Several mammalian formins participate in filopodial assembly, but the identity of the actin nucleation factor is not clear. Nucleation factor requirements differ between nonadherent and adherent cells. Arp2/3 complex is essential in suspension cells, whereas the situation is more complex in adherent cells.
Abstract
Filopodia are finger-like protrusions from the plasma membrane and are of fundamental importance to cellular physiology, but the mechanisms governing their assembly are still in question. One model, called convergent elongation, proposes that filopodia arise from Arp2/3 complex–nucleated dendritic actin networks, with factors such as formins elongating these filaments into filopodia. We test this model using constitutively active constructs of two formins, FMNL3 and mDia2. Surprisingly, filopodial assembly requirements differ between suspension and adherent cells. In suspension cells, Arp2/3 complex is required for filopodial assembly through either formin. In contrast, a subset of filopodia remains after Arp2/3 complex inhibition in adherent cells. In adherent cells only, mDia1 and VASP also contribute to filopodial assembly, and filopodia are disproportionately associated with focal adhesions. We propose an extension of the existing models for filopodial assembly in which any cluster of actin filament barbed ends in proximity to the plasma membrane, either Arp2/3 complex dependent or independent, can initiate filopodial assembly by specific formins.
INTRODUCTION
Two abundant actin-based structures in metazoan cells are lamellipodia and filopodia (Blanchoin et al., 2014; Skau and Waterman, 2015). Both structures consist of multiple actin filaments oriented with their fast-growing “barbed” ends toward the plasma membrane but are morphologically distinct. Lamellipodia are sheet-like protrusions containing short, dendritically branched actin filaments. In contrast, filopodia are finger-like protrusions (<200 nm diameter) containing longer, unbranched filaments. In both structures, it is the addition of actin monomers to barbed ends that leads to membrane protrusion.
In keeping with their differing morphologies, the assembly mechanisms for lamellipodia and filopodia also differ. Lamellipodial assembly is well characterized, with actin-related protein 2/3 (Arp2/3) complex being responsible for nucleating and cross-linking the dendritic network and barbed end–capping proteins terminating filament elongation (Blanchoin et al., 2014). In contrast, the details of filopodial assembly remain unclear, with two predominant models: convergent elongation and tip nucleation (Faix et al., 2009; Yang et al., 2009). The convergent elongation model proposes that filopodia arise directly from the lamellipodial dendritic network, dependent on Arp2/3 complex activity. A subset of these filaments is then reorganized by formins and/or Enabled/vasodilator-stimulated phosphoprotein (ENA/VASP) proteins through their abilities to block capping proteins and allow sustained elongation (Svitkina et al., 2003; Yang et al., 2007). Actin-bundling proteins, including fascin and possibly formins and/or ENA/VASP proteins (Harris and Higgs, 2006; Harris et al., 2010; Schirenbeck et al., 2006; Vignjevic et al., 2006; Block et al., 2008), further remodel elongating filaments into parallel bundles characteristic of filopodia.
The convergent elongation model has been supported by both in vitro studies (Vignjevic et al., 2003; Haviv et al., 2006) and cellular studies (Svitkina et al., 2003). However, there is also evidence for Arp2/3 complex–independent mechanisms of filopodial assembly (Steffen et al., 2006; Suraneni et al., 2012; Wu et al., 2012), prompting the tip nucleation model (Schirenbeck et al., 2006; Block et al., 2008; Faix et al., 2009; Yang and Svitkina, 2011). In this model, formin proteins serve as both actin nucleation and elongation factors to assemble filopodial actin filaments.
Biochemically, formin proteins have the ability to accelerate both actin nucleation and elongation, as well as to compete with capping proteins (Chesarone et al., 2010). Thus they are attractive candidates for filopodia assembly by either convergent elongation or tip nucleation. Indeed, a number of cellular studies suggest that formins participate in filopodial assembly, notably mammalian Diaphanous (mDia)1 (Sarmiento et al., 2008; Goh et al., 2011; Naj et al., 2013), mDia2 (Pellegrin and Mellor, 2005; Yang et al., 2007; Block et al., 2008; Harris et al., 2010), disheveled-associated activator of morphogenesis-1 (DAAM1; Matusek et al., 2008; Jaiswal et al., 2013), formin-like 2 (FMNL2; Block et al., 2012), and FMNL3 (Harris et al., 2010; Gauvin et al., 2015). Of interest, most of these formins (mDia2, DAAM1, FMNL2, and FMNL3) also have the ability to bundle actin filaments (Harris et al., 2006; Vaillant et al., 2008; Heimsath and Higgs, 2012; Jaiswal et al., 2013). However, it is unclear whether these formins serve as the nucleator in a tip nucleation–type model or use filaments nucleated by Arp2/3 complex.
Ena/VASP proteins have somewhat similar effects on actin as formins: elongation regulation, competitive inhibition of barbed end capping, and filament bundling (Bachmann et al., 1999; Bear et al., 2002; Barzik et al., 2005; Winkelman et al., 2014). Ena/VASP proteins have also been linked to filopodia assembly (Svitkina et al., 2003; Lebrand et al., 2004; Applewhite et al., 2007) but are not always present in filopodial tips (Yang et al., 2007; Barzik et al., 2014) and are also enriched at other sites, such as focal adhesions, stress fibers, and the lamellipodial leading edge (Reinhard et al., 1992, 1995; Bear et al., 2002; Gateva et al., 2014). It is unclear how formins and Ena/VASP work together in filopodia assembly (Yang et al., 2007; Homem and Peifer, 2009; Barzik et al., 2014; Bilancia et al., 2014).
In this study, we use a simple cellular system to examine fundamental features of formin-mediated filopodial assembly, using constitutively active constructs of the formins mDia2 and FMNL3. These data suggest that any cluster of actin filament barbed ends in proximity to the plasma membrane can serve as the starting material for formin-mediated filopodial assembly.
RESULTS
Filopodial assembly requires Arp2/3 complex in suspension cells
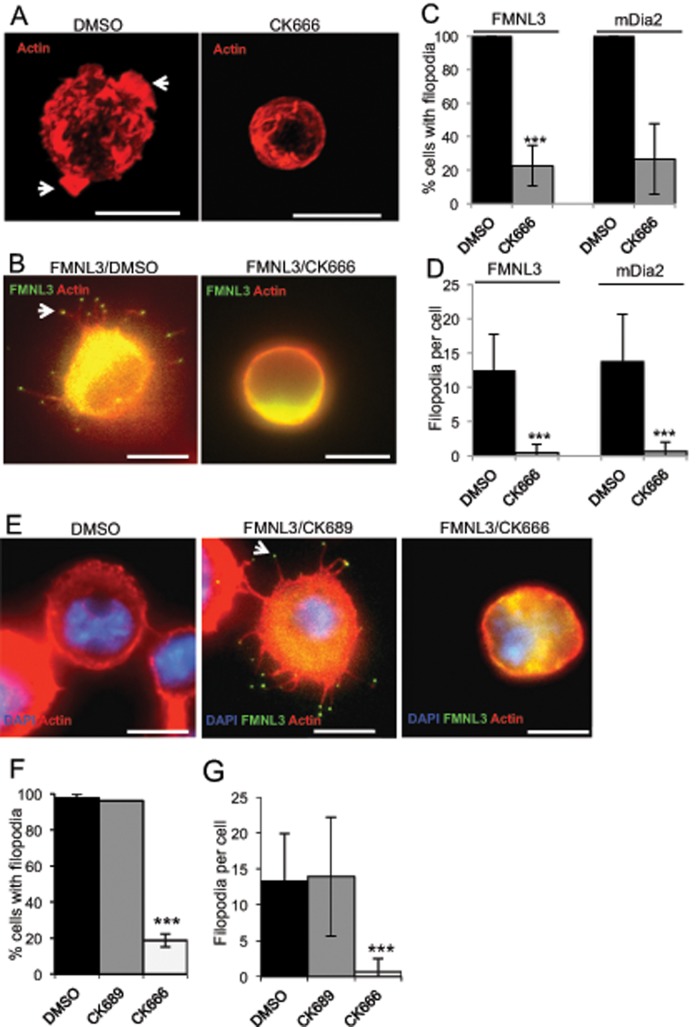
We used our previously developed Jurkat human T-cell system (Harris et al., 2010) to test whether Arp2/3 complex activity is necessary for filopodial assembly by constitutively active formins. Jurkat cells grow in suspension and display ruffles as their predominant cell surface feature (Majstoravich et al., 2004; Figure 1A). As we previously observed, transfection with a green fluorescent protein (GFP)–tagged construct containing the formin homology 1 (FH1) and FH2 domains of FMNL3 (GFP-FMNL3-FF) or one containing the FH1, FH2, and C-terminal domains of mDia2 (GFP-mDia2-FFC; diagram in Supplemental Figure S1A) causes surface conversion from ruffles to filopodia, with GFP enriched at filopodial tips (Figure 1B and Supplemental Figure S1B). GFP-fusion expression level does not influence filopodial number significantly within the transfection range analyzed (Supplemental Figure S1C). The small molecule CK666 effectively inhibits Arp2/3 complex in these cells, as judged by inhibition of ruffles in untransfected cells (Figure 1A). In formin-transfected cells, CK666 has a dramatic effect on filopodia, reducing the percentage of cells displaying filopodia by 77 and 73% and the number of filopodia per cell by 96 and 95% for FMNL3 and mDia2, respectively (Figure 1, C and D).
