Abstract
Liver transplantation is the ultimate treatment for severe hepatic failure to date. However, the limited supply of donor organs has severely hampered this treatment. So far, great potentials of using mesenchymal stem cells (MSCs) to replenish the hepatic cell population have been shown; nevertheless, there still is a lack of an optimal three-dimensional scaffold for generation of well-transplantable hepatic tissues. In this study, we utilized a cryo-chemical decellularization method which combines physical and chemical approach to generate acellular liver scaffolds (ALS) from the whole liver. The produced ALS provides a biomimetic three-dimensional environment to support hepatic differentiation of MSCs, evidenced by expression of hepatic-associated genes and marker protein, glycogen storage, albumin secretion, and urea production. It is also found that hepatic differentiation of MSCs within the ALS is much more efficient than two-dimensional culture in vitro. Importantly, the hepatic-like tissues (HLT) generated by repopulating ALS with MSCs are able to act as functional grafts and rescue lethal hepatic failure after transplantation in vivo. In summary, the cryo-chemical method used in this study is suitable for decellularization of liver and create acellular scaffolds that can support hepatic differentiation of MSCs and be used to fabricate functional tissue-engineered liver constructs.
Keywords: Acellular liver scaffolds, Mesenchymal stem cells, Hepatic-like tissues, Transplantation
1. Introduction
Liver diseases are one of the leading causes of death worldwide and account for approximately 1–2 million deaths per annum according to the World Health Organization. The only curative mode of management for end-stage chronic hepatic diseases is liver transplantation. However, the limited availability of donor organs for transplantation is a major issue in this context [1]. Alternative approaches, such as a stem cell-based regenerative medicine, offer possibilities that help to overcome problems related to the shortage of livers for transplantation [2]. In our previous study, we were able to successfully induce human mesenchymal stem cells (MSCs) to differentiate into hepatocyte-like cells using a two-step protocol in vitro [3]. MSCs possess significant potential for hepatic tissue engineering because autologous MSCs can easily be isolated and extensively scaled up [4,5]. In addition, the technique of differentiation of MSCs into hepatocyte-like cells using a two-dimensional (2D) culture system has been well established. However, it has been challenging to find a suitable environment that will allow these multipotent cells to develop toward a more specific state and allow the formation of an implant construct that is able to function well in vivo.
Tissue engineering is developing into an advanced area of research that aims at generating biological substitutes which can be used to repair, regenerate or even replace malfunctioning tissues [6]. Scaffolds play a crucial role in providing an appropriate biological environment that includes structural support for cell attachment and subsequent tissue development [7]. Therefore, the scaffold materials must have the chemical and physical properties that support the clinical usefulness of the final tissue. However, the selections of an ideal scaffold for hepatic tissue engineering remains an open question. Previous studies have utilized both synthetic (e.g. polylactic acid, polyglycolic acid, and polyethylene glycol hydrogels) [8–10] and natural (e.g. collagen, alginate and chitosan) [11–14] three-dimensional (3D) scaffolds that provide an environment supporting the maintenance and growth of hepatocytes. More recently, studies have shown that organ decellularization is an attractive strategy because it allows the creation of a naturally occurring 3D biomimetic scaffold that is available for tissue engineering [15,16]. Using this approach, organs such as heart [17], lung [18] and kidney [19] etc, are able to retain most of their native extracellular matrix proteins, bio-molecules and spatial organization. In addition, this 3D architecture provides an optimum vascular structure for oxygen and nutrient diffusion.
In this study, we aim to fabricate acellular liver scaffolds (ALS) from the whole liver using a cryo-chemical decellularized procedure, which is capable of preserving most of the major components of the native liver extracellular matrix and retaining the intact vascular framework [20,21]. Subsequently, by employing a previously developed hepatic differentiation platform [3], the ALS is used as a scaffold for MSCs to differentiate into hepatic-like cells; the functions of MSC-differentiated hepatic-like cells are compared with the traditional 2D cell culture system. Moreover, in vivo functionality of MSC-laden ALS is to be tested in a carbon tetrachloride (CCl4) induced fulminant hepatic failure mice models.
2. Materials and methods
2.1. Fabrication of ALS
The whole livers were harvested from Balb/c mice, which were purchased from the National Laboratory Animal Center (Taipei, Taiwan). After anesthesia with 2.5% Avertin (Sigma–Aldrich, 10 ml/kg body weight), a longitudinal abdominal incision was made in order to expose the liver. The portal vein was cannulated with Polyethylene Tubing-50 and attached to a peristaltic pump (EYELA, Japan). Deionized water was perfused through the portal vein at a rate of approximately 1 ml/min for 1 h and then the liver was frozen at −80 °C for 24 h. Next, the frozen livers were thawed at room temperature and perfused with deionizer water at 1 ml/min for 1 h. Subsequently, 1% Triton-X 100 (Sigma–Aldrich) plus 0.1% ammonium hydroxide (Sigma–Aldrich) in deionizer water was perfused throughout the livers to bring about decellularization and at the rate of 1 ml/min until the perfusate became clarity. Finally, prior to sterilization by gamma irradiation, the acellular liver scaffold that had been created by the above procedures was rinsed with sterile water to remove the remaining decellularization detergent.
2.2. Repopulation of ALS by MSCs
MSCs used in this study were isolated from the bone marrow of Balb/c mice. For repopulation of ALS, 10th- to 12th-passage cells, at 1.0–1.2 × 104 cells/cm2, were maintained in low-glucose Dulbecco's Modified Eagle Medium (Sigma–Aldrich) supplemented with 10% fetal bovine serum and 100 units of penicillin, 1000 units of streptomycin, and 2 mmol/L l-glutamine (Gibco BRL). MSCs were introduced into ALS via portal vein which was retained in ALS and cannulated with Polyethylene Tubing-50 ex vivo. Each ALS had culture medium circulated through it via the portal vein by the peristaltic pump (EYELA, Japan) at 1 ml/min for 1 h prior to recellularization. To allow total recellularization using 50 million cells, MSCs were infused into ALS using five steps at 10 min intervals; each step consisting of 10 million cells. After 40 min, the perfusate was collected, and cell viability and retention in the scaffold were determined. The flow rate of medium was used at 1 ml/min for culture of the HLT.
2.3. In vitro hepatic differentiation
To induce hepatic differentiation, MSCs were cultured in ALS and tissue culture dish. Differentiation was induced by treating the MSCs for 4 weeks with a 2-step protocol that we had previously reported [3]. Thereafter, medium replacements were performed twice weekly. Each of study group was following the above procedure for hepatic differentiation.
