Abstract
Purpose
The best screening practice for Lynch syndrome (LS) in endometrial cancer (EC) remains unknown. We sought to determine whether tumor microsatellite instability (MSI) typing along with immunohistochemistry (IHC) and MLH1 methylation analysis can help identify women with LS.
Patients and Methods
ECs from GOG210 patients were assessed for MSI, MLH1 methylation, and mismatch repair (MMR) protein expression. Each tumor was classified as having normal MMR, defective MMR associated with MLH1 methylation, or probable MMR mutation (ie, defective MMR but no methylation). Cancer family history and demographic and clinical features were compared for the three groups. Lynch mutation testing was performed for a subset of women.
Results
Analysis of 1,002 ECs suggested possible MMR mutation in 11.8% of tumors. The number of patients with a family history suggestive of LS was highest among women whose tumors were classified as probable MMR mutation (P = .001). Lynch mutations were identified in 41% of patient cases classified as probable mutation (21 of 51 tested). One of the MSH6 Lynch mutations was identified in a patient whose tumor had intact MSH6 expression. Age at diagnosis was younger for mutation carriers than noncarriers (54.3 v 62.3 years; P < .01), with five carriers diagnosed at age > 60 years.
Conclusion
Combined MSI, methylation, and IHC analysis may prove useful in Lynch screening in EC. Twenty-four percent of mutation carriers presented with ECs at age > 60 years, and one carrier had an MSI-positive tumor with no IHC defect. Restricting Lynch testing to women diagnosed at age < 60 years or to women with IHC defects could result in missing a substantial fraction of genetic disease.
INTRODUCTION
Endometrial cancer (EC) is the second most common malignancy in patients with Lynch syndrome (LS). Identifying patients with EC with LS benefits both those individuals already affected with cancer and their at-risk relatives. Estimates for LS frequency among patients with EC have ranged from 2% to 6%.1–5 A majority of Lynch families have mutations in MSH2, MLH1, MSH6, PMS2, or EPCAM. Mutation penetrance and expressivity are determined by which Lynch genes are defective and the nature of the mutations.6 MSH6 mutation confers a particular risk for EC and a relatively lower risk for colon cancers.7 International collaborative studies have led to screening recommendations reflecting the risks associated with the gene responsible for disease in a given family and age of cancer onset in relatives.7–9
The best practices for identifying LS are still being determined, with general consensus that many, if not all, patients with colon cancer or EC should be screened for LS.10–13 Tumor immunohistochemistry (IHC) is central to screening and has been widely adopted; however, Lynch screening in patients with EC presents challenges. Somatic or epigenetic inactivation of the MLH1 gene is a frequent event, and consequently, triage based on MLH1 methylation has been recommended.12 The higher frequency of MSH6 defects in EC and the distinct clinical features associated with MSH6 mutations also need to be considered in screening for LS in patients with EC. Later age of onset for Lynch mutation carriers, lower levels of tumor microsatellite instability (MSI), and differences in MSH6 mutation penetrance and expressivity compared with other Lynch genes must be considered as part of screening efforts.
In this study, we assessed tumor IHC, MSI, and MLH1 methylation analysis in a large cohort of patients with endometrioid EC enrolled onto an NRG Oncology and Gynecologic Oncology Group (GOG) trial to determine which test or combination of tests best predicts LS. Analyses were limited to endometrioid tumors, the most common histologic type of EC seen in LS.9
Each patient was classified as having either no defect in DNA mismatch repair (MMR), a sporadic epigenetic MMR defect, or probable MMR mutation based on tumor findings. Germline mutation testing was performed for a subset of patients considered to be possible mutation carriers based on tumor testing studies. Age at diagnosis, cancer family history, tumor, and Lynch testing (as appropriate) findings were compared for the three molecularly defined groups. Our analysis of 1,002 tumors illustrated that tumor screening for LS that includes MSI analysis identifies germline mutation carriers who would have gone untested based on IHC screening alone and that as many as 24% of mutation carriers were age > 60 years at the time of EC diagnosis.
PATIENTS AND METHODS
Patient Cohort and Clinical, Demographic, and Family History Data
Patients were investigated as part of the GOG8020 protocol. They were recruited to GOG210 (Molecular Staging Study of Endometrial Carcinoma; ClinicalTrials.gov identifier NCT00340808) during the so-called unrestricted enrollment period when all stages, grades, and histologic subtypes were eligible (2003 to 2007),14 after which eligibility was restricted to poor-prognosis tumors or tumors occurring among nonobese and nonwhite patients. Family history data were abstracted from the GOG210 questionnaire (family history section on cancers in first-degree relatives).14 Clinical reports and pathologic slides of tumors were centrally reviewed by the NRG/GOG Pathology Committee. Analyses were limited to endometrioid tumors, the most common histologic type seen in LS.9
Molecular Analysis of Tumors and Normal DNA
DNA preparation was carried out as previously described using Maxwell 16 (Promega, Madison, WI).15,16 Frozen tissues suitable for analysis were available for 611 patients, all reviewed by qualified pathologists to identify representative normal myometrium and high neoplastic cellularity (> 66%). Formalin-fixed tissues served as the source of DNA for 432 patient cases.
MSI testing was performed using a five-plex assay for the National Cancer Institute consensus markers.17 Alleles were detected using an ABI3130 analyzer and GeneMapper software (version 4.0; (Applied Biosystems, Foster City, CA). Tumors were classified as MSI high if novel alleles were seen at ≥ two loci. All instances of MSI with a single marker were confirmed with repeat polymerase chain reaction and classified as MSI low. MLH1 methylation was evaluated using pyrosequencing and/or combined bisulfite restriction analysis (COBRA).18 Primers and conditions are available on request. Finally, MSH6, MSH2, and MLH1 IHC was performed using whole-section slides; PMS2 was evaluated in a subset of patient cases.16,19,20 IHC staining was interpreted by a gynecologic pathologist (R.R.B.).
Normal DNA from 51 patient cases of probable mutation with sufficient high-quality DNA available were tested for LS mutations using ColoSeq (http://tests.labmed.washington.edu/COLOSEQ).21 Two additional DNA samples failed quality control assays for mutation testing. Patients considered probable carriers of Lynch mutations for whom normal tumor DNA yield or quality was inadequate were not tested. None of the IHC-normal MSI-low patient cases were considered for mutation testing.
Statistical Analysis
The patterns of cancer family history for the three molecularly defined patient groups were compared descriptively using contingency analyses. Ages were compared using Mann-Whitney tests. Pearson's correlation analysis was used to assess pyrosequencing methylation data (InStat3 software; GraphPad, La Jolla, CA).
RESULTS
Molecular Features of Tumors
MSI, IHC, and MLH1 methylation analysis was undertaken for 1,043 ECs. Overall, 28.4% of tumors (296 of 1,043) were MSI high, with only 29 MSI low (2.8%). Thirty-nine tumors failed MLH1 analysis, and three failed IHC (one failing both), leaving 1,002 tumors for further analysis. MLH1 methylation pyrosequencing was successful for 673 patient cases (67.2%), with COBRA used for the remainder. COBRA findings were 100% concordant for 86 tumors assessed by pyrosequencing. Methylation levels at the four CpG DNA sequences investigated were highly correlated (r2 = 0.98; Pearson's P < .001; primary data available on request). Tumors with ≥ 12% methylation at all four CpGs were classified as methylation positive.