FIGURE 1:
Arp2/3 complex inhibition effectively ablates formin-mediated filopodia in suspension cells. (A) Representative micrographs of untransfected Jurkat T-cells treated in suspension for 2 h with DMSO (left) or 200 μM Arp2/3 complex inhibitor CK666 (right) and stained with TRITC-phalloidin (red). Cells were fixed in suspension before adhesion to PLL-coated coverslips. Arrowheads represent actin-rich membrane ruffles in DMSO control. Scale bar, 10 μm. (B) Representative micrographs of GFP-FMNL3-FF–transfected Jurkat T-cells treated in suspension for 2 h with DMSO (left) or 200 μM CK666 (right). Similar micrographs for mDia2-FFC are shown in Supplemental Figure S1B. Cells were adhered to PLL-coated coverslips for 10 min before fixation. Actin is stained with TRITC-phalloidin (red), and GFP-FMNL3-FF is green. Arrowhead indicates FMNL3 enriched at filopodial tip. Filopodia number in examples: DMSO, 13; and CK666, 0. Scale bar, 10 μm. (C) Quantification of percentage of transfected cells displaying filopodia after indicated treatment. (D) Quantification of filopodia per transfected cell after indicated treatment. Results are pooled from three to five independent experiments, 55–80 cells (FMNL3) and 34–68 cells (mDia2). (E) Representative micrographs of 300.19 cells untransfected and treated with DMSO showing no filopodia (left) and transfected with GFP-FMNL3-FF and treated with 200 μM inactive CK689 control (middle) or 200 μM Arp2/3 complex inhibitor CK666 (right). All cells were treated in suspension for 2 h, followed by 10 min of adhesion onto PLL-coated coverslips before paraformaldehyde fixation. Actin is stained with TRITC-phalloidin (red) and DNA with DAPI (blue), and GFP-FMNL3-FF is green. Arrowhead indicates FMNL3 enriched at the filopodial tip. Filopodia number in examples: CK689, 15; and CK666, 0. Scale bar, 5 μm. (F) Quantification of percentage of transfected cells displaying filopodia after the indicated treatment. (G) Quantification of filopodia number per transfected cell treated as indicated. Results are pooled from three to five independent experiments (except for CK689, one experiment); 51–133 cells. Error bars, SD. ***p < 0.001.
CK666 has a similar effect on GFP-FMNL3-FF–induced filopodia in a second suspension cell line, 300.19 murine pre–B lymphoma cells, whereas an inactive analogue, CK689, has no effect (Figure 1E-G). We also confirmed these results using scanning electron microscopy (SEM; Supplemental Figure S2). Taken together, these data show that Arp2/3 complex is required for FMNL3 and mDia2-mediated filopodia in suspension cells, suggesting use of the convergent elongation model for their assembly.
A subset of filopodia persists upon Arp2/3 complex inhibition in adherent cells
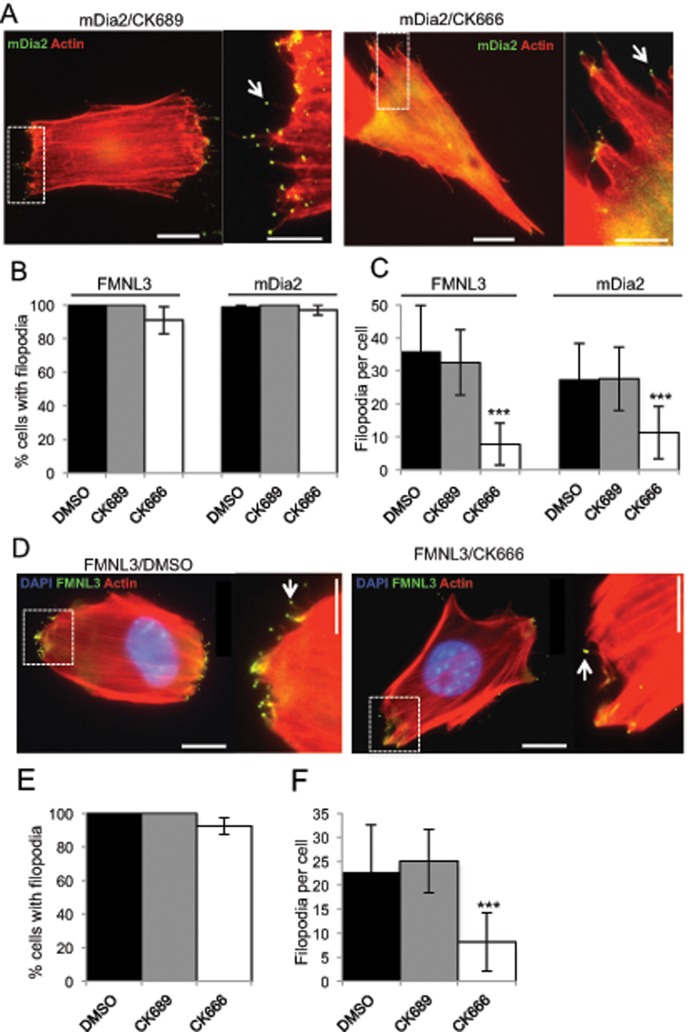
We also tested the effect of CK666 on filopodial assembly in adherent cells, initially using U2OS human osteosarcoma cells. As in suspension cells, transfection with either GFP-FMNL3-FF or GFP-mDia2-FFC induces filopodia with GFP enriched at their tips (Harris et al., 2010; Figure 2A). These structures are truly protrusive filopodia and not retraction fibers, as judged by live-cell analysis (Supplemental Figure S3C and Supplemental Movie S1). Although there is significant variability in the dynamics of these filopodia both within a single cell and between cells, the average filopodial lifetime is 264 ± 153 s (Supplemental Figure S3B), with an average maximal length of 1.9 ± 0.8 mm (Supplemental Figure S3E).
FIGURE 2:
Arp2/3 complex inhibition reduces formin-mediated filopodia in adherent cells. (A) Representative micrographs of U2OS cells transfected with GFP-mDia2-FFC and treated for 2 h with 200 μM CK689 (left) or 200 μM CK666 (right). Actin is stained with TRITC-phalloidin (red), and GFP-mDia2-FFC is green. Arrowheads indicate mDia2 enriched at filopodial tips. Filopodia number in insets: GFP/DMSO, 0; mDia2/CK689, 10; and mDia2/CK666, 5. Examples of untreated U2OS cells transfected with GFP alone in Supplemental Figure S3. (B) Quantification of percentage of transfected cells displaying filopodia after the indicated treatment. (C) Quantification of filopodia number on transfected cells treated as indicated. Results are pooled from three or four independent experiments, 24–37 cells (FMNL3) and 92–141 cells (mDia2). (D) Representative micrographs of NIH 3T3 cells transfected with GFP-FMNL3-FF and then treated for 2 h with DMSO (left) or 200 μM CK666 (right). Actin is stained with TRITC-phalloidin (red) and DNA with DAPI (blue), and GFP-FMNL3-FF is green. Arrowheads indicate FMNL3 enriched at filopodial tips. Filopodia numbers in insets: DMSO, 9; and CK666, 2. Scale bar, 10 μm (main image) and 5 μm (insets). (E) Quantification of percentage of transfected cells displaying filopodia after indicated treatment. (F) Quantification of filopodia number on each transfected cell after the indicated treatment. Results are pooled from three to five independent experiments; 48–118 cells. Scale bars: 10 μm (main image) and 5 μm (insets). Error bars, SD. ***p < 0.001.
In contrast to the results in suspension cells, CK666 only slightly reduces the percentage of cells possessing at least one filopodium (10% reduction for FMNL3 and 3% for mDia2; Figure 2B). However, the number of filopodia per cell is significantly reduced by CK666 treatment, with 76% reduction for FMNL3 and 58% reduction for mDia2 (Figure 2C). We also tested CK666 effects in a second adherent cell line, NIH 3T3 mouse fibroblasts. As in U2OS cells, CK666 does not cause a large reduction in the percentage of FMNL3-FF–transfected 3T3 cells displaying filopodia (8%), yet the number of filopodia per cell is significantly reduced (64%; Figure 2, D–F).
One possibility is that the filopodia remaining after CK666 treatment represent a stable subtype that does not turn over during the treatment period. To test this possibility, we examined the effect of CK666 on live cells, comparing dynamics before and after treatment. CK666 treatment decreases filopodial assembly rate by 75% (Supplemental Figure S3, D and F, and Supplemental Movie S2). The filopodia that do assemble in CK666-treated cells have shorter lifetimes than in the control condition (199 ± 105 s; Supplemental Figure S3B), whereas their average maximal length is similar (1.9 ± 0.7 mm; Supplemental Figure S3E). These results suggest that the filopodia that remain after Arp2/3 complex inhibition do not represent a stable population.
One possible explanation for the difference between suspension cells and adherent cells is that CK666 is only partially effective on adherent cells. To test this possibility, we used small interfering RNA (siRNA) to suppress the Arp2 subunit in U2OS cells (∼80% depletion; Supplemental Figure 4A), followed by CK666 treatment. Our rationale was that the combination siRNA/CK666 treatment should cause a larger reduction in filopodial number if CK666 alone only partially inhibits Arp2/3 complex. However, combined siRNA/CK666 treatment does not cause a further reduction in filopodia number (Figure 3A) or the percentage of cells displaying filopodia (Figure 3B) over CK666 alone, suggesting that CK666 inhibits the majority of active Arp2/3 complex. The situation is similar for both FMNL3 and mDia2. Micrographs representative of some of these conditions are shown in Supplemental Figure S4B.
FIGURE 3:
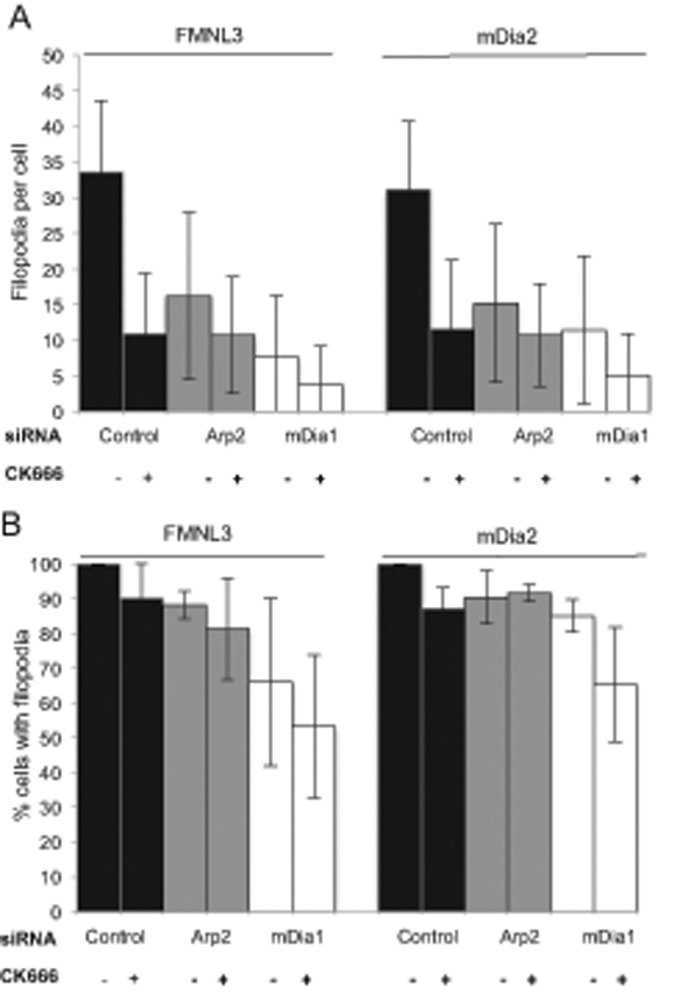
mDia1 depletion reduces formin-mediated filopodia in adherent cells. U2OS cells were transfected for 72 h with the indicated siRNA (control, mDia1 targeting, or Arp2 targeting). At 48 h after siRNA transfection, cells were transfected with either GFP-FMNL3-FF or GFP-mDia2-FFC and treated for 2 h with DMSO or 200 μM CK666. (A) Quantification of filopodia number per transfected cell after the indicated treatment. Results with CK666 differ from DMSO control with p < 0.001 for all but the Arp2 siRNA, mDia2-transfected results (p = 0.003). (B) Percentage of GFP-FMNL3-FF or GFP-mDia2-FFC transfected cells displaying filopodia after the indicated treatment. Results with CK666 differ from DMSO control with p > 0.1 for all conditions. Results are pooled from three of four independent experiments; 88–124 cells (FMNL3) and 79–127 cells (mDia2). Error bar, SD.