2.4. Histological analysis
Liver tissues were fixed in 3.7% formaldehyde (Sigma–Aldrich) in phosphate buffered saline (PBS, Gibco BRL) solution overnight. Samples were dehydrated in 30% sucrose in PBS overnight, embedded in O.C.T compound (Sakura), frozen quickly in liquid nitrogen and sectioned at 5 μm thickness. Sample sections were stained using Mayers hematoxylin (Sigma–Aldrich) and counterstained with Eosin-Y (Sigma–Aldrich). Stained sections were observed using an Olympus AX80 microscope (Olympus).
2.5. Scanning electron microscopy
Samples were fixed with 2% glutaraldehyde in PBS for 2 h and post-fixed with 1% osmium tetroxide (Electron Microscopy Sciences) for 1 h and rinsed three times in PBS. The tissue samples were dehydrated through a graded series of ethanol washes, beginning with 50% and progressing through 70%, 90% and 100% absolute ethanol, which were followed by critical point drying using an Emscope CPD 750. Samples were sputter coated with a 7-nm layer of gold-palladium (Cressington 108 sputter coater) and visualized at a voltage of 12 kV using a JEM 6335F field emission gun SEM (JEOL, Peabody, MA).
2.6. DNA content assay
DNA was extracted from a small piece of excised normal livers and ALS using the illustraTM tissue and cells genomic Prep Mini Spin Kit (GE Healthcare) according to the manufacturer's instructions. DNA concentration was determined by spectrophotometer.
2.7. Sirius red/fast green staining for collagen content
Sirius red/fast green solution was prepared using a saturated solution of picric acid (Sigma–Aldrich) in deionizer water (1.2 g per 100 ml) that contained 0.1 g of Direct Red (Sigma–Aldrich) and 0.1 g of Fast Green FCF (Sigma–Aldrich). The solution was allowed to stand for 15 min at room temperature before filtration. After immersing in deionizer water for 5 min, 0.2 ml Sirius Red/Fast Green solution was added to the frozen sections for 30 min. Sections were washed with deionizer water, scraped off the slides, incubated in 2 ml 0.1 N NaOH (Sigma–Aldrich) in methanol for 1 min, and centrifuged. The absorbance of the supernatant was determined at 605 and 540 nm. The value corresponding to 26% of the optical density at 605 nm was calculated, representing the contribution of Fast Green to the absorbance of Sirius Red at 540 nm. The above value was subtracted from the absorbance at 540 nm to obtain the corrected absorbance. Subsequently, the absorbance at 605 nm was divided by 2.08 and the corrected absorbance was divided by 38.4 to obtain the net amount of collagen and non-collagen protein in the section. Total protein was the sum of both values. Results were expressed as the ratio of collagen (μg) to total protein (mg) in order to remove any differences due to variations in the weight of the slices. All analyses were performed in triplicate.
2.8. RNA extraction
Total RNA was isolated using Trizol Reagent (Sigma–Aldrich) as described by the manufacturer's instructions. RNA concentration was determined by spectrophotometer at 260 nm. The purity of the RNA was determined by spectrophotometry at 280 nm and the samples were stored at −80 °C until further use. RNA samples (2 μg) were reverse transcribed to cDNA using Moloney murine leukemia virus reverse transcriptase (MMLV Reverse Transcriptase, Promega).
2.9. Real time RT-polymerase chain reaction
Real time quantitative PCR was performed using an ABI PRISM 7700 (Applied Biosystems) and analyzed using the accompanying software. Target genes that were regulated at the mRNA level during MSCs differentiation were pinpointed and then specific primers targeting these genes were designed using the Primer Express software (Applied Biosystems). DNA-intercalating SYBR green Master Mix reagent (Applied Biosystems) was used to detect the reverse transcribed PCR product. For the reverse-transcription polymerase chain reaction (RT-PCR), the following conditions were used: 50 °C for 2 min, 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 s and 60 °C for 1 min. Quantitative real-time RT-PCR (Q-PCR) was performed using the StepOnePlus™ Real-Time PCR System (Applied Biosystems). The alteration in gene expression was obtained using the Δ Δ Ct method in which all samples were first normalized against the level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) present in the same sample.
2.10. Periodic acid-schiff (PAS) staining
The differentiated cells were fixed with 3.7% formaldehyde and permeabilized with 0.1% Triton X-100 in PBS for 15 min. Samples were then oxidized in 1% periodic acid (Sigma–Aldrich) for 5 min, rinsed three times with deionizer water, treated with Schiff's reagent (Sigma–Aldrich) for 30 min, and then rinsed three times with deionizer water for 5–10 min. Finally, the samples were observed using an Olympus AX80 microscope (Olympus).
2.11. Immunofluorescence staining
The differentiated cells from 2D culture and ALS groups were fixed in 3.7% formaldehyde in PBS and permeabilized in 0.1% Triton X-100 for 15 min at room temperature. After permeabilization, the samples were blocked with 2% bovine serum albumin (BSA, Sigma–Aldrich) in PBS for 1 h and thereafter treated with primary antibodies overnight at 4 °C diluted in blocking buffer. The antibodies used in this study were mouse polyclonal anti-collagen type I (COL-I, 1:250 Abcam), mouse polyclonal anti-collagen type IV (COL-IV, 1:250 Abonva), mouse polyclonal anti-fibronectin (FN, 1:250 Abonva), mouse polyclonal anti-laminin (1:250 Abonva), mouse monoclonal anti-serum albumin (ALB, 1:250 R&D Systems), alpha-fetoprotein (AFP, 1:250 R&D System) and rabbit polyclonal anti-cytokeratin-19 (CK-19, 1:50 Abcam). Following three washes with PBS, the samples were incubated at room temperature for 1 h with goat anti-mouse Alexa fluor 594, goat anti-mouse Alexa fluor 488 and goat anti-rabbit Alexa fluor 488 conjugated secondary antibodies (Jackson ImmunoResearch). After incubation in the secondary antibody, the samples were then placed in PBS and incubated with 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI, 1:5000 Sigma–Aldrich) for 5 min to stain nuclear DNA. Finally, the samples were rinsed three times with PBS and mounted with mounting medium before imaging. Samples were imaged using an inverted Olympus AX80 microscope (Olympus).
2.12. Western blot analysis
Protein concentration was determined and 25 μg of protein was separated on 10% sodium dodecylsulfate/polyacrylamide gel electrophoresis and blotted onto a PVDF membrane (Bio-Rad Laboratories, Inc.). The membranes were blocked with HyBlock blocking buffer (HyCell Biotechnology, Inc.) for 30 min. The membrane was sequentially hybridized with the primary antibody. The antibodies used in this study were rabbit polyclonal anti-albumin (ALB, 1:100 Abcam), rabbit polyclonal anti-alpha-fetoprotein (AFP, 1:1000 Cell Signaling), rabbit polyclonal anti-cytokeratin 19 (CK19, 1:100 Abcam) and mouse monoclonal anti-beta-actin (1:2000 Sigma–Aldrich). Following three washes with TBST, the membrane were incubated at room temperature for 1 h with HRP-conjugated secondary antibodies (Sigma–Aldrich). After incubation in the secondary antibody, the membranes were washed three times with TBST. Protein intensity was determined with Immobilon Western (Millipore Corporation).
