Abstract
CRISPR-Cas9 represents a promising technology for genome editing, yet means of safe and efficient delivery remain to be fully realized. Here, we report a novel delivery vehicle to deliver the Cas9 protein and single-guide RNA simultaneously based on DNA nanoclews, yarn-like DNA nanoparticles synthesized by rolling circle amplification. The bio-inspired vehicles efficiently loaded Cas9/single-guide RNA complexes and delivered the complexes to the nuclei of human cells, allowing targeted gene disruptions while maintaining cell viability. Editing was most efficient when the DNA nanoclew sequence and the sgRNA guide sequence were partially complementary, offering a design rule for enhancing delivery. Overall, this strategy provides a versatile platform that could be adapted for delivering other DNA-binding proteins or for functional nucleic acids.
Keywords: drug delivery, DNA, nanoparticles, CRISPR-Cas9, genome editing
CRISPR-Cas9 has rapidly transitioned from an RNA-directed defense system in prokaryotes to a facile genome-editing technology.[1] The editing merely requires the Cas9 nuclease and an engineered single-guide RNA (sgRNA): the 20-nucleotide guide portion of the sgRNA recognizes complementary DNA sequences flanked by a protospacer-adjacent motif (PAM), and Cas9 cleaves the recognized DNA.[2] The double-stranded break is then repaired through non-homologous end joining (NHEJ) or homology-directed repair (HDR), allowing defined alterations to the targeted region.[3]
As CRISPR-Cas9 systems undergo further development toward human therapeutics, delivery poses the major challenge. Cas9 and the sgRNA have been overwhelmingly encoded within the DNA of plasmids of viral vectors.[4] However, this DNA can randomly integrate into the genome, potentially giving rise to cancer or other genetic diseases.[5] Furthermore, the template-driven nature of gene expression limits control over the total amount of Cas9 protein and sgRNAs, where excess dosing has been attributed to off-target cleavage.[6] One alternative is to deliver the Cas9/sgRNA ribonucleotprotein complex,[7] which enables greater control over its intracellular concentration and limits the timeframe in which editing can occur. However, delivering protein and RNA remains a central challenge in drug delivery.[8] Most protein therapeutics, such as enzymes,[9] antibodies[10] or transcription factors,[11] suffer from low stability and poor cell membrane permeability as a result of their fragile tertiary structures and large molecular sizes.[8] The strong negative charges of RNA therapeutics, including siRNA or miRNA, blocks them from diffusing across cell membrane and their susceptibility to endonuclease often requires chemical modification to prevent degradation.[12] Thus, devising an appropriate carrier to shield the protein and RNA from detrimental physiological environment and escort them simultaneously to cell nucleus is highly desirable.
Herein, we report a novel delivery vehicle for CRISPR-Cas9 based on biologically inspired yarn-like DNA nanoclew (NC) (Figure 1). The DNA NCs are synthesized by rolling circle amplification (RCA)[13] with palindromic sequences encoded to drive the self-assembly of nanoparticles. We previously demonstrated that the DNA NC could encapsulate the chemotherapeutic agent doxorubicin and drive its release based on environmental conditions.[14] Here, we hypothesized that the DNA NC can load and deliver the Cas9 protein together with an sgRNA for genome editing. Inspired by the ability of single stranded DNA (ssDNA) to base pair with the guide portion of the Cas9-bound sgRNA,[15] we designed the DNA NC to be partially complementary to the sgRNA. Following loading of the DNA NC with the Cas9/sgRNA complex, we applied a coating of the cationic polymer polyethylenimine (PEI) to help induce endosomal escape.[16] The Cas9/sgRNA complex delivered to the cytoplasm could then be transported into the nucleus via nuclear-localization-signal peptides fused to Cas9. We expected that the resulting delivery vehicle could form uniform particles and drive the formation of targeted insertions or deletions (indels) without measurable impact on cell viability.
Figure 1.

Schematic design of the DNA NC mediated CRISPR-Cas9 delivery system. (a) Preparation of Cas9/sgRNA/NC/PEI. I: The NC was synthesized by RCA and loaded with the Cas9/sgRNA complex through Watson-Crick base pairing; II: PEI was coated onto Cas9/sgRNA/NC for enhanced endosome escape. (b) Delivery of Cas9/sgRNA by the DNA NC based carrier to the nucleus of the cell for genome editing. I: Bind to cell membrane; II: Endocytosis; III: Endosome escape; IV; Transport into the nucleus; V: Search for target DNA locus in the chromosome and introduce double strand breaks for genome editing.