Roles for mDia1 and VASP in filopodial assembly in adherent cells
Our inability to abolish filopodia by Arp2/3 complex inhibition in adherent cells prompted us to ask whether other nucleation factors may play a role. We previously found that constitutively active constructs of the formin mDia1 did not induce filopodia (Harris et al., 2010). However, other studies have shown a role for mDia1 in filopodia assembly (Sarmiento et al., 2008; Goh et al., 2011; Naj et al., 2013), in addition to its roles in stress fiber and focal adhesion assembly (Watanabe et al., 1999; Hotulainen and Lappalainen, 2006; Zaoui et al., 2008; Fan et al., 2010; Rao et al., 2013; Burnette et al., 2014) in a variety of cell types. Given that a difference between adherent and suspension cells is the presence of stress fibers and focal adhesions, we postulated that mDia1 might contribute to filopodia assembly only in adherent cells.
We tested mDia1’s role by siRNA depletion in U2OS cells (∼87% mDia1 depletion; Supplemental Figure S5A). Depletion of mDia1 reduces filopodia number by a comparable amount to Arp2/3 complex inhibition (78% reduction for FMNL3-FF and 64% reduction for mDia2-FFC; Figure 3A), and reduces the percentage of cells possessing filopodia to a greater degree than Arp2/3 complex inhibition (34 and 15% decrease for FMNL3 and mDia2, respectively; Figure 3B). When combining mDia1 depletion with Arp2/3 inhibition (through CK666 treatment), both the number of filopodia per cell and the percentage of cells displaying filopodia are reduced further than with either treatment alone (Figure 3). However, a small population of filopodia persists under combined mDia1 and Arp2/3 complex inhibition. Micrographs representative of some of these conditions are shown in Supplemental Figure S5B. Collectively these data suggest that both mDia1 and Arp2/3 complex play roles in the majority of filopodial assembly in adherent cells.
We also examined the role of mDia1 in filopodia assembly in suspension cells. Of interest, mDia1 depletion does not reduce the percentage of cells with filopodia (100% of cells have filopodia; unpublished data), or the number of filopodia per cell (Figure 4B). However, mDia1 depletion does cause a significant increase in the number of “forked” filopodia (filopodia that originate from a common origin; Figure 4, A and C). These results suggest that the role of mDia1 in filopodial assembly is different between adherent and suspension cells.
FIGURE 4:
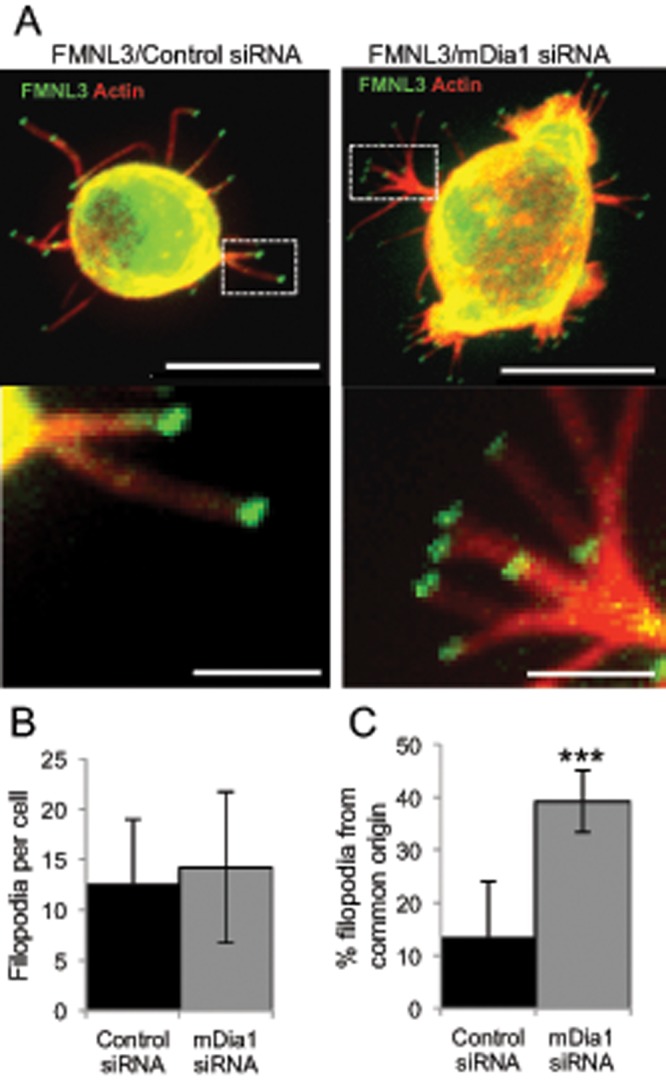
mDia1 depletion does not reduce FMNL3-mediated filopodia in Jurkat T-cells. (A) Representative images of Jurkat T-cells transfected for 72 h with control (left) or mDia1-targeting siRNA (right), followed by transfection with GFP-FMNL3-FF for the final 24 h. Cells were adhered to PLL-coated coverslips for 10 min before paraformaldehyde fixation. Actin is stained with TRITC-phalloidin (red), and FMNL3 is green. Insets provide examples of filopodia originating from a common base. Images are maximum intensity projections from 25 × 0.2–μm Z-slices. Scale bar, 10 μm (main image) and 2 μm (insets). (B) Quantification of filopodia per transfected cell after the indicated treatment. Results are pooled from four independent experiments; 104–118 cells. Error bar, SD. p = 0.09. (C) Quantification of percentage of filopodia that originate from a common base after the indicated treatment. Results are pooled from four independent experiments; 1317–1688 filopodia. Error bar, SD. ***p < 0.001.
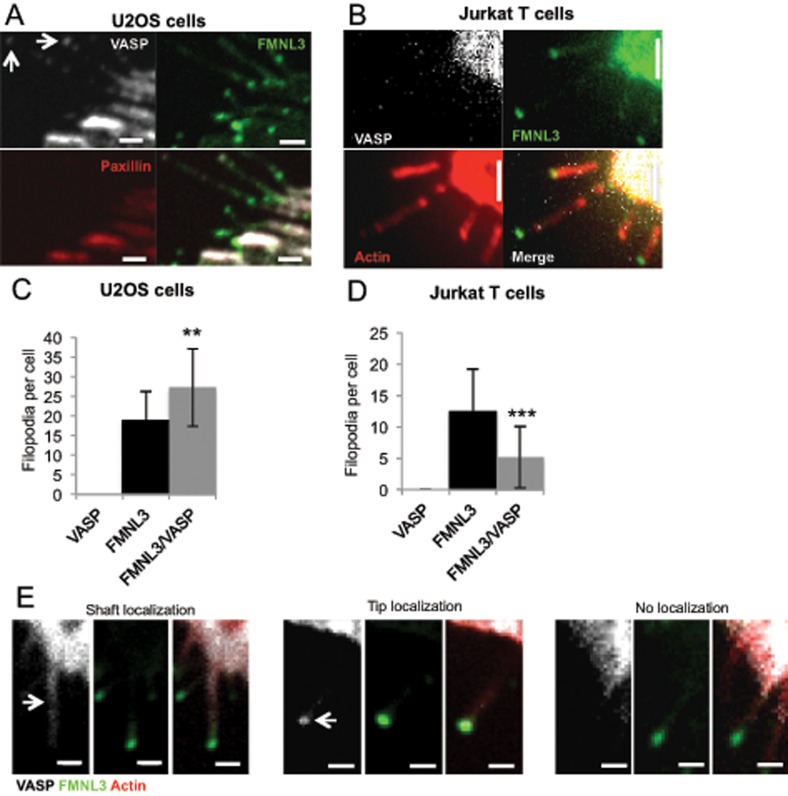
VASP is another actin assembly factor that has been linked to filopodia in many studies (Svitkina et al., 2003; Lebrand et al., 2004; Applewhite et al., 2007). We therefore examined the role of VASP in our system. In U2OS cells, endogenous VASP enriches at focal adhesions, but only enriches at the tips of a subset of filopodial tips, which appear to contain less GFP-FMNL3 (Figure 5A). In Jurkat cells, we find no specific localization of endogenous VASP in the cell body or in FMNL3-assembled filopodia (Figure 5B). Of interest, overexpression of mApple-VASP has opposite effects in suspension and adherent cells, decreasing the number of GFP-FMNL3-FF–containing filopodia in Jurkat cells by 44% but increasing these filopodia in U2OS cells by 59% (Figure 5, C and D). Despite this increase in U2OS filopodia, mApple-VASP is rarely enriched in filopodial tips (2.6%) or shafts (21.1%; n = 76 filopodia), whereas GFP-FMNL3-FF is present at 100% of filopodial tips (Figure 5E). These results suggest that VASP’s role in formin-mediated filopodial assembly differs between suspension and adherent cells and that its primary role might not be at filopodial tips.