2.13. Animal model
Non-obese diabetic severe combined immunodeficient (NOD-SCID) mice were purchased from National Laboratory Animal Center (Taipei, Taiwan). All animal experiments were performed with the approval of the Animal Care Committee of the Taipei Veterans General Hospital. Carbon tetrachloride (CCl4, Sigma–Aldrich) was dissolved in olive oil to give a 10% concentration and then administered to the animals by intraperitoneal injection. Treatment with a dose of 10 ml CCl4/kg body weight was tested for hepatotoxicity and lethality. Transplants were performed at 24 h after administration of CCl4.
2.14. Intrahepatic transplantation
Prior to transplantation, HLT was cut to generate 1 × 1 mm pieces in sterile PBS. The liver injured mice model was performed under general anesthesia (2.5% Avertin, 10 ml/kg intraperitoneally). After a mid-abdominal incision, the left lobe of the liver was exposed and a 2-mm incision was introduced into the lobe to make space for transplantation. The following groups of transplantation were carried out: (1) 200 μl of PBS (Placebo), (2) 2 × 106 undifferentiated MSCs suspended in 200 μl of PBS, (3) 2 × 106 MSC-derived hepatocyte-like cells (MDHs) obtained in 2D culture in 200 μl of PBS, (4) ALS only and (5) HLT containing 2 × 106 MSCs. Transplants in each group was directly embedded into the liver parenchyma and were secured by hemostatic mesh and the application of biological glue (Histoacryl, Sigma–Aldrich). The mid-abdominal incision then was sutured. Regular post-operative care was administered.
2.15. Biochemical analysis
Conditioned media from the differentiated cells cultured from both the 2D culture and ALS groups were collected after hepatic induction for four weeks; and stored at −20 °C until further assay. The various media were assessed for albumin secretion using an immunoperoxidase assay that was able to determine albumin (Immunology Consultants Laboratory Inc) and using a urea assay kit (BioVision) that was able to measure urea production; both assays were carried out according to manufacturer's recommendations.
2.16. Liver function tests
The serum concentrations of serum glutamyl oxaloacetic transaminase (SGOT), serum glutamyl pyruvic transaminase (SGPT), albumin and total bilirubin were determined using a SPOTCHEM SP-4410 automated dry chemistry analyzer system (Kyoto Daiichi-Kagaku, Kyoto) as per the manufacturer's instructions.
2.17. Immunohistochemistry
Samples were fixed in 3.7% formaldehyde in PBS solution overnight. These samples were then dehydrated in 30% sucrose in PBS overnight, which was followed by embedding in O.C.T compound. The embedded samples were frozen quickly in liquid nitrogen and then sectioned to 5 μm thickness. Staining was carried out according to the DakoCytomation EnVision + Dual Link System-HRP detection system protocol. Sections were treated with superblock and mouse-to-mouse block (ScyTek Laboratories) at each blocking step. Sections were then incubated with mouse monoclonal anti-albumin (ALB, 1:250 R&D Systems) antibody at 4 °C overnight, followed by staining with goat anti-mouse IgG antibody (Dako) for another 1 h at room temperature before DAB (Dako) was applied. Each slide was counterstained with hematoxylin.
2.18. Statistical analysis
KaplanMeier survival analysis was used to analyze the animal experiments and the results were statistically compared by log-rank tests. Data were performed as mean ± SD, and P-values less than 0.05 was considered to be statistically significant.
3. Results
3.1. Construction of the ALS
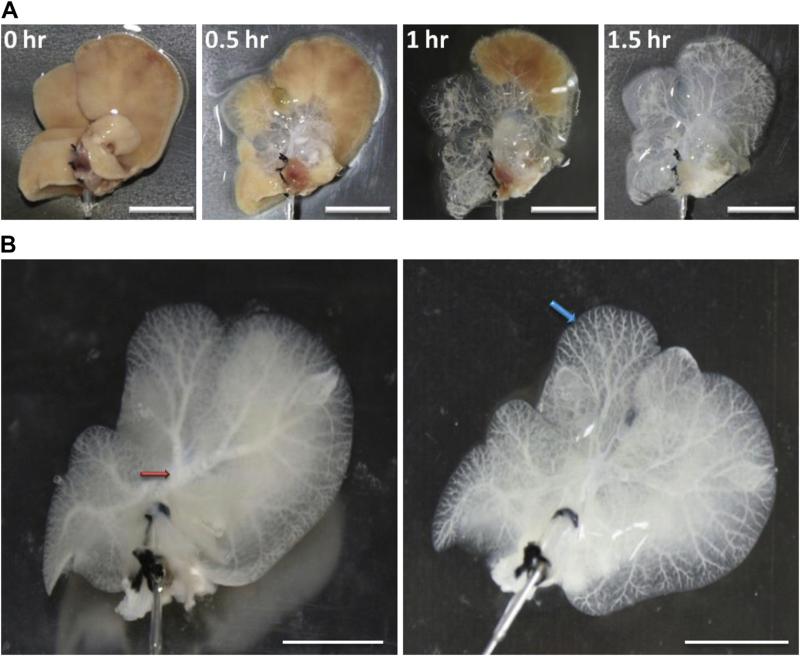
We carried out whole liver decellularization by our cryo-chemical method. This treatment effectively lyses cell membranes, disrupts intracellular organelles and removes most cellular debris from the tissue. During the decellularization process, a translucent acellular scaffold, which retained the complete shape of liver, was generated (Fig. 1A). The liver parenchyma became semi-transparent. The main and minor vascular structures were retained and were clearly visible when viewed under bright light (Fig. 1B).
Fig. 1.
