Abstract
Bacterial diversity and community structure of two maize varieties (white and yellow) during fermentation/steeping for ogi production, and the influence of spontaneous fermentation on mycotoxin reduction in the gruel were studied. A total of 142 bacterial isolates obtained at 24–96 h intervals were preliminarily identified by conventional microbiological methods while 60 selected isolates were clustered into 39 OTUs consisting of 15 species, 10 genera, and 3 phyla by 16S rRNA sequence analysis. Lactic acid bacteria constituted about 63% of all isolated bacteria and the genus Pediococcus dominated (white maize = 84.8%; yellow maize = 74.4%). Pediococcus acidilactici and Lactobacillus paraplantarum were found at all steeping intervals of white and yellow maize, respectively, while P. claussenii was present only at the climax stage of steeping white maize. In both maize varieties, P. pentosaceus was found at 24–72 h. Mycotoxin concentrations (μg/kg) in the unsteeped grains were: white maize (aflatoxin B1 = 0.60; citrinin = 85.8; cyclopiazonic acid = 23.5; fumonisins (B1/B2/B3) = 68.4–483; zearalenone = 3.3) and yellow maize (aflatoxins (B1/B2/M1) = 22.7–513; citrinin = 16,800; cyclopiazonic acid = 247; fumonisins (B1/B2/B3) = 252–1,586; zearalenone = 205). Mycotoxins in both maize varieties were significantly (p < 0.05) reduced across steeping periods. This study reports for the first time: (a) the association of L. paraplantarum, P. acidilactici, and P. claussenii with ogi production from maize, (b) citrinin occurrence in Nigerian maize and ogi, and (c) aflatoxin M1, citrinin and cyclopiazonic acid degradation/loss due to fermentation in traditional cereal-based fermented food.
Keywords: diversity, ecology, fermentation, food safety, maize, mycotoxins, ogi
Introduction
Maize (Zea mays L.) is a staple food in many parts of the world including sub-Saharan Africa (SSA). In Nigeria and some other West African countries, it is traditionally transformed by submerged fermentation to ogi – a complementary weaning food for infants and young children, convenient food for the sick, convalescent and elderly or quick breakfast mostly for those living in rural areas characterized by low income (Onyekwere et al., 1989; Steinkraus, 1996). Ogi is preferred by nearly 150 million West Africans (Oguntoyinbo and Narbad, 2012) mainly due to the ease of preparation of this gruel and the numerous associated nutritional benefits including high calorie, minerals, vitamins, and probiotic contents (Opere et al., 2012). The rich probiotic contents of ogi and other traditionally fermented cereal foods result from the indigenous beneficial microbial flora that play significant roles during cereal fermentation to yield the final product (Odunfa, 1985). Furthermore, fermentation provides a variety of foods and contributes to food preservation.
Fermentation during ogi production occurs in two distinct stages: (1) steeping of maize prior to obtaining ogi gruel and (2) souring of fermented ogi (Omemu, 2011). Several authors have reported on ogi production from various varieties of maize (white and yellow), from guinea corn, millet and sorghum after steeping for either 24, 48, 72, or 96 h (Odunfa and Adeyele, 1985; Teniola and Odunfa, 2002; Teniola et al., 2005; Adebayo and Aderiye, 2007; Adebayo-tayo and Onilude, 2008; Dike and Sanni, 2010; Omemu, 2011; Banwo et al., 2012; Oyedeji et al., 2013). Usual practice in the local/traditional setting, however, is that maize is steeped for at least 48 h and may extend to 96 h. In order to understand the ecology of species and promote biotechnology through beneficial strain selection for improved ogi production, microbial communities associated with spontaneous maize fermentation to ogi have been characterized and studied. The majority of the studies considered microbial diversity in mashed fermented maize grains (Teniola et al., 2005; Adebayo and Aderiye, 2007; Omemu, 2011) while others characterized fermenters from souring ogi samples (Odunfa and Adeyele, 1985; Teniola and Odunfa, 2002; Adebayo-tayo and Onilude, 2008; Oguntoyinbo et al., 2011; Banwo et al., 2012; Oguntoyinbo and Narbad, 2012); only Oyedeji et al. (2013) examined maize steep liquor of 24–72 h for fermenter diversity. From the aforementioned studies, Lactobacillus and Leuconostoc have been reported as the occurring genera of lactic acid bacteria (LAB) in maize steep liquor and fermented/souring ogi while Pediococcus was only reported in fermented/souring ogi.
Consumption of maize and maize-based food products is threatened by the presence of fungal toxins (e.g., aflatoxins, cyclopiazonic acid; fumonisins, and zearalenone; Warth et al., 2012; Abia et al., 2013a; Kayode et al., 2013; Adetunji et al., 2014a). Potential mycotoxin exposure through maize consumption has been identified to be higher in rural areas of developing countries where vulnerable consumers utilize broken and damaged kernels for diverse dietary purposes; such kernels are usually cheaper but could possibly be higher in mycotoxin content (Abia et al., 2013b; Njumbe Ediage et al., 2013; Adetunji et al., 2014b; Ezekiel et al., 2014). Efforts toward diversification of maize meals for enhanced nutritional benefits led to ogi production involving fermentation steps. There are suggestions that traditional fermentation can reduce mycotoxin transfer from grains to fermented foods whilst enhancing the nutrient content of food through the synthesis of protein, vitamins, and amino acids (Mokoena et al., 2005; Shetty and Jespersen, 2006; Juodeikiene et al., 2012).
Past studies with respect to ogi production focused mostly on studying microbial diversity and the roles of fermenters against natural microbial contaminants in ogi production (Odunfa and Adeyele, 1985; Teniola and Odunfa, 2002; Teniola et al., 2005; Adebayo and Aderiye, 2007; Adebayo-tayo and Onilude, 2008; Oguntoyinbo et al., 2011; Omemu, 2011; Banwo et al., 2012; Oguntoyinbo and Narbad, 2012; Oyedeji et al., 2013). There are also reports available on mycotoxin reduction mediated by single or combined cultures of LAB and other fermentation bacteria artificially inoculated into maize and other grains (Mokoena et al., 2005; Shetty and Jespersen, 2006; Oluwafemi and Da-Silva, 2009; Cho et al., 2010; Nyamete, 2013; Zhao et al., 2015). In spite of the aforementioned reports, there is no information on the influence of spontaneous fermentation mediated by autochthonous microbial communities interacting during ogi production on mycotoxin reduction in the gruel. Such information could provide important data to establish the relationship between fermentation and mycotoxin levels in ogi. This study therefore aims to discover the diversity and succession of bacteria during maize fermentation (steeping) into ogi, and to determine possible mycotoxin reduction due to fermentation influenced by indigenous microbial communities.
Materials and Methods
Source of Maize Grains and Preparation of Ogi Samples
Two maize varieties (white and yellow) were purchased from Ikenne market in Ogun State, Nigeria and used for this study. Grains of the white variety had been stored for less than one month while those of the yellow variety were approximately 6 months old in store. The grains were sorted to remove particulate matter and batched (750 g per batch) into the various groups based on the steeping durations (48, 72, and 96 h). Each batch was set up in triplicate, steeped in 1.5 L of clean tap water in large plastic bowls with lids and allowed to undergo spontaneous fermentation at ambient temperature. Steep liquor from the batches was taken at intervals of 24–96 h and bacteria associated with the steeping process were isolated and characterized. The pH of each steep liquor was determined at 24–96 h.
At the end of each steeping duration (48, 72, and 96 h), the remaining steep liquors were drained off and discarded. The fermented/softened maize grains were processed into ogi as previously described by Adebayo and Aderiye (2007). Ogi samples obtained from the various batches representing the steep durations were not left to sour but were immediately pressed to remove excess water and taken for mycotoxin analysis.
Isolation of Bacteria from Maize Steep Liquor
Serial dilutions of steep liquor obtained from maize fermentation batches were performed and used for isolation of aerobic and anaerobic bacteria on plate count agar (PCA, Oxoid CM 325, Unipath, Hampshire, England) and deMann Rogosa Sharpe (MRS, Oxoid CM 361) agar, respectively (Haddadin et al., 2004). The pour-plate method was used for inoculation. MRS agar was supplemented with sodium azide (0.01%, Labtech Chemicals, Chinchwad Gaon, Pune, India) to inhibit aerobic bacteria and facilitate the growth of LAB (Cho et al., 2013). PCA plates were incubated for 24 h at 35°C for isolation of mesophilic aerobic bacteria while MRS agar plates were incubated for 24–72 h at 35–45°C in an Anaerobic Gas-Pack system Jung et al. (2012). In order to ensure good representation of isolates, all colonies were sampled from plates containing fewer than 10 colonies or five distinct colonies were randomly selected in colony dense plates. Isolates obtained were purified by a two-time repeated streaking on fresh agar plates of PCA and MRS, and maintained on same agar slants at 4°C at the Microbiology Laboratory of Babcock University, Nigeria.
