlinear regression analysis.
Arabidopsis Mutant Lines Used and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants of ecotype Columbia and its knockout mutants in HAK5, AKT1 (Rubio et al., 2008), and CIPK23 (Nieves-Cordones et al., 2012), hak5-3, akt1-2, and cipk23-5, were used. In addition, the double mutants hak5-3 cipk23-5 and akt1-2 cipk23-5 as well as the triple mutant hak5-3 akt1-2 cipk23-5 were obtained by crossing of the corresponding single mutants. The three new mutant lines obtained here were checked by PCR on genomic DNA with the primers used to check the original single mutants as described elsewhere (Rubio et al., 2008; Nieves-Cordones et al., 2012).
Arabidopsis plants were grown hydroponically on a modified one-fifth-strength Hoagland solution that consisted of the macronutrients (mm) 1.4 Ca(NO3)2, 0.35 MgSO4, and 0.1 Ca(H2PO4)2 and the micronutrients (µm) 50 CaCl2, 12.5 H3BO3, 1 MnSO4, 1 ZnSO4, 0.5 CuSO4, 0.1 H2MoO4, 0.1 NiSO4, and 10 Fe-EDDHA. KCl was added as indicated for each case. Plants were grown in a controlled-conditions chamber (8-h-day/16-h-night cycle at 150 µmol m–2 s–1 light, 22°C, and 60% relative humidity).
Rb+ Depletion Experiments with Arabidopsis Plants
Plants of various genotypes were grown for 30 d in the conditions described above with a solution that contained 1.4 mm KCl. Then, plants were transferred to a solution with no KCl added and grown for 7 d to subject the plants to K+ starvation. Individual plants were transferred to tubes containing 30 mL of nutrient solution with no KCl added and supplemented with RbCl to a final concentration of 45 µm Rb+. Rb+ was used as a K+ analog, because when K+ is used, the K+ efflux that occurs during the 7 h of the experiment produced an underestimation of the K+ uptake (Rubio et al., 2008). Samples of the external solution were taken at different time points and had their Rb+ concentration determined by atomic emission spectrometry as indicated above. After 7 h of depletion experiment, plants were harvested, separated in roots and shoots, and dried in an oven at 65°C for 3 d. Dry weight of roots and shoots were determined, and then the plant material was acid digested with an HNO3:HClO4 (2:1) mix. K+ concentrations of acid-extracted plant material were determined by atomic emission spectrometry as indicated above.
Tissue K+ Concentration Determination
Plants grown under the K+ treatments indicated were separated into root and shoot. Then, the plant material was dried at 65°C for 4 d and weighed. When sufficient plant material was obtained for proper analysis, it was digested with HNO3:HClO4 (2:1, v/v), and the K+ concentrations were then determined in an inductively coupled plasma mass spectrometry by using an Iris Intrepid II inductively coupled plasma spectrometer (Thermo Electron Corporation). The data reported are the averages of four values per treatment, and error bars denote ses.
Expression and Purification of Fusion Proteins
The translational fusion GST-CIPK23(T190D,∆331) was purified from yeast as described (Chaves-Sanjuan et al., 2014). GST fusion proteins consisting of the N terminus (amino acids 1–94), internal loop (residues 123–182), or the C terminus (residues 530–784) of HAK5 were constructed by PCR amplification using the following primers pairs, BamHI_HAK5M1_F and XbaI_HAK5D94_R; BamHI_HAK5D123_F and XbaI_HAK5F182_R; and BamHI_HAK5K530_F and SalI_HAK5STOP_R, respectively. PCR primers incorporated restriction sites at the 5′ and 3′ ends, thereby allowing subcloning into the fusion protein vector pGEX4T1 or pGEX4T2 (GE Healthcare). Primer sequences are provided in Supplemental Table S1.
All GST fusion constructs were transformed into Escherichia coli Rosetta cells (Merck Millipore). A 10-mL overnight Luria-Bertani culture was transferred to a fresh 200 mL of 2x yeast extract tryptone medium and further cultured at 37°C until the A600 reached about 0.8. Recombinant protein expression was induced by 1 mm isopropyl β-d-thiogalactopyranoside for 4 h. The cells were harvested by centrifugation, and the pellets were resuspended in ice-cold phosphate-buffered saline buffer, pH 7.5, containing protease inhibitors (Sigma). Lysozyme (1 mg mL–1) was added to the suspension and incubated on ice with gentle shaking before sonication. The lysate was clarified by centrifugation, and the recombinant proteins were affinity purified by glutathione-Sepharose 4B (GE Healthcare). SDS-PAGE analysis was used to evaluate the protein composition of each preparation. Gels were stained with Coomassie Brilliant Blue R-250 (Sigma).
