Abstract
Dilated cardiomyopathy (DCM) is a major cause of sudden cardiac death and heart failure, and it is characterized by genetic and clinical heterogeneity, even for some patients with a very poor clinical prognosis; in the majority of cases, DCM necessitates a heart transplant. Genetic mutations have long been considered to be associated with this disease. At present, mutations in over 50 genes related to DCM have been documented. This study was carried out to elucidate the characteristics of gene mutations in patients with DCM. The candidate genes that may cause DCM include MYBPC3, MYH6, MYH7, LMNA, TNNT2, TNNI3, MYPN, MYL3, TPM1, SCN5A, DES, ACTC1 and RBM20. Using next-generation sequencing (NGS) and subsequent mutation confirmation with traditional capillary Sanger sequencing analysis, possible causative non-synonymous mutations were identified in ~57% (12/21) of patients with DCM. As a result, 7 novel mutations (MYPN, p.E630K; TNNT2, p.G180A; MYH6, p.R1047C; TNNC1, p.D3V; DES, p.R386H; MYBPC3, p.C1124F; and MYL3, p.D126G), 3 variants of uncertain significance (RBM20, p.R1182H; MYH6, p.T1253M; and VCL, p.M209L), and 2 known mutations (MYH7, p.A26V and MYBPC3, p.R160W) were revealed to be associated with DCM. The mutations were most frequently found in the sarcomere (MYH6, MYBPC3, MYH7, TNNC1, TNNT2 and MYL3) and cytoskeletal (MYPN, DES and VCL) genes. As genetic testing is a useful tool in the clinical management of disease, testing for pathogenic mutations is beneficial to the treatment of patients with DCM and may assist in predicting disease risk for their family members before the onset of symptoms.
Keywords: dilated cardiomyopathy, genetic testing, candidate genes, next-generation sequencing
Introduction
Dilated cardiomyopathy (DCM) is one of the most prevalent inherited cardiomyopathies and is known to be one of the leading causes of heart failure and sudden cardiac death, and typically necessitates a heart transplant. DCM is a genetically heterogeneous disease characterized by cardiac left ventricular enlargement and systolic dysfunction (1–3). In the general population, the prevalence of DCM is greater than 1 in 2,500 individuals (4). The majority of patients present with sporadic DCM (SDCM); however, genetic screening of first-degree relatives reveals that 20–35% of cases are familial DCM. Familial DCM is defined as SDCM in two or more closely related family members or, when involving a first-degree relative of a patient with SDCM (5,6).
DCM is primarily caused by pathogenic gene mutations inherited in a Mendelian autosomal dominant pattern (7–9). However, in a small number of cases, autosomal recessive, X-linked, or mitochondrial DNA inheritance characteristics have been observed (10–14). Currently, over 50 genes have been documented to cause DCM. Of these, the majority harbour missense mutations located in sarcomere proteins, including cardiac myosin-binding protein C (MYBPC3), cardiac alpha-myosin heavy chain (MYH6), cardiac beta-myosin heavy chain (MYH7), troponin T type 2 (cardiac) (TNNT2), troponin I type 3 (cardiac) (TNNI3), tropomyosin 1 (alpha) (TPM1), myosin light chain 3 (MYL3), and actin, alpha, cardiac muscle 1 (ACTC1). In addition, mutations in the nuclear envelope proteins of nuclear lamin A/C (LMNA); cytoskeletal proteins, including myopalladin (MYPN), desmin (DES) and vinculin (VCL); calcium/sodium-handling proteins of sodium channel, voltage gated, type V alpha subunit (SCN5A); desmosomal proteins, such as desmoplakin (DSP) and RNA-binding motif protein 20 (RBM20); as well as other types of genes are considered to be closely related to the occurrence of DCM (Table I) (4,15).
Table I.
Selected genes associated with inherited cardiomyopathy.
| Gene name | Protein name | Chromosome | Exons | OMIM | Inherited cardiomyopathy phenotype |
|---|---|---|---|---|---|
| ABCC9 | ATP-binding cassette, subfamily C, member 9 | 12p12.1 | 39 | 601439 | DCM |
| ACTN2 | Actinin, alpha 2 | 1q42-q43 | 21 | 102573 | HCM, DCM |
| CAV3 | Caveolin 3 | 3p25.3 | 2 | 601253 | DCM |
| DES | Desmin | 2q35 | 9 | 125660 | DCM, ARVC |
| LAMA4 | Laminin alpha 4 | 6q21 | 39 | 600133 | DCM |
| LAMP2 | Lysosome-associated membrane protein 2 | Xq24 | 10 | 309060 | HCM, DCM |
| LDB3 | LIM domain-binding 3 | 10q23.2 | 16 | 605906 | HCM, DCM |
| LMNA | Lamin A/C | 1q22 | 12 | 150330 | DCM, ARVC |
| MYBPC3 | Cardiac myosin-binding protein-C | 11p11.2 | 35 | 600958 | HCM, DCM |
| MYH6 | Myosin, heavy chain 6, cardiac muscle, alpha | 14q11.2-q12 | 39 | 160710 | HCM, DCM |
| MYH7 | Myosin, heavy chain 7, cardiac muscle, beta | 14q11.2-q12 | 40 | 160760 | HCM, DCM |
| MYL2 | Myosin light chain 2 | 12q23-q.24.3 | 7 | 160781 | HCM |
| MYL3 | Myosin light chain 3 | 3p21.2-p21.3 | 7 | 160790 | HCM |
| MYPN | Myopalladin | 10q21.1 | 20 | 608517 | DCM |
| PSEN2 | Presenilin 2 | 1q42.13 | 13 | 600759 | DCM |
| RBM20 | RNA-binding motif protein 20 | 10q25.2 | 14 | 613171 | DCM |
| SCN5A | Sodium channel, voltage-gated type V, alpha subunit | 3p22.2 | 29 | 600163 | DCM |
| SGCD | Delta-sarcoglycan | 5q33.3 | 9 | 601411 | DCM |
| TMPO | Thymopoietin | 12q23.1 | 10 | 188380 | DCM |
| TCAP | Telethonin | 17q12-q21.1 | 2 | 604488 | HCM, |
| TNNC1 | Troponin C type 1 (slow) | 3p21.1 | 6 | 191040 | HCM, DCM |
| TNNI3 | Troponin I, cardiac muscle | 19q13.4 | 8 | 191044 | HCM, DCM |
| TNNT2 | Troponin T, cardiac muscle | 1q32 | 17 | 191045 | HCM, DCM |
| TPM1 | Tropomyosin 1 alpha | 15q22.1-q22.2 | 10 | 191010 | HCM, DCM |
| VCL | Vinculin | 10q22.1-q23 | 22 | 193065 | HCM, DCM |
DCM, dilated cardiomyopathy; HCM, hypertrophic cardiomyopathy; ARVC, arrhythmogenic right ventricular cardiomyopathy.