Identification of Bacterial Isolates
Morphological and Biochemical Identification
For preliminary identification of isolates to genus level, cell morphology (Gram reaction, cell shape and arrangement) of all 142 selected colonies were examined under a phase contrast microscope (Olympus CX21FS1, Tokyo, Japan) after Gram staining. Simple biochemical tests (e.g., catalase production, motility, sugar fermentation, and acid and gas production in MRS broth) were also carried out on each presumed LAB isolate according to the method of Dykes (1994). Based on preliminary data obtained, 80 (21 aerobic and 59 anaerobic) of the 142 isolates were selected, preserved in nutrient broth (LAB ‘E’, LAB068, UK) supplemented with 40% glycerol (BDH, Poole, England) and sent to the Agricultural Research Council–Institute for Soil Climate and Water (ARC–ISCW), South Africa for molecular characterization.
Molecular Characterization of Isolates
Sixty (21 aerobic and 39 anaerobic) isolates were further selected based on further preliminary tests, for DNA extraction and molecular analysis to determine diversity and phylogeny of the isolates. Genomic DNA extraction was performed according to the protocol described in the ZR Soil Microbe DNA MiniPrep extraction kit (Zymo Research Corporation, Irvine, CA, USA) with modifications. DNA quantification was by the Qubit 2.0 Fluorometer (Q32867, Life Technologies, Grand Island, NY, USA) while verification of DNA quality was performed on 1.5% agarose gel electrophoresis prior to PCR.
All the isolated DNA sequences were used as template to amplify bacterial 16S rRNA genes. PCR amplification was performed with primers 341F (CCTACGGGAGGCAGCAG) and 907R (CCGTCAATTCCTTTAAGTTT). The amplicon sizes were ∼600 bp and were purified and sequenced using the Sanger sequence approach at Inqaba Biotech, South Africa, a commercial service provider. Bidirectional Sanger sequence reads were obtained by standard procedures and the contigs were assembled by the Bio-Edit sequence assembly program (Hall, 1999). Sequence data obtained have been submitted to GenBank under accession numbers: KR812487–KR812544 as well as BWS2E and FWS4B (Table 1).
Table 1.
Bacterial populations identified during steeping of maize for ogi production.
| Accession numbers | OTUs1 | Matched species (close relatives from NCBI) | Phylum |
|---|---|---|---|
| KR812487 | OTU2 | Lactobacillus paraplantarum (AJ306297) | Firmicutes |
| KR812488 | OTU4 | Pediococcus acidilactici (AJ305320) | Firmicutes |
| KR812489 | OTU4 | P. pentosaceus (AJ305321) | Firmicutes |
| KR812490 | OTU1 | P. acidilactici (AJ305320) | Firmicutes |
| KR812491 | OTU8 | P. pentosaceus (AJ305321) | Firmicutes |
| KR812492 | OTU2 | L. paraplantarum (AJ306297) | Firmicutes |
| KR812493 | OTU8 | P. acidilactici (AJ305320) | Firmicutes |
| KR812494 | OTU2 | L. paraplantarum (AJ306297) | Firmicutes |
| KR812495 | OTU8 | P. pentosaceus (AJ305321) | Firmicutes |
| KR812496 | OTU2 | L. paraplantarum (AJ306297) | Firmicutes |
| KR812497 | OTU8 | P. pentosaceus (AJ305321) | Firmicutes |
| KR812498 | OTU8 | P. pentosaceus (AJ305321) | Firmicutes |
| KR812499 | OTU7 | P. acidilactici (AJ305320) | Firmicutes |
| KR812500 | OTU3 | Pseudomonas hibiscicola (AB021405) | Proteobacteria |
| KR812501 | OTU4 | P. acidilactici (AJ305320) | Firmicutes |
| KR812502 | OTU8 | P. pentosaceus (AJ305321) | Firmicutes |
| KR812503 | OTU4 | P. acidilactici (AJ305320) | Firmicutes |
| KR812504 | OTU7 | P. acidilactici (AJ305320) | Firmicutes |
| KR812505 | OTU5 | Ps. hibiscicola (AB021405) | Proteobacteria |
| KR812506 | OTU8 | P. acidilactici (AJ305320) | Firmicutes |
| KR812507 | OTU6 | L. paraplantarum (AJ306297) | Firmicutes |
| KR812508 | OTU7 | P. acidilactici (AJ305320) | Firmicutes |
| KR812509 | OTU8 | P. pentosaceus (AJ305321) | Firmicutes |
| KR812510 | OTU8 | P. pentosaceus (AJ305321) | Firmicutes |
| KR812511 | OTU9 | Ps. hibiscicola (AB021405) | Proteobacteria |
| KR812512 | OTU10 | Bacillus thuringiensis (D16281) | Firmicutes |
| KR812513 | OTU16 | B. mycoides (AB021192) | Firmicutes |
| FWS4B | OTU11 | Enterobacter sp. (KF599041) | Proteobacteria |
| KR812514 | OTU12 | P. acidilactici (AJ305320) | Firmicutes |
| BWS2E | OTU13 | Moorella glycerini (U82327) | Firmicutes |
| KR812515 | OTU14 | B. mycoides (AB021192) | Firmicutes |
| KR812516 | OTU15 | P. pentosaceus (AJ305321) | Firmicutes |
| KR812517 | OTU16 | B. mycoides (AB021192) | Firmicutes |
| KR812518 | OTU16 | B. mycoides (AB021192) | Firmicutes |
| KR812519 | OTU16 | B. mycoides (AB021192) | Firmicutes |
| KR812520 | OTU16 | B. mycoides (AB021192) | Firmicutes |
| KR812521 | OTU8 | P. acidilactici (AJ305320) | Firmicutes |
| KR812522 | OTU17 | L. paraplantarum (AJ306297) | Firmicutes |
| KR812523 | OTU18 | P. acidilactici (AJ305320) | Firmicutes |
| KR812524 | OTU19 | P. acidilactici (AJ305320) | Firmicutes |
| KR812525 | OTU20 | P. acidilactici (AJ305320) | Firmicutes |
| KR812526 | OTU21 | P. acidilactici (AJ305320) | Firmicutes |
| KR812527 | OTU22 | Ps. hibiscicola (AB021405) | Proteobacteria |
| KR812528 | OTU23 | Ps. hibiscicola (AB021405) | Proteobacteria |
| KR812529 | OTU24 | Myroides marinus (GQ857652) | Bacteroidetes |
| KR812530 | OTU25 | Ps. hibiscicola (AB021405) | Proteobacteria |
| KR812531 | OTU26 | Alcaligenes faecalis (AJ242986) | Proteobacteria |
| KR812532 | OTU27 | Enterobacter sp. (KF588515) | Proteobacteria |
| KR812533 | OTU28 | Bordetella avium (AF177666) | Proteobacteria |
| KR812534 | OTU29 | P. pentosaceus (AJ305321) | Firmicutes |
| KR812535 | OTU30 | B. mycoides (AB021192) | Firmicutes |
| KR812536 | OTU31 | M. marinus (GQ857652) | Bacteroidetes |
| KR812537 | OTU32 | P. acidilactici (FJ905315) | Firmicutes |
| KR812538 | OTU33 | P. acidilactici (KT427449) | Firmicutes |
| KR812539 | OTU34 | Acinetobacter bereziniae (Z93443) | Proteobacteria |
| KR812540 | OTU35 | P. claussenii (NR_042232) | Firmicutes |
| KR812541 | OTU36 | Enterococcus faecium (AJ301830) | Firmicutes |
| KR812542 | OTU37 | B. bronchiseptica (U04948) | Proteobacteria |
| KR812543 | OTU38 | M. marinus (GQ857652) | Bacteroidetes |
| KR812544 | OTU39 | Ac. bereziniae (Z93443) | Proteobacteria |
1Operational Taxonomic Units.
The 16S rRNA gene sequences were first affiliated to bacterial taxa using SeqMatch on the Ribosomal Database Project (RDP) website (http://rdp.cme.msu.edu/index.jsp; Cole et al., 2009). Multiple sequence alignments and clustering into operational taxonomic units (OTUs) of the 60 sequences considered herein were performed with Mothur (Schloss et al., 2009), using a 1% dissimilarity level between OTUs (Table 1). The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987).
Matched sequences, one for each OTU, were later obtained from the GenBank using the accession numbers. These sequences alongside the OTU representatives were used to construct a library. All sequences were aligned using the multiple sequence alignment software, MAFFT version 7 (Katoh and Standley, 2013). A region of specific data matrice was built on sequence alignment and used to generate combined sequence alignment of multiple regions. Finally, Mega4 software was used to generate a phylogenetic tree consisting of representative OTUs and their close counterparts (matched sequences; Tamura et al., 2007), using the Aquifex aeolicus 16S rRNA gene as an outgroup sequence. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein, 1985).