Average methylation for 282 MSI patient cases was 61.2% (range, 0% to 97.2%). Mean methylation value of MSI-low tumors (17 assessed by pyrosequencing) was 10.3%, with only three classified as methylation positive. Forty-eight of 265 MSI-high tumors (18.1%) lacked methylation. Average methylation for 391 microsatellite stable (MSS) tumors assessed by pyroseqeuncing was 4.58% (range, 0% to 92.1%); 21 methylated tumors (mean methylation, 37.8%) expressed MLH1. COBRA confirmed methylation in 10 of 10 tumors tested.
The combined molecular data were used to assign tumors to one of three molecular classes: 617 (61.6%) were classified as MMR normal (no MSI, no IHC defect), 266 (26.5%) as sporadic epigenetic MMR defective (MSI positive, methylation, and absent MLH1), and 119 (11.9%) as probable MMR mutation (absence of MLH1 methylation and MSI and/or combined MSI and IHC defect).
Family Cancer History for Lynch-Associated Tumors and Relationship With Tumor MMR Status
Family history data were available for 938 of 1,002 patient cases with molecularly characterized tumors. Clinicopathologic and demographic features are listed in Appendix Table A1 (online only). Most patients were white (90.4%) and had early-stage and low-grade disease, with a mean age of 62.1 years (range, 25 to 100 years) and body-mass index of 35 kg/m2 (range, 16.6 to 82.8 kg/m2).
Thirty-eight percent of tumors had features indicative of defective DNA MMR (Table 1). MLH1 methylation and tumor MSI were seen in 253 patient cases (70%). A majority of the additional 107 tumors with MMR defects considered probable mutation had MSI (MSI high, n = 79; MSI low, n = 20; MSS, n = 8). The most frequent IHC defects were combined MSH2 and MSH6 loss and MSH6 loss alone (22 and 21 instances, respectively). All 22 tumors lacking both MSH2 and MSH6, consistent with an MSH2 mutation, were MSI high. One, G838 T, had MLH1 methylation (31.5%) but expressed MLH1. Among the tumors that lacked MSH6 only, 15 were MSI high, one was MSI low, and five were MSS. Eighteen tumors (MSI high, n = 15; MSS, n = 3) failed to express both MLH1 and PMS2, suggestive of MLH1 mutation. All nine tumors that lacked PMS2 (with expression of other three MMR proteins) were MSI high. Thirty-three tumors (3.5% of entire cohort) had MSI but no IHC defect; 19 of these were MSI low. Finally, there were four MSI-high tumors for which ≥ one IHC marker failed, resulting in an uncertain class of defect (Table 1).
Table 1.
Molecular Characteristics of Endometrioid Endometrial Cancers for Women With Available Family History Data (n = 938)
| Characteristic | No. |
|---|---|
| MMR Status | |
| Normal | 578 |
| Defective | 360 |
| Type of MMR Defect | |
| Sporadic epigenetic | 253 |
| Methylated MLH1, absent MLH1 expression | |
| MSI high | 249* |
| MSI low | 4* |
| Probable MMR mutations (unmethylated) | 107 |
| MSH2 and MSH6 absent† | 22 |
| MSI high | 22 |
| MSH6 only absent | 21 |
| MSI high | 15 |
| MSI low | 1 |
| MSS | 5 |
| MLH1 and PMS2 absent | 18 |
| MSI high | 15 |
| MSS | 3 |
| PMS2 only absent | 9 |
| MSI high | 9 |
| No IHC defect | 33 |
| MSI high | 14 |
| MSI low | 19 |
| Mixed or uncertain | 4 |
| MSI high | 4‡ |
Abbreviations: IHC, immunohistochemistry; MMR, mismatch repair; MSI, microsatellite instability; MSS, microsatellite stability.
Thirteen patient cases expressed MLH1, MSH6, and MSH2 and had extensive MSI and methylation. Eleven of 13 did not express PMS2, consistent with MLH1 false-positive staining. One had uncertain MLH1 and PMS2 staining. One expressed all four MMR proteins. These 13 patient cases were considered sporadic epigenetic, along with one MSI-low patient case with scattered foci expressing MLH1.
One tumor had MLH1 methylation but still expressed MLH1 protein.
Three patient cases with failure for ≥ one IHC marker, and one tumor with mixed IHC abnormalities.
A total of 347 Lynch-associated cancers (LACs) were reported among 6,615 relatives of the 938 probands, with 13 relatives having two LACs (Table 2). The most common LAC was colon (females, n = 78; males, n = 64), followed by endometrial or reproductive system and ovarian cancers (n = 70 and 36, respectively). There was a significant excess of affected female relatives (χ2 P < .001), largely attributable to gynecologic cancers. Nearly twice as many cancers were in mothers than sisters and daughters. The 19 reported female reproductive system cancers (mothers, n = 15; sisters, n = 3; daughter, n = 1) were considered endometrial for these analyses.
Table 2.
Lynch-Associated Cancers Reported in First-Degree Relatives of Probands With Endometrial Cancer (n = 938)
| Relative | No. (%) | No. of Cancers (No. Diagnosed at Age < 50 years) |
|||
|---|---|---|---|---|---|
| Colon | Endometrial | Ovarian | Other* | ||
| Mother† | 854 (13) | 56 (3) | 38 (10) | 23 (7) | 28 (1) |
| Father‡ | 760 (11) | 40 (4) | — | — | 38 (5) |
| Sister§ | 1,473 (22) | 19 (6) | 25 (11) | 10 (4) | 12 (9) |
| Brother‖ | 1,466 (22) | 24 (7) | — | — | 15 (3) |
| Daughter¶ | 1,009 (15) | 3 (2) | 7 (7) | 3 (3) | 4 (2) |
| Son | 1,053 (16) | 0 | — | — | 2 |
NOTE. No data for: 85 mothers, 180 fathers, 29 sisters, 41 brothers, 19 daughters, and 45 sons.
Other Lynch-associated cancers included stomach, hepatobiliary system, small bowel, renal pelvis or ureter, glioblastoma or brain, pancreas, and female reproductive tract.
Seven mothers with ≥ two cancers.
One father with ≥ two cancers.
Three sisters with two cancers.
One brother with two cancers.
One daughter with two cancers.
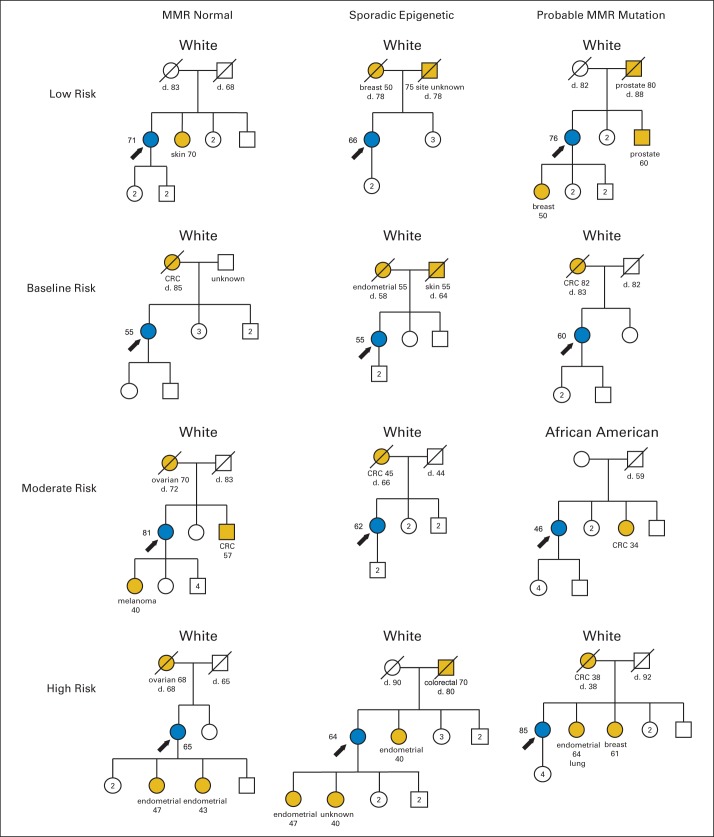
Each proband was assigned to one of four Lynch cancer family history risk groups based on number and age of onset of LACs in first-degree relatives (familial risk classes listed in Table 3). A total of 658 women (70.1%) reported no relatives with LACs and were considered to have low familial risk for LS (Table 3). There were 235 probands who reported a single relative with an LAC; of those, 181 were considered to have baseline risk (single relative with one LAC diagnosed at age ≥ 50 years). Forty-five probands had one relative with an early-onset LAC (considered moderate risk), and another 22 had > two affected relatives, for a total of 67 with moderate risk (7.1% of cohort). Thirty-two probands (3.4%) had high familial risk for LS (nine had single relative with double primary cancer; remainder had ≥ two relatives with early-onset and/or double primary LACs). Overall, 10.6% of probands had elevated (moderate or high) familial risk. Representative pedigrees of the four risk classes are presented in Figure 1. Proband age at diagnosis was not associated with familial risk class.