To demonstrate the DNA NC-mediated delivery of CRISPR-Cas9, we first selected the well-characterized and most extensively applied Streptococcus pyogenes Cas9.[17] Recombinant Cas9 fused with N-terminal and C-terminal nuclear localization signals[18] was purified following overexpression in Escherichia coli (Figure S1 in the SI) and incubated with one of two sgRNAs: one designed to target a sequence within the enhanced green fluorescent protein (EGFP) gene flanked by an NGG PAM, and the other control sgRNA (cgRNA) designed not to appreciably target any DNA sequence in EGFP or the human genome (Figure S2a). We confirmed that the resulting Cas9/sgRNA complex was active in vitro based on cleavage of a linearized plasmid encoding the EGFP gene, but only in the presence of Cas9 and the EGFP-targeting sgRNA (Figure S2b).
We next generated the DNA NC to bind the Cas9/sgRNA complex. The DNA template for RCA was designed to encode 12 nucleotides complementary to the 5’ end of the sgRNA (NC-12) along with the palindromic repeat that drives self-assembly (Table S1). The rationale was that the complementary sequence would promote base pairing between the DNA NC and the Cas9/sgRNA complex, thereby forming a strong but reversible interaction. To form the nanoparticle consisting of Cas9, sgRNA, NC-12, and PEI (Cas9/sgRNA/NC-12/PEI), Cas9 and the sgRNA were incubated together, followed by the addition of the NC-12, and then the addition of PEI. Measuring the zeta potential at each assembly step showed that the positively charged Cas9 (+19.3 ± 3.8 mV) became negatively charged with the addition of sgRNA (−19.4 ± 3.7 mV) and then NC-12 (−28.6 ± 5 mV), which was reverted to positive charge upon the addition of PEI (+18.6 ± 4.1 mV) (Figure 2a, S3). Dynamic light scattering analysis (Figure 2b, S4), atomic force microscopy (Figure 2c, S4) and transmission electron microscopy (Figure 2d) revealed that the Cas9/sgRNA/NC-12/PEI nanoparticles were uniformly sized with a hydrodynamic size of ~56 nm. Interestingly, the fully assembled particle was more compact and uniformly sized than the NC-12 nanoclew and the Cas9/sgRNA/NC-12 complex, potentially due to offsetting the dispersing charges. To assess the co-localization of each component, we applied confocal laser scanning microscopy (CLSM) to image nanoparticles comprised of Cas9 labeled with Alexa Fluor 647 (AF647), the sgRNA, the NC-12 stained with Hoechst 33342, and PEI conjugated with FITC. Imaging revealed consistent co-localization of all dyes (Figure S5), confirming the stable assembly of Cas9/sgRNA/NC-12/PEI.
Figure 2.

Particle characterization of Cas9/sgRNA/NC-12/PEI. (a) Monitoring zeta potential of the Cas9/sgRNA/NC-12/PEI assembly process. Bars represent mean ± SD (n = 3). (b) Hydrodynamic size distribution of Cas9/sgRNA/NC-12/PEI. (c) AFM image and d) TEM image of Cas9/sgRNA/NC-12/PEI with scale bars of 400 nm and 100 nm, respectively.
We further investigated the ability of the particles to deliver Cas9/sgRNA into cultured cells. As a model, we used an established U2OS cell line that constitutively expresses a destabilized form of EGFP (U2OS.EGFP).[6b] CLSM, a technique with depth selectivity for analyzing subcellular location of delivered drugs,[14, 19] was first applied to evaluate the localization of the Cas9/sgRNA/NC-12/PEI nanoparticles containing the AF647-labeled Cas9 (Figure 3a, S6). Over the course of six hours, the labeled Cas9 first binds to the cell surface, then enters the cytosol, and finally localizes to the nuclei as indicated by the colocalization of the red fluorescence signal from AF647-Cas9 with the blue fluorescent signal of stained nuclei. To elucidate the mechanism of internalization, we applied inhibitors of different endocytosis pathways[19b] and measured the relative uptake of the Cas9/sgRNA/NC-12/PEI nanoparticles containing AF647-labeled Cas9. Flow cytometry analysis revealed that the inhibitors methyl-β-cyclodextrin (MCD) and amiloride (AMI) imparted the greatest reduction in Cas9 uptake (Figure 3b), suggesting that the particles were mainly internalized through lipid rafts and macropinocytosis.[19b] Furthermore, we evaluated the impact of the nanoparticles on cell viability. TO-PRO-3 live/dead assay[7a] demonstrated no measurable impact on viability even at high concentrations (200 nM) of Cas9 (Figure 3c).