FIGURE 5:
Effect of VASP on formin-mediated filopodial assembly differs between suspension and adherent cells. (A, B) Endogenous VASP (white) staining by immunofluorescence microscopy of U2OS cells (A) or Jurkat cells (B) transfected with GFP-FMNL3-FF (green). Red for U2OS cells is endogenous paxillin and for Jurkat cells indicates actin filaments (TRITC-phalloidin). Arrows indicate two filopodia with some degree of VASP at tips. Scale bar, 2 μm. (C) Quantification of filopodia number per transfected U2OS cell after transfection with mApple-VASP, GFP-FMNL3-FF, or mApple-VASP and GFP-FMNL3-FF; 20–25 cells. Error bar, SD. **p < 0.01. (D) Quantification of filopodia number per transfected Jurkat cell after transfection with mApple-VASP, GFP-FMNL3-FF, or mApple-VASP and GFP-FMNL3-FF; 30–87 cells. Error bar, SD. ***p < 0.001. (E) Fluorescence micrographs of mApple-VASP (white), GFP-FMNL3-FF (green), and BFP-Lifeact (red) in U2OS cells, showing examples of VASP enrichment in filopodial shaft (left), VASP enrichment in filopodial tip (middle), or no VASP enrichment in filopodia (right). Scale bar, 1 μm.
A population of FMNL3-mediated filopodia associates with focal adhesions
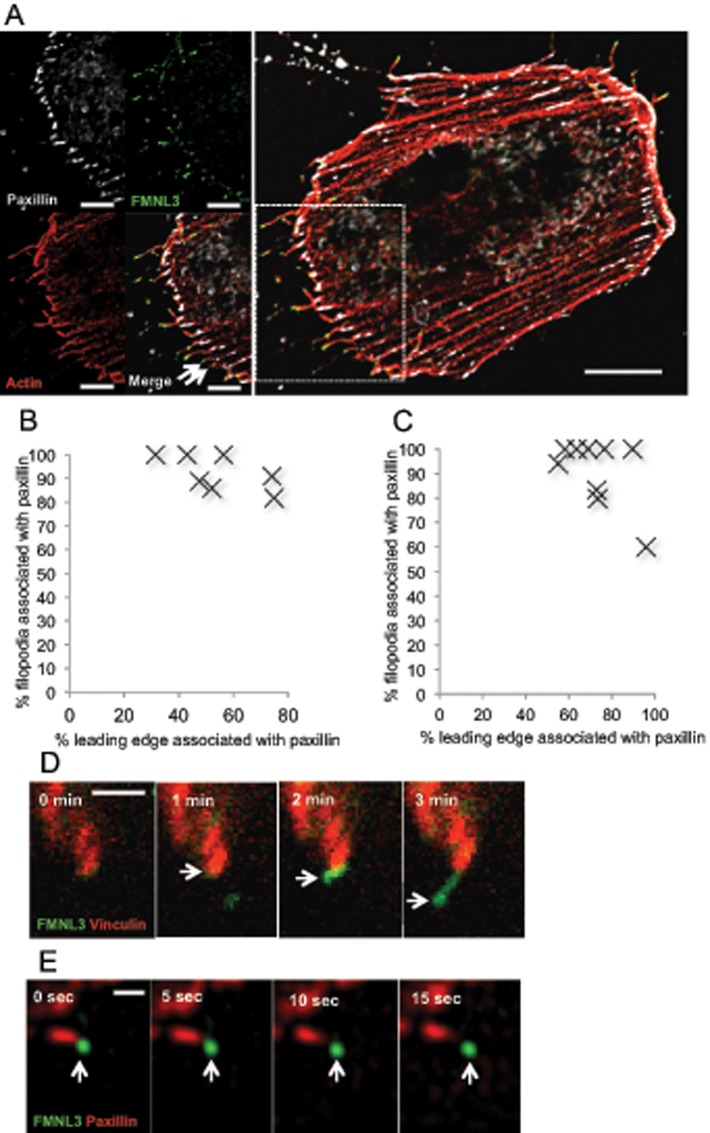
The effects of mDia1 and VASP in adherent cells raise the possibility that some filopodia might require actin filaments that are associated with focal adhesions. We investigated the relationship between GFP-FMNL3-FF–mediated filopodia and focal adhesions by staining fixed cells for paxillin and actin filaments and scoring filopodia/focal adhesion association at the leading edge. Although the focal adhesion density at the leading edge varies, a disproportionately high percentage of filopodia originates at focal adhesions, even in cells for which focal adhesion density is low (Figure 6, A and B). We also imaged live cells cotransfected with GFP-FMNL3-FF and either mApple-vinculin (Figure 6D and Supplemental Movie S3) or mRFP-paxillin (Figure 6E and Supplemental Movie S4). Puncta of GFP-FMNL3 appear to extend directly from the focal adhesion in both cases. We quantified the percentage of focal adhesion–associated filopodia from confocal movies and found a similar trend as in fixed cells (Figure 6C). These results suggest that a population of filopodia emanates directly from focal adhesions in these cells.
FIGURE 6:
A subset of FMNL3-FF–mediated filopodia associates with focal adhesions. (A) Representative micrograph of the leading-edge region of a U2OS cell cotransfected with RFP-paxillin (white), BFP-Lifeact (red), and GFP-FMNL3-FF (green). Images are maximum intensity projections from 10 × 0.2–μm Z-slices. Arrowheads indicate two filopodia that are not associated with focal adhesions in this example. Scale bar, 0.5 μm. (B) Quantification of percentage of FMNL3-FF–enriched filopodia that are associated with focal adhesions (judged from fixed cells stained with anti-paxillin) as a function of percentage of focal adhesion coverage at the leading edge; 7 cells. See Materials and Methods for description of quantification. (C) Quantification of focal adhesion–associated filopodia from confocal movies similar to D. (D) Time-lapse montage of FMNL3 punctum emanating from a focal adhesion (individual time points from Supplemental Movie S3). U2OS cells cotransfected with mApple-vinculin (red) and GFP-FMNL3-FF (green). mApple and GFP micrographs were acquired sequentially (<1 s between colors) every 30 s. Scale bar, 2 μm. (E) Structured illumination microscopy time-lapse montage of FMNL3 punctum emanating from a focal adhesion (individual time points from Supplemental Movie S4). U2OS cells cotransfected with RFP-paxillin (red) and GFP-FMNL3 (green). RFP and GFP micrographs were acquired sequentially (<1 s between colors) every 5 s. Scale bar, 0.5 μm.
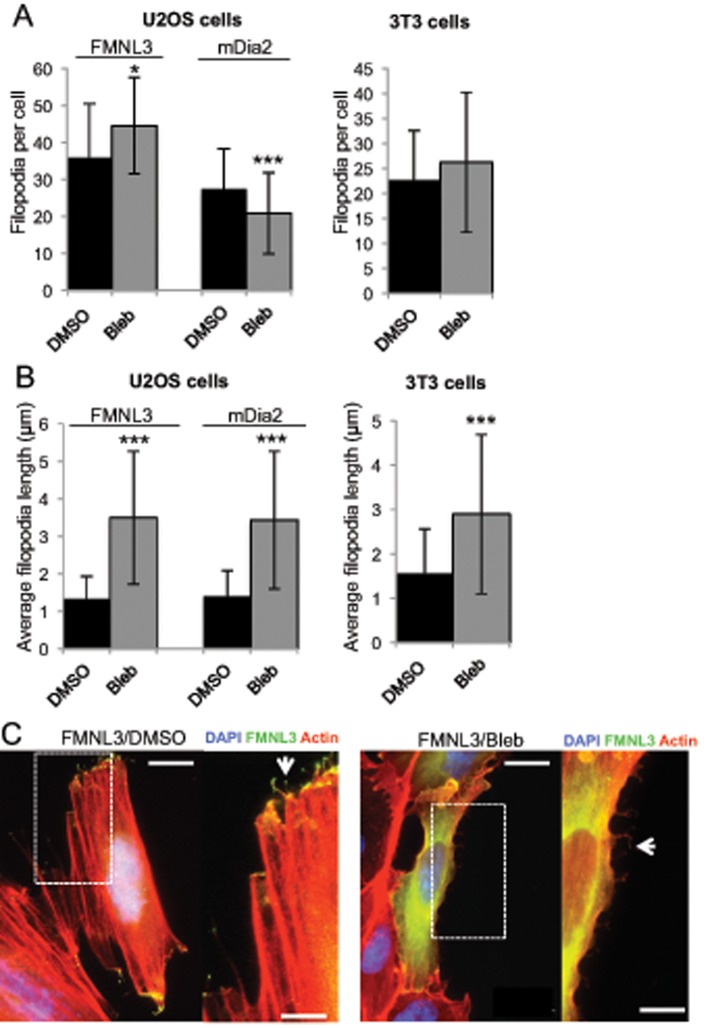
The myosin II inhibitor blebbistatin causes a reduction in focal adhesions and stress fibers (Hotulainen and Lappalainen, 2006; Aratyn-Schaus et al., 2011). We tested whether blebbistatin treatment would influence filopodia assembly in U2OS and 3T3 cells. Surprisingly, blebbistatin does not reduce filopodial number (Figure 7A), despite causing a significant reduction in stress fibers. We observe, however that blebbistatin causes a significant increase in filopodial length in both U2OS and 3T3 cells (Figure 7B). This effect is likely due to the function of myosin II on retrograde flow and has been observed in other systems (Medeiros et al., 2006; Shin et al., 2014). An interesting feature is that, upon blebbistatin treatment, filopodia appear to emanate throughout the cell periphery, whereas they are largely confined to the protruding ends of untreated cells (Figure 7C).
FIGURE 7:
Myosin II inhibition does not reduce filopodia number but increases filopodial length. U2OS cells or NIH 3T3 cells were transfected with GFP-FMNL3-FF (both cell types) or GFP-mDia2-FFC (U2OS only) and treated for 2 h with DMSO or 50 μM blebbistatin. (A) Quantification of filopodia number per transfected cell treated as indicated. Results are pooled from three or four independent experiments; 31–103 U2OS cells, 83–117 NIH 3T3 cells. Error bar, SD. *p < 0.05, ***p < 0.001. (B) Quantification of filopodial length in GFP-FMNL3-FF– or GFP-mDia2-FFC–transfected cells. Results are pooled from three or four independent experiments; 40–95 filopodia in U2OS cells, 297–423 filopodia in 3T3 cells. Error bar, SD. ***p < 0.001. (C) Representative micrographs of U2OS cells transfected with GFP-FMNL3 and treated with DMSO (left) or 50 μM blebbistatin (right). Scale bar, 10 μm (main image) and 5 μm (insets).
