Abstract
In single particle reconstruction (SPR) from cryo-electron microscopy (EM), the 3D structure of a molecule needs to be determined from its 2D projection images taken at unknown viewing directions. Zvi Kam showed already in 1980 that the autocorrelation function of the 3D molecule over the rotation group SO(3) can be estimated from 2D projection images whose viewing directions are uniformly distributed over the sphere. The autocorrelation function determines the expansion coefficients of the 3D molecule in spherical harmonics up to an orthogonal matrix of size (2l + 1) × (2l + 1) for each l = 0,1,2,…. In this paper we show how techniques for solving the phase retrieval problem in X-ray crystallography can be modified for the cryo-EM setup for retrieving the missing orthogonal matrices. Specifically, we present two new approaches that we term Orthogonal Extension and Orthogonal Replacement, in which the main algorithmic components are the singular value decomposition and semidefinite programming. We demonstrate the utility of these approaches through numerical experiments on simulated data.
Index Terms: Cryo-electron microscopy, 3D reconstruction, single particle analysis, ab-initio modelling, autocorrelation, spherical harmonics, polar decomposition, semidefinite programming, convex relaxation
1. Introduction
SPR from cryo-EM is an increasingly popular technique in structural biology for determining 3D structures of macro-molecular complexes that resist crystallization [1, 2, 3]. In the basic setup of SPR, the data collected are 2D projection images of ideally assumed identical, but randomly oriented, copies of a macromolecule. In cryo-EM, the sample of molecules is rapidly frozen in a thin layer of vitreous ice, and maintained at liquid nitrogen temperature throughout the imaging process [4]. The electron microscope provides a top view of the molecules in the form of a large image called a micrograph. The projections of the individual particles can be picked out from the micrograph, resulting in a set of projection images. Datasets typically range from 104 to 105 projection images whose size is roughly 100 × 100 pixels.
Mathematically, ignoring the effects of the microscope's contrast transfer function and noise, a 2D projection image I : ℝ2 → ℝ corresponding to rotation R is given by the integral of the Coulomb potential ϕ : ℝ3 → ℝ that the molecule induces
| (1) |
where r = (x, y, z)T. The 3D reconstruction problem in cryo-EM is a non-linear inverse problem in which ϕ needs to be estimated from multiple noisy discretized projection images of the form (1) for which the rotations are unknown.
Radiation damage limits the maximum allowed electron dose. As a result, the acquired 2D projection images are extremely noisy with poor signal-to-noise ratio (SNR). Estimating ϕ and the unknown rotations at very low SNR is a major challenge.
The 3D reconstruction problem is typically solved by guessing an initial structure and then performing an iterative refinement procedure, where iterations alternate between estimating the rotations given a structure and estimating the structure given rotations [1, 5, 6]. When the particles are too small and images too noisy, the final result of the refinement process depends heavily on the choice of the initial model, which makes it crucial to have a good initial model. If the molecule is known to have a preferred orientation, then it is possible to find an ab-initio 3D structure using the random conical tilt method [7, 8]. There are two known approaches to ab initio estimation that do not involve tilting: the method of moments [9, 10], and common-lines based methods [11, 12, 13].
Using common-lines based approaches, [14] was able to obtain three-dimensional ab-initio reconstructions from real microscope images of large complexes that had undergone only rudimentary averaging. However, researchers have so far been unsuccessful in obtaining meaningful 3D ab-initio models directly from raw images that have not been averaged, especially for small complexes.
We present here two new approaches for ab-initio modelling that are based on Kam's theory [15] and that can be regarded as a generalization of the molecular replacement method from X-ray crystallography to cryo-EM. The only requirement for our methods to succeed is that the number of collected images is large enough for accurate estimation of the covariance matrix of the 2D projection images.
2. Kam's Theory and the Orthogonal Matrix Retrieval Problem
Kam showed [15] using the Fourier projection slice theorem (see, e.g., [16, p. 11]) that if the viewing directions of the projection images are uniformly distributed over the sphere, then the autocorrelation function of the 3D volume with itself over the rotation group SO(3) can be directly computed from the covariance matrix of the 2D images. Let ϕ̂ : ℝ3 → ℂ be the 3D Fourier transform of ϕ and consider its expansion in spherical coordinates
| (2) |
where k is the radial frequency and are the real spherical harmonics. Kam showed that
| (3) |
can be estimated from the covariance matrix of the 2D projection images. For images sampled on a Cartesian grid, each matrix Cl is of size K × K, where K is the maximum frequency (dictated by the experimental setting). In matrix notation, eq.(3) can be rewritten as
| (4) |
where Al is a matrix size K × (2l + 1) whose m'th column is Alm. The factorization (4) of Cl, also known as the Cholesky decomposition, is not unique: If Al satisfies (4), then Al U also satisfies (4) for any (2l + 1) × (2l + 1) unitary matrix U (i.e., UU* = U*U = I).