Figure 3.

a) CLSM images of U2OS.EGFP cells incubated with Cas9/sgRNA/NC-12/PEI for 1 h, 2 h, 4 h and 6 h (Cas9 and sgRNA concentrations at 100 nM). Green for EGFP, red for Cas9 stained with AF647 and blue for nuclei stained with Hoechst 33342. Scale bar is 10 µm. b) Relative Cas9/sgRNA/NC-12/PEI uptake by U2OS.EGFP cells in the presence of different endocytosis inhibitors (Cas9 and sgRNA concentrations at 100 nM). **P<0.01 as compared to the control group. Bars represent mean ± SD (n = 3). c) In vitro cell viability of U2OS.EGFP cells treated with Cas9/sgRNA/NC-12/PEI and Cas9/sgRNA/PEI by flow cytometry. The cells were stained with TO-PRO-3 live/dead stain after the treatment and analyzed by flow cytometry. Bars represent mean ± SD (n = 3).
Based on the evidence that the Cas9/sgRNA would reach cell nucleus, we next evaluated the extent to which Cas9/sgRNA could drive the formation of indels through targeted DNA cleavage and repair by the endogenous NHEJ pathway. By targeting the coding region of EGFP, most indels would shift the reading frame, thereby preventing proper EGFP expression. To evaluate the impact on EGFP expression, we incubated cells with the particles containing the EGFP-targeting sgRNA (Cas9/sgRNA/NC-12/PEI, Figure 4a) or the non-targeting cgRNA (Cas9/cgRNA/NC-12/PEI, Figure S7). Fluorescence microscopy and flow cytometry analysis revealed that the sgRNA reduced fluorescence in ~36% of the cells, whereas the cgRNA had a negligible effect in comparison to untreated cells. We also evaluated particles prepared with only Cas9, sgRNA, and PEI; these particles reduced fluorescence in only 5% of the cells, demonstrating the importance of the DNA NC for effective delivery. To assess whether the reduction in fluorescence was attributed to indel formation, we applied the SURVEYOR assay that quantifies the frequency of mutations within an amplified target region.[3] The assay revealed mutation frequencies of 28% and 1.5% for cells treated with Cas9/sgRNA/NC-12/PEI and Cas9/sgRNA/PEI (Figure 4b), respectively, closely paralleling the flow cytometry analysis. We also subcloned the amplified target region of cells incubated with the Cas9/sgRNA/NC-12/PEI nanoparticles. Sanger sequencing of 20 clones revealed 7 clones with typical indels within the PAM or the sequence complementary to the sgRNA guide (Figure S7), confirming the genetic disruption of EGFP expression by CRISPR-Cas9.[3] One-time treatment with the DNA NC mediated Cas9/sgRNA delivery system lead to higher editing efficacy than the cell-penetrating peptides (CPPs) based vector (9.7%) if the variation of cell line and targeted locus were not taken into account.[7b] Although the cationic lipid/anionic EGFP based delivery strategy showed higher editing efficacy (80%),[7a] lipid-vehicles are often hampered by serum instability, which could be alleviated by polymer-based carriers.[8, 20]
Figure 4.

Genome editing by Cas9/sgRNA delivered by DNA NC (8 µg/mL) coated with PEI (10 µg/mL). a) Fluorescent microscope images and flow cytometry analysis of U2OS.EGFP cells treated with Cas9/sgRNA/PEI and Cas9/sgRNA/NC-12/PEI (Cas9 and sgRNA concentrations at 100 nM). Green represents EGFP and blue represents nuclei stained with Hoechst 33342. Scale bar is 100 µm. b) T7EI assay of U2OS.EGFP cells treated with Cas9/gRNA/NC-12/PEI and Cas9/gRNA/PEI. c) EGFP disruption assay of Cas9/gRNA delivered by different DNA NCs. Percentages of EGFP negative cells after treating with Cas9/sgRNA/NC-23/PEI, Cas9/sgRNA/NC-12/PEI, Cas9/sgRNA/NC-0/PEI and Cas9/sgRNA/PEI at different Cas9/sgRNA molar ratios were profiled. Bars represent mean ± SD (n = 3).