Phosphorylation Assays
Substrate recombinant proteins (approximately 200 ng) were subjected to phosphorylation by the CIPK23(T190D/Δ331) protein kinase (approximately 200 ng) in 30 μL of buffer (20 mm Tris⋅HCl, pH 7.5, 5 mm MgCl2, and 1 mm dithiothreitol). Reactions were started by adding ATP (0.2 mm with 1 μCi of [γ-32P]ATP), which was incubated at 30°C for 30 min, and stopped with 10 μL of 4× SDS/PAGE sample buffer. Aliquots were then resolved by SDS/PAGE, and the gel was exposed to x-ray films.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Fresh weight of shoots and roots of the Arabidopsis wild type and mutant lines grown in a range of K+ concentrations.
Supplemental Figure S2. K+ concentrations of shoots of Arabidopsis plants grown in a range of K+ concentrations.
Supplemental Table S1. Primers used in this work.
Glossary
- GST
glutathione S-transferase
- cDNA
complementary DNA
Footnotes
This work was supported by the Ministerio de Enonomía y Competitividad, Spain (grant nos. BFU2012–35060 to J.M.P., BIO2012–36533 to F.J.Q., and AGL2012–33504 to F.R., which were cofinanced by the European Regional Development Fund) and an Formación Profesorado Universitario predoctoral contract from the Ministerio de Educación, Cultura y Deporte (to R.R.).
Articles can be viewed without a subscription.
References
- Alemán F, Nieves-Cordones M, Martínez V, Rubio F (2009) Differential regulation of the HAK5 genes encoding the high-affinity K+ transporters of Thellungiella halophila and Arabidopsis thaliana. Environ Exp Bot 65: 263–269 [Google Scholar]
- Alemán F, Nieves-Cordones M, Martínez V, Rubio F (2011) Root K+ acquisition in plants: the Arabidopsis thaliana model. Plant Cell Physiol 52: 1603–1612 [DOI] [PubMed] [Google Scholar]
- Bañuelos MA, Klein RD, Alexander-Bowman SJ, Rodríguez-Navarro A (1995) A potassium transporter of the yeast Schwanniomyces occidentalis homologous to the Kup system of Escherichia coli has a high concentrative capacity. EMBO J 14: 3021–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O, Waadt R, Steinhorst L, Held K, Kudla J (2010) CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J 61: 211–222 [DOI] [PubMed] [Google Scholar]
- Bohnert HJ, Gong Q, Li P, Ma S (2006) Unraveling abiotic stress tolerance mechanisms: getting genomics going. Curr Opin Plant Biol 9: 180–188 [DOI] [PubMed] [Google Scholar]
- Brunelli JP, Pall ML (1993) A series of yeast shuttle vectors for expression of cDNAs and other DNA sequences. Yeast 9: 1299–1308 [DOI] [PubMed] [Google Scholar]
- Buschmann PH, Vaidyanathan R, Gassmann W, Schroeder JI (2000) Enhancement of Na+ uptake currents, time-dependent inward-rectifying K+ channel currents, and K+ channel transcripts by K+ starvation in wheat root cells. Plant Physiol 122: 1387–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero F, Botella MA, Rubio L, Fernández JA, Martínez V, Rubio F (2012) A Ca2+-sensitive system mediates low-affinity K+ uptake in the absence of AKT1 in Arabidopsis plants. Plant Cell Physiol 53: 2047–2059 [DOI] [PubMed] [Google Scholar]
- Chaves-Sanjuan A, Sanchez-Barrena MJ, Gonzalez-Rubio JM, Moreno M, Ragel P, Jimenez M, Pardo JM, Martinez-Ripoll M, Quintero FJ, Albert A (2014) Structural basis of the regulatory mechanism of the plant CIPK family of protein kinases controlling ion homeostasis and abiotic stress. Proc Natl Acad Sci USA 111: E4532–E4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NH, Pittman JK, Zhu JK, Hirschi KD (2004) The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J Biol Chem 279: 2922–2926 [DOI] [PubMed] [Google Scholar]
- Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim BG, Lee SC, Kudla J, Luan S (2007) Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J 52: 223–239 [DOI] [PubMed] [Google Scholar]
- Chérel I, Lefoulon C, Boeglin M, Sentenac H (2014) Molecular mechanisms involved in plant adaptation to low K+ availability. J Exp Bot 65: 833–848 [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122 [DOI] [PubMed] [Google Scholar]
- Corratgé-Faillie C, Jabnoune M, Zimmermann S, Véry AA, Fizames C, Sentenac H (2010) Potassium and sodium transport in non-animal cells: the Trk/Ktr/HKT transporter family. Cell Mol Life Sci 67: 2511–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuéllar T, Pascaud F, Verdeil JL, Torregrosa L, Adam-Blondon AF, Thibaud JB, Sentenac H, Gaillard I (2010) A grapevine Shaker inward K+ channel activated by the calcineurin B-like calcium sensor 1-protein kinase CIPK23 network is expressed in grape berries under drought stress conditions. Plant J 61: 58–69 [DOI] [PubMed] [Google Scholar]