Recent technical advances have allowed high-throughput next-generation sequencing (NGS) to overcome the limitations of traditional capillary Sanger sequencing (16,17). NGS has previously been used for genetic diagnoses in clinical settings (18). In theory, genetic testing can identify patients at risk of developing DCM prior to the onset of clinical symptoms. The most frequently used Sanger sequencing technique is, however, cumbersome and expensive. Comparatively, NGS techniques are cost-efficient and generate a large amount of data in one reaction (19,20). Moreover, the molecular genetics data of patients with DCM are insufficient in China and have not been reported previously in relation to the Yunnan population. To provide new insight into the genetic profile of DCM in the Yunnan population in China, in the current study, we employed NGS screening to investigate the major DCM-causing genes in patients diagnosed with the disease. To the best of our knowledge, this is the first description of a gene mutation profile of patients with DCM from Yunnan in southwestern China. In view of the fact that genetic testing is a useful indicator for the clinical management of disease, testing for pathogenic mutations may be beneficial to the treatment of patients with DCM and may be used to predict disease risk for their family members prior to the onset of symptoms.
Materials and methods
Subjects and clinical evaluation
All subjects (n=21; 15 male, 6 female; median age at onset, 48.7 (±11.7) years; age range, 26–73 years) were recruited from southern and western Yunnan, China, and had been previously diagnosed with DCM in accordance with previously published guidelines (21). At the clinical interview, physical examinations, invasive examinations and echocardiography (ECG) were performed to determine the phenotype of each SDCM [left ventricular ejection fraction (LVEF) <50%; left ventricular end-diastolic diameter (LVED) >112% of the predicted value]. Written informed consent and additional clinical information were obtained from each patient with DCM. This study was approved by the Institutional Ethics Committee of the First People's Hospital of Yunnan Province (Affiliated Hospital of Kunming University of Science and Technology) and complied with the principles of the Declaration of Helsinki.
Candidate gene sequencing
Peripheral whole blood lymphocyte samples (2 ml) from each patient were collected in Vacutainer tubes coated with EDTA (BD Biosciences, Franklin Lakes, NJ, USA) and stored at 4°C until DNA extraction. Genomic DNA was extracted from each blood sample using a commercially available genomic AxyPrep DNA miniprep kit (Axygen Biosciences, Union City, CA, USA) following the manufacturer's instructions. A total of 25 genes associated with DCM (Table I) were selected as candidate genes. The amplicons were captured using a custom-designed multi-target gene library (Agilent Technology, Santa Clara, CA, USA) to achieve sufficient sequences covering the genes of interest, and the amplicons were then sequenced using a Genome Analyzer IIx (Illumina, Inc., San Diego, CA, USA).
Sequence alignment and variant calling
Low-quality reads were discarded following an initial inspection with FASTX-tools (http://hannonlab.cshl.edu/fastx_toolkit/index.html), and qualified sequences were aligned together with the human reference genome assembly (GRCh37/hg19) in Burrows-Wheeler Aligner, Smith-Waterman Alignment (BWA-SW, version 0.5.9) and Sequence Alignment/Map (SAM; version 0.1.16) tools software package, as previously described (22,23). Sequence calls for variants were identified using Variant caller (version 4.2.0), as previously described (24). Following variant detection, the identified variants (splice, stop loss, synonymous, non-synonymous, insertion, or deletion variants) were annotated with the ANNOVAR annotation tool or using online Ion Reporter software (25; https://ionreporter.lifetechnologies.com/ir/secure/home.html).