Analysis of Maize and Ogi Samples for Mycotoxins
Prior to batching the maize grains for steeping and ogi production (see section on Source of maize grains and preparation of ogi samples), 500 g subsample of the grains was randomly taken, milled and quartered. A quarter (125 g) of the milled sample was taken for mycotoxin analysis and triplicate maize samples were analyzed. About 80 g subsample of each pressed ogi (see section on Source of maize grains and preparation of ogi samples) was also taken and quartered. The maize and ogi samples were immediately shipped on dry ice to IFA-Tulln, Austria for mycotoxin analysis. All samples were kept at –20°C at IFA-Tulln until mycotoxin analysis. Mycotoxin analysis of maize and ogi samples was performed by liquid chromatography tandem mass spectrometry (LC–MS/MS).
Five grams of each homogenized representative maize or ogi sample was weighed into a 50 ml polypropylene tube (Sarstedt, Germany) and extracted with 15 ml acetonitrile/water/acetic acid (79:20:1, v/v/v) for 90 min on a GFL 3017 rotary shaker (GFL, Burgwedel, Germany). Extracts were diluted in extraction solvent and injected into the LC instrument as described in detail by Malachova et al. (2014). Mycotoxins and other microbial metabolites described by Malachova et al. (2014) were screened using a QTrap 5500 LC-MS/MS System (Applied Biosystems, Foster City, CA, USA) equipped with a TurboV electrospray ionization (ESI) source and a 1290 Series UHPLC System (Agilent Technologies, Waldbronn, Germany). Chromatographic separation was performed at 25°C on a Gemini® C18-column, 150 mm × 4.6 mm i.d., 5 μm particle size, equipped with a C18 security guard cartridge, 4 mm × 3 mm i.d. (all from Phenomenex, Torrance, CA, USA). Positive analyte identification was confirmed by the acquisition of two MS/MS transitions which yielded 4.0 identification points according to commission decision 2002/657/EC. Furthermore, the LC retention time and the intensity ratio of the two MRM transitions agreed with the related values of an authentic standard within 0.1 min and 30% rel., respectively. Further details relating to spiking, recoveries and additional LC–MS/MS parameters are as reported in our previous papers (Warth et al., 2012; Abia et al., 2013a).
Estimation of Mycotoxin Reduction in Ogi Due to Fermentation
In order to estimate percentage reduction of each mycotoxin due to fermentation influenced by fermenter diversity, the percentage difference between mycotoxin levels in the grain and final product (ogi) was calculated, taking into consideration the sum of mycotoxin levels lost due to other processes involved in ogi production (e.g., washing of grains, mashing and sieving of slurry, and discarding of pomace). Details of the influence of steeping and processing practices on reduction of mycotoxins and other microbial metabolites during ogi production will be described elsewhere (Okeke et al., manuscript in preparation).
Data Analysis
SPSS 15.0 for Windows (SPSS, Inc., Chicago, IL, USA) was used for data analyses of (a) occurrence of fermenter species, and (b) mycotoxin reduction levels. Means were separated by the Duncan’s Multiple Range test and tested for significance by one-way analysis of variance at α = 0.05.
Results and Discussion
Bacterial Diversity During Steeping of Maize for Ogi Production
A total of 142 bacterial isolates were obtained from the steeping processes of both maize varieties; 73 and 69 isolates from white and yellow varieties, respectively. The isolates obtained on PCA (n = 51) represented aerobic or facultatively anaerobic species while those on MRS agar (n = 91) were LAB and represented obligate or facultative anaerobic homofermentative cocci or heterofermentative cocci and rods. Preliminary identification tests of all the 142 isolates suggested the LAB isolates were Gram-positive, catalase negative and non-motile (Odunfa and Adeyele, 1985; Teniola and Odunfa, 2002; Teniola et al., 2005) and belonged to Lactobacillus and Pediococcus, while other bacteria belonged to Bacillus, Enterococcus and a range of rod-shaped bacteria.
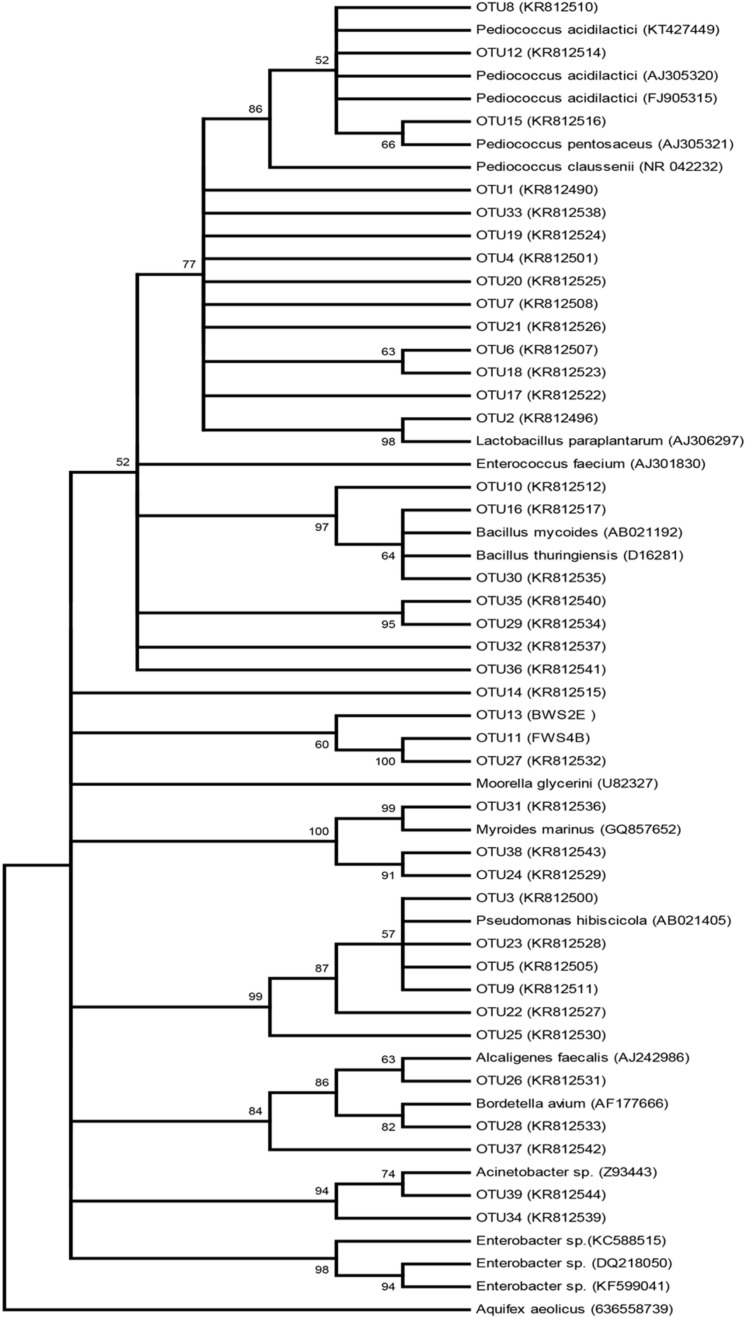
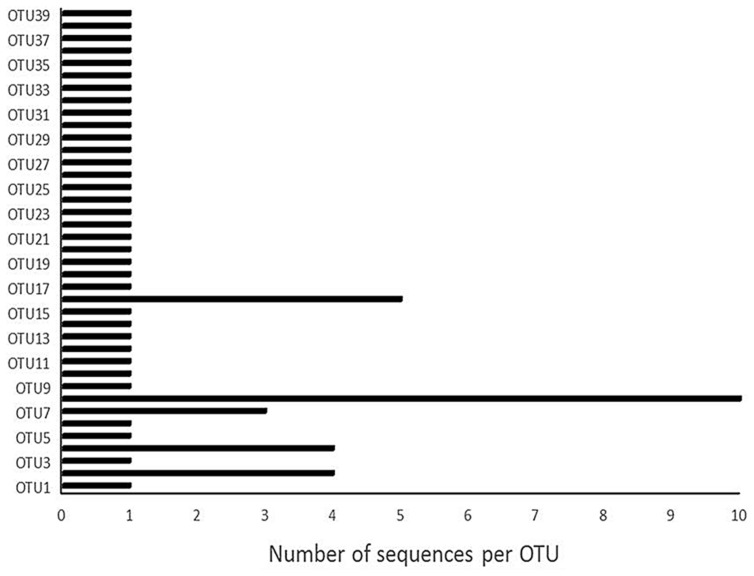
Our choice of cultivation techniques aided the discrimination of living and dead microorganisms that are able to participate in the fermentation process. However, to ensure proper identification of the bacterial isolates, molecular techniques were employed. Molecular characterization of the isolates clustered them into 39 OTUs representing three phyla, eight families, 10 genera and 15 species (Table 1, Figures 1 and 2). The families (Alcaligenaceae, Bacillaceae, Enterobacteriaceae, Enterococcaceae, Flavobacteriaceae, Lactobacillaceae, Moraxellaceae, and Xanthomonadaceae) are not shown in the tables or figures.
FIGURE 1.