Since ϕ, the electric potential induced by the molecule, is real-valued, its Fourier transform ϕ̂ satisfies , or equivalently, . Together with properties of the real spherical harmonics, it follows that Alm(k) (and therefore Al) is real for even l and purely imaginary for odd l. Then Al is unique up to a (2l + 1) × (2l + 1) orthogonal matrix Ol ∈ O(2l + 1), where
| (5) |
Originally, 2l + 1 functions of the radial frequency are required for each l in order to completely characterize ϕ. With the additional knowledge of Cl the parameter space is reduced to O(2l + 1). We refer to the problem of recovering the missing orthogonal matrices O0, O1, O2,… as the orthogonal matrix retrieval problem in cryo-EM.
2.1. Analogy with X-ray crystallography
The orthogonal matrix retrieval problem is akin to the phase retrieval problem in X-ray crystallography. In crystallography, the measured diffraction patterns contain information about the modulus of the 3D Fourier transform of the structure but the phase information is missing and needs to be obtained by other means. Notice that in crystallography, the particle's orientations are known but the phases of the Fourier coefficient are missing, while in electron microscopy, the projection images contain phase information but the orientations of the particles are missing. Kam's theory converts the cryo-EM problem to one akin to the phase retrieval problem in crystallography. From a mathematical standpoint, the phase retrieval problem in crystallography is perhaps more challenging than the orthogonal matrix retrieval problem in cryo-EM, because in crystallography each Fourier coefficient is missing its phase, while in cryo-EM only a single orthogonal matrix is missing per several radial components.
3. Orthogonal Extension (OE)
A classical solution to the missing phase problem in crystallography is molecular replacement, which relies upon the existence of a previously solved structure which is similar to the unknown structure from which the diffraction data is obtained. The structure is then estimated using the Fourier magnitudes from the diffraction data with the phases from the homologous structure. We mimic this approach in cryo-EM, by grafting the orthogonal matrices of the already resolved similar structure onto the unknown structure.
Let ϕ be the unknown structure, and suppose ψ is a known homologous structure, whose 3D Fourier transform ψ̂ has the following expansion in spherical harmonics
| (6) |
We can obtain the auto-correlation matrices Cl from the cryo-EM images of the unknown structure ϕ using Kam's method. Let Fl be any matrix satisfying , determined from the Cholesky decomposition of Cl. Then
| (7) |
where Ol ∈ O(2l + 1). Requiring Al ≈ Bl, in orthogonal extension we determine Ol as the solution to the least squares problem
| (8) |
where ‖·‖F denotes the Frobenius norm.
Although the orthogonal group is non-convex, there is a closed form solution to (8) (see, e.g., [17]) given by
| (9) |
where
| (10) |
is the singular value decomposition (SVD) of . Thus, we estimate Al by
| (11) |
In analogy with crystallography, the phase information from the resolved homologous structure appends the experimentally measured intensity information (Fl). We note that other magnitude correction schemes have been used in crystallography. For example, setting the magnitude to be twice the magnitude from the desired structure minus the magnitude from the known structure, has the desired effect of properly weighting the difference between the two structures, but also the undesired effect of doubling the noise level. The cryo-EM analog in this case would be estimating Al by
| (12) |
4. Orthogonal Replacement (OR)
We move on to describe Orthogonal Replacement, our approach for resolving structures for which there does not exist a homologous structure. Suppose ϕ(1) and ϕ(2) are two unknown structures for which we have cryo-EM images. We assume that their difference Δϕ = ϕ(2) – ϕ(1) is known. This can happen, for example, when an antibody fragment of a known structure binds to a protein. We have two sets of cryo-EM images, one from the protein alone, ϕ(1) and another from the protein plus the antibody, ϕ(2). Let be the matrices computed from the sample covariance matrices of the 2D projection images of ϕ(i), (i = 1, 2). Let be any matrix satisfying . We have , where . The matrices need to be determined for i = 1, 2 and l = 0, 1, 2,…. The difference is known from the 3D Fourier transform of the binding structure Δϕ. We have
| (13) |
4.1. Relaxation to a Semidefinite Program
The least squares problem
| (14) |
is a non-convex optimization problem with no closed form solution. We find using convex relaxation in the form of semidefinite programming (SDP). We first homogenize (13) by introducing a slack unitary variable and consider the augmented linear system
| (15) |
If the triplet is a solution to (15), then the pair is a solution to the original linear system (13). The corresponding least squares problem
| (16) |
is still non-convex. But it can be relaxed to an SDP. Let Q ∈ ℝ3(2l+1) × 3(2l+1) be a symmetric matrix, which can be expressed as a 3 × 3 block matrix with block size 2l + 1, and the ij'th block is given by
| (17) |
It follows that Q is positive semidefinite (denoted Q ≥ 0). Moreover, the three diagonal blocks of Q are Qii = I (i = 1, 2, 3) and rank(Q) = 2l + 1. The cost function in (16) is quadratic in , so it is linear in Q. The problem can be equivalently rewritten as
| (18) |
over Q ∈ ℝ3(2l+1) × 3(2l+1), subject to Qii = I, rank(Q) = 2l + 1 and Q ≥ 0, where the matrix W can be written in terms of . Here, we have only one non-convex constraint – the rank constraint. Upon dropping the rank constraint we arrive at an SDP that can be solved efficiently in polynomial time in l. We extract the orthogonal matrices from the decomposition (17) of Q. If the solution matrix Q has rank greater than 2l + 1 (which is possible since we dropped the rank constraint), then we employ the rounding procedure of [18].