Molecular genetic analysis
The potential pathogenic role of variants in the coding region was focused on in the present study, i.e. only the statistical variation in the coding region. Putative pathogenic mutations were considered to be pathogenic based on the following criteria (26–29): i) the mutation has been reported to be associated with the disease phenotype in the reference or Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/docs/login.html); ii) the mutation has a minor allele frequency (MAF) of <1% in the NCBI dbSNP Build 137 (http://www.ncbi.nlm.nih.gov/projects/SNP), the 1000 Genomes Project (http://www.1000genomes.org/), and the National Heart, Lung, and Blood Institute Exome Sequencing Project (https://esp.gs.washington.edu/drupal/) databases; iii) the protein structure and function was significantly altered, and the amino acid was highly conserved across a number of species; iv) the mutation was analyzed and shown to be pathogenic using the PolyPhen-2, SIFT, or MutationTaster (30–32) algorithms; or v) novel mutation or variants of uncertain significance had to be absent from unrelated and healthy controls which were matched for ethnicity. The mutation nomenclature was based on the nucleotide reference sequences: MYPN, NM_032578; TNNT2, NM_001001432; MYH7, NM_000257; DES, NM_001927; VCL, NM_014000.2; MYH6, NM_002471.3; RBM20, NM_001134363.1; TNNC1, NM_003280.2; MYBPC3, NM_000256; and NM_000258.2 for MYL3.
Validation of selected mutations
All of the putative pathogenic mutations were reconfirmed by traditional capillary Sanger sequencing (ABI 3130 Genetic Analyzer; Life Technology, Carlsbad, CA, USA) of the PCR product for all suspected samples, using primers for preliminary determined mutations (Table II). The amplification of each mutation fragment was undertaken using primeSTAR GXL DNA polymerase (Takara, Otsu, Japan, code no. R050A). PCR conditions consisted of 1 cycle of 96°C for 5 min; 30 cycles of 96°C for 3 min, 54°C or 57°C for 30 sec, 68°C for 1 min (200 bp<fragment size<1000 bp) or 2 min (1000 bp<fragment size<2000 bp); 1 extension cycle of 68°C for 5 min and holding at 16°C. Control alleles (n=200) were taken from 100 unrelated healthy subjects with normal phenotypes (matched for gender, age and ethnicity) to exclude the possibility of rare polymorphisms of the novel mutations and variants of uncertain significance.
Table II.
PCR primers for amplification of the mutation sites.
| Gene symbol | Nucleotide changes | Exon | Primer (5′→3′) | Annealing temperature (°C) |
|---|---|---|---|---|
| MYPN | c.1888G>A | 10 | Sense: ACTCAGCCAACTCTACCACCAACC | 54 |
| Antisense: ACTGGAGGGGGCTCTTTCACG | ||||
| TNNT2 | c.539G>C | 12 | Sense: ACCTTCTCCCTATGCACACCT | 57 |
| Antisense: CACAGCAGCTGGGAATCTCTT | ||||
| MYH7 | c.77C>T | 3 | Sense: AGCCAGCTTCTGCTCACTCCAG | 57 |
| Antisense: GCCACTTGTAAGGGTTGACGGT | ||||
| DES | c.1157G>A | 6 | Sense: CAGGAGATGATGGAATACCGACAC | 54 |
| Antisense: ACAGAAATGGACCACCCAGCAC | ||||
| VCL | c.625A>T | 6 | Sense: CAGCAGGATACGCCATTCAGAGT | 57 |
| Antisense: GACCAGAGCAGCAGCAACACA | ||||
| RBM20 | c.3545G>A | 13 | Sense: GCAGAGAGAGGTACAGTGTGAAG | 57 |
| Antisense: AAGTCTATGGGAAGATTAGGGGTTT | ||||
| MYBPC3 | c.3371G>T | 31 | Sense: AGAGGCTCTCGGCATCAGGAAG | 54 |
| Antisense: ACATAGATGCCCCCGTCAAAGG | ||||
| TNNC1 | c.8A>T | 1 | Sense: GGGATAAACTTGATACGAACTCTG | 57 |
| Antisense: GAGGAAGAGATGAAAAGAGAAAGG | ||||
| MYH6 | c.3139C>T | 24 | Sense: TTTTGCTCTCTGTAGTTCCTCAC | 54 |
| Antisense: TGCTCATCCTCAATCTTACTGTTC | ||||
| MYH6 | c.3758C>T | 27 | Sense: ATGCCTTCTCTCTCTGTCTGCC | 54 |
| Antisense: GTGAAATCATTGAGGGAGCGTT | ||||
| MYBPC3 | c.478C>T | 4 | Sense: GCCCTTCAGTCTCAGCTTTTAGC | 54 |
| Antisense: CTTGAGCCCTTAGCCCTGATACT | ||||
| MYL3 | c.377A>G | 4 | Sense: AAAGTGCCTCGCGATGGTAGTTTG | 57 |
| Antisense: GTCTGCCATTGAGGCTCCCTAATT |
Statistical analysis
Statistical analysis was used to summarize the clinical characteristics of the subjects; SPSS 16.0 software was used. The mean values ± standard deviation (SD) are shown when comparing the presence and absence of mutation groups. T-test results of independent samples with p-values <0.05 were considered to indicate statistically significant differences.
Results
Study population
Molecular screening was performed on a cohort of 21 unrelated patients in Yunnan, who were recruited with an original clinical diagnosis of DCM. Of these patients, 15 were male and 6 were female, with a median age at onset of 48.7 (±11.7) years, and an age range of 26–73 years. ECG revealed enlarged heart chambers [LVED = 68.9±10.6; left ventricular end systolic diameter (LVESD) = 58.5±11.4] and a low LVEF (LVEF = 31.4±10.9); interventricular septal thickness and left ventricular posterior wall thickness were 9.3 and 8.6 mm, respectively. The left atrium size was 45.7 mm. The clinical characteristics of the patients with DCM are presented in Table III.