DISCUSSION
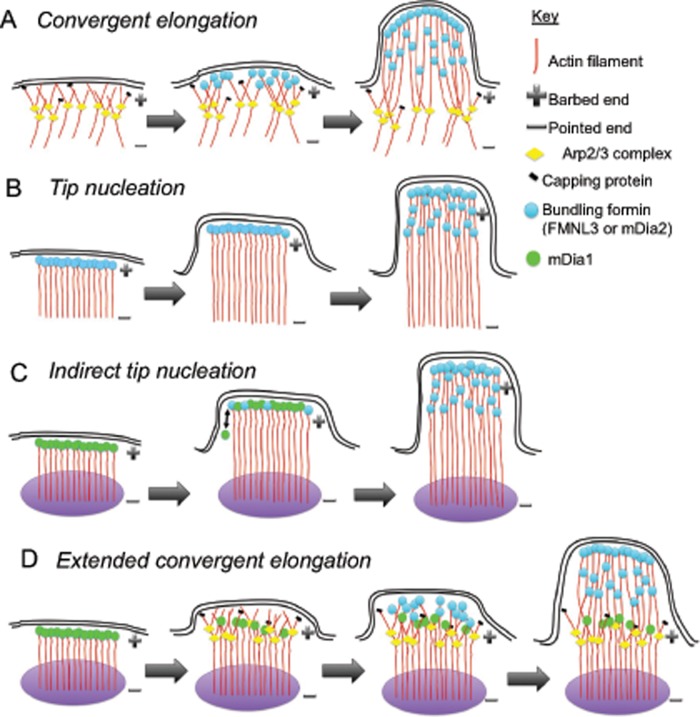
In this work, we use a relatively simple system, expression of constitutively active formin proteins, to ask fundamental questions about filopodial assembly. Collectively our results show that Arp2/3 complex is an important nucleation factor in formin-mediated filopodial assembly. However, we also show that a population of filopodia remains after Arp2/3 complex inhibition in adherent cells and that filopodia frequently assemble from focal adhesions. We therefore propose a modification to existing assembly models: filopodia can assemble from any collection of barbed ends in proximity to the plasma membrane, not just from the lamellipodial network. These filaments might be nucleated by a number of mechanisms, including Arp2/3 complex–nucleated dendritic network (convergent elongation; Figure 8A), formin-nucleated filaments (tip nucleation; Figure 8B), formin-nucleated filaments in which the nucleating formin is displaced by a bundling formin (indirect tip nucleation; Figure 8C), and a sequential combination of formin- and Arp2/3 complex–mediated nucleation (extended convergent elongation; Figure 8D).
FIGURE 8:
Models for filopodial assembly. Models A (convergent elongation) and B (tip nucleation) were proposed previously. We propose models C and D from our results in this work. (C) Indirect tip nucleation model. A nucleation factor (e.g., mDia1) assembles new filaments at a specific site near the leading edge (purple oval), such as a focal adhesion. During filament elongation, the barbed end–bound nucleation factor is displaced by a bundling formin (e.g., FMNL3 or mDia2), which allows further elongation, as well as bundling, to drive productive filopodial growth. (D) Extended convergent elongation model. Actin filaments are assembled at a specific site (purple oval) by a nucleation factor such as mDia1 and serve as mother filaments for Arp2/3 complex–mediated nucleation. Bundling formins such as FMNL3 and mDia2 then mediate convergent elongation on the Arp2/3 complex–nucleated barbed ends. We do not depict VASP in these models because its role is still unclear, but it could in principle be enhancing the initial assembly of filaments in models A, C, and D.
Much of the uncertainty surrounding filopodial assembly concerns the identity of the actin nucleation involved (Chhabra and Higgs, 2007; Faix et al., 2009; Harris et al., 2010). In principle, formin proteins can serve as both nucleation and elongation factors, making the tip nucleation model intuitively attractive. In practice, however, at least two filopodia-assembling formins, FMNL3 and DAAM1, are extremely poor actin-nucleating proteins in vitro (Heimsath and Higgs, 2012; Jaiswal et al., 2013), suggesting that these formins might not necessarily nucleate filopodial actin filaments.
Arp2/3 complex clearly plays a major role in filopodial assembly in all cells we analyzed, suggesting that convergent elongation is an important assembly mechanism. In adherent cells, however, a population of filopodia remains upon Arp2/3 complex inhibition, suggesting the existence of other assembly mechanisms. We find that inhibition of another potential nucleation factor, mDia1, reduces filopodia in adherent cells. Other studies have linked mDia1 with filopodia (Sarmiento et al., 2008; Goh et al., 2011, 2012; Naj et al., 2013), but its specific function has been unclear.
What is the mechanistic explanation for both Arp2/3 complex and mDia1 acting in filopodial assembly in adherent cells? The simplest scenario is that two independent pathways exist by which filopodia could assemble from either linear mDia1-generated filaments (Figure 8C) or branched Arp2/3 complex–generated filaments (Figure 8A). There has long been evidence for distinct actin networks existing at the leading edge of adherent cells, one being the Arp2/3 complex–assembled dendritic network and the other being less defined. In addition to ventral stress fibers, there may be a network of longer filaments ventral to the lamellipodium in some cells (Ponti et al., 2004; Giannone et al., 2007; Urban et al., 2010). Yet another population of filaments called dorsal stress fibers originates at focal adhesions (Hotulainen and Lappalainen, 2006; Burnette et al., 2014).
We favor dorsal stress fibers as a possible second source of filaments for filopodial assembly. Current evidence suggests that dorsal stress fibers consist of uniformly oriented actin bundles that require mDia1 for assembly but do not contain significant myosin II (Hotulainen and Lappalainen, 2006; Oakes et al., 2012; Burnette et al., 2014). These findings agree well with our results correlating the majority of filopodia with focal adhesions, as well as with the filopodial inhibition we observe upon mDia1 suppression but not upon myosin II inhibition. Of interest, myosin II inhibition did result in the loss of filopodial polarity (Figure 7C). Another source of filaments might be the cortical actin network, since this network requires both Arp2/3 complex and mDia1 (Bovellan et al., 2014). However, the lack of effect of mDia1 suppression in Jurkat cells, which should have a cortical actin network, argues against this possibility.
An argument against the existence of two independent filopodial nucleation pathways is the fact that inhibition of either Arp2/3 complex or mDia1 individually results in >50% reduction in filopodia number, whereas one might expect the sum of these effects to be no more than 100% if in independent pathways. A second possibility is that mDia1 and Arp2/3 complex act together in the same pathway to supply filaments that can be used by bundling formins such as FMNL3 and mDia2. For example, Arp2/3 complex could elaborate a branched network from mDia1-assembled dorsal stress fibers, which can be organized into filopodia by bundling formins (Figure 8D).
Why does a population of filopodia remain after inhibition of both mDia1 and Arp2/3 complex in adherent cells? Possibilities include the following: 1) there is a low level of tip nucleation activity directly by FMNL3 or mDia2 (Figure 8B), 2) other nucleating activities occur near the cortex, either other formins or the WH2-containing proteins Spire or Cordon Bleu, and 3) residual mDia1 protein and/or Arp2/3 complex activity might be sufficient to maintain a low degree of filopodial assembly.
Our results show that VASP’s role on FMNL3-mediated filopodia differs between suspension and adherent cells. In neither cell type do we find consistent VASP enrichment at filopodial tips. The role of Ena/VASP proteins in filopodia assembly has been unclear. Ena/VASP has been implicated in filopodial assembly in several cellular systems (Han et al., 2002; Svitkina et al., 2003; Schirenbeck et al., 2006; Kwiatkowski et al., 2007) and is found at the tips of or along the shafts of filopodia in some systems (Lanier et al., 1999; Rottner et al., 1999; Schirenbeck et al., 2006; Gates et al., 2007). VASP interacts directly with filopodia-assembling formin proteins in Dictyostelium (Schirenbeck et al., 2006), Drosophila (Homem and Peifer, 2009), and mammals (Grosse et al., 2003; Barzik et al., 2014) and might potentiate elongation of formin-mediated filopodia (Barzik et al., 2014). In addition, elegant studies in Drosophila suggest that the functional interplay between VASP and formins is intricate and has major consequences for development (Homem and Peifer, 2009; Bilancia et al., 2014).
However, it is not clear in all systems that VASP contributes directly to filopodial assembly. VASP also localizes to other areas, such as focal adhesions (Reinhard et al., 1992; Bear et al., 2000, 2002; Grosse et al., 2003), dorsal stress fibers (Gateva et al., 2014), and the leading edge in general (Rottner et al., 1999). A recent study in adherent cells showed that VASP rarely enriched at filopodial tips in the presence of constitutively active mDia2 but did increase the lifetime of these filopodia (Barzik et al., 2014). These results are similar to ours and might suggest that at least part of VASP’s effect on filopodia could be due to the VASP population in the focal adhesions, perhaps either through increased assembly of dorsal stress fibers (Gateva et al., 2014) or through increased substrate adhesion (Schafer et al., 2009).
The behavior of full-length formins might differ somewhat from our simple system, which uses formin constructs lacking the N-terminal regulatory region. In both mDia and FMNL family proteins, this region contains a Rho GTPase–binding domain, autoinhibitory DID domain (sometimes referred to as FH3), and a dimerization domain (Wallar et al., 2006; Kuhn et al., 2015) and participates in cellular localization through interactions with other proteins (Seth et al., 2006; Brandt et al., 2007). The interaction and possible competition between endogenous formins and the expressed formin constructs at filopodial tips might also contribute to the effects observed here. Further studies with full-length formins in more defined endogenous formin backgrounds will be useful in testing our proposed models.
A final question pertains to how our simple system relates to other filopodial systems. In cell lines in which Arp2/3 complex subunits are either suppressed (Steffen et al., 2006; Gomez et al., 2007; Wu et al., 2012) or ablated (Suraneni et al., 2012), assembly of endogenous filopodia still occurs. These filopodia appear to be enriched in mDia1 and mDia2 (Suraneni et al., 2012) and are formin dependent (Suraneni et al., 2015). These studies provide evidence for an Arp2/3 complex–independent filopodial assembly mechanism. Given the variety of filopodia-like structures that exists, it is certainly possible that multiple variations on either convergent elongation or tip nucleation might be used. In addition, there may be formin-independent mechanisms of filopodial assembly. Our study shows that a diverse array of actin filament sources, not simply the lamellipodial network, is capable of serving as a precursor for formin-mediated filopodia.