Then we asked how complementarity between the DNA NC and the sgRNA impacted the efficacy of Cas9-driven genome editing. To address this, we generated two additional variants of the DNA NC with 0 or 23 nucleotides complementary to the sgRNA (designated as NC-0 and NC-23, respectively). Agarose gel electrophoresis confirmed that NC-0 and NC-23 yielded similar molecular weight distributions as NC-12 and were resistant to Cas9/sgRNA degradation (Figure S9). Subjecting the resulting particles to the U2OS.EGFP cells revealed that NC-12 yielded the highest fraction of EGFP negative cells (Figure 4c). This trend was upheld for different molar ratios of Cas9 and the sgRNA, where the 1:1 standard stoichiometry of the Cas9/sgRNA complex yielded the greatest activity. Altogether, these results suggest that partial complementarity between the sgRNA and the NC are important for efficient delivery, which may be attributed to the need for balancing Cas9/sgRNA loading and release.
We further evaluated the in vivo EGFP disruption potency of Cas9/sgRNA delivered by NC-12 using U2OS.EGFP tumor bearing mice as models. 10 days after intratumoral injection, ~25% the U2OS.EGFP cells in the frozen tumor sections near the site of injection lost EGFP expression in the Cas9/sgRNA/NC-12/PEI treated mice, while the tumors in the untreated group or the group treated with Cas9/cgRNA/NC-12/PEI did not show any loss of EGFP signal (Figure 5, S10).
Figure 5.
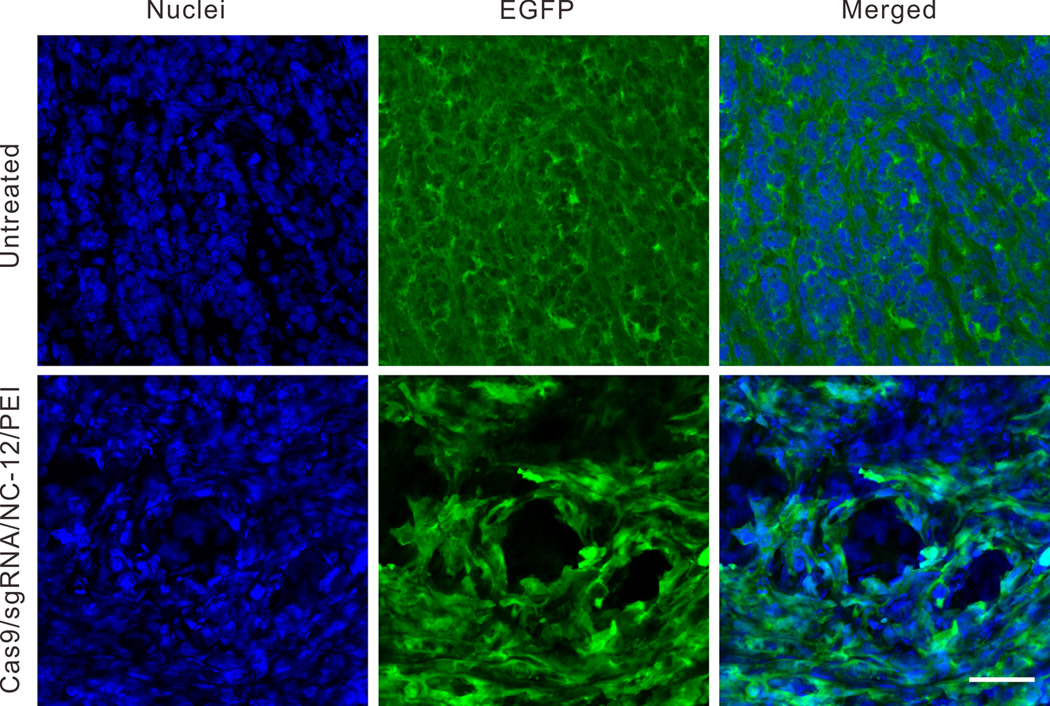
In vivo delivery of Cas9/sgRNA into U2OS.EGFP xenograft tumors in nude mice. Tumor sections were collected 10 days after intratumoral injection of Cas9/sgRNA/NC-12/PEI. The EGFP was stained by FITC conjugated GFP antibody and nuclei were stained with Hoechst 33342. Scale bar is 50 µm.