Table 3.
Familial Risk, Proband Age, and Tumor MMR Status for Patient Cases of Endometrioid Endometrial Cancer (n = 938)
| Tumor MMR Status | Familial Risk*† |
P | Age of Probands Median (range) | P | |||
|---|---|---|---|---|---|---|---|
| Low | Baseline | Moderate‡ | High | ||||
| MMR normal | 427 | 99 | 36 | 16 | < .001§ | 60 (25-91) | < .001‖ |
| Sporadic epigenetic | 169 | 58 | 20 | 6 | 65 (36-100) | ||
| Probable mutation | 62 | 24 | 11 | 10 | 59 (35-87) | ||
Abbreviations: LAC, Lynch-associated cancer; MMR, mismatch repair.
Familial risk classification: low, no relative with LAC; baseline, single relative with one LAC diagnosed at age > 50 years; moderate, one relative with two LACs and/or diagnosed at young age; high, ≥ two relatives with LACs and/or diagnosed at young age.
Mean age (range) of four risk groups: low, 62 (25-91); baseline, 63 (37-89); moderate, 61 (30-81); and high, 62 years (43-100).
Fifty-six had a single relative who either had early-onset cancer (n = 46) or double primary LACs (n = 10).
χ2 test.
Kruskal-Wallis test.
Fig 1.
Two-generation pedigrees representative of familial risk group for women whose tumors classified as mismatch repair (MMR) normal, sporadic epigenetic MMR defect, or probable MMR mutation. Blue symbols indicate histologically confirmed endometrioid endometrial cancer. Gold symbols represent reported cancers. Age at diagnosis and at death (d) given when known. CRC, colorectal cancer.
Tumor MMR status was associated with familial risk (χ2 P = .001; Table 3). Among the 107 probands whose tumors were classified as having probable MMR mutation, 21 (19.6%) had moderate or high familial risk for LS. Among probands classified as having sporadic MMR defect, only 26 (10.2%) had moderate or high familial risk, and only 9% of probands (52 of 578) whose tumors had normal MMR had moderate or high risk. Proband age at diagnosis was different for the groups (Kruskal-Wallis P < .001; Table 3). There was no difference between the MMR normal and probable mutation groups (mean age, 61.2 v 59.9 years), whereas women whose tumors had sporadic epigenetic MMR defects (silencing of MLH1) were older (mean age, 65.4 years; Mann-Whitney P < .001 for both comparisons).
Germline Mutations in MMR Genes
Forty-seven germline DNA samples from probands whose tumors were classified as probable mutation and for whom family history data were available were tested for mutations in MLH1, MSH6, MSH2, and PMS2 using ColoSeq.21 The MSI, MLH1 methylation, IHC, and predicted molecular defect information is listed in Table 4. Nineteen germline mutations were identified (40.4% of those tested). One woman had a variant of uncertain significance (VUS).
Table 4.
Tumor and ColoSeq Findings for Women With Tumors Classified As Having Probable Genetic MMR Defects
| Predicted Gene Defect* | Mutation Identified | Proband Age (years) | Risk Category | MSI Status | PREMM1,2,6 Risk Score (%) |
|||
|---|---|---|---|---|---|---|---|---|
| Overall | MLH1 | MSH2 | MSH6 | |||||
| MSH2 | ||||||||
| G494 T | MSH2 c.1853delC, p.P618Hfs*17 | 52 | High | High | 27.8 | 8.8 | 14.5 | 4.5 |
| G839 T | MSH2 c.1861C>T, p.R621* | 53 | Moderate | High | 22.7 | 7.3 | 12.5 | 3.0 |
| G194 T | MSH2 del ex11 | 35 | Low | High | 5.9 | 1.1 | 2.7 | 2.1 |
| G930 T | MSH2 c.229_230delAG, p.S77Cfs*4 | 57 | Low | High | 5.4 | 1.1 | 1.7 | 2.6 |
| G1116 T | MSH2 del ex 1-6 | 55 | Baseline | High | 13.9 | 2.6 | 3.0 | 8.4 |
| G734 T | MSH2 c.1226_1227delAG, p.Q409Rfs*7 | 46 | High | High | 33.4 | 10.7 | 18.8 | 4.0 |
| G119 T | — | 54 | Baseline | High | 8.0 | 1.2 | 2.5 | 4.3 |
| G800 T | — | 83 | Low | High | 5.3 | 1.1 | 1.0 | 3.2 |
| G838 T | — | 53 | Low | High | 5.4 | 1.1 | 1.8 | 2.5 |
| G669 T | — | 69 | Low | High | 5.3 | 1.1 | 1.3 | 2.9 |
| G1148 T | — | 54 | Baseline | High | 8.0 | 1.2 | 2.5 | 4.3 |
| G1166 T | — | 55 | Low | High | 5.4 | 1.1 | 1.8 | 2.6 |
| G531 T | — | 61 | Low | High | 5.3 | 1.1 | 1.5 | 2.7 |
| G209 T | — | 54 | Baseline | High | 5.4 | 1.1 | 1.8 | 2.5 |
| MSH6 | ||||||||
| G778 T | MSH6 c.3768T>G, p.Y1256* | 51 | Low | High | 5.5 | 1.1 | 1.9 | 2.5 |
| G783 T | MSH6 c.892C>T, p.R298* | 53 | High | High | 20.1 | 2.8 | 5.8 | 11.5 |
| G852 T | MSH6 c.3332_3335dup, p.D1112Efs*2 | 54 | Low | High | 5.4 | 1.1 | 1.8 | 2.5 |
| G573 T | MSH6 c.3939_3957dupTCAAAAGGGACATAGAAAA, p.A1320Sfs*5 | 55 | Baseline | High | 5.4 | 1.1 | 1.7 | 2.6 |
| G31 T | MSH6 c.3013C>T, p.R1005* | 45 | Low | High | 5.6 | 1.1 | 2.2 | 2.3 |
| G1064 T | MSH6 c.3991C>T, p.R1331* | 61 | Moderate | High | 13.8 | 4.7 | 6.0 | 3.1 |
| G697 T | MSH6 c.3202C>T, p.R1068* | 55 | Moderate | High | 21.0 | 5.3 | 6.8 | 8.9 |
| G705 T | — | 59 | Low | High | 5.4 | 1.1 | 1.6 | 2.7 |
| G1171 T | — | 68 | Low | MSS | 5.3 | 1.1 | 1.3 | 2.9 |
| G116 T | — | 65 | Low | High | 5.3 | 1.1 | 1.4 | 2.8 |
| G117 T | — | 74 | Baseline | MSS | 7.9 | 1.2 | 1.6 | 5.1 |
| G562 T | — | 84 | Low | MSS | 5.3 | 1.1 | 1.0 | 3.2 |
| PMS2 | ||||||||
| G480 T | PMS2 c.736_741delCCCCCTinsTGTGTGTGAAG, p.P246_P247Ffs*7 | 57 | Baseline | High | 14.0 | 2.6 | 4.0 | 7.4 |
| G212 T | PMS2 del ex8 | 85 | High | High | 37.0 | 11.1 | 9.1 | 16.7 |
| G236 T | MLH1 c.191A>G, p.N64S | 61 | Low | High | 5.3 | 1.1 | 1.5 | 2.7 |
| G717 T | — | 70 | Low | High | 5.3 | 1.1 | 1.3 | 2.9 |
| G174 T | — | 59 | Low | High | 5.4 | 1.1 | 1.6 | 2.7 |
| G262 T | — | 54 | Low | High | 5.4 | 1.1 | 1.8 | 2.5 |
| G206 T | — | 64 | Moderate | High | 7.9 | 1.2 | 2.0 | 4.7 |
| No IHC defect or epitope stable | ||||||||
| G25 T | MSH6 c.393delAC, p.V131fs*2 | 56 | Low | High | 5.4 | 1.1 | 1.7 | 2.6 |
| G894 T | — | 55 | Baseline | High | 14.0 | 2.6 | 3.8 | 7.6 |
| G920 T | — | 50 | Low | High | 5.5 | 1.1 | 1.9 | 2.4 |
| G983 T | — | 76 | Low | High | 5.3 | 1.1 | 1.1 | 3.1 |