Table III.
Clinical characteristics of patients with dilated cardiomyopathy according to diagnosis.
| Characteristic | Cardiac screening (%) | Genetic testing
|
P-value | |
|---|---|---|---|---|
| Mutation absence: 9 (42.8%) | Mutation presence: 12 (57.2%) | |||
| Gender, n (%) | ||||
| Male | 15 (71.5) | 7 (77.8) | 8 (66.6) | |
| Female | 6 (28.5) | 2 (22.2) | 4 (33.4) | N/A |
| Age at onset (years) | 48.7±11.7 | 57.1±9.1 | 42.3±9.3 | 0.002a |
| IVST (mm) | 9.3±1.9 | 10.2±1.6 | 8.6±1.7 | N/A |
| LVED (mm) | 68.9±10.6 | 74.4±12.6 | 64.8±6.6 | N/A |
| LVESD (mm) | 58.5±11.4 | 65.5±12.4 | 53.3±7.4 | N/A |
| LVPWT (mm) | 8.6±1.5 | 8.3±0.7 | 8.8±1.8 | N/A |
| LA (mm) | 45.7±9.5 | 50.3±10.8 | 42.3±7.1 | N/A |
| LVEF (%) | 31.4±10.9 | 26.6±9.0 | 35.0±11.1 | 0.078 |
IVST, interventricular septal thickness; LVED, left ventricular end-diastolic diameter; LVESD, left ventricular end systolic diameter; LVPWT, left ventricular posterior wall thickness; LA, left atrium; LVEF, left ventricular ejection fraction; N/A, not applicable.
Statistically significant, p<0.05.
Identification of DCM-related mutations
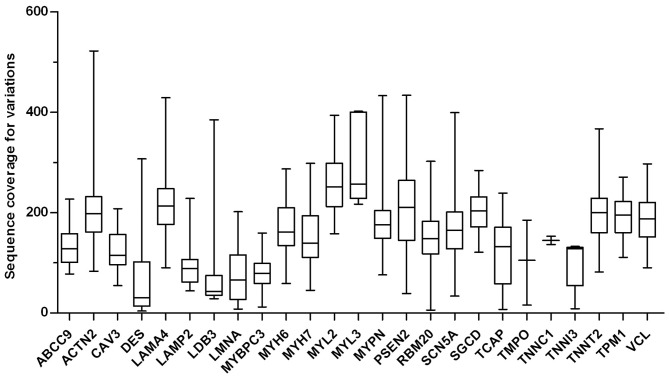
Sequencing of the major DCM-causing genes (Table I) was performed. Subsequently, the short reads mapped onto the reference genome (GRCh37/hg19) were annotated with the ANNOVAR annotation tool or Ion Reporter software. All successfully mapped sequence reads were analyzed to detect sequence variants, including non-synonymous, synonymous, insertion, and deletion variants. In all patients, the mean read depth of coverage for all target gene variants was 170.8X (range, 4 to 522X; Fig. 1). Altogether, 243 missense mutations (data available upon request) were identified with an average of 12 variants found per patient with DCM. On the other hand, the potential of the variations were detected in the coding region (CDS) of the target genes in 21 patients with DCM (data available upon request).
Figure 1.
Sequence coverage for mutations within selected target genes. A box plot of the distribution of the coverage on mutations from the target genes is shown, ranging from 4 to 522X in 21 consecutive patients with DCM. The mean coverage was 172.4X.
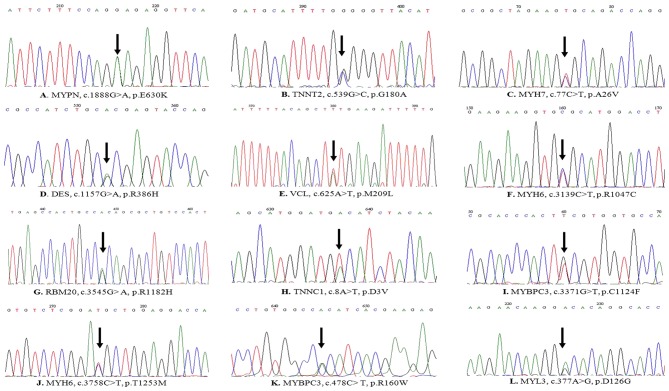
The variants were selected from most of the synonymous or non-synonymous variants and compared with the reported data from NCBI dbSNP (MAF>0.01). Finally, 12 possibly pathogenic heterozygous mutations from the candidate genes were revealed (Table IV), and these mutations were confirmed using traditional capillary Sanger sequencing (Fig. 2). Of these, 2 mutations, p.A26V in MYH7 and p.R160W in MYBPC3, have been previously documented (33), and 3 variants of uncertain significance (VCL, p.M209L; RBM20, p.R1128H; MYH6, p.T1253M) in dbSNP have been documented in the 1000 Genomes Project database at MAF = 0.0001, 0.0049, and 0, respectively, in the Han Chinese population from Beijing. Furthermore, it is worth noting that 7 novel mutations (MYPN, p.E630K; TNNT2, p.G180A; DES, p.R386H; MYBPC3, p.C1124F; MYH6, p.R1047C; MYL3, p.D126G; and TNNC1, p.D3V) are not listed in the NCBI dbSNP database, 1000 Genomes Project, the Human Gene Mutation database, or Exome Sequencing Project database. These 7 novel mutations were absent in the healthy control samples, and the corresponding altered amino acids of the 7 novel mutations and 3 variants of uncertain significance were highly conserved across a number of species. This indicates that these altered amino acids share related protein functions (data available upon request). Finally, based on the online analysis using MutationTaster, PolyPhen2 and SIFT, these mutations were predicted to be disease-causing, not tolerated, or probably damaging (Table IV). These results suggest that the 7 novel mutations and 3 variants of uncertain significance were highly associated with DCM in the patients studied.