MATERIALS AND METHODS
Cell culture
Human Jurkat T lymphocytes (American Type Culture Collection, Manassas, VA) were grown in RPMI 1640 (Life Technologies, Carlsbad, CA) supplemented with 5% fetal bovine serum (FBS; Atlanta Biologicals, Norcross, GA), 2 mg/ml glucose, 1 mM sodium pyruvate (Life Technologies), and 50 μM β-mercaptoethanol (Life Technologies). Murine 300.19 pre–B lymphoma cells (a gift from Geoffrey Kansas, Northwestern Medical School, Chicago, IL) were grown in RPMI supplemented with 5% FBS and 50 μM β-mercaptoethanol. Both Jurkat and 300.19 cells were maintained in suspension at concentrations of <1 million cells/ml. Mouse NIH 3T3 cells (a gift from Gregg Gundersen, Columbia University, New York, NY) and cultured in DMEM with 4.5 g/l glucose, l-glutamine, sodium pyruvate, and 10% neonatal calf serum (NCS; Atlanta Biologicals). U2OS human osteosarcoma cells (a gift from Duane Compton, Geisel School of Medicine, Dartmouth College, Hanover, NH) and cultured in DMEM with 4.5 g/l glucose, l-glutamine, sodium pyruvate, and 10% NCS (Atlanta Biologicals).
DNA transfections
For 300.19 and 3T3 cells, DNA constructs were introduced by electroporation. Briefly, cells were washed twice in phosphate-buffered saline (PBS) and resuspended to 12.5 × 106 cells/ml in PBS. Cells, 400 μl, were mixed with 20 μg of plasmid DNA in a 4-mm gap electroporation cuvette (BTX Harvard Apparatus) and electroporated at 250 V and 975-μF capacitance in a Gene Pulser II (Bio-Rad, Hercules, CA). Prewarmed medium (1 ml) was added, and cell debris was removed. For 3T3 cells, cells were moved to 4-ml prewarmed medium, centrifuged at 300 × g for 5 min, and seeded onto glass coverslips at 1 × 105 cells/ml. Cells were incubated for 24 h before treatment and fixation. For 300.19 cells, cells were added to 6 ml of prewarmed medium and left at 37°C/5% CO2 for 6 h. Cells were centrifuged at 300 × g for 5 min, and the cell pellet was brought up in 1 ml of prewarmed medium. A 0.5-ml amount of suspension was placed on a 12-mm round coverslip coated with 0.01% poly-l-lysine (PLL; >300,000 molecular weight; Sigma-Aldrich, St. Louis, MO) and incubated for 10 min before fixation.
For U2OS cells, DNA constructs were introduced using JetPrime transfection reagent (PolyPlus, Illkirch, France); a total of 0.5 μg of each plasmid DNA was used for all transfections. Cells were incubated for 24 h before treatment and fixation.
For Jurkat T-cells, DNA constructs were introduced using the Amaxa Nucleofector Kit (Lonza, Cologne, Germany). Before nucleoporation, 1 × 106 cells were centrifuged at 300 × g for 10 min, resuspended in 100 μl of Nucleofector solution V, and mixed with 2 μg of plasmid. Resuspended cells were nucleoporated using the program X-001 (Nucleofection 2b Device), and 500 μl of full medium was added. Cells were transferred to a 12-well plate containing 1 ml of prewarmed medium and incubated at 37°C/5% CO2 for up to 8 h before treatment and fixation.
The following expression constructs were used: pEGFP-C1 FMNL3-FF (Harris et al., 2010), pEGFP-C1 mDia2-FFC (Harris et al., 2010), mApple VASP-5 (Addgene; a gift from Michael Davidson, Florida State University, Tallahassee, FL), mApple vinculin (a gift from Clare Waterman, National Institutes of Health, Bethesda, MD), mRFPn paxillin (a gift from Scott Blystone, Upstate Medical University, Syracuse, NY), F-tractin mCherry fusion (a gift from Clare Waterman, National Institutes of Health) and mTagBFP2-Lifeact7 (a gift from Michael Davidson).
Inhibitor treatment
Cells were treated with inhibitors for 2 h at 37°C/5% CO2 (unless otherwise stated). All inhibitors were diluted from dimethyl sulfoxide (DMSO) stocks (Sigma-Aldrich) to the following concentrations in full cell medium: CK666 or CK689 to 200 μM (Calbiochem, Billerica, MA) and blebbistatin to 50 μM (Sigma-Aldrich).
siRNA treatment
For U2OS cells, 50 nM each siRNA oligonucleotide was introduced using Lipofectamine RNAiMAX following the manufacturer’s instructions. Cells were analyzed 72 h posttransfection. For Jurkat T-cells, 1 μM each siRNA oligonucleotide was introduced using the Amaxa Nucleofector Kit V. Cells were analyzed 72 h posttransfection. Oligonucleotides for siRNA included human Arp2 (IDT, Integrated DNA Technologies, Coralville, IA; CGAUCAAUGUUAGAAUUAACUACC), human mDia1 (Ambion, Waltham, MA; GGAGUUACGAUAGCCGGAA), and negative control (IDT; CGUUAAUCGCGUAUAAUACGCGUAT).
Western blotting
Cells were extracted 72 h posttransfection. U2OS cells were scraped with 100 μl of 2× DB (50 mM Tris-HCl, pH 6.8, 2 mM EDTA, 20% glycerol, 0.8% SDS, 0.02% bromophenol blue, 1000 mM NaCl, 4 M urea). Jurkat T-cells (∼1 × 106 cells) were resuspended in 100 μl of 2× DB. Proteins were separated by 7.5% SDS–PAGE and transferred to a polyvinylidene fluoride membrane (Millipore, Billerica, MA). The membrane was blocked with TBS-T (20 mM Tris-HCl, pH 7.6, 136 mM NaCl, and 0.1% Tween-20) containing 3% bovine serum albumin (Research Organics, Cleveland, OH) for 1 h and then incubated with primary antibody at 4°C overnight. After washing with TBS-T, the membrane was incubated with horseradish peroxidase–conjugated secondary antibody (Bio-Rad) for 2 h at room temperature. Protein signal was detected by chemiluminescence, and Western band intensity was quantified using ImageJ (National Institutes of Health, Bethesda, MD). Primary antibodies used were anti-mDia1 produced in chicken (Aves Labs, Tigard, OH) using amino acids 748–1175 of mouse mDia1 as antigen, followed by affinity purification against the same antigen; anti–inverted formin-2 (Ramabhadran et al., 2011); anti-tubulin (DM1-α; Sigma-Aldrich); and anti-Arp2 (H-84; Santa Cruz Biotechnology, Dallas, TX).
Immunofluorescence microscopy
300.19 and Jurkat T-cells were fixed on 12-mm round coverslips by removal of medium and addition of 4% paraformaldehyde in PBS for at least 1 h. Before fixation, 300.19 and Jurkat T-cells were allowed to settle on coverslips precoated with 0.1% PLL for 10 min at 37°C. To fix Jurkat T-cells in suspension, 3 × 106 cells were fixed in 4% paraformaldehyde in PBS (1 ml) for at least 1 h at room temperature. Fixed cells were resuspended in PBS (100 μl/coverslip) and then allowed to settle onto coverslips precoated with 0.1% PLL for 10 min at 37°C, followed by a further 10 min of incubation in 4% paraformaldehyde.
U2OS and 3T3 cells were fixed on 12-mm round coverslips by removal of medium and addition of 4% formaldehyde in PBS for 20 min.
For staining, all cells were washed in 3× PBS before permeabilization with 0.1% Triton X-100 in PBS for 15 min. Cells were washed 3× with PBS and incubated in blocking buffer (10% calf serum, 0.1% saponin, 0.02% sodium azide in PBS) for 30 min. Cells were stained with combinations of the following: primary antibody (described later), 1 mM tetramethylrhodamine isothiocyanate (TRITC)–phalloidin (Sigma-Aldrich), and 0.4 ng/μl 4′,6-diamidino-2-phenylindole (DAPI) in block solution diluted 1:10 in PBS for 1 h at room temperature. Cells were incubated with primary treatment for 1 h, followed by 3× PBS washes and incubation in secondary antibody for 1 h. Cells were washed 3× with PBS and mounted on polyvinyl alcohol–1,4-diazabicyclo[2.2.2]octane.
Images were acquired using a Nikon TE-2000E microscope with either the 60×/1.4 numerical aperture (NA) or 100×/1.4 NA objective, a Roper CoolSnap camera, and Nikon Elements software. Alternatively, images where maximum intensity projections are shown (see figure legends) were acquired using a Wave FX spinning-disk confocal microscope (Quorum Technologies, Sussex, UK; on a Nikon Eclipse microscope) with Z-stacks of 0.2-μm sections.
Primary antibodies were monoclonal anti-paxillin (610569; BD Biosciences, San Jose, CA) and monoclonal anti-VASP 9A2 (3132; Cell Signaling, Danvers, MA). Secondary antibodies were Alexa Fluor 405–conjugated anti-mouse immunoglobulin G (IgG; Invitrogen, Carlsbad, CA), Texas red anti-mouse IgG (Vector Laboratories, Burlingame, CA), and Alexa Fluor 405–conjugated anti-rabbit IgG (Invitrogen).
Live-cell microscopy
For live-cell acquisition, coverslips were mounted onto a flow chamber in live-cell medium (DMEM minus phenol red, 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 10% NCS) and then onto a Wave FX spinning-disk confocal microscope (Quorum Technologies, Guelph, Canada; on a Nikon Eclipse microscope) with Bionomic Controller (20/20 Technology, Montreal, Canada), and temperature-controlled stage set to 37°C. After equilibration to temperature for 10 min, cells were imaged through the 60×/1.4 NA Plan Apo objective (Nikon USA, Melville, NY) using the 403-nm laser and 435/485 filter for blue fluorescent protein (BFP), the 491-nm laser and 525/20 filter for GFP, and the 561-nm laser and 593/40 filter for monomeric red fluorescent protein (mRFP)/mApple. For Supplemental Movies S1, S2, and S4, GFP and mRFP/mApple micrographs were acquired sequentially (< 1 s between channels) at 5-s intervals, For Supplemental Movie S3, GFP and mApple micrographs were acquired sequentially (<1 s between channels) at 30-s intervals. Inhibitor was added 2 h before live-cell analysis and was kept in cell medium during acquisition.