| G234 T | — | 67 | Low | High | 5.3 | 1.1 | 1.4 | 2.8 |
| MLH1 | ||||||||
| G146 T | MLH1 c.34insG, p.G12fs*17 | 46 | Moderate | High | 19.8 | 6.6 | 10.5 | 2.6 |
| G805 T | — | 62 | Low | High | 5.3 | 1.1 | 1.5 | 2.7 |
| G345 T | — | 60 | Low | High | 5.4 | 1.1 | 1.6 | 2.7 |
| G1117 T | — | 50 | Low | High | 5.5 | 1.1 | 1.9 | 2.4 |
| G118 T | — | 58 | Baseline | High | 7.9 | 1.2 | 2.3 | 4.4 |
| G510 T | — | 65 | High | High | 45.7 | 21.5 | 21.5 | 2.7 |
| Uncertain staining | ||||||||
| G1063 T | MSH6 c.3261delC, p.F1088Sfs*2 | 55 | Moderate | High | 21.6 | 8.3 | 10.5 | 2.9 |
| G359 T† | Variant of uncertain significance MSH6 c.2057G>A, p.G686D | 53 | Moderate | High | 8.0 | 1.2 | 2.5 | 4.3 |
| G677 T | — | 57 | Baseline | High | 7.1 | 1.5 | 2.7 | 2.9 |
Abbreviations: IHC, immunohistochemistry; MMR, mismatch repair; MSI, microsatellite instability.
Based on IHC and MSI findings; all tumors unmethylated for MLH1 except for G838 T.
Variant of uncertain significance not considered mutation.
On the basis of the nine MSH6, six MSH2, two PMS2, and two MLH1 germline mutations identified, we estimated the rate of LS at 4.4%. However, when the frequency of each class of predicted defect was considered, the overall minimum rate for LS was 3.89% (Appendix Table A2, online only). It is noteworthy that the largest single group of predicted mutations was those with no IHC defect (n = 33; 3.5% of entire cohort; Table 1). Among these patient cases, most women had MSI-low tumors; none were tested for mutations. The single mutation identified in the no–IHC defect group was in MSH6, and one additional MSH6 mutation was detected in a patient whose tumor was MSI high but for whom IHC classification was uncertain.
For the 47 probands assessed for mutations, PREMM1,2,6 gave overall risk predictions for LS ranging from 5.3% to 45.7% (Table 4).22 Only 13 probands were assigned risk > 10%. Eleven of 13 had Lynch mutations, and among the 34 with risk < 10%, eight had mutations. The sensitivity of the PREMM1,2,6 prediction model was 58% and specificity 93% in this molecularly high-risk selected cohort.
Our ColoSeq mutation testing included four probands whose family history data were unavailable. Three carried germline mutations: one each in MSH2, MSH6, and PMS2; one had a PMS2 VUS (Appendix Table A3, online only). Unexpectedly, both patient cases with PMS2 variants (mutation and VUS) had IHC defects consistent with an MSH2 mutation (absent MSH2 and MSH6). On the basis of three mutations identified, we estimate approximately one in 300 patients with EC carry a PMS2 mutation, consistent with IHC predictions for colorectal cancer.23–26 With the additional MSH6 mutation (10 total), MSH6 remains the most frequent cause of LS. Mutation carriers were younger than noncarriers (54.3 v 62.3; Mann-Whitney P < .01).
Molecular Features of Tumors and MMR Germline Mutations
MSH6 was the most frequently mutated Lynch gene in our cohort (Table 4). Tumors from nine MSH6 mutation carriers were MSI high; the number of MSI events in MSH6 MSI-high tumors was, however, fewer than that for tumors from women with MSH2, MLH1, and PMS2 mutations (P < .001; Appendix Table A4, online only). Mononucleotide repeats (BAT26 and BAT25) accounted for most MSI events, with only four of nine MSH6 carriers' tumors showing a dinucleotide change. It was noteworthy that for the 19 MSI-low tumors with no IHC defect, 16 had dinucleotide, and only three had mononucleotide repeat MSI.
DISCUSSION
Our analysis of endometrioid ECs from GOG210 provides an estimate of 3.89% frequency for LS, consistent with other large population-based series.2,12 The frequency of LS may be higher because of the fact that only 5% of the cohort (51 of 1,002) had germline mutation testing, and some women with prior colorectal cancers would have been excluded from GOG210. The GOG210 protocol was, however, amended on September 18, 2006, to allow for patients with prior malignancies. Given that metachronous cancers are a hallmark of LS, and EC is a second malignancy in approximately 50% of patients with LS, it is probable some Lynch patient cases were excluded.27
Combined, IHC and MLH1 methylation of tumors identified Lynch patient cases that would not have been considered for mutation testing if only IHC and methylation analysis were used for initial screening for referral for genetic testing. One patient, G25, had an MSI-high tumor that expressed all four MMR proteins and carried a germline MSH6 mutation. IHC findings were inconclusive for a second MSH6 mutation carrier, G1063. MSH2 and MSH6 staining was uncertain for both and reported as “favor positive,” but on the basis of tumor MSI status, we undertook mutation analysis. Considering the testing was limited to < 50% of the patients with probable MMR mutation, we estimate approximately one in 150 women with ECs have LS with tumors that do not have IHC defects (Appendix Table A4). We note that some tumors with IHC defects lacked MSI (Appendix Table A4).
Another important and clinically relevant finding is that Lynch mutations are seen at appreciable frequency in patients with EC diagnosed at age > 60 years. Five mutation carriers (MSH6, n = 3; MLH1, n = 1; PMS2, n = 1) were identified among the 17 women age > 60 years tested for germline mutations (Table 4; Appendix Table A4). Thirty-two women with tumors that had IHC defects or were MSI high but lacked MLH1 methylation were diagnosed at age > 60 years (3.2% of cohort; 938 had family history data; 64 lacked family data). On the basis of these data, we estimate 0.94% of women diagnosed with EC at age > 60 years have LS. Overall, this represents 24% of Lynch patient cases presenting with EC.