Phylogenetic tree illustrating major bacterial taxa identified during steeping of maize for ogi production and their relatives obtained from the Seqmatch (RDP).
FIGURE 2.
Operational Taxonomic Units (OTUs) and number of sequences per OTU.
Among the identified OTUs were four distinct LAB species – L. paraplantarum, P. acidilactici, P. claussenii, and P. pentosaceus (Table 1, Figure 2). Similar spectra and even more species of LAB excluding L. paraplantarum and P. claussenii have been previously reported in ogi made from a range of cereals including guinea-corn, maize, millet, and sorghum (Teniola and Odunfa, 2002; Teniola et al., 2005; Adebayo and Aderiye, 2007; Adebayo-tayo and Onilude, 2008; Oguntoyinbo et al., 2011; Omemu, 2011; Banwo et al., 2012; Oguntoyinbo and Narbad, 2012). L. plantarum and various species of Lactococcus and Leuconostoc were previously reported in maize steep liquor at 24–72 h by Oyedeji et al. (2013). However, this first report of L. paraplantarum and absence of L. plantarum, Lactococcus, and Leuconostoc in our study may have been mainly due to selective/biased isolation/subculturing influenced by culture-dependent methods, especially when L. plantarum was reported to be predominant during ogi production by previous authors who employed molecular tools for species identification (Oguntoyinbo et al., 2011; Banwo et al., 2012; Oguntoyinbo and Narbad, 2012). L. paraplantarum was previously found in beer (Curk et al., 1996) and subsequently in koko sour water from millet in Ghana (Lei and Jakobsen, 2004).
Among the Pediococcus species we found, P. pentosaceus had previously been identified in kenkey and white maize grains steeped for 24 h for ogi (Halm et al., 1993; Teniola et al., 2005) while P. acidilactici was also found in kenkey and during maize fermentation for masa (Oyeyiola, 1990; Halm et al., 1993; Fowoyo and Ogunbanwo, 2010; Sanni and Adesulu, 2013) and recently, in sorghum ogi (Banwo et al., 2012). Our study is therefore the first report on association of P. acidilactici and P. claussenii with ogi production from maize. Additional isolates obtained in the present study belonged to Acinetobacter, Alcaligenes, Bacillus, Bordetella, Enterobacter, Enterococcus, Moorella, Myroides, and Pseudomonas (Table 1). These genera are known soil bacteria, human, and animal pathogens, and are of no value to fermentation except for Bacillus, Enterobacter, and Enterococcus which had been detected during food fermentations in Africa and Asia (Abriouel et al., 2006; Chiang et al., 2006; Oguntoyinbo et al., 2010; Oguntoyinbo and Narbad, 2012).
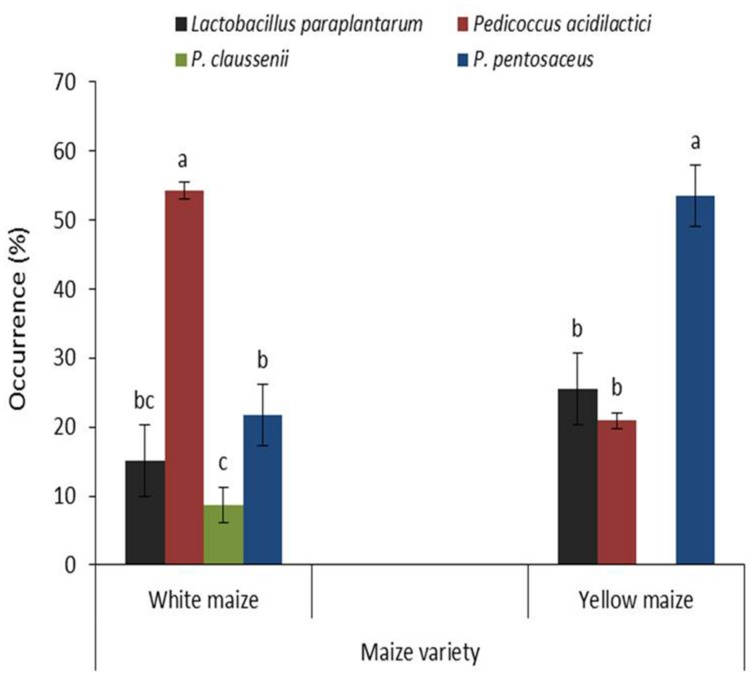
Maize varietal specific distribution of the LAB species is shown in Figure 3. Overall, LAB constituted 62.3–63.0% of all isolated bacteria from steep water of both maize varieties; Pediococcus dominated (84.8% in white maize and 74.4% in yellow maize) while the occurrence of Lactobacillus was 15.2 and 25.6% in white and yellow maize, respectively. The frequencies of the LAB isolates by OTU cluster in each maize variety are given here: white maize (Pediococcus = 20.5%, Lactobacillus = 5.1%); yellow maize: (Pediococcus = 15.4%, Lactobacillus = 2.6%). In terms of species distribution, Pediococcus acidilactici (54.3%) and P. pentosaceus (53.5%) significantly (p < 0.05) predominated over other species in steep water of white and yellow maize varieties, respectively, while P. claussenii (8.7%) was found only in white maize (Figure 3). Our findings contrast with previous reports on several species of Lactobacillus (e.g., L. delbrueckii subsp. bulgaricus, L. fermentum, and L. plantarum) predominating in ogi, it’s fermentation processes, or in other traditional foods such as fermenting cassava for garri, raw milk for nono, massa, and wara (Johansson et al., 1995; Teniola and Odunfa, 2002; Adebayo and Aderiye, 2007; Adebayo-tayo and Onilude, 2008; Fowoyo and Ogunbanwo, 2010; Oguntoyinbo et al., 2011; Omemu, 2011; Banwo et al., 2012; Oguntoyinbo and Narbad, 2012; Obinna-Echem et al., 2013; Oyedeji et al., 2013). This emphasizes the need for more elaborate efforts toward studying the diversity of beneficial natural flora of maize and exploitation of such bacteria for traditional food processes.
FIGURE 3.
Occurrence of lactic acid bacteria in steep liquor of two varieties of maize grains fermented for 94 h. Vertical lines on bars indicate the standard error of mean (α = 0.05). Bars with different alphabets are significantly different by DMRT at α = 0.05.
Bacterial Succession During Steeping of Maize for Ogi Production
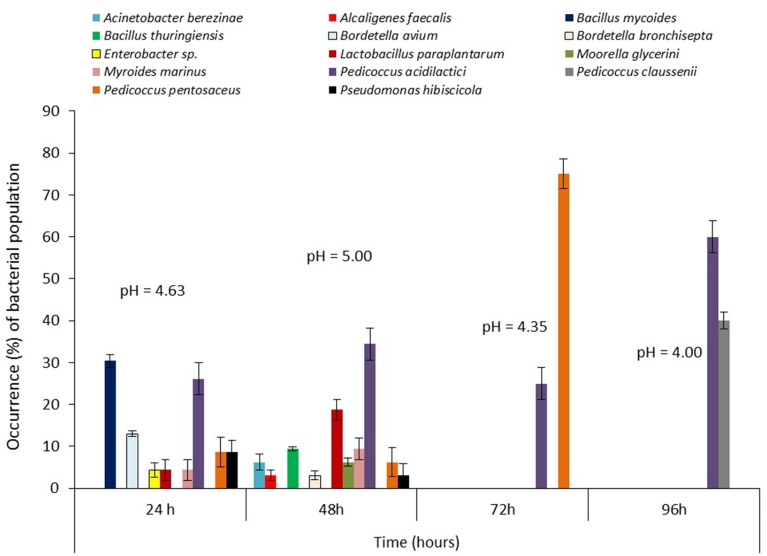
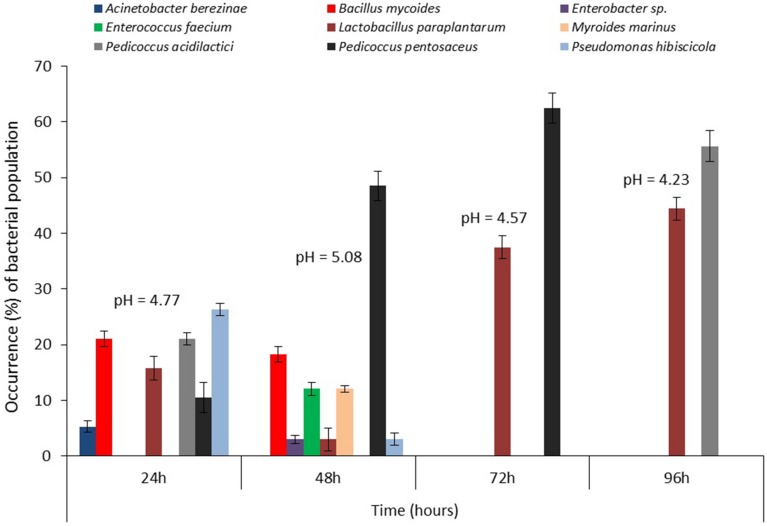
Molecular analysis strongly indicated succession among the bacterial communities during the steeping/fermentation process of both maize varieties (Figures 4 and 5). Succession patterns differed from previous reports (Teniola et al., 2005; Adebayo and Aderiye, 2007; Omemu, 2011; Oyedeji et al., 2013) and also in the two maize varieties though the LAB isolates dominated through the successional periods. For the white maize, P. acidilactici occurred at all stages of steeping [pioneered at 24 h (occurrence = 26.1%) and climaxed at 96 h (occurrence = 60%)] while L. paraplantarum (4.4–18.8%) and P. pentosaceus (6.3–75%) were detected only at seral stages (24–48 h and 24–72 h, respectively; Figure 4). Pediococcus claussenii (40%) was detected only at the climax stage. In addition, the non-LAB species (3.1–30.4%) were prominent only at the early stages (24–48 h) with Bacillus mycoides (30.4%) dominating at the pioneer stage. In the yellow maize, L. paraplantarum was detected from pioneer (occurrence = 15.8%) to climax (occurrence = 44.4%) of steeping duration. Pediococcus pentosaceus (10.5–62.5%) co-pioneered with L. paraplantarum, P. acidilactici (21.1%), and the non-LAB species (5.3–26.3%), extending to 72 h before disappearing from the community while P. acidilactici was subsequently detected at climax where it dominated (occurrence = 55.6%). Clearly, a wide range of beneficial, opportunistic and potentially pathogenic microbial communities were present and actively interacting in the microenvironment.