Superresolution three-dimensional structured illumination microscopy images were acquired on a DeltaVision OMX V4 (GE Healthcare, Waterbury, VT) equipped with a 60×/1.42 NA PlanApo oil immersion objective (Olympus, Center Valley, PA), 405-, 488-, 568-, and 642-nm solid-state lasers (100 mW), and sCMOS cameras (pco.edge). Image stacks of 1 μm with 0.125-μm-thick Z-sections and 15 images per optical slice (3 angles and 5 phases) were acquired using immersion oil with a refractive index of 1.524. Images were reconstructed using Wiener filter settings of 0.005 and optical transfer functions measured specifically for each channel with SoftWoRx 6.1.3 (GE Healthcare) to obtain superresolution images with a twofold increase in resolution both axially and laterally. Images from different color channels were registered using parameters generated from a gold grid registration slide (GE Healthcare) and SoftWoRx 6.1.3.
Quantification methods
NIS-Elements software was used for all analysis.
For quantification of fixed-cell phenotypes, FMNL3/mDia2-dependent filopodia were quantified on cells transfected with GFP-FMNL3-FF or GFP-mDia2-FFC and stained with TRITC-phalloidin. Filopodia were positive when GFP was found at the tip of an actin-stained thin protrusion (width of protrusion at or near the resolution limit, ∼200 nm) from the cell body. Filopodial length was measured from the central point of the tip-localized GFP punctum to the interface between the protrusion and the cell body.
For quantification of GFP fluorescence intensity versus filopodia number, total GFP fluorescence intensity of 10 randomly selected cells for GFP-FMNL3-FF–transfected Jurkat T-cells was normalized by subtracting background fluorescence from areas devoid of cells. Filopodia number per cell was counted manually and plotted as a function of GFP fluorescence intensity. Results are shown in Supplemental Figure S1C.
For focal adhesion quantification, GFP-FMNL3-FF–transfected U2OS cells were stained for paxillin (anti-paxillin) and actin filaments (TRITC-phalloidin). Leading-edge length was measured by tracing the edge of the cell body (following the phalloidin signal). Filopodia were excluded from the trace. Focal adhesion coverage was calculated by measuring the width of paxillin staining at the leading edge; the sum of all widths was divided by the total leading-edge length to obtain the percentage of focal adhesion coverage. Maximum intensity projections of confocal-acquired images were used to trace the phalloidin signal back from the GFP-labeled filopodial tip to the cell body. Analysis of live-cell filopodia and focal adhesion association was done in a similar manner. U2OS cells were cotransfected with BFP-Lifeact, RFP-paxillin, and GFP-FMNL3-FF.
Quantification of filopodia lifetimes and maximum filopodial length was done in a 10-μm region of the leading edge from time-lapse series of 10–15 min. Quantification of the filopodia birth rate was done in a 10-μm region of the leading edge in a 5-min acquisition time.
Electron microscopy
The 300.19 cells were fixed for 1–2 h in 3.5% glutaraldehyde in 100 mM NaPO4, pH 7.4. Suspension cells were adhered to PLL coating by allowing fixed cells to settle onto coverslips (as described for immunofluorescence), followed by refixing. Coverslips were washed with 20 mM sodium phosphate and stained for 1 h with 1% osmium tetroxide (Electron Microscopy Sciences, Hatfield, PA) diluted in 100 mM sodium phosphate (pH 6.0). Coverslips were washed once in water and dehydrated sequentially with 30, 50, 70, 95, and 100% ethanol (>10-min incubation in each). Coverslips were washed 2 × 5 min with 1:1 and 1:2 dilutions of ethanol:hexamethyldisilazane (HMDS; Sigma-Aldrich) and then 3 × 20 min with 100% HMDS. Coverslips were dried overnight in a desiccator under vacuum. Before viewing, coverslips were adhered to aluminum SEM stubs (Electron Microscopy Sciences) and coated with 3 nm of osmium tetroxide using an OPC-60 Osmium Plasma Coater (SPI Supplies, West Chester, PA). Cells were imaged in SE mode with an FEI XL-30 FEG ESEM using an accelerating voltage of 5 kV and spot size of 3. Quantification of SEM images was done using low-magnification images (1000×) to evaluate percentage of cells with filopodia. Six randomly selected fields of view with 76–108 cells were counted. From these images, filopodia per cell number was evaluated from those cells that contained at least one filopodium.
Supplementary Material
Acknowledgments
We thank Nick Deakin, Christopher Turner, and Clare Waterman for plasmids, Guillaume Charras for helpful advice on cortical actin, and Sam Besly for putting it all together. This work was supported by National Institutes of Health Grants GM109965 and GM069818 to H.N.H.
Abbreviations used:
- Arp
actin-related protein
- DAAM1
disheveled-associated activator of morphogenesis-1
- Ena/VASP
Enabled/vasodilator-stimulated phosphoprotein
- FF
formin homology 1–formin homology 2
- FFC
FF C-terminus
- FH
formin homology
- FMNL
formin-like
- mDia
mammalian Diaphanous.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-09-0626) on October 7, 2015.
REFERENCES
- Applewhite DA, Barzik M, Kojima S-I, Svitkina TM, Gertler FB, Borisy GG. Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol Biol Cell. 2007;18:2579–2591. doi: 10.1091/mbc.E06-11-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aratyn-Schaus Y, Oakes PW, Gardel ML. Dynamic and structural signatures of lamellar actomyosin force generation. Mol Biol Cell. 2011;22:1330–1339. doi: 10.1091/mbc.E10-11-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann C, Fischer L, Walter U, Reinhard M. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J Biol Chem. 1999;274:23549–23557. doi: 10.1074/jbc.274.33.23549. [DOI] [PubMed] [Google Scholar]
- Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, Schafer DA. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem. 2005;280:28653–28662. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzik M, McClain LM, Gupton SL, Gertler FB. Ena/VASP regulates mDia2-initiated filopodial length, dynamics, and function. Mol Biol Cell. 2014;25:2604–2619. doi: 10.1091/mbc.E14-02-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Loureiro JJ, Libova I, Fassler R, Wehland J, Gertler FB. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 2000;101:717–728. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, Gertler FB. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Bilancia CG, Winkelman JD, Tsygankov D, Nowotarski SH, Sees JA, Comber K, Evans I, Lakhani V, Wood W, Elston TC, et al. Enabled negatively regulates diaphanous-driven actin dynamics in vitro and in vivo. Dev Cell. 2014;28:394–408. doi: 10.1016/j.devcel.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- Block J, Breitsprecher D, Kuhn S, Winterhoff M, Kage F, Geffers R, Duwe P, Rohn JL, Baum B, Brakebusch C, et al. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr Biol. 2012;22:1005–1012. doi: 10.1016/j.cub.2012.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block J, Stradal TE, Hanisch J, Geffers R, Kostler SA, Urban E, Small JV, Rottner K, Faix J. Filopodia formation induced by active mDia2/Drf3. J Microsc. 2008;231:506–517. doi: 10.1111/j.1365-2818.2008.02063.x. [DOI] [PubMed] [Google Scholar]
- Bovellan M, Romeo Y, Biro M, Boden A, Chugh P, Yonis A, Vaghela M, Fritzsche M, Moulding D, Thorogate R, et al. Cellular control of cortical actin nucleation. Curr Biol. 2014;24:1628–1635. doi: 10.1016/j.cub.2014.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol. 2007;178:193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette DT, Shao L, Ott C, Pasapera AM, Fischer RS, Baird MA, Der Loughian C, Delanoe-Ayari H, Paszek MJ, Davidson MW, et al. A contractile and counterbalancing adhesion system controls the 3D shape of crawling cells. J Cell Biol. 2014;205:83–96. doi: 10.1083/jcb.201311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Faix J, Breitsprecher D, Stradal TE, Rottner K. Filopodia: complex models for simple rods. Int J Biochem Cell Biol. 2009;41:1656–1664. doi: 10.1016/j.biocel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Fan L, Pellegrin S, Scott A, Mellor H. The small GTPase Rif is an alternative trigger for the formation of actin stress fibers in epithelial cells. J Cell Sci. 2010;123:1247–1252. doi: 10.1242/jcs.061754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates J, Mahaffey JP, Rogers SL, Emerson M, Rogers EM, Sottile SL, Van Vactor D, Gertler FB, Peifer M. Enabled plays key roles in embryonic epithelial morphogenesis in Drosophila. Development. 2007;134:2027–2039. doi: 10.1242/dev.02849. [DOI] [PubMed] [Google Scholar]
- Gateva G, Tojkander S, Koho S, Carpen O, Lappalainen P. Palladin promotes assembly of non-contractile dorsal stress fibers through VASP recruitment. J Cell Sci. 2014;127:1887–1898. doi: 10.1242/jcs.135780. [DOI] [PubMed] [Google Scholar]
- Gauvin TJ, Young LE, Higgs HN. The formin FMNL3 assembles plasma membrane protrusions that participate in cell-cell adhesion. Mol Biol Cell. 2015;26:467–477. doi: 10.1091/mbc.E14-07-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G, Dubin-Thaler BJ, Rossier O, Cai Y, Chaga O, Jiang G, Beaver W, Dobereiner HG, Freund Y, Borisy G, Sheetz MP. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 2007;128:561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh WI, Lim KB, Sudhaharan T, Sem KP, Bu W, Chou AM, Ahmed S. mDia1 and WAVE2 proteins interact directly with IRSp53 in filopodia and are involved in filopodium formation. J Biol Chem. 2012;287:4702–4714. doi: 10.1074/jbc.M111.305102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh WI, Sudhaharan T, Lim KB, Sem KP, Lau CL, Ahmed S. Rif-mDia1 interaction is involved in filopodium formation independent of Cdc42 and Rac effectors. J Biol Chem. 2011;286:13681–13694. doi: 10.1074/jbc.M110.182683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Kumar K, Medeiros RB, Shimizu Y, Leibson PJ, Billadeau DD. Formins regulate the actin-related protein 2/3 complex-independent polarization of the centrosome to the immunological synapse. Immunity. 2007;26:177–190. doi: 10.1016/j.immuni.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse R, Copeland JW, Newsome TP, Way M, Treisman R. A role for VASP in RhoA-Diaphanous signalling to actin dynamics and SRF activity. EMBO J. 2003;22:3050–3061. doi: 10.1093/emboj/cdg287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y-H, Chung CY, Wessels D, Stephens S, Titus MA, Soll DR, Firtel RA. Requirement of a vasodilator-stimulated phosphoprotein family member for cell adhesion, the formation of filopodia, and chemotaxis in dictyostelium. J Biol Chem. 2002;277:49877–49887. doi: 10.1074/jbc.M209107200. [DOI] [PubMed] [Google Scholar]