MSH6 mutations accounted for half of Lynch patient cases in our series, confirming earlier reports that MSH6 is a major cause of LS among families ascertained through EC probands.2,3 Among relatives of the 938 probands with family history data, ECs were almost as frequent as colon cancers among female relatives (Table 2), which could reflect genetic and nongenetic risk factors.28,29 It is noteworthy that 11% of probands whose tumors were classified as having probable MMR mutation reported ≥ one relative with EC, compared with 6.7% for the rest of the cohort (Appendix Table A5, online only).
Cancer family risk (our categories or PREMM1,2,6 scores) did not reliably predict germline mutation, and several mutation carriers had no history of LACs in relatives (Table 4), confirming reports that family history fails to identify Lynch carriers.30–33 As noted, some women with a previous history of cancer were excluded from the GOG210 study.
Universal germline Lynch testing for patients with EC is cost prohibitive, given the low incidence of Lynch mutations in the general population, and despite nearly two decades of research, best approaches in triage for Lynch testing remains uncertain.31,34 Personal and family histories of cancer lack sensitivity because of variable penetrance and expressivity of the different LS genes and alleles and because of the lack of informativity for patients with EC from small families or for those women with limited knowledge of their biologic relatives. IHC screening identifies many ECs with MMR defects associated with epigenetic silencing of MLH1 that are not the result of inherited Lynch mutations. Buchanan et al12 highlighted the importance of MLH1 methylation analysis in tumors to triage patient cases for Lynch screening in EC. Whereas colon cancers with somatic or epigenetic inactivation of MLH1 frequently have BRAF mutations, and presence of BRAF mutation is used clinically in triage, no such marker exists for EC. Our study confirms the high frequency of epigenetic silencing of MLH1 (sporadic epigenetic MMR defect), with 27% of cancers having MLH1 methylation and MSI (Table 1). As recommended by Buchanan et al, we considered these patient cases to represent sporadic or epigenetic MMR defects; however, we did not test for germline methylation in this group.12 Thus, germline epimutation cannot be excluded. In fact, 26 probands with MSI-positive methylated tumors had moderate or high familial risk (Table 3). The six probands with high familial risk (example shown in Fig 1) had a history consistent with inherited MLH1 epimutation.35,36 We tested the normal DNA from these probands, and all were unmethylated. This finding is consistent with the low incidence of germline epimutation.
In summary, our analysis of a large cohort of endometrioid ECs points to the importance of combined IHC, methylation, and MSI tumor typing in Lynch screening and the need to evaluate women diagnosed at age > 60 years. Our data strongly suggest all women with endometrioid EC should undergo LS screening that includes MMR protein IHC combined with MSI and MLH1 methylation analysis. Because nonendometrioid and mixed-histology tumors were not evaluated, we are unable to predict the overall benefit of combined IHC, MSI, and MLH1 methylation in women with less common histologies that are also seen in women with LS mutations. Prospective studies will clarify the utility of IHC, MSI, and MLH1 methylation analysis in these patients and in the EC population in general.
Acknowledgment
We thank Erika Crouch, MD, and Neha Dahiya, MD (Washington University School of Medicine), for their assistance with immunohistochemistry, Tom Walsh for development of mutation testing methods, and Ms Mary McNulty for her support in biospecimen processing.
Appendix
The following institutions participated in this study: Roswell Park Cancer Institute, University of Alabama at Birmingham, Duke University Medical Center, Abington Memorial Hospital, Walter Reed Army Medical Center, Wayne State University, University of Minnesota Medical School, Northwestern University, University of Mississippi, University of Colorado–Anschutz Cancer Pavilion, University of California at Los Angeles, Fred Hutchinson Cancer Research Center, Penn State Milton S. Hershey Medical Center, University of Cincinnati, University of North Carolina, University of Iowa Hospitals and Clinics, University of Texas Southwestern Medical Center, Indiana University Medical Center, Wake Forest University Health Sciences, University of California Medical Center at Irvine–Orange Campus, Magee Women's Hospital–University of Pittsburgh Medical Center, University of New Mexico, Cleveland Clinic Foundation, State University of New York at Stony Brook, Washington University School of Medicine, Cooper Hospital/University Medical Center, Columbus Cancer Council/Ohio State University, University of Massachusetts Memorial Health Care, Fox Chase Cancer Center, Women's Cancer Center of Nevada, University of Oklahoma Health Sciences Center, University of Virginia, University of Chicago, Mayo Clinic, Case Western Reserve University, Moffitt Cancer Center and Research Institute, Yale University, University of Wisconsin Hospital, Women and Infants' Hospital of Rhode Island, Hospital of Central Connecticut at New Britain General, Gynecologic Oncology of West Michigan, and Community Clinical Oncology Program.
Table A1.
Clinicopathologic and Demographic Characteristics of GOG210 Endometrioid Endometrial Cancers Investigated
| Characteristic | No. (%) |
|---|---|
| Race | |
| White | 848 (90.4) |
| African American | 55 (5.9) |
| Asian | 17 (1.8) |
| Other | 7 (0.7) |
| Unknown/not specified | 11 (1.2) |
| Grade | |
| 1 | 383 (40.8) |
| 2 | 408 (43.5) |
| 3 | 147 (15.7) |
| Stage | |
| I | 702 (74.8) |
| II | 88 (9.4) |
| III | 129 (13.8) |
| IV | 19 (2.0) |
| Age (mean, range)* | 62 (25-100) |
| BMI (mean, range) | 35 (16.6-82.8) |
Abbreviation: BMI, body-mass index.
At time of hysterectomy.
Table A2.
Estimated Frequencies of Germline Mutations
| Predicted Gene Defect* | No. (%) | No. Tested | No. Mutation Positive (%) | Predicted Mutation Frequency (%) |
|---|---|---|---|---|
| MSH6 | 21 (2.2) | 12 | 7 (58.3) | 1.31 |
| MSH2 | 22 (2.3) | 14 | 6 (42.9) | 1.01 |
| PMS2 | 9 (1.0) | 7 | 3 (42.9)† | 0.41† |
| MLH1 | 18 (1.9) | 6 | 1 (16.7) | 0.32 |
| Unknown (no IHC defect)‡ | 33 (3.5) | 5 | 1 (20)§ | 0.70 |
| Uncertain | 4 (0.4) | 3 | 1 (33.3)§ | 0.14 |
Abbreviation: IHC, immunohistochemistry; MSI, microsatellite instability.
Based on MSI, IHC, and MLH1 methylation.
Two PMS2 mutations and one MLH1 mutation.
Only MSI-high patient cases were tested, and as such, we cannot accurately predict mutation rate for this group.
MSH6 mutation.
Table A3.
Tumor and ColoSeq Findings for Additional Women With Tumors Classified As Having Probable Genetic MMR Defects But No Family History Data Unavailable
| Predicted Gene Defect | Mutation Identified | Proband Age (years) | MSI Status |
|---|---|---|---|
| MSH2 | |||
| G979 T | MSH2 del ex 1-6 | 61 | High (four of five markers) |
| MSH2 | |||
| G199 T | PMS2 p.Arg153Glufs*48 | 28 | High (four of five markers) |
| MSH2 | |||
| G728 T* | Variant of uncertain significance PMS2 c.241G>A, p.E81K | 44 | High (five of five markers) |
| MSH6 | |||
| G1051 T | MSH6 c.1969delC, p.Q657Rfs*6 | 62 | Low (BAT26 only) |
Abbreviations: MMR, mismatch repair; MSI, microsatellite instability.