FIGURE 4.
Changes in bacterial community structure and pH during steeping of white maize grains for ogi production.
FIGURE 5.
Changes in bacterial community structure and pH during steeping of yellow maize grains for ogi production.
The presence and dominance of LAB species such as L. paraplantarum, P. acidilactici, and P. pentosaceus at various steeping times suggest that these and other previously reported LAB species not found in our study are the primary and influential bacterial species involved in the fermentation of maize to ogi. These LAB species may therefore be exploited as probiotics and potential starter cultures. The fluctuations of the pH from 4.63 through 5.00 to 4.00 in white maize and from 4.77 through 5.08 to 4.23 in yellow maize steep liquors indicate fluctuations in acid production and release into the microenvironment (Adebayo and Aderiye, 2007; Oyedeji et al., 2013). Furthermore, the extremely low pH (4.00) environment at 96 h of white maize fermentation created by P. acidilactici may have led to the entry and stabilization of P. claussenii in the succession as this LAB species has only been implicated in spoilage of fermented beverage (beer) due to diacetyl production (Bergsveinson et al., 2012). It was obvious that the non-LAB isolates were excluded from the environment marked by higher acidity (after 48 h) due to intense competition from LAB isolates capable of producing antibacterial compounds such as hydrogen peroxide, diacetyl, and bacteriocins during the process of sugar utilization to release acids (Teniola and Odunfa, 2002; Ogunbanwo et al., 2003, 2004; Adebayo and Aderiye, 2007; Dike and Sanni, 2010; Oyedeji et al., 2013), though these chemical products were not studied here.
Occurrence of Major Mycotoxins in White and Yellow Maize Grain
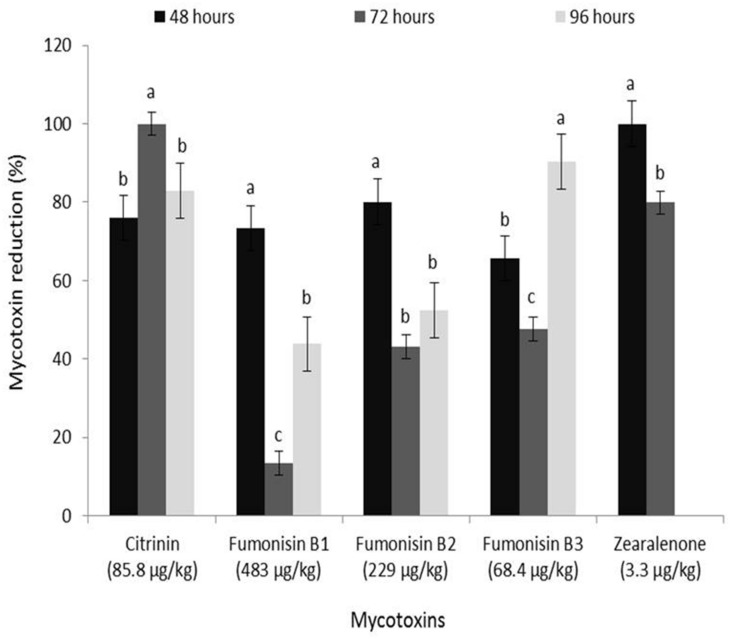
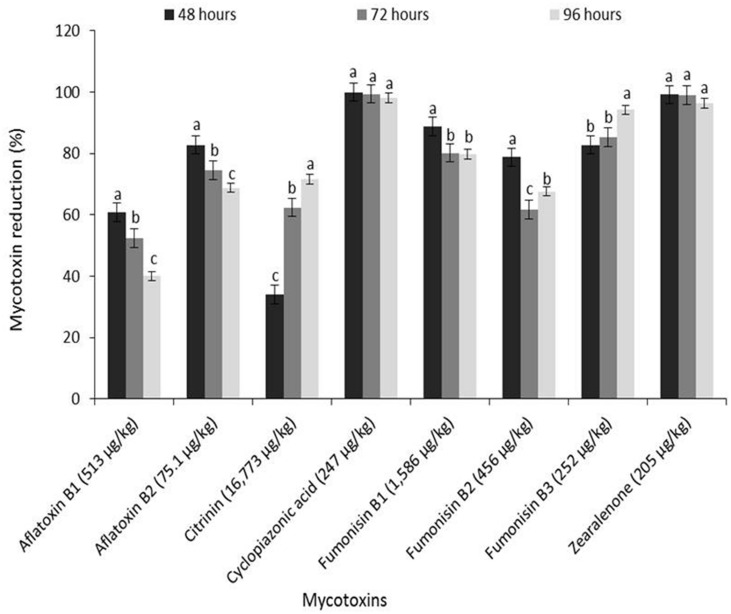
The concentrations of mycotoxins quantified in unsteeped grains of the two maize varieties are shown in Figures 6 and 7. Seven mycotoxins [aflatoxin B1 (AFB1) = 0.60 μg/kg; citrinin (CIT) = 85.8 μg/kg; cyclopiazonic acid (CPA) = 23.5 μg/kg; fumonisin B1 (FB1) = 483 μg/kg; fumonisin B2 (FB2) = 229 μg/kg; fumonisin B3 (FB3) = 68.4 μg/kg; zearalenone (ZEN) = 3.3 μg/kg] were found in the white grain (Figure 6), while nine mycotoxins [AFB1 = 513 μg/kg; aflatoxin B2 (AFB2) = 75.1 μg/kg; aflatoxin M1 (AFM1) = 22.7 μg/kg; CIT = 16,800 μg/kg; CPA = 247 μg/kg; FB1 = 1,586 μg/kg; FB2 = 456 μg/kg; FB3 = 252 μg/kg; ZEN = 205 μg/kg] occurred in the yellow variety (Figure 7). Mycotoxin levels in the yellow grains stored for about six months were at least twofold higher than the levels in the white variety barely stored for a month. Citrinin, a mycotoxin produced by Aspergillus and Penicillium, is reported here for the first time in Nigerian maize and ogi (see section on Reduction of mycotoxins in ogi influenced by fermentation) though it has previously been reported in maize from India and fermented maize from Ghana (Vella et al., 1995; Janardhana et al., 1999). The spectrum and levels of the other mycotoxins reported in this study, especially the increased levels in stored yellow maize grain, are similar to those previously reported across sub-Saharan Africa. This provides further evidence that mycotoxin contamination of maize and especially their accumulation under poor storage conditions remain a major food safety challenge warranting urgent attention in many countries in SSA (Udoh et al., 2000; Kankolongo et al., 2009; Ncube et al., 2011; Warth et al., 2012; Abia et al., 2013a; Adetunji et al., 2014a).
FIGURE 6.
Reduction (%) of mycotoxins in freshly fermented ogi due to fermentation during steeping of white maize grains. Concentrations given on x-axis indicate mycotoxin levels in raw maize grains before steeping. Cyclopiazonic acid (23.5 μg/kg) was reduced by 100% at all time intervals. Vertical lines on bars indicate the standard error of mean (α = 0.05). Bars with different alphabets are significantly different by DMRT at α = 0.05.
FIGURE 7.
Reduction (%) of mycotoxins in freshly fermented ogi due to fermentation during steeping of yellow maize grains. Concentrations given on x-axis indicate mycotoxin levels in raw maize grains before steeping. Aflatoxin M1 (22.7 μg/kg) was reduced by 100% at all time intervals. Vertical lines on bars indicate the standard error of mean (α = 0.05). Bars with different alphabets are significantly different by DMRT at α = 0.05.