In summary, we have demonstrated a novel delivery vehicle to achieve targeted genome editing with CRISPR-Cas9. Our DNA NC-based delivery system represents, to our knowledge, the first example of a polymeric nanoparticle for the delivery of CRISPR-Cas9. The DNA NC pre-organized the Cas9/sgRNA into nanoparticles and increased the charge densities of the core in the core-shell assembly, which may have acted to stabilize the nanoparticle.[7a, 21] Partial complementarity between the DNA nanoclew and the sgRNA guide sequence promoted the greatest extent of gene editing, potentially due to balancing binding and release of the Cas9/sgRNA complex by the nanoclew. Future implementation of the delivery vehicles may focus on attaching cell-specific targeting ligands,[22] engineering the environmentally responsive release of the CRISPR-Cas9,[14, 23] modifying the sequence of DNA NC to incorporate multiple sgRNAs for multiplexed editing, or employing the DNA NC or packaged DNA sequences as templates for homology-directed repair. The same NC architecture could also be used to incorporate other functional DNA-binding proteins, such as transcription factors, zinc-finger nucleases, and TALE nucleases, as well as other functional or protein-coding RNAs. The potential immunogenicity associated with DNA NCs should be further investigated for clinical translation.[24]
Supplementary Material
Footnotes
This work was supported by the grants from NC TraCS, NIH’s Clinical and Translational Science Awards (CTSA, NIH grant 1UL1TR001111) at UNC-CH to Z.G. and from NSF (MCB-1452902) to C.L.B. We thank Dr. Elizabeth Loboa and Dr. Glenn Walker for providing experimental facilities. We acknowledge Dr. J Keith Joung from Massachusetts General Hospital for providing the U2OS.EGFP cell line. We acknowledge the use of the Analytical Instrumentation Facility (AIF) at NC State, which is supported by the State of North Carolina and NSF.
Contributor Information
Wujin Sun, Joint Department of Biomedical Engineering, University of North Carolina at Chapel Hill and North Carolina State University, Raleigh, NC 27695, USA, and Division of Molecular Pharmaceutics and Center for Nanotechnology in Drug Delivery, Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Wenyan Ji, Joint Department of Biomedical Engineering, University of North Carolina at Chapel Hill and North Carolina State University, Raleigh, NC 27695, USA, and Division of Molecular Pharmaceutics and Center for Nanotechnology in Drug Delivery, Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Jordan M. Hall, Department of Chemical and Biomolecular Engineering, North Carolina State University, Raleigh, NC 27695-7905, USA
Quanyin Hu, Joint Department of Biomedical Engineering, University of North Carolina at Chapel Hill and North Carolina State University, Raleigh, NC 27695, USA, and Division of Molecular Pharmaceutics and Center for Nanotechnology in Drug Delivery, Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Chao Wang, Joint Department of Biomedical Engineering, University of North Carolina at Chapel Hill and North Carolina State University, Raleigh, NC 27695, USA, and Division of Molecular Pharmaceutics and Center for Nanotechnology in Drug Delivery, Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Prof. Dr. Chase L. Beisel, Email: cbeisel@ncsu.edu, Department of Chemical and Biomolecular Engineering, North Carolina State University, Raleigh, NC 27695-7905, USA.
Prof. Dr. Zhen Gu, Email: zgu@email.unc.edu, Joint Department of Biomedical Engineering, University of North Carolina at Chapel Hill and North Carolina State University, Raleigh, NC 27695, USA, and Division of Molecular Pharmaceutics and Center for Nanotechnology in Drug Delivery, Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; Department of Medicine, University of North Carolina School of Medicine, Chapel Hill, NC 27599, USA.