Table IV.
Summary of the disease mutations found in DCM patients.
| Patient no. | Gene | Gender | Nucleotide change | Amino acid change | SIFT | Ploy Phen-2 | Mutation Taster | Previously reported |
|---|---|---|---|---|---|---|---|---|
| DCM-1 | MYPN | M | c.1888G>A | p.E630K | T | B | DC | NO |
| DCM-10 | TNNT2 | M | c.539G>C | p.G180A | T | PD | DC | NO |
| DCM-12 | MYH7 | F | c.77C>T | p.A26V | NA | NA | NA | YES |
| DCM-21 | DES | F | c.1157G>A | p.R386H | NT | PD | DC | NO |
| DCM-29 | VCL | M | c.625A>T | p.M209L | T | B | DC | VUS |
| DCM-33 | MYH6 | M | c.3139C>T | p.R1047C | NT | PD | DC | NO |
| DCM-45 | RBM20 | M | c.3545G>A | p.R1182H | NT | B | DC | VUS |
| DCM-46 | TNNC1 | M | c.8A>T | p.D3V | NT | PD | DC | NO |
| DCM-7 | MYBPC3 | F | c.3371G>T | p.C1124F | T | B | DC | NO |
| DCM-55 | MYH6 | M | c.3758C>T | p.T1253M | T | PD | DC | VUS |
| DCM-11 | MYBPC3 | F | c.478C>T | p.R160W | NA | NA | NA | YES |
| DCM-13 | MYL3 | M | c.377A>G | p.D126G | NA | B | DC | NO |
NA, not applicable; T, tolerated; NT, not tolerated; B, benign; PD, probably damaging; DC, disease causing; VUS, variant of uncertain significance; DCM, dilated cardiomyopathy.
Figure 2.
(A-L) Sanger sequencing showing gene mutations observed in the present study in patients with DCM. Mutation sites are indicated by arrows.
Association of genotype and phenotype
To elucidate the association of genotype and phenotype in patients with DCM, a comparison of the clinical characteristics, such as age at diagnosis and LVEF, was performed between groups of patients with and without gene mutations (Table III). No statistically significant difference in LVEF was observed in the patients with DCM based on the presence or absence of genetic mutations (P=0.078>0.05). However, it was demonstrated that patients with DCM harbouring DCM-associated mutations had been diagnosed with their condition at a significantly younger age than patients with DCM without the mutations (57.1±9.1 vs. 42.3±9.3 years; P=0.002<0.05; Table III).
Mutant spectra in patients with DCM
As shown in Table IV, there were 12 probands (patients with DCM) harbouring one mutation (12/21, ~57.2%), including the 7 novel mutations, 3 variants of uncertain significance, and 2 previously reported mutations. From the patients with DCM carrying genetic variants (8 male, 4 female), the highest frequency of mutations was detected in sarcomere genes (8/12, ~66.7%; MYH7, MYBPC3, MYH6, TNNT2, TNNC1 and MYL3), followed by cytoskeletal genes (3/12, ~25%; MYPN, DES and VCL) and other types of genes (1/12, ~8.3%; RBM20). A small number of mutated genes were identified with a high frequency in the patients with DCM. To the best of our knowledge, the mutated genes, MYBPC3 and MYH6, were the most frequently observed genes with 2 out of 12 patients harbouring mutations in these two genes. However, the other mutated genes were only found in 1 patient each.
Discussion
DCM is primarily caused by pathogenic genetic mutations. For example, it has been suggested that mutations in genes encoding contractile proteins result in functional changes and lead to contractile dysfunction of cardiomyocytes (34,35). The majority of patients with DCM exhibit autosomal dominant genetic disease, although there are several reported cases with recessive, X-linked, and other patterns of inheritance (11,12,14,15). Thus, the Heart Rhythm Society/European Heart Rhythm Association expert consensus statement recommends the performance of genetic testing on patients with DCM. As regards clinical intervention for patients with DCM, there is still no effective treatment, aside from heart transplants. The identification of the molecular genetic basis of the disease is important for determining appropriate strategies for DCM prevention and management before the onset of symptoms.