4.2. Exact Recovery and Resolution Limit
We have the following theoretical guarantee on recovery of using the SDP relaxation in the noiseless case:
Theorem 1. Assume that are elementwise sampled from i.i.d. Gaussian N(0, 1), and K > 2l + 1, then the SDP method recovers almost surely.
The proof of Theorem 1 is beyond the scope of this paper and is deferred to a separate publication. Theorem 1 shows that the SDP method almost achieves the theoretical information limit, since by counting the degrees of freedom in (13) it is impossible to recover if K < 2l. Indeed, the number of free parameters associated with an orthogonal matrix in O(2l + 1) is l(2l + 1), while the number of equations in (13) is K(2l + 1). This introduces a natural resolution limit on structures that can be resolved. Only angular frequencies for which can be determined using OR.
5. Numerical Experiments
We present the results of numerical experiments on simulated images (109 × 109 pixels) of the Kv1.2 potassium channel complex (Fig. 1 A and B) with clean and noisy projection images. The experiments were performed in MATLAB in UNIX environment on an Intel (R) Xeon(R) X7542 with 2 CPUs, having 6 cores each, running at 2.67 GHz, and with 256 GB RAM in total. To solve the SDP we used the MATLAB package CVX [19], and to compute the covariance matrix of the 2D images we used the steerable PCA procedure [20].
Fig. 1.
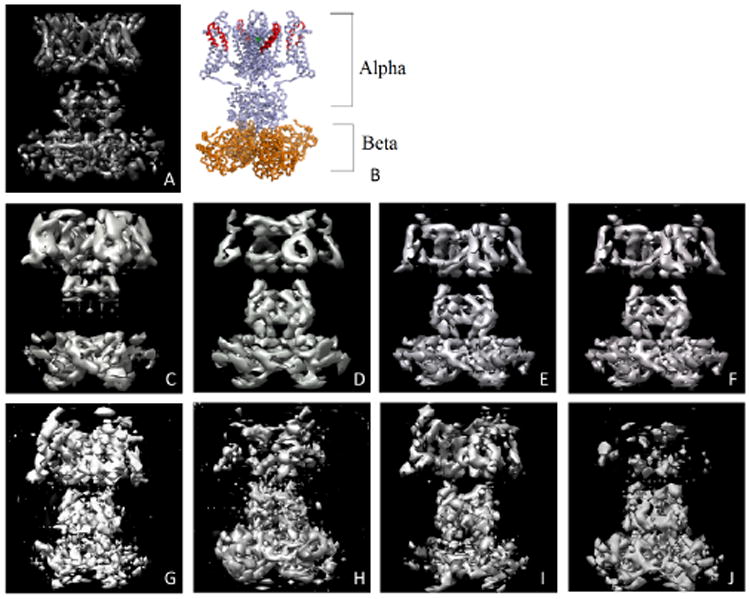
Kv1.2 potassium channel: A) Volume visualization in UCSF Chimera [21]. B) Image from Protein Data Bank Japan (PDBj). C through F show reconstructions from clean images - C) OE with α4 known, D) OE with β4 known, E) OR with α4 known, and F) OE with β4 known. G through J show reconstructions from noisy images using OR - G) SNR=0.7 with α4 known, H) SNR=0.7 with β4 known, I) SNR=0.35 with α4 known, and J) SNR=0.35 with β4 known.