Reduction of Mycotoxins in Ogi Influenced by Fermentation
Estimated percentage reductions of mycotoxins in white and yellow ogi due to fermentation are shown in Figures 6 and 7. Estimates were based on percentage differences between mycotoxin levels in the grain and ogi, taking into consideration the sum of mycotoxin levels lost due to other processes involved in ogi production. Details on the influence of steeping and processing practices on reduction of mycotoxins and other microbial metabolites during ogi production will be described elsewhere (Okeke et al., manuscript in preparation). The level of AFB1 (0.60 μg/kg) in white maize was very low to determine reduction, hence this was excluded from the percentage reduction estimations. On the other hand, CPA and AFM1 were reduced to levels below the limits of detection (LOD; CPA: <25 μg/kg; AFM1: <0.4 μg/kg, equivalent to 100% reduction) by fermentation of white and yellow maize, respectively, into ogi at all time intervals. Additionally, CIT and ZEN in white maize were completely lost (levels detected in ogi were <LOD; CIT ≤ 2.5 μg/kg, ZEN ≤ 0.05 μg/kg) during steeping for 72 and 48 h, respectively, while levels of CPA in yellow maize completely diminished (<LOD) at 48 h of steeping (Figures 6 and 7). This is the first report of AFM1, CIT and CPA degradation/loss due to fermentation in any traditional cereal-based fermented food product.
There were significant (p < 0.05) differences in the percentage reductions of mycotoxins across steeping time intervals (48–96 h) for both maize varieties though no time-dependent reduction of the mycotoxins was recorded for the white maize (Figure 6). For the yellow maize, percentage reduction of CIT and FB3 significantly (p < 0.05) increased as steeping time increased while for other toxins (AFB1, AFB2, FB1, and FB2) percentage reduction dropped significantly (p < 0.05) as steeping time increased from 48 to 96 h (Figure 7). Reduction levels of CPA (100–98.1%) and ZEN (99.2–96.4%) dropped insignificantly from 48 to 96 h. Generally, steeping of maize for 48 h drastically reduced fumonisin (FB1, FB2, and FB3) levels in both varieties (white maize: 65.7–80.1%; yellow maize: 78.7–88.8%) as well as aflatoxins (AFB1: 60.8%; AFB2: 82.8%) and ZEN (99.2%) in the yellow variety.
The extremely high levels of AFB1, CIT, CPA, FB1, and ZEN reported in raw maize samples in this study raise questions about the safety of consuming maize-based foods contaminated by mycotoxins. These mycotoxins exert diverse individual or synergistic toxicological effects (hepatotoxicity, nephrotoxicity, genotoxicity, teratogenicity, and immunotoxicity) on human and animal systems (Vella et al., 1995; Janardhana et al., 1999; Bondy and Pestka, 2000; Council for Agricultural Science Technology [Cast], 2003; Flajs and Peraica, 2009). However, the remarkable reduction of all the mycotoxins especially after 48 h of steeping proves that fermentation mediated by natural maize flora reduces mycotoxin levels of ogi, thus making it a safer food for consumption than its parent maize material. For fermentation to be very effective in producing foods with the safest levels of mycotoxins it is important to ensure that mycotoxin control begins from the field through post-harvest (storage and handling); this will lead to having unsteeped maize with low mycotoxin levels.
Some mycotoxins (e.g., AFB1, FB1, and ZEN) have been shown to be degraded to various extents by fermentation bacteria or bio-transformed during fermentation processes (Mokoena et al., 2005; Shetty and Jespersen, 2006; Oluwafemi and Da-Silva, 2009; Cho et al., 2010; Nyamete, 2013; Ezekiel et al., 2015; Zhao et al., 2015) and we observed same. However, our study contradicts the reports of Fandohan et al. (2005) and Mokoena et al. (2006) who reported very low reduction levels of 18–28% for AFB1 under spontaneous fermentation conditions and suggested increased but insignificant toxin reduction with prolonged fermentation time. The higher reduction (>80%) observed for ZEN at all time intervals in contrast to the lower levels (∼45%) previously reported by Zhao et al. (2015) when strains of L. plantarum were used to remove ZEN from MRS medium, may indicate that mycotoxin degradation during fermentation may either be strain specific or require synergistic interaction of more than one species/strain. The fact that non-LAB species (e.g., Bacillus subtilis) has been implicated in ZEN degradation (up to 99% of 1 mg/kg after 24 h) in liquid medium (Cho et al., 2010) supports our findings of higher reduction at 48 h in both maize varieties. At 48 h, bacterial population was highest in both maize varieties (occurrence: white maize = 32/73, yellow maize = 33/69) and species diversity in the microenvironment included LAB and non-LAB species including numerous colonies of Bacillus species. In our study, we did not find any bio-transformation product of ZEN (e.g., β-zearalenol – a less estrogenic form) in contrast to reports from Mizutani et al. (2011) and Ezekiel et al. (2015) which showed that fermentation, especially submerged, leads to β-zearalenol formation. An assumption may be that the product released was further degraded by specialized bacteria – a finding that requires further investigation.
Citrinin and FB3 reduction levels increased with prolonged fermentation time while levels of other mycotoxins (including AFB1) reduced. This may be linked to specialization by bacteria – it is most likely that the LAB isolates which dominated the latter stages of the successional chain were fully responsible for CIT and FB3 degradation or detoxification; this needs to be proven. However, the binding of mycotoxins (e.g., aflatoxins) to the surface of Lactobacillus species (Haskard et al., 2001; Oluwafemi and Da-Silva, 2009) or reformation of aflatoxins, for example, in increased acidic conditions as explained by Kpodo et al. (1996) may have been the underlying factor of the lower reduction of some mycotoxins as fermentation days extended. Cho et al. (2010) and Zhao et al. (2015) suggested that the viable cell count of bacteria influences toxin detoxification, while Guan et al. (2005) reported the involvement of bacterial peptidoglycans in binding foreign matter in order to confer protection against infections. In view of these suggestions and the fact that Cho et al. (2010) and other authors reported the involvement of non-LAB species and some fungi in mycotoxin detoxification, we propose that mycotoxins (excluding aflatoxins) whose reduction levels decreased with prolonged fermentation were most likely due to more diverse bacteria with mycotoxin-binding capacity existing in the early stages of succession. As one moves up the successional chain toward climax, the diversity of bacteria in the microenvironment reduces which influences effective binding of mycotoxins; hence decreased reduction levels. Our reports on reduction levels of aflatoxins being decreased with increased fermentation time fully agrees with the idea on acidification of the environment which interferes with aflatoxin reduction since pH of the steep liquor dropped drastically (Kpodo et al., 1996).
Conclusion
The combined application of culture-dependent and molecular tools in this study has provided verified information on the community structure of bacteria playing successive roles during steeping of maize for ogi production. This has also translated into tracing the relationship between bacterial diversity and mycotoxin reduction under natural fermentation of maize to ogi. Mycotoxin levels in the unsteeped maize grains which were quite high were drastically and significantly reduced during the steeping/fermentation process mediated by diverse bacterial communities including fermenters. In view of our findings, ogi may be a relatively safe food for human consumption in terms of mycotoxin contamination. The present study identified knowledge gaps worth investigating and, therefore, propose the following: (a) a comprehensive study of the diversity and successional pattern of bacteria/microbes from steeping to souring, and how these influence mycotoxin transfer from raw maize to soured ogi; (b) study to understand which specific bacterial/microbial populations degrade or detoxify specific mycotoxins under natural fermentation conditions using labeled mycotoxins and metabolomics tools. This will enhance selection of appropriate starter cultures capable of optimal fermentation while at the same time, detoxifying mycotoxins.
Author Contributions
CNE conceived and designed the study. COE, MS, CCN, and SKD contributed to the design of the study. CAO, CCN, and MS carried out the experiment/lab work in Nigeria, South Africa and Austria. CNE, COE, SKD, RAA, and RK supervised the experiment/lab work. CNE, CCN, MS, and RAA analyzed the data. CAO, CNE, and CCN drafted the manuscript. All authors critically revised, fine-tuned, and approved the final draft manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful to the South African National Research Foundation Thuthuka grant # 841688 as well as the ARC-ISCW facility for molecular analyses.