Features of the whole liver scaffolds before and after decellularization. (A) Phase contrast microscopic analysis during perfusion with the decellularization reagents via the portal vein at 0 h, 0.5 h, 1 h and 1.5 h. (B) The main (red arrow) and branch (blue arrow) vascular structures can be seen to have remained intact after decellularization. Scale bar: 10 mm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Biochemical composition of the ALS
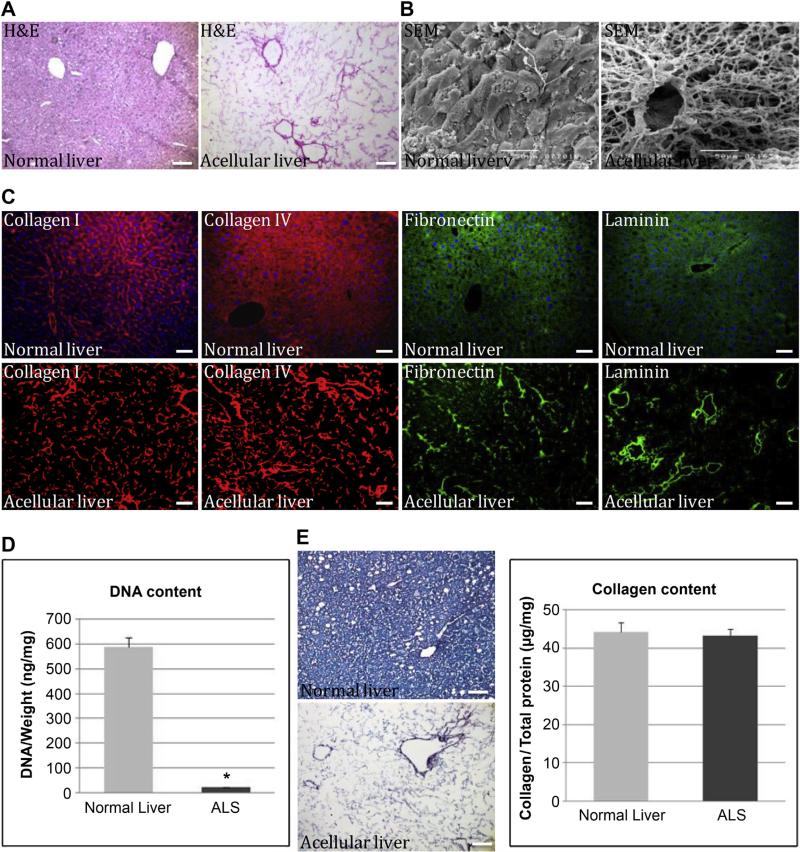
Histochemical staining was carried out to further characterize the composition of the ALS. H&E staining showed the presence of pink eosinophilic staining that is typical of collagen; however, no basophilic staining, which is a typical of cellular nuclear material, was detected (Fig. 2A). Scanning electron microscopy (SEM) images confirmed the presence of large vessels composed of collagen fibers within the ALS (Fig. 2B). We also observed free spaces that were about the size of hepatocytes. This cobweb of acellular material, consisting of 10–20-μm diameter gaps in the major extracellular matrix (ECM), was thought to be the remnants of the mouse hepatocytes that were removed by decellularization (Fig. 2B). Immunostaining for four ECM proteins, type I collagen, type IV collagen, fibronectin and laminin, indicated that the structural components and basement membrane composition of the ECM had been retained and were similar to that of a normal liver (Fig. 2C). No DAPI-positive nuclei (blue) were found in the ALS, which confirmed the absence of cellular material (Fig. 2C). DNA content in normal livers (588.3 ± 37.5 ng/mg) was significantly decreased when compared with ALS (22.9 ± 1.0 ng/mg) (P = 0.000697) (Fig. 2D). In addition, Sirius Red/Fast Green staining was used to confirm the above results, and also revealed the preservation of collagen within the intact liver scaffold. When quantitatively measured, the relative collagen content in the ALS (43.2 ± 1.8 μg/mg) was found by colorimetric assay to show no significant difference when compared with normal livers (44.3 ± 2.4 μg/mg) (P = 0.289) (Fig. 2E).
Fig. 2.
Ultra-structural analysis of normal and acellular livers. (A) H&E staining showed the cellular material and proteinous extracellular matrix in acellular liver section. (B) Scanning electron micrograph of normal and acellular livers at 500× magnification showing vessels composed of collagen networks together with free spaces of approximately hepatocyte size. (C) Immunofluorescent staining showed various ECM components, namely type I collagen (red), type IV collagen (red), fibronectin (green) and laminin (green). All samples were counterstained with DAPI. (D) DNA content in liver scaffold after decellularization was normalized to initial weight of the sample (P = 0.00697). (E) Analysis and quantification of the collagen content of acellular liver scaffolds were performed using Sirius Red/Fast Green assay (P = 0.289). Data are presented as a mean ± SD from three independent experiments as histograms (*, P < 0.05). Scale bar: 100 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Recellularization of MSCs within the whole ALS
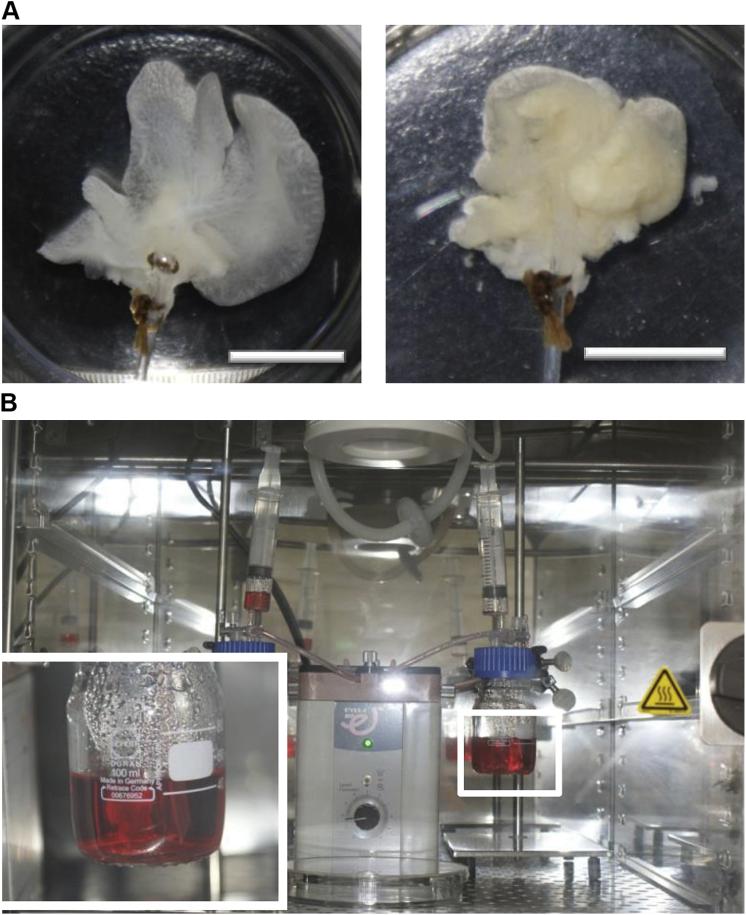
A representative image of the construct before and after recellularization process was shown (Fig. 3A). In order to accelerate gas and nutrient exchange, a simple perfusion system at a low flow rate was used for the in vitro culture of the HLT (Fig. 3B).
Fig. 3.
Recellularization of ALS with MSCs. (A) The representative photograph of ALS before and after repopulation with MSCs. (B) The perfusion culture system used for MSCs repopulation and hepatic differentiation over 4 weeks via dynamic circulation in incubator. Scale bar: 10 mm.