The present study aimed to assess the prevalence of known DCM-related genes and the association between the results of cardiac screening and genetic testing on patients from Yunnan, China. The screening of candidate genes (Table I) in 21 patients with DCM was performed using NGS. Mutations were found in more than half of the DCM patients (12/21, ~57.2%). In comparison with previous studies (36–38), pathogenic mutations were present at a higher frequency in Yunnan DCM patients than in patients from other populations. Moreover, pathogenic mutations were not detected in 9 patients with DCM, due to localisation of the mutations to other untested genes, as has been previously suggested (39). MYH7-p.A26V (40,41) and MYBPC3-p.R160W (32,42) have been reported previously in different patient groups with hypertrophic cardiomyopathy (HCM). However, these mutations (MYH7-p.A26V and MYBPC3-p.R160W) were originally found in patients with DCM, once again illustrating the clinical and genetic heterogeneity of patients with DCM. Of these two mutations, MYH7-p.A26V results in an amino acid substitution located in the myosin heavy chain affecting different functional domains of the head or the head-rod junction of MYH7 (43). MYBPC3-p. R160W results in changes in the charge of the altered amino acid in the immunoglobulin-like domain, and this mutation is associated with both HCM (32,42) and DCM.
Notably, the 7 novel mutations, p.E630K in MYPN, p.G180A in TNNT2, p.R1047C in MYH6, p.D3V in TNNC1, p.R386H in DES, p.C1124F in MYBPC3, and p.D126G in MYL3, and the three variants of uncertain significance (RBM20, p.R1182H; MYH6, p.T1253M; and VCL, p.M209L) were not found in the genomes of 100 healthy controls with matched age, gender, and geographical region (data available upon request). A cross-species alignment of those sequences showed that their corresponding amino acids were highly evolutionarily conserved (data available upon request). Online bioinformatics software (PolyPhen-2, SIFT and MutationTaster) was used to predict the functional effects of the altered proteins in the DCM patients, resulting fromin? the seven novel mutations and three variants of uncertain significance. The results of the present analysis implied that these altered amino acids damage protein functions, as these mutations were predicted to localise to the functional region of the proteins (data available upon request).
Of the patients with DCM recruited for this study harbouring identified mutations, mutations in sarcomere genes and cytoskeletal genes were the most common, with a prevalence of ~66.7% (8/12) and ~25% (3/12), respectively (18). Of the tested genes in the present study, sarcomere (MYBPC3, MYH7, MYH6, TNNT2, TNNC1 and MYL3) and cytoskeletal genes (MYPN, DES and VCL) were the most frequently mutated. It is worth noting that MYH6 (p.R1047C and p.T1253M) and MYBPC3 (p.R160W and p.C1124F) gene mutations were found in two patients; the other gene mutations (MYH7, TNNT2, TNNC1, MYL3, MYPN, DES, RBM20 and VCL) were identified in one patient each. Comparatively, MYH6 and MYBPC3 gene mutations had not occurred at such a high frequency in previously documented DCM patients (2,44,37). This implies that genetic testing on sarcomere and cytoskeletal genes is a valuable diagnostic tool for individuals at a high risk for DCM. However, due to the limitations caused by the small sample size in this study, it is necessary to increase the sample size in future studies to confirm the genetic basis of SDCM.
In the current study, the prevalence and distribution of disease genes, the spectrum of gene mutations, and clinical features were first reported in DCM patients in Yunnan Province, China. We found a mutation in 57.2% of the tested DCM patients, a total of 12 non-synonymous mutations, and of these, seven novel mutations, were identified using NGS. MYBPC3 and MYH6, both sarcomere protein-encoding genes, were the most commonly identified. Therefore, our results indicate that targeted gene sequencing is a feasible approach to the identification of pathogenic mutations in DCM patients. Compared with whole genome sequencing, whole exome sequencing, and Sanger sequencing, NGS screening is efficient, fast, and cost-effective (45,46). We suggest that genetic testing on DCM patients thus provides the most effective means to identify at-risk family members, particularly those whose clinical features are mild or ambiguous. It is also important for early diagnostic clinical evaluation and the better management of family members at risk for DCM.
However, there were several limitations to the present study. The only information obtained for the control subjects was age, gender, ethnicity and certain clinical characteristics. Only 21 patients with DCM were recruited and screened, thus resulting in a small sample size. Thus, additional independent studies with a larger sample size are warranted in order to confirm our results. In the current study, we have only described the gene mutations of DCM patients; therefore, further studies on the mechanisms of novel mutations are necessary in order to elucidate the disease mechanisms at the level of cell or transgenic animal models.
Acknowledgments
The authors acknowledge the contributions of the participating patients in the dilated cardiomyopathy registry, and the authors would like to thank the staff of The First Hospital of Yunnan Province for providing their support. The present study was supported by Major Program of Applied Basic Research of Yunnan Province, China (grant no. 2013FC007).