Variants of uncertain significance not considered mutations.
Table A4.
MSI Events in Patient Cases Classified As Probable Genetic Disease (n = 107)
| Predicted Gene Defect* | Mutation Identified | D17S250 Status | BAT25 Status | D5S346 Status | BAT26 Status | D2S123 Status | Total No. of MSI Events |
|---|---|---|---|---|---|---|---|
| MSH2 | |||||||
| G494 T | MSH2 c.1853delC, p.P618Hfs*17 | MSI | MSI | MSI | MSI | MSI | 5 |
| G839 T | MSH2 c.1861C>T, p.R621* | MSI | MSI | MSI | MSI | MSI | 5 |
| G194 T | MSH2 del ex11 | MSI | MSI | MSI | MSI | MSI | 5 |
| G930 T | MSH2 c.229_230delAG, p.S77Cfs*4 | MSI | MSI | MSI | AI | MSI | 4 |
| G1116 T | MSH2 del ex 1-6 | MSI | NI | MSI | MSI | MSI | 4 |
| G734 T | MSH2 c.1226_1227delAG, p.Q409Rfs*7 | MSI | MSI | MSI | MSI | MSI | 5 |
| G119 T | — | MSI | MSI | MSI | MSI | MSI | 5 |
| G800 T | — | MSI | MSI | MSI | MSI | MSI | 5 |
| G838 T | — | MSI | NI | MSI | MSI | MSI | 4 |
| G669 T | — | MSI | MSI | MSI | MSI | MSI | 5 |
| G1148 T | — | MSI | MSI | MSI | MSI | MSI | 4 |
| G1166 T | — | MSI | MSI | MSI | MSI | MSI | 5 |
| G531 T | — | MSI | MSI | MSI | MSI | MSI | 4 |
| G209 T | — | ND | MSI | MSI | MSI | AI | 4 |
| Not tested | |||||||
| G71 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G78 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G170 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G351 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G485 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G820 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G850 T | MSI | MSI | MSI | MSI | NL | 4 | |
| G1210T | MSI | MSI | MSI | MSI | MSI | 5 | |
| MSH6 | |||||||
| G778 T | MSH6 c.3768T>G, p.Y1256* | LOH | MSI | NL | MSI | LOH | 2 |
| G783 T | MSH6 c.892C>T, p.R298* | NL | MSI | NI | MSI | NL | 2 |
| G852 T | MSH6 c.3332_3335dup, p.D1112Efs*2 | NL | MSI | NI | MSI | NL | 2 |
| G573 T | MSH6 c.3939_3957dupTCAAAAGGGACATAGAAAA, p.A1320Sfs*5 | MSI | MSI | MSI | MSI | NL | 4 |
| G31 T | MSH6 c.3013C>T, p.Arg1005* | NL | NI | MSI | MSI | MSI | 3 |
| G1064 T | MSH6 c.3991C>T, p.R1331* | MSI | MSI | NL | MSI | NL | 2 |
| G697 T | MSH6 c.3202C>T, p.R1068* | NL | MSI | NL | MSI | AI | 2 |
| G705 T | — | NL | MSI | NI | MSI | MSI | 3 |
| G116 T | — | NI | NI | NL | MSI | MSI | 2 |
| G1171 T | — | NL | NI | NL | NI | NI | 0 |
| G117 T | — | LOH | NI | LOH | NI | NL | 0 |
| G562 T | — | NL | NI | NL | NI | NL | 0 |
| Not tested | |||||||
| G429 T | NI | MSI | NL | MSI | MSI | 3 | |
| G703 T | NI | MSI | NI | MSI | NI | 2 | |
| G868 T | MSI | NI | MSI | MSI | NL | 3 | |
| G968 T | MSI | MSI | NL | MSI | NL | 3 | |
| G993 T | NI | MSI | NL | MSI | MSI | 3 | |
| G1093 T | NL | MSI | NI | MSI | NI | 2 | |
| G1126 T | NI | MSI | NI | NI | NL | 1 | |
| G257 T | NL | NI | NI | NI | NL | 0 | |
| G766 T | NL | NI | NI | NI | NL | 0 | |
| PMS2 | |||||||
| G480 T | PMS2 c.736_741delCCCCCTinsTGTGTGTGAAG, p.P246_P247Ffs*7 | MSI | MSI | MSI | MSI | MSI | 5 |
| G212 T | PMS2 del ex8 | NL | MSI | MSI | MSI | MSI | 4 |
| G236 T | MLH1 c.191A>G, p.Asn64Ser | MSI | MSI | MSI | MSI | MSI | 5 |
| G717 T | — | MSI | MSI | NL | MSI | MSI | 4 |
| G174 T | — | MSI | MSI | MSI | MSI | MSI | 5 |
| G262 T | — | MSI | MSI | MSI | MSI | MSI | 5 |
| G206 T | — | MSI | MSI | MSI | MSI | NL | 4 |
| Not tested | |||||||
| G184 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G890 T | NI | NI | MSI | MSI | MSI | 3 | |
| No IHC defect/epitope stable | |||||||
| G25 T | MSH6 c.393delAC, p.Val131fsX2 | MSI | NI | NL | MSI | NL | 2 |
| G894 T | — | MSI | MSI | MSI | MSI | MSI | 5 |
| G920 T | — | ND | MSI | MSI | NI | MSI | 4 |
| G983 T | — | MSI | NI | MSI | NI | MSI | 3 |
| G234 T | — | MSI | MSI | NL | NI | LOH | 2 |
| Not tested | |||||||
| G3 T | MSI | MSI | MSI | MSI | AI or MSI | 5 | |
| G52 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G182 T | NL | MSI | MSI | NI | NL | 2 | |
| G233 T | MSI | MSI | NL | MSI | LOH | 3 | |
| G388 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G647 T | MSI | MSI | MSI | MSI | NL | 4 | |
| G893 T | MSI | NI | NI | NI | MSI | 2 | |
| G908 T | MSI | NI | MSI | MSI | NL | 3 | |
| G1182 T | NI | MSI | MSI | NI | NL | 2 | |
| G13 T | MSI | NI | NL | NI | NL | 1 | |
| G20 T | MSI | NI | NL | NI | NL | 1 | |
| G64 T | MSI | NI | NL | NI/AI | NL | 1 | |
| G122 T | NL | MSI | NI | NI | NL | 1 | |
| G216 T | MSI | NL | NL | NL | NL | 1 | |
| G380 T | NL | MSI | NL | NI | NI | 1 | |
| G466 T | MSI | NI | NL | NI | NL | 1 | |
| G478 T | NL | NI | NL | NI | MSI | 1 | |
| G507 T | MSI | NI | NL | NI | NL | 1 | |
| G522 T | NL | NI | MSI | NI | NL | 1 | |
| G569 T | NL | NI | NL | NI | MSI | 1 | |
| G720 T | NI | NI | MSI | NI | NI | 1 | |
| G933 T | MSI | NI | NL | NI | NL | 1 | |
| G957 T | NI | MSI | NI | NI | NI | 1 | |
| G970 T | NI | NI | NL | NI | MSI | 1 | |
| G1030 T | NI | NI | MSI | NI | NI | 1 | |
| G1042 T | NL | NI | MSI | NI | NI | 1 | |
| G1160 T | MSI | NL | NI | NI | NI | 1 | |
| G1211 T | NI | NI | MSI | NI | NI | 1 | |
| MLH1 | |||||||
| G146 T | MLH1 c.34insG, p.Gly12fsX17 | LOH | MSI | MSI | MSI | NI | 3 |
| G805 T | — | MSI | MSI | MSI | MSI | MSI | 5 |
| G345 T | — | MSI | NI | MSI | NI | MSI | 3 |
| G1117 T | — | MSI | MSI | MSI | MSI | MSI | 5 |
| G118 T | — | MSI | MSI | MSI | MSI | MSI | 5 |
| G510 T | — | MSI | MSI | MSI | MSI | MSI | 5 |
| Not tested | |||||||
| G85 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G465 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G683 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G769 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G823 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G854 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G878 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G917T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G926 T | MSI | MSI | MSI | MSI | MSI | 5 | |
| G139 T | NL | NI | NL | NI | NL | 0 | |
| G354 T | NL | NI | NL | NI | NL | 0 | |
| G708 T | NL | NI | NI | NI | NL | 0 | |
| Uncertain staining | |||||||
| G1063 T | MSH6 c.3261delC, p.F1088Sfs*2 | NL | MSI | NL | MSI | MSI | 3 |
| G359 T† | Variant of uncertain significance MSH6 c.2057G>A, p.Gly686Asp | MSI | MSI | MSI | MSI | MSI | 5 |
| G677 T | — | MSI | NI | NI | MSI | MSI | 3 |
| Not tested | |||||||
| G369 T | NI | MSI | NL | MSI | MSI | 3 |
Abbreviations: AI, allelic imbalance; IHC, immunohistochemistry; LOH, loss of heterozygosity; MSI, microsatellite instability; NI, not informative and no evidence of MSI; NL, no loss (informative).