Kv1.2 is a dumbbell-shaped particle consisting of two subunits - a small β4 subunit and a larger α4 subunit, connected by a central connector. We performed experiments using OE and OR, assuming one of the subunits (e.g., α4) is known, while the other is unknown. In the case of OR, we additionally used projection images of the unknown subunit.
5.1. Clean and Noisy Projections
We reconstruct the structure from both clean and noisy projection images. The reconstruction of Kv1.2 obtained from clean images using OE and OR is shown in Fig. 1 C through F. We used the true Cl matrices for the known subunit, and a maximum l of 30. We tested OR to reconstruct Kv1.2 from noisy projections at various values of SNR. A sample projection image at different values of SNR is shown in Fig. 2.
Fig. 2.
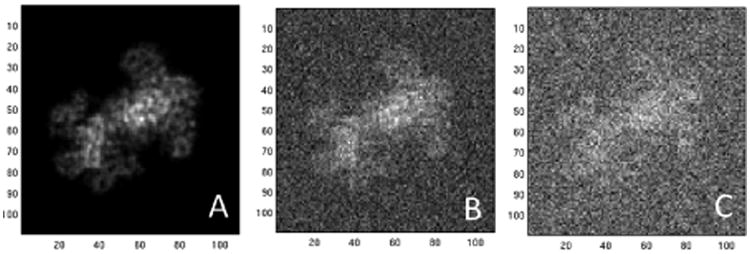
Projection images at different values of SNR: A) Clean image, B) SNR=0.7, and C) SNR=0.35.
The Cl matrices were estimated from the noisy projection images. In Fig. 1 G through J we show the reconstructions obtained from 10000 projections using OR at SNR=0.7, and from 40000 projections using OR at SNR=0.35. In our simulations with 10000 images, it takes 416 seconds to perform steerable PCA, 194 seconds to calculate the Cl matrices using the maximum l as 30, and the time to solve the SDP as a function of l ranges from 5 seconds for l = 5 to 194 seconds for l = 30.
5.2. Comparison between OE and OR
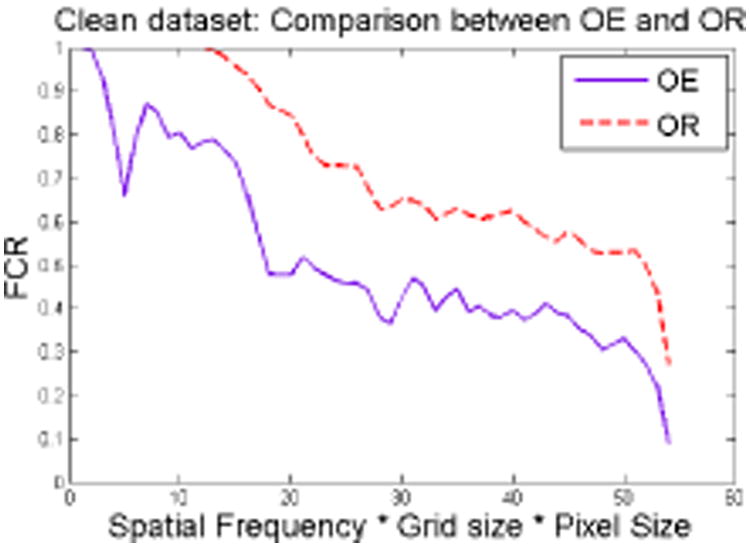
We quantify the ‘goodness’ of the reconstruction using the Fourier Cross Resolution (FCR) [22]. In Fig. 3 we show the FCR curves for the reconstruction from the β4 complex using OE and OR. The additional information in OR, from the projection images of α4, results in a better reconstruction, as seen from the FCR curve. The Kv1.2 complex has C4 symmetry, which reduces the rank of the Cl matrices. Our experiment thus benefits from the reduced size of the orthogonal matrices to be recovered.
Fig. 3. FCR curve for reconstruction from β4 (clean images).
6. Summary
We presented two new approaches based on Kam's theory for ab-initio modelling of macromolecules for SPR from cryo-EM. Ab-initio modelling of small complexes is a challenging problem in cryo-EM because it is difficult to detect common lines between noisy projection images at low SNR. Our methods only require reliable estimation of the covariance matrix of the projection images which can be met even at low SNR if the number of images is sufficiently large. In future work we plan to estimate the covariance matrix when images are not centered, and to apply our methods to experimental datasets.