References
- 1.Taylor MR, Carniel E, Mestroni L. Cardiomyopathy, familial dilated. Orphanet J Rare Dis. 2006;1:27. doi: 10.1186/1750-1172-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hershberger RE, Siegfried JD. Update 2011: clinical and genetic issues in familial dilated cardiomyopathy. J Am Coll Cardiol. 2011;57:1641–1649. doi: 10.1016/j.jacc.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michels VV, Moll PP, Miller FA, Tajik AJ, Chu JS, Driscoll DJ, Burnett JC, Rodeheffer RJ, Chesebro JH, Tazelaar HD. The frequency of familial dilated cardiomyopathy in a series of patients with idiopathic dilated cardiomyopathy. N Engl J Med. 1992;326:77–82. doi: 10.1056/NEJM199201093260201. [DOI] [PubMed] [Google Scholar]
- 4.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 5.Zhao L, Xu JH, Xu WJ, Yu H, Wang Q, Zheng HZ, Jiang WF, Jiang JF, Yang YQ. A novel GATA4 loss-of-function mutation responsible for familial dilated cardiomyopathy. Int J Mol Med. 2014;33:654–660. doi: 10.3892/ijmm.2013.1600. [DOI] [PubMed] [Google Scholar]
- 6.Zhou W, Zhao L, Jiang JQ, Jiang WF, Yang YQ, Qiu XB. A novel TBX5 loss-of-function mutation associated with sporadic dilated cardiomyopathy. Int J Mol Med. 2015;36:282–288. doi: 10.3892/ijmm.2015.2206. [DOI] [PubMed] [Google Scholar]
- 7.Xu L, Zhao L, Yuan F, Jiang WF, Liu H, Li RG, Xu YJ, Zhang M, Fang WY, Qu XK, et al. GATA6 loss-of-function mutations contribute to familial dilated cardiomyopathy. Int J Mol Med. 2014;34:1315–1322. doi: 10.3892/ijmm.2014.1896. [DOI] [PubMed] [Google Scholar]
- 8.Zhang XL, Dai N, Tang K, Chen YQ, Chen W, Wang J, Zhao CM, Yuan F, Qiu XB, Qu XK, et al. GATA5 loss-of-function mutation in familial dilated cardiomyopathy. Int J Mol Med. 2015;35:763–770. doi: 10.3892/ijmm.2014.2050. [DOI] [PubMed] [Google Scholar]
- 9.Yuan F, Qiu XB, Li RG, Qu XK, Wang J, Xu YJ, Liu X, Fang WY, Yang YQ, Liao DN. A novel NKX2-5 loss-of-function mutation predisposes to familial dilated cardiomyopathy and arrhythmias. Int J Mol Med. 2015;35:478–486. doi: 10.3892/ijmm.2014.2029. [DOI] [PubMed] [Google Scholar]
- 10.Hershberger RE, Morales A, Siegfried JD. Clinical and genetic issues in dilated cardiomyopathy: a review for genetics professionals. Genet Med. 2010;12:655–667. doi: 10.1097/GIM.0b013e3181f2481f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy RT, Mogensen J, Shaw A, Kubo T, Hughes S, McKenna WJ. Novel mutation in cardiac troponin I in recessive idiopathic dilated cardiomyopathy. Lancet. 2004;363:371–372. doi: 10.1016/S0140-6736(04)15468-8. [DOI] [PubMed] [Google Scholar]
- 12.Roh JI, Cheong C, Sung YH, Lee J, Oh J, Lee BS, Lee JE, Gho YS, Kim DK, Park CB, et al. Perturbation of NCOA6 leads to dilated cardiomyopathy. Cell Rep. 2014;8:991–998. doi: 10.1016/j.celrep.2014.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Man E, Lafferty KA, Funke BH, Lun KS, Chan SY, Chau AK, Chung BH. NGS identifies TAZ mutation in a family with X-linked dilated cardiomyopathy. BMJ Case Rep. 2013;2013 doi: 10.1136/bcr-2012-007529. pii: bcr2012007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charron P, Arad M, Arbustini E, Basso C, Bilinska Z, Elliott P, Helio T, Keren A, McKenna WJ, Monserrat L, et al. European Society of Cardiology Working Group on Myocardial and Pericardial Diseases: Genetic counselling and testing in cardiomyopathies: a position statement of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J. 2010;31:2715–2726. doi: 10.1093/eurheartj/ehq271. [DOI] [PubMed] [Google Scholar]
- 15.Posafalvi A, Herkert JC, Sinke RJ, van den Berg MP, Mogensen J, Jongbloed JD, van Tintelen JP. Clinical utility gene card for: dilated cardiomyopathy (CMD) Eur J Hum Genet. 2013;21:21. doi: 10.1038/ejhg.2012.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 17.Schuster SC. Next-generation sequencing transforms today's biology. Nat Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 18.Ware SM. Genetic diagnosis in pediatric cardiomyopathy: clinical application and research perspectives. Prog Pediatr Cardiol. 2011;31:99–102. doi: 10.1016/j.ppedcard.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biswas A, Rao VR, Seth S, Maulik SK. Next generation sequencing in cardiomyopathy: towards personalized genomics and medicine. Mol Biol Rep. 2014;41:4881–4888. doi: 10.1007/s11033-014-3418-9. [DOI] [PubMed] [Google Scholar]
- 20.Punetha J, Hoffman EP. Short read (next-generation) sequencing: a tutorial with cardiomyopathy diagnostics as an exemplar. Circ Cardiovasc Genet. 2013;6:427–434. doi: 10.1161/CIRCGENETICS.113.000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mestroni L, Maisch B, McKenna WJ, Schwartz K, Charron P, Rocco C, Tesson F, Richter A, Wilke A, Komajda M. Guidelines for the study of familial dilated cardiomyopathies. Collaborative Research Group of the European Human and Capital Mobility Project on Familial Dilated Cardiomyopathy. Eur Heart J. 1999;20:93–102. doi: 10.1053/euhj.1998.1145. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 1000 Genome Project Data Processing Subgroup: The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tester DJ, Ackerman MJ. Genetic testing for potentially lethal, highly treatable inherited cardiomyopathies/channelopathies in clinical practice. Circulation. 2011;123:1021–1037. doi: 10.1161/CIRCULATIONAHA.109.914838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maron BJ, Semsarian C. Prevention of sudden death for patients with cardiomyopathies another step forward. J Am Coll Cardiol. 2012;59:501–502. doi: 10.1016/j.jacc.2011.10.869. [DOI] [PubMed] [Google Scholar]