- Epstein E, Rains DW, Elzam OE (1963) Resolution of dual mechanisms of potassium absorption by barley roots. Proc Natl Acad Sci USA 49: 684–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Becker D, Vosloh D, Gambale F, Palme K, Rehers M, Anschuetz U, Dreyer I, Kudla J, Hedrich R (2009) Heteromeric AtKC1middle dotAKT1 channels in Arabidopsis roots facilitate growth under K+-limiting conditions. J Biol Chem 284: 21288–21295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierth M, Mäser P, Schroeder JI (2005) The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol 137: 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Qiu QS, Quintero FJ, Pardo JM, Ohta M, Zhang C, Schumaker KS, Zhu JK (2004) Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement for its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis thaliana. Plant Cell 16: 435–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Song CP, Gong D, Halfter U, Zhu JK (2002) A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev Cell 3: 233–244 [DOI] [PubMed] [Google Scholar]
- Held K, Pascaud F, Eckert C, Gajdanowicz P, Hashimoto K, Corratgé-Faillie C, Offenborn JN, Lacombe B, Dreyer I, Thibaud JB, et al. (2011) Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res 21: 1116–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CH, Frommer WB (2014) Fluorescent sensors for activity and regulation of the nitrate transceptor CHL1/NRT1.1 and oligopeptide transporters. eLife 3: e01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Ho CH, Tsay YF (2010) Nitrate, ammonium, and potassium sensing and signaling. Curr Opin Plant Biol 13: 604–610 [DOI] [PubMed] [Google Scholar]
- Hong JP, Takeshi Y, Kondou Y, Schachtman DP, Matsui M, Shin R (2013) Identification and characterization of transcription factors regulating Arabidopsis HAK5. Plant Cell Physiol 54: 1478–1490 [DOI] [PubMed] [Google Scholar]
- Honsbein A, Sokolovski S, Grefen C, Campanoni P, Pratelli R, Paneque M, Chen Z, Johansson I, Blatt MR (2009) A tripartite SNARE-K+ channel complex mediates in channel-dependent K+ nutrition in Arabidopsis. Plant Cell 21: 2859–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Wang W, Deng K, Li H, Zhang Z, Zhang L, Chu C (2015) MicroRNA399 is involved in multiple nutrient starvation responses in rice. Front Plant Sci 6: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JY, Shin R, Schachtman DP (2009) Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21: 607–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermeier F, Armengaud P, Seditas TJ, Danku J, Salt DE, Amtmann A (2014) Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 26: 1480–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Ciani S, Schachtman DP (2010) A peroxidase contributes to ROS production during Arabidopsis root response to potassium deficiency. Mol Plant 3: 420–427 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Ruzicka D, Shin R, Schachtman DP (2012) The Arabidopsis AP2/ERF transcription factor RAP2.11 modulates plant response to low-potassium conditions. Mol Plant 5: 1042–1057 [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Krouk G, Crawford NM, Coruzzi GM, Tsay YF (2010) Nitrate signaling: adaptation to fluctuating environments. Curr Opin Plant Biol 13: 266–273 [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan WZ, Kim BG, Li L, Cheong YH, Pandey GK, Lu G, Buchanan BB, Luan S (2007) A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc Natl Acad Sci USA 104: 15959–15964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh RA, Wyn Jones RG (1984) A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant-cell. New Phytol 97: 1–13 [Google Scholar]
- Li J, Long Y, Qi GN, Li J, Xu ZJ, Wu WH, Wang Y (2014) The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell 26: 3387–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kim BG, Cheong YH, Pandey GK, Luan S (2006) A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc Natl Acad Sci USA 103: 12625–12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SH, Kuo HF, Canivenc G, Lin CS, Lepetit M, Hsu PK, Tillard P, Lin HL, Wang YY, Tsai CB, et al. (2008) Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20: 2514–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markadieu N, Delpire E (2014) Physiology and pathophysiology of SLC12A1/2 transporters. Pflugers Arch 466: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner P. (2012) Marschner’s Mineral Nutrition of Higher Plants, Ed 3 Academic Press, San Diego [Google Scholar]
- Martínez-Cordero MA, Martínez V, Rubio F (2004) Cloning and functional characterization of the high-affinity K+ transporter HAK1 of pepper. Plant Mol Biol 56: 413–421 [DOI] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Nieves-Cordones M, Caballero F, Martínez V, Rubio F (2012) Disruption of the Arabidopsis thaliana inward-rectifier K+ channel AKT1 improves plant responses to water stress. Plant Cell Physiol 53: 423–432 [DOI] [PubMed] [Google Scholar]