- Harris ES, Gauvin TJ, Heimsath EG, Higgs HN. Assembly of filopodia by the formin FRL2 (FMNL3) Cytoskeleton (Hoboken) 2010;67:755–772. doi: 10.1002/cm.20485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ES, Higgs HN. Biochemical analysis of mammalian formin effects on actin dynamics. Methods Enzymol. 2006;406:190–214. doi: 10.1016/S0076-6879(06)06015-0. [DOI] [PubMed] [Google Scholar]
- Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem. 2006;281:14383–14392. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- Haviv L, Brill-Karniely Y, Mahaffy R, Backouche F, Ben-Shaul A, Pollard TD, Bernheim-Groswasser A. Reconstitution of the transition from lamellipodium to filopodium in a membrane-free system. Proc Natl Acad Sci USA. 2006;103:4906–4911. doi: 10.1073/pnas.0508269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimsath EG, Jr, Higgs HN. The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends. J Biol Chem. 2012;287:3087–3098. doi: 10.1074/jbc.M111.312207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem CC, Peifer M. Exploring the roles of diaphanous and enabled activity in shaping the balance between filopodia and lamellipodia. Mol Biol Cell. 2009;20:5138–5155. doi: 10.1091/mbc.E09-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal R, Breitsprecher D, Collins A, Corrêa IR, Xu M-Q, Goode BL. The formin Daam1 and fascin directly collaborate to promote filopodia formation. Curr Biol. 2013;23:1373–1379. doi: 10.1016/j.cub.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn S, Erdmann C, Kage F, Block J, Schwenkmezger L, Steffen A, Rottner K, Geyer M. The structure of FMNL2-Cdc42 yields insights into the mechanism of lamellipodia and filopodia formation. Nat Commun. 2015:6, 7088. doi: 10.1038/ncomms8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski AV, Rubinson DA, Dent EW, Edward van Veen J, Leslie JD, Zhang J, Mebane LM, Philippar U, Pinheiro EM, Burds AA, et al. Ena/VASP is required for neuritogenesis in the developing cortex. Neuron. 2007;56:441–455. doi: 10.1016/j.neuron.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Lanier LM, Gates MA, Witke W, Menzies AS, Wehman AM, Macklis JD, Kwiatkowski D, Soriano P, Gertler FB. Mena is required for neurulation and commissure formation. Neuron. 1999;22:313–325. doi: 10.1016/s0896-6273(00)81092-2. [DOI] [PubMed] [Google Scholar]
- Lebrand C, Dent EW, Strasser GA, Lanier LM, Krause M, Svitkina TM, Borisy GG, Gertler FB. Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of netrin-1. Neuron. 2004;42:37–49. doi: 10.1016/s0896-6273(04)00108-4. [DOI] [PubMed] [Google Scholar]
- Majstoravich S, Zhang J, Nicholson-Dykstra S, Linder S, Friedrich W, Siminovitch KA, Higgs HN. Lymphocyte microvilli are dynamic, actin-dependent structures that do not require Wiskott-Aldrich syndrome protein (WASp) for their morphology. Blood. 2004;104:1396–1403. doi: 10.1182/blood-2004-02-0437. [DOI] [PubMed] [Google Scholar]
- Matusek T, Gombos R, Szécsényi A, Sánchez-Soriano N, Czibula Á, Pataki C, Gedai A, Prokop A, Raskó I, Mihály J. Formin proteins of the DAAM subfamily play a role during axon growth. J Neurosci. 2008;28:13310–13319. doi: 10.1523/JNEUROSCI.2727-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros NA, Burnette DT, Forscher P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat Cell Biol. 2006;8:215–226. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- Naj X, Hoffmann A-K, Himmel M, Linder S. The formins FMNL1 and mDia1 regulate coiling phagocytosis of Borrelia burgdorferi by primary human macrophages. Infect Immun. 2013;81:1683–1695. doi: 10.1128/IAI.01411-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes PW, Beckham Y, Stricker J, Gardel ML. Tension is required but not sufficient for focal adhesion maturation without a stress fiber template. J Cell Biol. 2012;196:363–374. doi: 10.1083/jcb.201107042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrin S, Mellor H. The Rho family GTPase Rif induces filopodia through mDia2. Curr Biol. 2005;15:129–133. doi: 10.1016/j.cub.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- Ramabhadran V, Korobova F, Rahme GJ, Higgs HN. Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol Biol Cell. 2011;22:4822–4833. doi: 10.1091/mbc.E11-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MV, Chu P-H, Hahn KM, Zaidel-Bar R. An optogenetic tool for the activation of endogenous diaphanous-related formins induces thickening of stress fibers without an increase in contractility. Cytoskeleton (Hoboken) 2013;70:394–407. doi: 10.1002/cm.21115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Halbrugge M, Scheer U, Wiegand C, Jockusch BM, Walter U. The 46/50 kDa phosphoprotein VASP purified from human platelets is a novel protein associated with actin filaments and focal contacts. EMBO J. 1992;11:2063–2070. doi: 10.1002/j.1460-2075.1992.tb05264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Jouvenal K, Tripier D, Walter U. Identification, purification, and characterization of a zyxin-related protein that binds the focal adhesion and microfilament protein VASP (vasodilator-stimulated phosphoprotein) Proc Natl Acad Sci USA. 1995;92:7956–7960. doi: 10.1073/pnas.92.17.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K, Behrendt B, Small JV, Wehland J. VASP dynamics during lamellipodia protrusion. Nat Cell Biol. 1999;1:321–322. doi: 10.1038/13040. [DOI] [PubMed] [Google Scholar]
- Sarmiento C, Wang W, Dovas A, Yamaguchi H, Sidani M, El-Sibai M, Desmarais V, Holman HA, Kitchen S, Backer JM, et al. WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. J Cell Biol. 2008;180:1245–1260. doi: 10.1083/jcb.200708123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer C, Borm B, Born S, Mohl C, Eibl EM, Hoffmann B. One step ahead: role of filopodia in adhesion formation during cell migration of keratinocytes. Exp Cell Res. 2009;315:1212–1224. doi: 10.1016/j.yexcr.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Schirenbeck A, Arasada R, Bretschneider T, Stradal TEB, Schleicher M, Faix J. The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc Natl Acad Sci USA. 2006;103:7694–7699. doi: 10.1073/pnas.0511243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A, Otomo C, Rosen MK. Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLalpha and mDia1. J Cell Biol. 2006;174:701–713. doi: 10.1083/jcb.200605006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin EY, Lee CS, Yun CY, Won SY, Kim HK, Lee YH, Kwak SJ, Kim EG. Non-muscle myosin II regulates neuronal actin dynamics by interacting with guanine nucleotide exchange factors. PLoS One. 2014;9:e95212. doi: 10.1371/journal.pone.0095212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skau CT, Waterman CM. Specification of architecture and function of actin structures by actin nucleation factors. Annu Rev Biophys. 2015;44:285–310. doi: 10.1146/annurev-biophys-060414-034308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A, Faix J, Resch GP, Linkner J, Wehland J, Small JV, Rottner K, Stradal TE. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol Biol Cell. 2006;17:2581–2591. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P, Fogelson B, Rubinstein B, Noguera P, Volkmann N, Hanein D, Mogilner A, Li R. A mechanism of leading edge protrusion in the absence of Arp2/3 complex. Mol Biol Cell. 2015;26:901–912. doi: 10.1091/mbc.E14-07-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P, Rubinstein B, Unruh JR, Durnin M, Hanein D, Li R. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J Cell Biol. 2012;197:239–251. doi: 10.1083/jcb.201112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban E, Jacob S, Nemethova M, Resch GP, Small JV. Electron tomography reveals unbranched networks of actin filaments in lamellipodia. Nat Cell Biol. 2010;12:429–435. doi: 10.1038/ncb2044. [DOI] [PubMed] [Google Scholar]
- Vaillant DC, Copeland SJ, Davis C, Thurston SF, Abdennur N, Copeland JW. Interaction of the N- and C-terminal auto-regulatory domains of FRL2 does not inhibit FRL2 activity. J Biol Chem. 2008;283:33750–33762. doi: 10.1074/jbc.M803156200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG. Role of fascin in filopodial protrusion. J Cell Biol. 2006;174:863–875. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D, Yarar D, Welch MD, Peloquin J, Svitkina T, Borisy GG. Formation of filopodia-like bundles in vitro from a dendritic network. J Cell Biol. 2003;160:951–962. doi: 10.1083/jcb.200208059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallar BJ, Stropich BN, Schoenherr JA, Holman HA, Kitchen SM, Alberts AS. The basic region of the diaphanous-autoregulatory domain (DAD) is required for autoregulatory interactions with the diaphanous-related formin inhibitory domain. J Biol Chem. 2006;281:4300–4307. doi: 10.1074/jbc.M510277200. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol. 1999;1:136–143. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- Winkelman JD, Bilancia CG, Peifer M, Kovar DR. Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. Proc Natl Acad Sci USA. 2014;111:4121–4126. doi: 10.1073/pnas.1322093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:2624–2645. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Hoelzle M, Disanza A, Scita G, Svitkina T. Coordination of membrane and actin cytoskeleton dynamics during filopodia protrusion. PLoS One. 2009;4:e5678. doi: 10.1371/journal.pone.0005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Svitkina T. Filopodia initiation: focus on the Arp2/3 complex and formins. Cell Adh Migr. 2011;5:402–408. doi: 10.4161/cam.5.5.16971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaoui K, Honoré S, Isnardon D, Braguer D, Badache A. Memo–RhoA–mDia1 signaling controls microtubules, the actin network, and adhesion site formation in migrating cells. J Cell Biol. 2008;183:401–408. doi: 10.1083/jcb.200805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.