- 28.Chiu C, Tebo M, Ingles J, Yeates L, Arthur JW, Lind JM, Semsarian C. Genetic screening of calcium regulation genes in familial hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2007;43:337–343. doi: 10.1016/j.yjmcc.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 29.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 30.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 31.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 32.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang S, Zou Y, Fu C, Xu X, Wang J, Song L, Wang H, Chen J, Wang J, Huan T, Hui R. Worse prognosis with gene mutations of beta-myosin heavy chain than myosin-binding protein C in Chinese patients with hypertrophic cardiomyopathy. Clin Cardiol. 2008;31:114–118. doi: 10.1002/clc.20151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson P, Griffiths PJ, Watkins H, Redwood CS. Dilated and hypertrophic cardiomyopathy mutations in troponin and alpha-tropomyosin have opposing effects on the calcium affinity of cardiac thin filaments. Circ Res. 2007;101:1266–1273. doi: 10.1161/CIRCRESAHA.107.156380. [DOI] [PubMed] [Google Scholar]
- 35.Mirza M, Marston S, Willott R, Ashley C, Mogensen J, McKenna W, Robinson P, Redwood C, Watkins H. Dilated cardiomyopathy mutations in three thin filament regulatory proteins result in a common functional phenotype. J Biol Chem. 2005;280:28498–28506. doi: 10.1074/jbc.M412281200. [DOI] [PubMed] [Google Scholar]
- 36.Mller DV, Andersen PS, Hedley P, Ersbll MK, Bundgaard H, Moolman-Smook J, Christiansen M, Kber L. The role of sarcomere gene mutations in patients with idiopathic dilated cardiomyopathy. Eur J Hum Genet. 2009;17:1241–1249. doi: 10.1038/ejhg.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hershberger RE, Norton N, Morales A, Li D, Siegfried JD, Gonzalez-Quintana J. Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3:155–161. doi: 10.1161/CIRCGENETICS.109.912345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hershberger RE, Parks SB, Kushner JD, Li D, Ludwigsen S, Jakobs P, Nauman D, Burgess D, Partain J, Litt M. Coding sequence mutations identified in MYH7, TNNT2, SCN5A, CSRP3, LBD3, and TCAP from 313 patients with familial or idiopathic dilated cardiomyopathy. Clin Transl Sci. 2008;1:21–26. doi: 10.1111/j.1752-8062.2008.00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lakdawala NK, Funke BH, Baxter S, Cirino AL, Roberts AE, Judge DP, Johnson N, Mendelsohn NJ, Morel C, Care M, et al. Genetic testing for dilated cardiomyopathy in clinical practice. J Card Fail. 2012;18:296–303. doi: 10.1016/j.cardfail.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu W, Liu W, Hu D, Zhu T, Ma Z, Yang J, Xie W, Li C, Li L, Yang J, et al. Mutation spectrum in a large cohort of unrelated Chinese patients with hypertrophic cardiomyopathy. Am J Cardiol. 2013;112:585–589. doi: 10.1016/j.amjcard.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 41.Matsushita Y, Furukawa T, Kasanuki H, Nishibatake M, Kurihara Y, Ikeda A, Kamatani N, Takeshima H, Matsuoka R. Mutation of junctophilin type 2 associated with hypertrophic cardiomyopathy. J Hum Genet. 2007;52:543–548. doi: 10.1007/s10038-007-0149-y. [DOI] [PubMed] [Google Scholar]
- 42.Zou Y, Wang J, Liu X, Wang Y, Chen Y, Sun K, Gao S, Zhang C, Wang Z, Zhang Y, et al. Multiple gene mutations, not the type of mutation, are the modifier of left ventricle hypertrophy in patients with hypertrophic cardiomyopathy. Mol Biol Rep. 2013;40:3969–3976. doi: 10.1007/s11033-012-2474-2. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Xu SJ, Zhou H, Wang LJ, Hu B, Fang F, Zhang XM, Luo YW, He XY, Zhuang SW, et al. A novel mutation of the beta myosin heavy chain gene responsible for familial hypertrophic cardiomyopathy. Clin Cardiol. 2009;32:E16–E21. doi: 10.1002/clc.20520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carniel E, Taylor MR, Sinagra G, Di Lenarda A, Ku L, Fain PR, Boucek MM, Cavanaugh J, Miocic S, Slavov D, et al. Alpha-myosin heavy chain: a sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Circulation. 2005;112:54–59. doi: 10.1161/CIRCULATIONAHA.104.507699. [DOI] [PubMed] [Google Scholar]
- 45.Herman DS, Hovingh GK, Iartchouk O, Rehm HL, Kucherlapati R, Seidman JG, Seidman CE. Filter-based hybridization capture of subgenomes enables resequencing and copy-number detection. Nat Methods. 2009;6:507–510. doi: 10.1038/nmeth.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sikkema-Raddatz B, Johansson LF, de Boer EN, Almomani R, Boven LG, van den Berg MP, van Spaendonck-Zwarts KY, van Tintelen JP, Sijmons RH, Jongbloed JD, Sinke RJ. Targeted next-generation sequencing can replace Sanger sequencing in clinical diagnostics. Hum Mutat. 2013;34:1035–1042. doi: 10.1002/humu.22332. [DOI] [PubMed] [Google Scholar]