References
- Abia W. A., Warth B., Sulyok M., Krska R., Tchana A. N., Njobeh P. B., et al. (2013a). Determination of multi-mycotoxin occurrence in cereals, nuts and their products in Cameroon by liquid chromatography tandem mass spectrometry (LC–MS/MS). Food Control 31 438–453. 10.1016/j.foodcont.2012.10.006 [DOI] [Google Scholar]
- Abia W. A., Warth B., Sulyok M., Krska R., Tchana A. N., Njobeh P. B., et al. (2013b). Bio-monitoring of mycotoxin exposure in Cameroon using a urinary multi-biomarker approach. Food Chem. Toxicol. 62 927–934. 10.1016/j.fct.2013.10.003 [DOI] [PubMed] [Google Scholar]
- Abriouel H., Omar N. B., Lopez R. L., Martinez-Canamero M., Keleke S., Galvez A. (2006). Culture-independent analysis of the microbial composition of the African traditional fermented foods poto poto and dégué by using three different DNA extraction methods. Int. J. Food Microbiol. 111 228–233. 10.1016/j.ijfoodmicro.2006.06.006 [DOI] [PubMed] [Google Scholar]
- Adebayo C. O., Aderiye B. I. (2007). Ecology and antibacteria potential of lactic acid bacteria associated with cereals and cassava. Res. J. Microbiol. 2 426–435. 10.3923/jm.2007.426.435 [DOI] [Google Scholar]
- Adebayo-tayo B. C., Onilude A. A. (2008). Screening of lactic acid bacteria strains isolated from some Nigerian fermented foods for EPS production. World Appl. Sci. J. 4 741–747. [Google Scholar]
- Adetunji M. C., Atanda O. O., Ezekiel C. N., Sulyok M., Warth B., Beltran E., et al. (2014a). Fungal and bacterial metabolites of stored maize (Zea mays, L.) from five agro-ecological zones of Nigeria. Mycotoxin Res. 30 89–102. 10.1007/s12550-014-0194-2 [DOI] [PubMed] [Google Scholar]
- Adetunji M. C., Atanda O. O., Ezekiel C. N., Dipeolu A. O., Uzochukwu S. V. A., Oyedepo J., et al. (2014b). Distribution of mycotoxins and risk assessment of maize consumers in five agro-ecological zones of Nigeria. Eur. Food Res. Technol. 239 287–296. 10.1007/s00217-014-2221-0 [DOI] [Google Scholar]
- Banwo K., Sanni A. I., Tan H., Tian Y. (2012). Phenotypic and genotypic characterization of lactic acid bacteria isolated from some Nigerian traditional fermented foods. Food Biotechnol. 26 124–142. 10.1080/08905436.2012.670831 [DOI] [Google Scholar]
- Bergsveinson J., Pittet V., Ziola B. (2012). RT-qPCR analysis of putative beer-spoilage gene expression during growth of Lactobacillus brevis BSO 464 and Pediococcus claussenii ATCC BAA-344T in beer. Appl. Microbiol. Biotechnol. 96 461–470. 10.1007/s00253-012-4334-3 [DOI] [PubMed] [Google Scholar]
- Bondy G. S., Pestka J. J. (2000). Immunomodulation by fungal toxins. J. Toxicol. Environ. Health B Crit. Rev. 3 109–143. 10.1080/109374000281113 [DOI] [PubMed] [Google Scholar]
- Chiang Y. W., Chye F. Y., Mohd Ismail A. (2006). Microbial diversity and proximate composition of tapal, a Sabah’s fermented beverage. Malays. J. Microbiol. 2 1–6. [Google Scholar]
- Cho K. J., Kang J. S., Cho W. T., Lee C. H., Ha J. K., Song K. B. (2010). In vitro degradation of zearalenone by Bacillus subtilis. Biotechnol. Lett. 32 1921–1924. 10.1007/s10529-010-0373-y [DOI] [PubMed] [Google Scholar]
- Cho Y. H., Hong S. M., Kim C. H. (2013). Isolation and characterization of lactic acid bacteria from kimchi, Korean traditional fermented food to apply into fermented dairy products. Korean J. Food Sci. Anim. Resour. 33 75–82. 10.5851/kosfa.2013.33.1.75 [DOI] [Google Scholar]
- Cole J. R., Wang Q., Cardenas E., Fish J., Chai B., Farris R. J., et al. (2009). The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37 141–145. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Council for Agricultural Science Technology [Cast] (2003). Mycotoxins: Risk in Plants, Animals, and Human Systems. CAST Task Force Report 2003 No. 139 Ames, IA: CAST. [Google Scholar]
- Curk M. C., Hubert J. C., Bringel F. (1996). Lactobacillus paraplantarum sp. nov., a new species related to Lactobacillus plantarum. Int. J. Syst. Bacteriol. 46 595–598. 10.1099/ijsem.0.000636 [DOI] [PubMed] [Google Scholar]
- Dike K. S., Sanni A. I. (2010). Influence of starter culture of lactic acid bacteria on the shelf life of agidi; an indigenous fermented cereal product. Afr. J. Biotechnol. 9 7922–7927. 10.5897/AJB09.1203 [DOI] [Google Scholar]
- Dykes G. A. (1994). Bacteriocins: ecological and evolutionary significance. Trends Ecol. Evol. 10 186–189. 10.1016/S0169-5347(00)89049-7 [DOI] [PubMed] [Google Scholar]
- Ezekiel C. N., Abia W. A., Ogara M. I., Sulyok M., Warth B., Krska R. (2015). Fate of mycotoxins in two popular traditional cereal-based beverages (kunu-zaki and pito) from rural Nigeria. LWT Food Sci. Technol. 60 137–141. 10.1016/j.lwt.2014.08.018 [DOI] [Google Scholar]
- Ezekiel C. N., Warth B., Ogara I. M., Abia W. A., Ezekiel V. C., Atehnkeng J., et al. (2014). Mycotoxin exposure in rural residents in northern Nigeria: a pilot study using multi-urinary biomarkers. Environ. Int. 66 138–145. 10.1016/j.envint.2014.02.003 [DOI] [PubMed] [Google Scholar]
- Fandohan P., Zoumenou D., Hounhouigan D. J., Marasas W. F. O., Wingfield M. J., Hell K. (2005). Fate of aflatoxins and fumonisins during the processing of maize into food products in Benin. Int. J. Food Microbiol. 98 249–259. 10.1016/j.ijfoodmicro.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39 783–791. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Flajs D., Peraica M. (2009). Toxicological properties of citrinin. Arh. Hig. Rada Toksikol. 60 457–464. 10.2478/10004-1254-60-2009-1992 [DOI] [PubMed] [Google Scholar]
- Fowoyo P. T., Ogunbanwo S. T. (2010). Phenotypic diversity of lactic acid bacteria isolated from massa; a fermented maize dough. Afr. J. Microbiol. Res. 4 2682–2691. [Google Scholar]
- Guan R., Wang Q., Sunberg E. J., Mariuzza R. A. (2005). Crystal structure of human peptidoglycan recognition protein S (PGRP-S) at 1.70 A resolution. J. Mol. Biol. 347 683–691. 10.1016/j.jmb.2005.01.070 [DOI] [PubMed] [Google Scholar]
- Haddadin M. S. Y., Awaisheh S. S., Robinson R. K. (2004). The production of yoghurt with probiotic bacteria isolated from infants in Jordan. Pak. J. Nutr. 3 290–293. 10.3923/pjn.2004.290.293 [DOI] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acid Symp. Ser. 41 95–98. [Google Scholar]
- Halm M., Lillie A., Sorensen A. K., Jakobsen M. (1993). Microbiological and aromatic characteristics of fermented maize doughs for Kenkey production in Ghana. Int. J. Food Microbiol. 19 135–143. 10.1016/0168-1605(93)90179-K [DOI] [PubMed] [Google Scholar]
- Haskard C. A., El–Nezami H. S., Kankaanää P. E., Salminen S., Ahohas J. T. (2001). Surface binding of aflatoxin B1 by lactic acid bacteria. Appl. Environ. Microbiol. 67 3086–3091. 10.1128/AEM.67.7.3086-3091.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janardhana G. R., Raveesha K. A., Shetty H. S. (1999). Mycotoxin contamination of maize grains grown in Karnataka (India). Food Chem. Toxicol. 37 863–886. 10.1016/S0278-6915(99)00067-8 [DOI] [PubMed] [Google Scholar]
- Johansson M. L., Sanni A., Lonner C., Molin G. (1995). Phenotypically based taxonomy using API 50CH of lactobacilli from Nigerian ogi, and the occurrence of starch fermenting strains. Int. J. Food Microbiol. 25 159–168. 10.1016/0168-1605(94)00096-O [DOI] [PubMed] [Google Scholar]
- Jung S. H., Joung W. P., Cho I. J., Lee N. K., Yeo I. C., Kim B. Y., et al. (2012). Characterization of lactic acid bacteria isolated from sauce-type kimchi. Prev. Nutr. Food Sci. 17 217–222. 10.3746/pnf.2012.17.3.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juodeikiene G., Basinskiene L., Bartkiene E., Matusevicius P. (2012). “Mycotoxin decontamination aspects in food, feed and renewables using fermentation processes,” in Structure and Function of Food Engineering, ed. Eissa A. A. (Rijeka: InTech; ), 171–204. [Google Scholar]