3.4. In vitro functional analysis of the hepatic differentiation of the MSCs in 2D and within the ALS
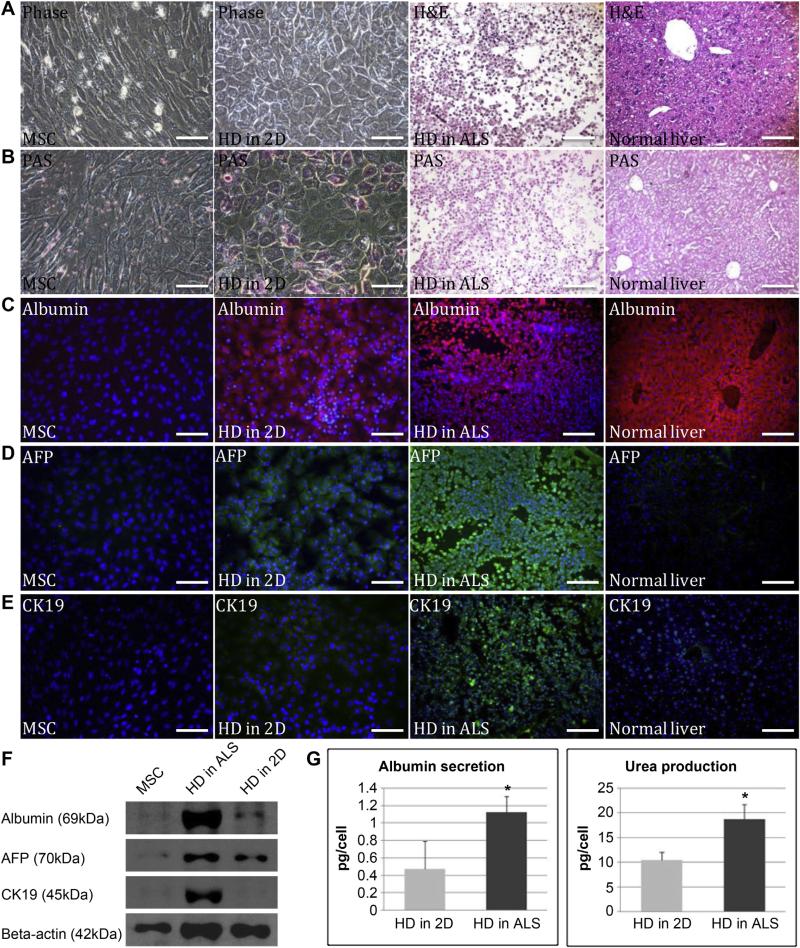
A comparison of gene expression during hepatic differentiation of MSCs within the ALS (HD in ALS) and 2D culture (HD in 2D) were carried out by qRT-PCR. To do this, the transcription levels of various hepatic development associated genes, namely hepatocyte nuclear factor 1 alpha (HNF1A), hepatocyte nuclear factor 1 beta (HNF1B), hepatocyte nuclear factor 3 beta (HNF3B), hepatocyte nuclear factor 4 alpha (HNF4A), alpha-fetoprotein (AFP), cytokeratin-19 (CK19), glucose-6-phosphatase (G6P), tyrosine aminotransferase (TAT), GATA binding protein 4 (GATA4), and epithelial cell adhesion molecule (EpCAM) were examined and found the gene expression levels of differentiating MSCs within the ALS after four weeks of hepatic induction were significantly higher than the those after four weeks of hepatic induction that were in culture dish (Fig. 4). Furthermore, we observed that the MSCs had altered their morphology from spindle to cuboidal shape after four weeks hepatic induction in 2D culture, while the cells in the ALS had dispersed into the parenchyma sites (Fig. 5A). Periodic Acid-Schiff (PAS) staining revealed that after hepatic differentiation in the ALS, there was higher glycogen storage capability than after HD in 2D (Fig. 5B). Immunofluorescence analysis was then used to reveal that the expression of various hepatic progenitor markers, namely albumin, alpha fetoprotein (AFP) and cytokeratin-19 (CK19). High AFP and CK19 expression observed from the cells in ALS may represent the phenotype of hepatic progenitors (Fig. 5C– E). Similarly, it was found that the expression levels of these proteins in the MSCs were higher in the ALS compared to the MSCs in monolayer culture after four weeks of induction (Fig. 5F). To evaluate metabolic activity, we quantified albumin secretion and urea production by the MSC-derived differentiated cells using the 2D system and the ALS after four weeks of hepatic induction. The cumulative albumin level of the differentiated cells in the ALS (1.13 ± 0.18 pg/cell) was statistically significantly higher than in 2D culture system (0.48 ± 0.32 pg/cell) over a 3 day culture period (P = 0.0148). The urea concentration in the media produced by differentiated cells in the ALS (18.74 ± 3.05 pg/cell) was remarkably higher that of the 2D culture (10.37 ± 1.71 pg/cell) media over the same 3 day period (P = 0.0194) (Fig. 5G). Taken together, the above results clearly demonstrate the superiority of the ALS scaffold over 2D culture in terms of supporting the hepatic differentiation of MSCs.
Fig. 4.
Hepatic-related genes expression analysis by real-time PCR. Comparison of gene transcription levels after four weeks of hepatic induction between culturing in culture dish (HD in 2D) and ALS (HD in ALS). Statistically significant differences relative to levels of undifferentiated MSCs using the 2D approach, which were arbitrarily set to 1.0, are indicated.
Fig. 5.