Based on IHC and MSI findings; all tumors unmethylated for MLH1 except for G838 T.
Variant of uncertain significance not considered mutation.
Table A5.
Lynch-Associated Cancers Reported in First-Degree Relatives by Molecular Group
| Molecular Tumor Classification | No. of Probands | No. Reporting Cancer |
||||
|---|---|---|---|---|---|---|
| Colon | Endometrial | Ovarian | Other | None | ||
| Probable MMR mutation | 107 | 28 | 12 | 4 | 16 | 62 |
| Sporadic | 253 | 37 | 21 | 11 | 36 | 169 |
| MMR normal | 578 | 70 | 35 | 21 | 53 | 427 |
Abbreviation: MMR, mismatch repair.
Support information appears at the end of this article.
Authors' disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
Clinical trial information: NCT00340808.
Support
Supported by Grant No. P50 CA134254 from the National Institutes of Health/National Cancer Institute; by Barnes-Jewish Hospital and Siteman Cancer Center; by Ohio State University James Comprehensive Cancer Center; and by National Cancer Institute grants to Gynecologic Oncology Group (GOG) Administrative Office (No. CA 27469), GOG Statistical Office (No. CA 37517), NRG Oncology Group (No. 1 U10 CA180822), and GOG Tissue Bank (No. U10 CA27469, U24 CA114793, and U10 CA180868).
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Disclosures provided by the authors are available with this article at www.jco.org.
AUTHOR CONTRIBUTIONS
Conception and design: Paul J. Goodfellow, Feng Gao, Matthew A. Powell, David Mutch
Administrative support: Paul J. Goodfellow
Provision of study materials or patients: Lisa M. Landrum, Michael L. Pearl, Matthew A. Powell, David Mutch
Collection and assembly of data: Caroline C. Billingsley, Heather A. Lankes, Shamshad Ali, Russell J. Broaddus, Nilsa Ramirez, Colin C. Pritchard, Alexis S. Chassen, Luke V. Simmons, Amy P. Schmidt, Louise A. Brinton, Lisa M. Landrum, Melissa A. Geller, Paul A. DiSilvestro, Michael L. Pearl, Shashikant B. Lele
Data analysis and interpretation: Caroline C. Billingsley, Shamshad Ali, David E. Cohn, Russell J. Broaddus, Colin C. Pritchard, Heather Hampel, Amy P. Schmidt, Floor Backes, Michael L. Pearl, Matthew A. Powell, Richard J. Zaino
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Combined Microsatellite Instability, MLH1 Methylation Analysis, and Immunohistochemistry for Lynch Syndrome Screening in Endometrial Cancers From GOG210: An NRG Oncology and Gynecologic Oncology Group Study
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Paul J. Goodfellow
No relationship to disclose
Caroline C. Billingsley
No relationship to disclose
Heather A. Lankes
No relationship to disclose
Shamshad Ali
No relationship to disclose
David E. Cohn
No relationship to disclose
Russell J. Broaddus
No relationship to disclose
Nilsa Ramirez
No relationship to disclose
Colin C. Pritchard
No relationship to disclose
Heather Hampel
Honoraria: Quest Diagnostics, Invitae Genetics, Prevention Genetics
Research Funding: Myriad Genetic Laboratories
Alexis S. Chassen
No relationship to disclose
Luke V. Simmons
No relationship to disclose
Amy P. Schmidt
No relationship to disclose
Feng Gao
No relationship to disclose
Louise A. Brinton
No relationship to disclose
Floor Backes
No relationship to disclose
Lisa M. Landrum
No relationship to disclose
Melissa A. Geller
Consulting or Advisory Role: OvaGene Oncology
Research Funding: TESARO
Paul A. DiSilvestro
Honoraria: Hologic
Speakers' Bureau: Hologic
Research Funding: AstraZeneca, TESARO, Genentech, Janssen
Michael L. Pearl
Consulting or Advisory Role: Ethicon
Patents, Royalties, Other Intellectual Property: Vitatex
Shashikant B. Lele
No relationship to disclose
Matthew A. Powell
Honoraria: Roche/Genentech, AstraZenca
Consulting or Advisory Role: Roche/Genentech, Arnotherapeutics, Eisai
Speakers' Bureau: Roche/Genentech, AstraZeneca
Richard J. Zaino
Consulting or Advisory Role: Repros, Roche Diagnostics
David Mutch
No relationship to disclose
REFERENCES
- 1.Ollikainen M, Abdel-Rahman WM, Moisio AL, et al. Molecular analysis of familial endometrial carcinoma: A manifestation of hereditary nonpolyposis colorectal cancer or a separate syndrome? J Clin Oncol. 2005;23:4609–4616. doi: 10.1200/JCO.2005.06.055. [DOI] [PubMed] [Google Scholar]
- 2.Hampel H, Frankel W, Panescu J, et al. Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res. 2006;66:7810–7817. doi: 10.1158/0008-5472.CAN-06-1114. [DOI] [PubMed] [Google Scholar]
- 3.Goodfellow PJ, Buttin BM, Herzog TJ, et al. Prevalence of defective DNA mismatch repair and MSH6 mutation in an unselected series of endometrial cancers. Proc Natl Acad Sci U S A. 2003;100:5908–5913. doi: 10.1073/pnas.1030231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egoavil C, Alenda C, Castillejo A, et al. Prevalence of Lynch syndrome among patients with newly diagnosed endometrial cancers. PLoS One. 2013;8:e79737. doi: 10.1371/journal.pone.0079737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferguson SE, Aronson M, Pollett A, et al. Performance characteristics of screening strategies for Lynch syndrome in unselected women with newly diagnosed endometrial cancer who have undergone universal germline mutation testing. Cancer. 2014;120:3932–3939. doi: 10.1002/cncr.28933. [DOI] [PubMed] [Google Scholar]
- 6.Lynch HT, Snyder CL, Shaw TG, et al. Milestones of Lynch syndrome: 1895-2015. Nat Rev Cancer. 2015;15:181–941. doi: 10.1038/nrc3878. [DOI] [PubMed] [Google Scholar]
- 7.Baglietto L, Lindor NM, Dowty JG, et al. Risks of Lynch syndrome cancers for MSH6 mutation carriers. J Natl Cancer Inst. 2010;102:193–201. doi: 10.1093/jnci/djp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasen HF, Abdirahman M, Brohet R, et al. One to 2-year surveillance intervals reduce risk of colorectal cancer in families with Lynch syndrome. Gastroenterology. 2010;138:2300–2306. doi: 10.1053/j.gastro.2010.02.053. [DOI] [PubMed] [Google Scholar]