- Nieves-Cordones M, Martinez-Cordero MA, Martinez V, Rubio F (2007) An NH4+-sensitive component dominates high-affinity K+ uptake in tomato plants. Plant Sci 172: 273–280 [Google Scholar]
- Nieves-Cordones M, Miller AJ, Alemán F, Martínez V, Rubio F (2008) A putative role for the plasma membrane potential in the control of the expression of the gene encoding the tomato high-affinity potassium transporter HAK5. Plant Mol Biol 68: 521–532 [DOI] [PubMed] [Google Scholar]
- Pandey G, Kanwar P, Pandey A (2014) Global Comparative Analysis of CBL-CIPK Gene Families in Plants. Springer International Publishing, New York [Google Scholar]
- Pyo YJ, Gierth M, Schroeder JI, Cho MH (2010) High-affinity K+ transport in Arabidopsis: AtHAK5 and AKT1 are vital for seedling establishment and postgermination growth under low-potassium conditions. Plant Physiol 153: 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Hampton CR, Shin R, Barkla BJ, White PJ, Schachtman DP (2008) The high affinity K+ transporter AtHAK5 plays a physiological role in planta at very low K+ concentrations and provides a caesium uptake pathway in Arabidopsis. J Exp Bot 59: 595–607 [DOI] [PubMed] [Google Scholar]
- Quan R, Lin H, Mendoza I, Zhang Y, Cao W, Yang Y, Shang M, Chen S, Pardo JM, Guo Y (2007) SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19: 1415–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero FJ, Martinez-Atienza J, Villalta I, Jiang X, Kim WY, Ali Z, Fujii H, Mendoza I, Yun DJ, Zhu JK, et al. (2011) Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc Natl Acad Sci USA 108: 2611–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XL, Qi GN, Feng HQ, Zhao S, Zhao SS, Wang Y, Wu WH (2013) Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J 74: 258–266 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Navarro A, Ramos J (1984) Dual system for potassium transport in Saccharomyces cerevisiae. J Bacteriol 159: 940–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio F, Alemán F, Nieves-Cordones M, Martínez V (2010) Studies on Arabidopsis athak5, atakt1 double mutants disclose the range of concentrations at which AtHAK5, AtAKT1 and unknown systems mediate K uptake. Physiol Plant 139: 220–228 [DOI] [PubMed] [Google Scholar]
- Rubio F, Fon M, Ródenas R, Nieves-Cordones M, Alemán F, Rivero RM, Martínez V (2014) A low K+ signal is required for functional high-affinity K+ uptake through HAK5 transporters. Physiol Plant 152: 558–570 [DOI] [PubMed] [Google Scholar]
- Rubio F, Nieves-Cordones M, Alemán F, Martínez V (2008) Relative contribution of AtHAK5 and AtAKT1 to K+ uptake in the high-affinity range of concentrations. Physiol Plant 134: 598–608 [DOI] [PubMed] [Google Scholar]
- Rubio F, Santa-Maria GE, Rodríguez-Navarro A (2000) Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol Plant 109: 34–43 [Google Scholar]
- Rudnick G, Krämer R, Blakely RD, Murphy DL, Verrey F (2014) The SLC6 transporters: perspectives on structure, functions, regulation, and models for transporter dysfunction. Pflugers Arch 466: 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzer S, Böhm J, Krol E, Shabala L, Kreuzer I, Larisch C, Bemm F, Al-Rasheid KAS, Shabala S, Rennenberg H, et al. (2015) Calcium sensor kinase activates potassium uptake systems in gland cells of Venus flytraps. Proc Natl Acad Sci USA 112: 7309–7314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Schachtman DP (2004) Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc Natl Acad Sci USA 101: 8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhorst L, Kudla J (2013) Calcium and reactive oxygen species rule the waves of signaling. Plant Physiol 163: 471–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Tachihara K, Tomitori H, Kashiwagi K, Igarashi K (2005) Characteristics of the polyamine transporter TPO1 and regulation of its activity and cellular localization by phosphorylation. J Biol Chem 280: 9646–9652 [DOI] [PubMed] [Google Scholar]
- Vastermark A, Wollwage S, Houle ME, Rio R, Saier MH Jr (2014) Expansion of the APC superfamily of secondary carriers. Proteins 82: 2797–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DJ, Leigh RA, Miller AJ (1996) Potassium homeostasis in vacuolate plant cells. Proc Natl Acad Sci USA 93: 10510–10514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Garvin DF, Kochian LV (2002) Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol 130: 1361–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PJ, Karley AJ (2010) Potassium. In Hell R, Mendel RR, eds, Cell Biology of Metals and Nutrients, Vol 17 Springer, Heidelberg, pp 199–224 [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360 [DOI] [PubMed] [Google Scholar]