In vitro analysis of liver function markers. (A) Phase of hepatic differentiation of MSCs using the 2D and ALS approaches compared to normal liver. (B) PAS staining showed the glycogen storage capability after four weeks of hepatic differentiation using the 2D and ALS approaches. (C)(D)(E) Immunofluorescence analysis of albumin (red), AFP (green) and CK19 (green) expression in undifferentiated MSCs and after four weeks of hepatic differentiation using the 2D and ALS approaches. (F) Western blot analysis showed the expression of albumin, AFP and CK19 in undifferentiated MSCs and after four weeks of hepatic differentiation using the 2D and ALS approaches. (G) Albumin secretion (P = 0.015) and urea production (P = 0.019) after hepatic differentiation for four weeks using the 2D and ALS approaches. Data are presented as means ± SD from three independent experiments (*, P < 0.05). Scale bar: 100 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. Transplantation of HLT in chemical liver injured mice
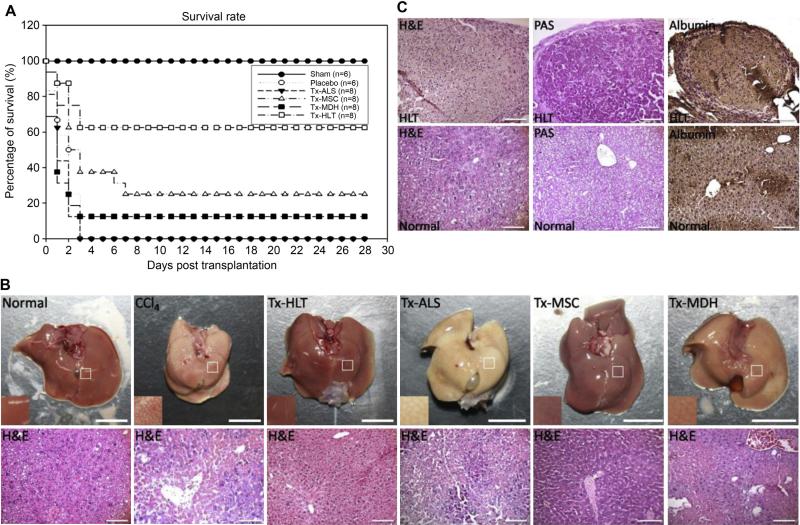
A fulminant hepatic failure mouse model involving the intraperitoneal injection of CCl4, which was known to induce hepatotoxicity by oxidative damage, cellular necrosis and extensive vacuolar degeneration, was used to evaluate the therapeutic potential of grafts. This involved the treatment of mice with placebo (PBS), ALS only, undifferentiated MSCs, MSC-derived hepatocyte-like cells using 2D approach and HLT. All the animals either infused with placebo or had received the ALS only had died of hepatic failure; 2 out of 8 mice survived by transplantation of MSCs; 1 out of 8 mice survived by transplantation of MDHs; 5 out of 8 mice survived by transplantation of HLT (Fig. 6A). The survival rate was significantly higher in this group (P = 0.017). These results suggested that the transplantation of HLT significantly prolonged the survival of mice that had received a lethal dose of CCl4 compared to transplantation of ALS only, MSCs, MDHs and placebo. Biochemical analysis showed a dramatic increase after 24 h in the concentrations of marker proteins such as albumin, serum glutamic oxaloacetic transaminase (SGOT), serum glutamic pyruvic transaminase (SGPT) and total bilirubin (TB) in the serum of the CCl4 treated mice, which confirms that CCl4 liver damage had been induced. Liver functions were improved after the transplantation of HLT, and recovery was completely achieved at 7 days after transplantation (Table 1). The appearance and histological analysis of the liver revealed the presence of serious liver necrosis after CCl4 treatment and necrosis was attenuated by transplantation of HLT compared with transplantation of ALS with no cells, MSCs, MDHs and placebo (Fig. 6B). After transplantation of HLT for four weeks, we harvested the transplanted site of liver to validate. Histochemical staining revealed that the HLT indeed engrafted into the region of lobule. In addition, in the liver failure mice at four weeks after transplantation, it was found that the HLT grafts possessed functional hepatic abilities, as well as exhibiting glycogen storage and albumin production (Fig. 6C).
Fig. 6.
Analysis of liver function after transplantation. (A) Survival curves of the NOD-SCID mice that had undergone intrahepatic transplantation with placebo (n = 6), ALS (n = 8) (P = 0.447), MSCs (n = 8) (P = 0.177), MDHs (n = 8) (P = 0.446) and HLT (n = 8) (P = 0.017) at 24 h after administration of 10 ml/kg CCl4. (B) Appearance and histological sections of liver from normal mice, CCl4-treated mice, HLT, ALS, MSCs and MDHs recipient mice after four weeks of transplantation. Scale bar: 10 mm (top panels); 100 μm (bottom panels). (C) Morphology and functionality of the HLT graft after transplantation in comparison to normal liver as demonstrated by PAS staining and immunohistological staining for albumin. Scale bar: 100 μm.
Table 1.
Liver function analysis of NOD-SCID mice that were administered 10 ml/kg CCl4 and intrahepatic transplantation with hepatic-like tissue.
| Albumin (g/dL) | SGOT (IU/L) | SGPT (IU/L) | TB (mg/dL) | |
|---|---|---|---|---|
| Normal | 2.8 ± 0.12 | 73 ± 12 | 13 ± 3.0 | 0.5 ± 0.2 |
| Post CCl4 day 1 (Transplantation) | 3.2 ± 0.058 | > 1000 | > 1000 | 1.4 ± 0.20 |
| Post IH-HLT day 3 | 2.8 ± 0.21 | 316 ± 146 | 218 ± 121 | 0.7 ± 0.2 |
| Post IH-HLT day 7 | 2.8 ± 0.058 | 97 ± 18 | 27 ± 15 | 0.2 ± 0.1 |
| Post IH-HLT day 14 | 2.9 ± 0.10 | 73 ± 5.7 | 16 ± 4.2 | 0.4 ± 0.2 |
| Post IH-HLT day 21 | 2.7 ± 0.058 | 58 ± 14 | 12 ± 2.9 | 0.2 ± 0.1 |
| Post IH-HLT day 28 | 2.7 ± 0.10 | 72 ± 2.1 | 17 ± 1.5 | 0.2 ± 0.0 |
NOTE. Data are presented as mean ± SD of three determinations.
IH, intrahepatic transplantation; HLT, hepatic-like tissue; SGOT, Serum glutamic oxaloacetic transaminase; SGPT, serum glutamic pyruvic transaminase; TB, total bilirubin.
4. Discussion
The challenge of tissue engineering is the supply of gas and nutrients into the cells seeded in the scaffolds [7,22]. Indeed, decellularization provides a new milestone in the engineering of a bioartificial liver [23]. In order to prepare an ideal liver scaffold, we adopt a sequential approach consisting of a combination of physical and chemical methods to decellularize the whole liver [20,21,24–27]. In our method, deionized water is used instead of PBS for washing to maintain most of the collagen within the ECM, followed by sequential freezing and thawing to physically disrupt cell membranes and to avoid disruptions of ECM ultrastructures. Subsequently, chemical treatment with Triton X-100 and ammonium hydroxide is used to facilitate the decellularized process. The reason of using Triton X-100 is that it is a relatively mild nonionic detergent and allows better extracellular matrix retention compared to SDS [24]. Furthermore, a unique perfusion system is generated to promote continuous flow and diffusion of chemical reagents to remove cellular components during the decellularization process of the whole liver. Finally, 50 million MSCs are seeded into the matrix, which is roughly the number of hepatocytes present in a native mouse liver. This is done via a multistep infusion strategy that gives the greatest seeding efficiency and facilitates cell repopulation [28]. The decellularized liver by the cryo-chemical decellularization method also preserves the intact vascular tree (Fig. 1), which is favorable for recellularization and revascularization after transplantation.