- 9.Lu KH, Daniels M. Endometrial and ovarian cancer in women with Lynch syndrome: Update in screening and prevention. Fam Cancer. 2013;12:273–277. doi: 10.1007/s10689-013-9664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoffel EM, Mangu PB, Gruber SB, et al. Hereditary colorectal cancer syndromes: American Society of Clinical Oncology clinical practice guideline endorsement of the familial risk-colorectal cancer—European Society for Medical Oncology clinical practice guidelines. J Clin Oncol. 2015;33:209–217. doi: 10.1200/JCO.2014.58.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lancaster JM, Powell CB, Chen LM, et al. Society of Gynecologic Oncology statement on risk assessment for inherited gynecologic cancer predispositions. Gynecol Oncol. 2015;136:3–7. doi: 10.1016/j.ygyno.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Buchanan DD, Tan YY, Walsh MD, et al. Tumor mismatch repair immunohistochemistry and DNA MLH1 methylation testing of patients with endometrial cancer diagnosed at age younger than 60 years optimizes triage for population-level germline mismatch repair gene mutation testing. J Clin Oncol. 2014;32:90–100. doi: 10.1200/JCO.2013.51.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu KH, Ring KL. One size may not fit all: The debate of universal tumor testing for Lynch syndrome. Gynecol Oncol. 2015;137:2–3. doi: 10.1016/j.ygyno.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 14.Brinton LA, Felix AS, McMeekin DS, et al. Etiologic heterogeneity in endometrial cancer: Evidence from a Gynecologic Oncology Group trial. Gynecol Oncol. 2013;129:277–284. doi: 10.1016/j.ygyno.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zighelboim I, Schmidt AP, Gao F, et al. ATR mutation in endometrioid endometrial cancer is associated with poor clinical outcomes. J Clin Oncol. 2009;27:3091–3096. doi: 10.1200/JCO.2008.19.9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dewdney SB, Rimel BJ, Thaker PH, et al. Aberrant methylation of the X-linked ribosomal S6 kinase RPS6KA6 (RSK4) in endometrial cancers. Clin Cancer Res. 2011;17:2120–2129. doi: 10.1158/1078-0432.CCR-10-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 18.Whitcomb BP, Mutch DG, Herzog TJ, et al. Frequent HOXA11 and THBS2 promoter methylation, and a methylator phenotype in endometrial adenocarcinoma. Clin Cancer Res. 2003;9:2277–2287. [PubMed] [Google Scholar]
- 19.Djordjevic B, Barkoh BA, Luthra R, et al. Relationship between PTEN, DNA mismatch repair, and tumor histotype in endometrial carcinoma: Retained positive expression of PTEN preferentially identifies sporadic non-endometrioid carcinomas. Mod Pathol. 2013;26:1401–1412. doi: 10.1038/modpathol.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruegl AS, Djordjevic B, Batte B, et al. Evaluation of clinical criteria for the identification of Lynch syndrome among unselected patients with endometrial cancer. Cancer Prev Res (Phila) 2014;7:686–697. doi: 10.1158/1940-6207.CAPR-13-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pritchard CC, Smith C, Salipante SJ, et al. ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J Mol Diagn. 2012;14:357–366. doi: 10.1016/j.jmoldx.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kastrinos F, Steyerberg EW, Mercado R, et al. The PREMM(1,2,6) model predicts risk of MLH1, MSH2, and MSH6 germline mutations based on cancer history. Gastroenterology. 2011;140:73–81. doi: 10.1053/j.gastro.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leenen CH, van Lier MG, van Doorn HC, et al. Prospective evaluation of molecular screening for Lynch syndrome in patients with endometrial cancer ≤ 70 years. Gynecol Oncol. 2012;125:414–420. doi: 10.1016/j.ygyno.2012.01.049. [DOI] [PubMed] [Google Scholar]
- 24.Clendenning M, Senter L, Hampel H, et al. A frame-shift mutation of PMS2 is a widespread cause of Lynch syndrome. J Med Genet. 2008;45:340–345. doi: 10.1136/jmg.2007.056150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Truninger K, Menigatti M, Luz J, et al. Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer. Gastroenterology. 2005;128:1160–1171. doi: 10.1053/j.gastro.2005.01.056. [DOI] [PubMed] [Google Scholar]
- 26.ten Broeke SW, Brohet RM, Tops CM, et al. Lynch syndrome caused by germline PMS2 mutations: Delineating the cancer risk. J Clin Oncol. 2015;33:319–325. doi: 10.1200/JCO.2014.57.8088. [DOI] [PubMed] [Google Scholar]
- 27.Lu KH, Dinh M, Kohlmann W, et al. Gynecologic cancer as a “sentinel cancer” for women with hereditary nonpolyposis colorectal cancer syndrome. Obstet Gynecol. 2005;105:569–574. doi: 10.1097/01.AOG.0000154885.44002.ae. [DOI] [PubMed] [Google Scholar]
- 28.Cook LS, Nelson HE, Stidley CA, et al. Endometrial cancer and a family history of cancer. Gynecol Oncol. 2013;130:334–339. doi: 10.1016/j.ygyno.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bharati R, Jenkins MA, Lindor NM, et al. Does risk of endometrial cancer for women without a germline mutation in a DNA mismatch repair gene depend on family history of endometrial cancer or colorectal cancer? Gynecol Oncol. 2014;133:287–292. doi: 10.1016/j.ygyno.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Backes FJ, Hampel H, Backes KA, et al. Are prediction models for Lynch syndrome valid for probands with endometrial cancer? Fam Cancer. 2009;8:483–487. doi: 10.1007/s10689-009-9273-5. [DOI] [PubMed] [Google Scholar]
- 31.Barzi A, Sadeghi S, Kattan MW, et al. Comparative effectiveness of screening strategies for lynch syndrome. J Natl Cancer Inst. 2015;20:107. doi: 10.1093/jnci/djv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan YY, McGaughran J, Ferguson K, et al. Improving identification of lynch syndrome patients: A comparison of research data with clinical records. Int J Cancer. 2013;132:2876–2883. doi: 10.1002/ijc.27978. [DOI] [PubMed] [Google Scholar]
- 33.Mercado RC, Hampel H, Kastrinos F, et al. Performance of PREMM(1,2,6), MMRpredict, and MMRpro in detecting Lynch syndrome among endometrial cancer cases. Genet Med. 2012;14:670–680. doi: 10.1038/gim.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gould-Suarez M, El-Serag HB, Musher B, et al. Cost-effectiveness and diagnostic effectiveness analyses of multiple algorithms for the diagnosis of Lynch syndrome. Dig Dis Sci. 2014;59:2913–2926. doi: 10.1007/s10620-014-3248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward RL, Dobbins T, Lindor NM, et al. Identification of constitutional MLH1 epimutations and promoter variants in colorectal cancer patients from the Colon Cancer Family Registry. Genet Med. 2013;15:25–35. doi: 10.1038/gim.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pineda M, Mur P, Iniesta MD, et al. MLH1 methylation screening is effective in identifying epimutation carriers. Eur J Hum Genet. 2012;20:1256–1264. doi: 10.1038/ejhg.2012.136. [DOI] [PMC free article] [PubMed] [Google Scholar]