- Kankolongo M. A., Hell K., Nawa I. N. (2009). Assessment for fungal, mycotoxin and insect spoilage in maize stored for human consumption in Zambia. J. Sci. Food Agric. 89 1366–1375. 10.1002/jsfa.3596 [DOI] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 772–778. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayode O. F., Sulyok M., Fapohunda S. O., Ezekiel C. N., Krska R., Oguntona C. R. B. (2013). Mycotoxins and fungal metabolites in groundnut– and maize-based snacks from Nigeria. Food Addit. Contam. 6 294–300. 10.1080/19393210.2013.823626 [DOI] [PubMed] [Google Scholar]
- Kpodo K., Sorensen A. K., Jakobsen M. (1996). The occurrence of mycotoxins in fermented maize products. Food Chem. 56 147–153. 10.1016/0308-8146(95)00155-7 [DOI] [Google Scholar]
- Lei V., Jakobsen M. (2004). Microbiological characterization and probiotic potential of koko and koko sour water, African spontaneously fermented millet porridge and drink. J. Appl. Microbiol. 96 384–397. 10.1046/j.1365-2672.2004.02162.x [DOI] [PubMed] [Google Scholar]
- Malachova A., Sulyok M., Beltran E., Berthiller F., Krska R. (2014). Optimization and validation of a quantitative liquid chromatography–tandem mass spectrometric method covering 295 bacterial and fungal metabolites including all regulated mycotoxins in four model food matrices. J. Chromatogr. A 1363 145–156. 10.1016/j.chroma.2014.08.037 [DOI] [PubMed] [Google Scholar]
- Mizutani K., Nagatomi Y., Mochizuki N. (2011). Metabolism of zearalenone in the course of beer fermentation. Toxins 3 134–141. 10.3390/toxins3020134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokoena M. P., Chelule P. K., Ggaleni N. (2005). Reduction of fumonisin B1 and zearalenone by lactic acid bacteria in fermented maize meal. J. Food Prot. 68 2095–2099. [DOI] [PubMed] [Google Scholar]
- Mokoena M. P., Chelule P. K., Ggaleni N. (2006). The toxicity and decreased concentration of aflatoxin B1 in natural lactic acid fermented maize meal. J. Appl. Microbiol. 100 773–777. 10.1111/j.1365-2672.2006.02881.x [DOI] [PubMed] [Google Scholar]
- Ncube E., Flett B. C., Waalwijk C., Viljoen A. (2011). Fusarium spp. and levels of fumonisins in maize produced by subsistence farmers in South Africa. South Afr. J. Sci. 107 33 10.4102/sajs.v107i1/2.367 [DOI] [Google Scholar]
- Njumbe Ediage E., Di Mavungu J. D., Song S. Q., De Saeger S. (2013). Multimycotoxin analysis in urines to assess infant exposure: a case study in Cameroon. Environ. Int. 5 50–59. 10.1016/j.envint.2013.04.002 [DOI] [PubMed] [Google Scholar]
- Nyamete F. A. (2013). Potential of Lactic Acid Fermentation in Reducing Aflatoxin B1 and Fumonisin B1 in Tanzanian Maize-Based Complementary Gruel. Ph.D. thesis, Michigan State University, East Lansing, MI. [Google Scholar]
- Obinna-Echem P. C., Kuri V., Beal J. (2013). Evaluation of the microbial community, acidity and proximate composition of akamu, a fermented maize food. J. Sci. Food Agric. 94 331–340. 10.1002/.jsfa.6264 [DOI] [PubMed] [Google Scholar]
- Odunfa S. A. (1985). “African fermented foods,” in Microbiology of Fermented Foods Vol. 2 ed. Wood B. J. (London: Elsevier; ), 155–191. [Google Scholar]
- Odunfa S. A., Adeyele S. (1985). Microbiological changes during the traditional production of ogi-baba, a West African fermented sorghum gruel. J. Cereal Sci. 3 173–180. 10.1016/S0733-5210(85)80027-8 [DOI] [Google Scholar]
- Ogunbanwo S. T., Sanni A. I., Onilude A. A. (2003). Characterization of bacteriocin produced by Lactobacillus plantarum F1 and Lactobacillus brevis OG1. Afr. J. Biotechnol. 2 219–227. 10.5897/AJB2003.000-1045 [DOI] [Google Scholar]
- Ogunbanwo S. T., Sanni A. I., Onilude A. A. (2004). Effect of bacteriocinogenic Lactobacillus spp. on the shelf life of fufu, a traditional fermented cassava product. World J. Microbiol. Biotechnol. 20 57–63. 10.1023/B:WIBI.0000013312.10120.2a [DOI] [Google Scholar]
- Oguntoyinbo F. A., Huch M., Cho G. S., Schillinger U., Holzapfel W. H., Sanni A. I., et al. (2010). Diversity of Bacillus species isolated from Okpehe, a traditional fermented soup condiment from Nigeria. J. Food Prot. 73 870–878. [DOI] [PubMed] [Google Scholar]
- Oguntoyinbo F. A., Narbad A. (2012). Molecular characterization of lactic acid bacteria and in situ amylase expression during traditional fermentation of cereal foods. Food Microbiol. 31 254–262. 10.1016/j.fm.2012.03.004 [DOI] [PubMed] [Google Scholar]
- Oguntoyinbo F. A., Tourlomousis P., Gasson M. J., Narbad A. (2011). Analysis of bacterial communities of traditional fermented West African cereal foods using culture independent methods. Int. J. Food Microbiol. 145 205–210. 10.1016/j.ijfoodmicro.2010.12.025 [DOI] [PubMed] [Google Scholar]
- Oluwafemi F., Da-Silva F. A. (2009). Removal of aflatoxins by viable and heat-killed Lactobacillus species isolated from fermented maize. J. Appl. Biosci. 16 871–876. [Google Scholar]
- Omemu A. M. (2011). Fermentation dynamics during production of ogi, a Nigerian fermented cereal porridge. Rep. Opin. 3 8–17. [Google Scholar]
- Onyekwere O. O., Akinrele I. A., Koleoso O. A. (1989). “Industrialization of ogi fermentation,” in Industrialization of Indigenous Fermented Foods Vol. 33 ed. Steinkraus K. H. (New York, NY: Marcel Dekker Inc; ), 329–362. [Google Scholar]
- Opere B., Aboaba E. O., Ugoji O. O., Iwalokun B. A. (2012). Estimation of nutritive value, organoleptic properties and consumer acceptability of fermented cereal gruel (ogi). Adv. J. Food Sci. Technol. 4 1–8. [Google Scholar]
- Oyedeji O., Ogunbanwo S. T., Onilude A. A. (2013). Predominant lactic acid bacteria involved in the traditional fermentation of fufu and ogi, two Nigerian fermented food products. Food Nutr. Sci. 4 40–46. 10.4236/fns.2013.411A006 [DOI] [Google Scholar]
- Oyeyiola G. P. (1990). Microbiological and biochemical changes during the fermentation of maize (Zea mays) grains formasa production. World J. Microbiol. Biotechnol. 6 171–177. 10.1007/BF01200938 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Sanni A. I., Adesulu A. T. (2013). Microbiological and physico-chemical changes during fermentation of maize for masa production. Afr. J. Microbiol. Res. 7 4355–4362. 10.5897/AJMR12.1362 [DOI] [Google Scholar]
- Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: open source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty P. H., Jespersen L. (2006). Saccharomyces cerevisiae and lactic acid bacteria as potential mycotoxin decontamination agents. Trends Food Sci. Technol. 17 48–55. 10.1007/BF02946747 [DOI] [Google Scholar]
- Steinkraus K. H. (1996). Handbook of Indigenous Fermented Foods, 2nd Edn New York, NY: Marcel Dekker Inc. [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Teniola O. D., Holzapfel W. H., Odunfa S. A. (2005). Comparative assessment of fermentation techniques useful in the processing of ogi. World J. Microbiol. Biotechnol. 21 39–43. 10.1007/s11274-004-1549-1 [DOI] [Google Scholar]
- Teniola O. D., Odunfa S. A. (2002). Microbial assessment and quality evaluation of ogi during spoilage. World J. Microbiol. Biotechnol. 18 731–737. 10.1023/A:1020426304881 [DOI] [Google Scholar]
- Udoh J. M., Ikotun T., Cardwell K. F. (2000). Storage structures and aflatoxin content of maize in five agroecological zones of Nigeria. J. Stored Prod. Res. 36 187–201. 10.1007/s12550-014-0194-2 [DOI] [PubMed] [Google Scholar]
- Vella F., Kpodo K., Sorensen A. K., Jakobsen M. (1995). The occurrence of mycotoxins in fermented maize products. Food Chem. 56 147–153. [Google Scholar]
- Warth B., Parich A., Atehnkeng J., Bandyopadhyay R., Schuhmacher R., Sulyok M., et al. (2012). Quantitation of mycotoxins in food and feed from Burkina Faso and Mozambique using a modern LC–MS/MS multitoxin method. J. Agric. Food Chem. 60 9352–9363. 10.1021/jf302003n [DOI] [PubMed] [Google Scholar]
- Zhao L., Jin H., Lan J., Zhang R., Ren H., Zhang X., et al. (2015). Detoxification of zearalenone by three strains of Lactobacillus plantarum from fermented food in vitro. Food Control 54 158–164. 10.1016/j.foodcont.2015.02.0 [DOI] [Google Scholar]