Previous studies have suggested that decellularized liver is able to maintain the physiological functioning of primary hepatocytes or precursor cells [23,29], Because of the shortage of hepatocyte from donor livers, we use MSCs, which possess the potential to differentiate into functional hepatocyte-like cells [5,13]. In addition, the use of MSCs can also bypasses the ethical controversies [30]. Our previous study has shown that MSCs can differentiate into hepatocyte-like cells in 2D culture dish by a defined protocol [3]. In this study, we further demonstrate that hepatic differentiation of MSCs in our 3D liver scaffolds is superior to the conventional 2D culture. The ALS generated in this study is able to efficiently promote hepatic differentiation of MSCs, implying its high efficiency in mass transfer.
A previous study has shown that decellularized liver matrix can indeed promote hepatic differentiation of MSCs in vitro [31]. In our study, we use a more effective liver decellularization process which can maintain inherent structure and matrix but can also eliminate the cellular debris (Fig. 2), and preserve the intact vascular architecture, which will facilitate cell seeding and nutrients exchange (Fig. 1). Also, we maximize the use of the whole organ perfusion culture to allow continuous flow and optimize mass transfer (Fig. 3), thus enhancing the hepatic differentiation of MSCs. Most importantly, the in vivo function of our HLT has been demonstrated by transplanting into in site injured livers. The advantage of directly transplanting the graft into the injured liver is that loss of cells can be minimized compared to intravenous injection [31].
To investigate the effect of ALS on MSCs under hepatic differentiation, we have cultured MSCs in tissue culture dish and ALS with the same hepatic induction medium and further validated the expression of liver development genes [32,33]. Up-regulation of hepatic-specific transcription factors (HNF1A, HNF1B, HNF3B and HNF4A), hepatic progenitor marker protein (AFP and CK19), liver-associated enzyme (G6P and TAT), transcription factor of fetal liver (GATA4) and epithelial marker (EpCAM) is found in MSCs cultured in ALS and the expression level is significantly higher than in 2D culture (Fig. 4). This result suggests that the liver scaffold perform the independent inductive potential for the differentiation of MSCs into the hepatic lineage.
According to our result, MSCs in whole ALS after four weeks of hepatic induction express high levels of AFP and CK19 (Fig. 5), suggesting that these cells, at least some of them are in the progenitor state. Previous studies have indicated that hepatic progenitor cells (or fetal liver cells) possess greater regenerate potential than fully matured hepatocytes in experimental liver failure models [34,35]; therefore, MSCs may be a better source for HLT compared to mature hepatocytes. It have also been shown that transplantation of fetal liver fragments or implants are functional at the intrahepatic implantation site [36]. Our findings clearly indicate that orthotopic intrahepatic transplantation of HLT in vivo can not only preserve liver function but can also potentially contribute to the restoration of liver function in the animal model of lethal fulminant hepatic failure (Fig. 6).
5. Conclusions
Our study proves that ALS made by cryo-chemical decellularization methods is able to effectively remove the cellular materials from the liver, whilst maintain the physical structure and the ECM components. ALS fabricated in this method promotes the differentiation of MSCs into functional hepatocyte-like cells under the whole organ perfusion culture system. HLT generated by re-seeding of MSCs into ALS severe as a transplantation source to rescue lethal fulminant hepatic failure. In conclusion, HLT generated by decellularization with cryo-chemical method by re-seeding of MSCs restore liver functions after transplantation. Such approach can be further translated into clinical application to overcome the current need of donor liver shortage.
Acknowledgments
This work was supported in part by the UST-UCSD International Center of Excellence in Advanced Bio-engineering sponsored by the Taiwan National Science Council I-RiCE Program under Grant Number: NSC101-2911-I-009-101. The authors also acknowledge financial support from the Taipei Veterans General Hospital (VGH102E1-007, VGH102C-001), the National Science Council, Taiwan (NSC102-2120-M-010-002, NSC100-2314-B-010-030-MY3, and NSC102-2321-B-010-008), and the Ministry of Economic Affairs, Taiwan (102-EC-17-A-17-S1-203). This study was also supported by a grant from Ministry of Education, Aiming for the Top University Plan.
Footnotes
Conflict of interest
There is no conflict of interest in the manuscript.
References
- 1.Brown RS., Jr Live donors in liver transplantation. Gastroenterology. 2008;134:1802–13. doi: 10.1053/j.gastro.2008.02.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalgetty DM, Medine CN, Iredale JP, Hay DC. Progress and future challenges in stem cell-derived liver technologies. Am J Physiol Gastrointest Liver Physiol. 2009;297:241–8. doi: 10.1152/ajpgi.00138.2009. [DOI] [PubMed] [Google Scholar]
- 3.Lee KD, Kuo TK, Whang-Peng J, Chung YF, Lin CT, Chou SH, et al. In vitro hepatic differentiation of human mesenchymal stem cells. Hepatology. 2004;40:1275–84. doi: 10.1002/hep.20469. [DOI] [PubMed] [Google Scholar]
- 4.Dai LJ, Li HY, Guan LX, Ritchie G, Zhou JX. The therapeutic potential of bone marrow-derived mesenchymal stem cells on hepatic cirrhosis. Stem Cell Res. 2009;2:16–25. doi: 10.1016/j.scr.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Kuo TK, Hung SP, Chuang CH, Chen CT, Shih YR, Fang SC, et al. Stem cell therapy for liver disease: parameters governing the success of using bone marrow mesenchymal stem cells. Gastroenterology. 2008;134:2111–21. doi: 10.1053/j.gastro.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatia SK. Tissue engineering for clinical applications. Biotechnol J. 2010;5:1309–23. doi: 10.1002/biot.201000230. [DOI] [PubMed] [Google Scholar]
- 7.Chan BP, Leong KW. Scaffolding in tissue engineering: general approaches and tissue-specific considerations. Eur Spine J. 2008;17:467–79. doi: 10.1007/s00586-008-0745-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ranucci CS, Moghe PV. Polymer substrate topography actively regulates the multicellular organization and liver-specific functions of cultured hepatocytes. Tissue Eng. 1999;5:407–20. doi: 10.1089/ten.1999.5.407. [DOI] [PubMed] [Google Scholar]