Acknowledgments
This research was partially supported by Award Number R01GM090200 from the NIGMS and Award Number N66001-13-1-4052 from DARPA.
References
- 1.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies : Visualization of Biological Molecules in Their Native State: Visualization of Biological Molecules in Their Native State. Oxford University Press; USA: 2006. [Google Scholar]
- 2.Kühlbrandt W. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- 3.Bai X, McMullan G, Scheres SHW. How cryoem is revolutionizing structural biology. Trends in Biochemical Sciences. doi: 10.1016/j.tibs.2014.10.005. in press. [DOI] [PubMed] [Google Scholar]
- 4.Wang L, Sigworth FJ. Cryo-EM and single particles. Physiology (Bethesda) 2006;21:13–18. doi: 10.1152/physiol.00045.2005. [DOI] [PubMed] [Google Scholar]
- 5.van Heel M, Gowen B, Matadeen R, Orlova EV, Finn R, Pape T, Cohen D, Stark H, Schmidt R, Patwardhan A. Single particle electron cryo-microscopy: Towards atomic resolution. Q Rev Biophys. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- 6.Penczek P, Renka R, Schomberg H. Gridding-based direct Fourier inversion of the three-dimensional ray transform. J Opt Soc Am A. 2004;21:499–509. doi: 10.1364/josaa.21.000499. [DOI] [PubMed] [Google Scholar]
- 7.Verschoor A, Radermacher M, Wagenknecht T, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50s ribosomal subunit of escherichia coli. Journal of Microscopy. 1987;146(2):113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 8.Verschoor A, Radermacher M, Wagenknecht T, Frank J. Three-dimensional structure of the large ribosomal subunit from Escherichia coli. EMBO J. 1987;6(4):1107–14. doi: 10.1002/j.1460-2075.1987.tb04865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goncharov AB. Integral geometry and three-dimensional reconstruction of randomly oriented identical particles from their electron microphotos. Acta Applicandae Mathematicae. 1988;11:199–211. [Google Scholar]
- 10.Salzman DB. A method of general moments for orienting 2D projections of unknown 3D objects. Comput Vision Graph Image Process. 1990 May;50(2):129–156. [Google Scholar]
- 11.Vainstein BK, Goncharov AB. Dokl Acad Sci USSR. 5. Vol. 287. Soviet Physics Doklady; 1986. Determining the spatial orientation of arbitrarily arranged particles given their projections; pp. 1131–1134. English translation. Vol. 31, p.278. [Google Scholar]
- 12.Van Heel M. Angular reconstitution: a posteriori assignment of projection directions for 3d reconstruction. Ultramicroscopy. 1987;21(2):111–123. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 13.Singer A, Shkolnisky Y. Three-dimensional structure determination from common lines in cryo-EM by eigenvectors and semidefinite programming. SIAM Journal on Imaging Sciences. 2011;4:543–572. doi: 10.1137/090767777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Z, Singer A. Rotationally invariant image representation for viewing direction classification in cryo-EM. Journal of Structural Biology. 2014;186(1):153–166. doi: 10.1016/j.jsb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kam Z. The reconstruction of structure from electron micrographs of randomly oriented particles. Journal of Theoretical Biology. 1980;82(1):15–39. doi: 10.1016/0022-5193(80)90088-0. [DOI] [PubMed] [Google Scholar]
- 16.Natterer F. The Mathematics of Computerized Tomography. Classics in Applied Mathematics. SIAM: Society for Industrial and Applied Mathematics; 2001. [Google Scholar]
- 17.Keller JB. Closest unitary, orthogonal and Hermitian operators to a given operator. Mathematics Magazine. 1975;48(4):192–197. [Google Scholar]
- 18.Bandeira AS, Singer A, Spielman DA. A Cheeger inequality for the graph connection Laplacian. SIAM Journal on Matrix Analysis and Applications. 2013;34(4):1611–1630. [Google Scholar]
- 19.Grant M, Boyd S. CVX: Matlab software for disciplined convex programming, version 2.1. 2014 Mar; http://cvxr.com/cvx.
- 20.Zhao Z, Singer A. Fourier Bessel rotational invariant eigenimages. J Opt Soc Am A. 2013 May;30(5):871–877. doi: 10.1364/JOSAA.30.000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 22.Penczek PA. Chapter three - resolution measures in molecular electron microscopy. In: Jensen Grant J., editor. Methods in Enzymology. Vol. 482. Academic Press; 2010. pp. 73–100. Cryo-EM, Part B: 3-D Reconstruction. [DOI] [PMC free article] [PubMed] [Google Scholar]