References
- 1.Hsu Patrick D, Lander Eric S, Zhang F. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Platt Randall J, Chen S, Zhou Y, Yim Michael J, Swiech L, Kempton Hannah R, Dahlman James E, Parnas O, Eisenhaure Thomas M, Jovanovic M, Graham Daniel B, Jhunjhunwala S, Heidenreich M, Xavier Ramnik J, Langer R, Anderson Daniel G, Hacohen N, Regev A, Feng G, Sharp Phillip A, Zhang F. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotterman MA, Schaffer DV. Nat. Rev. Genet. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. Nat. Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen Z-Y, Liu DR. Nat. Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, Kim H. Genome Res. 2014 doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu Z, Biswas A, Zhao M, Tang Y. Chem. Soc. Rev. 2011;40:3638–3655. doi: 10.1039/c0cs00227e. [DOI] [PubMed] [Google Scholar]
- 9.Gerngross TU. Nat. Biotechnol. 2004;22:1409–1414. doi: 10.1038/nbt1028. [DOI] [PubMed] [Google Scholar]
- 10.Sliwkowski MX, Mellman I. Science. 2013;341:1192–1198. doi: 10.1126/science.1241145. [DOI] [PubMed] [Google Scholar]
- 11.Wade M, Li Y-C, Wahl GM. Nat. Rev. Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Nat. Rev. Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 13.a) Ali MM, Li F, Zhang Z, Zhang K, Kang D-K, Ankrum JA, Le XC, Zhao W. Chem. Soc. Rev. 2014;43:3324–3341. doi: 10.1039/c3cs60439j. [DOI] [PubMed] [Google Scholar]; b) Hu R, Zhang X, Zhao Z, Zhu G, Chen T, Fu T, Tan W. Angew. Chem. 2014;126:5931–5936. doi: 10.1002/anie.201400323. Angew. Chem. Int. Ed.2014, 53, 5821–5826. [DOI] [PubMed] [Google Scholar]; c) Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT. Nat. Mater. 2012;11:316–322. doi: 10.1038/nmat3253. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Lee HY, Jeong H, Jung IY, Jang B, Seo YC, Lee H, Lee H. Adv. Mater. 2015;27:3513–3517. doi: 10.1002/adma.201500414. [DOI] [PubMed] [Google Scholar]; e) Macfarlane RJ, Thaner RV, Brown KA, Zhang J, Lee B, Nguyen ST, Mirkin CA. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14995–15000. doi: 10.1073/pnas.1416489111. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Jones MR, Seeman NC, Mirkin CA. Science. 2015;347 doi: 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]
- 14.Sun W, Jiang T, Lu Y, Reiff M, Mo R, Gu Z. J. Am. Chem. Soc. 2014;136:14722–14725. doi: 10.1021/ja5088024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varkouhi AK, Scholte M, Storm G, Haisma HJ. J. Control. Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Doudna JA, Charpentier E. Science. 2014;346 doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 18.Fujii W, Kawasaki K, Sugiura K, Naito K. Nucleic Acids Res. 2013;41:e187. doi: 10.1093/nar/gkt772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Bolte S, CordeliÈRes FP. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]; b) Mo R, Jiang T, DiSanto R, Tai W, Gu Z. Nat. Commun. 2014;5:3364. doi: 10.1038/ncomms4364. [DOI] [PubMed] [Google Scholar]
- 20.Hsu CYM, Uludag H. Nat. Protocols. 2012;7:935–945. doi: 10.1038/nprot.2012.038. [DOI] [PubMed] [Google Scholar]
- 21.Hong CA, Eltoukhy AA, Lee H, Langer R, Anderson DG, Nam YS. Angew. Chem. 2015;127:6844–6848. doi: 10.1002/anie.201412493. Angew. Chem. Int. Ed.2015, 54, 6740–6744. [DOI] [PubMed] [Google Scholar]
- 22.a) Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nat. Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]; b) Chow EK-H, Ho D. Sci. Transl. Med. 2013;5:216rv214. doi: 10.1126/scitranslmed.3005872. [DOI] [PubMed] [Google Scholar]
- 23.Zetsche B, Volz SE, Zhang F. Nat. Biotechnol. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.a) Tian J, Avalos AM, Mao S-Y, Chen B, Senthil K, Wu H, Parroche P, Drabic S, Golenbock D, Sirois C, Hua J, An LL, Audoly L, La Rosa G, Bierhaus A, Naworth P, Marshak-Rothstein A, Crow MK, Fitzgerald KA, Latz E, Kiener PA, Coyle AJ. Nat Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]; b) Sun W, Gu Z. Biomater. Sci. 2015;3:1018–1024. doi: 10.1039/c4bm00459k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