- 9.Kim SS, Utsunomiya H, Koski JA, Wu BM, Cima MJ, Sohn J, et al. Survival and function of hepatocytes on a novel three-dimensional synthetic biodegradable polymer scaffold with an intrinsic network of channels. Ann Surg. 1998;228:8–13. doi: 10.1097/00000658-199807000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Underhill GH, Chen AA, Albrecht DR, Bhatia SN. Assessment of hepatocellular function within PEG hydrogels. Biomaterials. 2007;28:256–70. doi: 10.1016/j.biomaterials.2006.08.043. [DOI] [PubMed] [Google Scholar]
- 11.Sugimoto S, Harada K, Shiotani T, Ikeda S, Katsura N, Ikai I, et al. Hepatic organoid formation in collagen sponge of cells isolated from human liver tissues. Tissue Eng. 2005;11:626–33. doi: 10.1089/ten.2005.11.626. [DOI] [PubMed] [Google Scholar]
- 12.Takimoto Y, Dixit V, Arthur M, Gitnick G. De novo liver tissue formation in rats using a novel collagen-polypropylene scaffold. Cell Transplant. 2003;12:413e, 21. doi: 10.3727/000000003108746966. [DOI] [PubMed] [Google Scholar]
- 13.Seo SJ, Choi YJ, Akaike T, Higuchi A, Cho CS. Alginate/galactosylated chitosan/ heparin scaffold as a new synthetic extracellular matrix for hepatocytes. Tissue Eng. 2006;12:33–44. doi: 10.1089/ten.2006.12.33. [DOI] [PubMed] [Google Scholar]
- 14.Cheng N, Wauthier E, Reid LM. Mature human hepatocytes from ex vivo differentiation of alginate-encapsulated hepatoblasts. Tissue Eng Part A. 2008;14:1–7. doi: 10.1089/ten.a.2007.0131. [DOI] [PubMed] [Google Scholar]
- 15.Badylak SF, Taylor D, Uygun K. Whole-organ tissue engineering: decellularization and recellularization of three-dimensional matrix scaffolds. Annu Rev Biomed Eng. 2011;13:27–53. doi: 10.1146/annurev-bioeng-071910-124743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song JJ, Ott HC. Organ engineering based on decellularized matrix scaffolds. Trends Mol Med. 2011;17:424–32. doi: 10.1016/j.molmed.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, et al. Perfusiondecellularized matrix: using nature's platform to engineer a bioartificial heart. Nat Med. 2008;14:213–21. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 18.Ott HC, Clippinger B, Conrad C, Schuetz C, Pomerantseva I, Ikonomou L, et al. Regeneration and orthotopic transplantation of a bioartificial lung. Nat Med. 2010;16:927–33. doi: 10.1038/nm.2193. [DOI] [PubMed] [Google Scholar]
- 19.Song JJ, Guyette JP, Gilpin SE, Gonzalez G, Vacanti JP, Ott HC. Regeneration and experimental orthotopic transplantation of a bioengineered kidney. Nat Med. 2013;19:646–51. doi: 10.1038/nm.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baptista PM, Siddiqui MM, Lozier G, Rodriguez SR, Atala A, Soker S. The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology. 2011;53:604–17. doi: 10.1002/hep.24067. [DOI] [PubMed] [Google Scholar]
- 21.Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011;32:3233–43. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiegel HC, Kneser U, Kluth D, Metzger R, Till H, Rolle U. Development of hepatic tissue engineering. Pediatr Surg Int. 2009;25:667–73. doi: 10.1007/s00383-009-2389-8. [DOI] [PubMed] [Google Scholar]
- 23.Uygun BE, Soto-Gutierrez A, Yagi H, Izamis ML, Guzzardi MA, Shulman C, et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat Med. 2010;16:814–20. doi: 10.1038/nm.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert TW, Sellaro TL, Badylak SF. Decellularization of tissues and organs. Biomaterials. 2006;27:3675–83. doi: 10.1016/j.biomaterials.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 25.Kajbafzadeh AM, Javan-Farazmand N, Monajemzadeh M, Baghayee A. Determining the optimal decellularization and sterilization protocol for preparing a tissue scaffold of a human-sized liver tissue. Tissue Eng Part C Methods. 2013;19:642–51. doi: 10.1089/ten.TEC.2012.0334. [DOI] [PubMed] [Google Scholar]
- 26.Lang R, Stern MM, Smith L, Liu Y, Bharadwaj S, Liu G, et al. Three-dimensional culture of hepatocytes on porcine liver tissue-derived extracellular matrix. Biomaterials. 2011;32:7042–52. doi: 10.1016/j.biomaterials.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Olausson M, Patil PB, Kuna VK, Chougule P, Hernandez N, Methe K, et al. Transplantation of an allogeneic vein bioengineered with autologous stem cells: a proof-of-concept study. Lancet. 2012;380:230–7. doi: 10.1016/S0140-6736(12)60633-3. [DOI] [PubMed] [Google Scholar]
- 28.Soto-Gutierrez A, Zhang L, Medberry C, Fukumitsu K, Faulk D, Jiang H, et al. A whole-organ regenerative medicine approach for liver replacement. Tissue Eng Part C Methods. 2011;17:677–86. doi: 10.1089/ten.tec.2010.0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Cui CB, Yamauchi M, Miguez P, Roach M, Malavarca R, et al. Lineage restriction of human hepatic stem cells to mature fates is made efficient by tissue-specific biomatrix scaffolds. Hepatology. 2011;53:293–305. doi: 10.1002/hep.24012. [DOI] [PubMed] [Google Scholar]
- 30.Keating A. Mesenchymal stromal cells: new directions. Cell Stem Cell. 2012;10:709–16. doi: 10.1016/j.stem.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 31.Ji R, Zhang N, You N, Li Q, Liu W, Jiang N, et al. The differentiation of MSCs into functional hepatocyte-like cells in a liver biomatrix scaffold and their transplantation into liver-fibrotic mice. Biomaterials. 2012;33:8995–9008. doi: 10.1016/j.biomaterials.2012.08.058. [DOI] [PubMed] [Google Scholar]
- 32.Kinoshita T, Miyajima A. Cytokine regulation of liver development. Biochim Biophys Acta. 2002;1592:303–12. doi: 10.1016/s0167-4889(02)00323-3. [DOI] [PubMed] [Google Scholar]
- 33.Duncan SA. Mechanisms controlling early development of the liver. Mech Dev. 2003;120:19–33. doi: 10.1016/s0925-4773(02)00328-3. [DOI] [PubMed] [Google Scholar]
- 34.Oertel M, Menthena A, Dabeva MD, Shafritz DA. Cell competition leads to a high level of normal liver reconstitution by transplanted fetal liver stem/ progenitor cells. Gastroenterology. 2006;130:507–20. doi: 10.1053/j.gastro.2005.10.049. [DOI] [PubMed] [Google Scholar]
- 35.Yovchev MI, Grozdanov PN, Zhou H, Racherla H, Guha C, Dabeva MD. Identification of adult hepatic progenitor cells capable of repopulating injured rat liver. Hepatology. 2008;47:636–47. doi: 10.1002/hep.22047. [DOI] [PubMed] [Google Scholar]
- 36.Katchman H, Tal O, Eventov-Friedman S, Shezen E, Aronovich A, Tchorsh D, et al. Embryonic porcine liver as a source for transplantation: advantage of intact liver implants over isolated hepatoblasts in overcoming homeostatic inhibition by the quiescent host liver. Stem Cells. 2008;26:1347–55. doi: 10.1634/stemcells.2007-0631. [DOI] [PubMed] [Google Scholar]