Abstract
Aberrant microRNA (miRNA) expression is a defining feature of human malignancy. Specific miRNAs have been identified as promoters or suppressors of metastatic progression. These miRNAs control metastasis through divergent or convergent regulation of metastatic gene pathways. Some miRNA regulatory networks govern cell-autonomous cancer phenotypes, while others modulate the cell-extrinsic composition of the metastatic microenvironment. The use of small RNAs as probes into the molecular and cellular underpinnings of metastasis holds promise for the identification of candidate genes for potential therapeutic intervention.
Introduction
Epithelial carcinomas are the predominant cancer types that kill humans by metastasizing from their tissues of origin to distal organs. Nevertheless, the cell-biological pathways regulating the multi-step metastatic cascade had not been adequately characterized until recently1-3. Several technological advances during the last decade, however, have greatly deepened our molecular and conceptual understanding of this process. The development of whole-genome profiling approaches has permitted global analysis of the cancer transcriptome, whereas the availability of RNA-interference tools and human cancer xenograft models have facilitated functional testing of specific genes in metastasis. A key concept that emerged from early mouse studies was that metastatic colonization requires the actions of many gene products4-6. These functional findings were corroborated by transcriptomic studies of human breast cancers, which revealed large sets of genes to be recurrently overexpressed in primary tumours that metastasize7. An outstanding question in the field at that time was how such concerted pro-metastatic gene expression states were attained. In recent years, small non-coding RNAs, or miRNAs, have emerged as a class of post-transcriptional regulators that displays a pervasive role in this type of coordinated gene expression control8-13. In this review, we provide a conceptual overview of the molecular and cellular processes governed by endogenous miRNAs during metastatic progression.
Basics of miRNA biogenesis and action
Although the majority of human miRNAs reside in intergenic regions or within the introns of genes, exonic miRNAs have also been described14. Transcription of a single or of multiple concatenated miRNA-coding sequences is mediated by RNA polymerase II, and less frequently by RNA polymerase III, yielding a primary miRNA transcript or pri-miRNA15,16. Mammalian pri-miRNAs are processed through a series of cleavage steps15-18. Initially, the RNA-binding protein DGCR8 recognizes and binds to double-stranded hairpin structures embedded within the pri-miRNA. This interaction allows the ribonuclease Drosha, which forms a complex with DGCR8, to cleave the pri-miRNA at the hairpin junction site, releasing a roughly 70-nt long stem-loop product known as the pre-miRNA15-17. Association of the pre-miRNA with Exportin-5 and its co-factor Ran-GTP, facilitates its nuclear export15,16. Once in the cytoplasm, the ribonuclease Dicer bound to its co-factor, the RNA-binding protein TARBP2, cleaves the pre-miRNA, yielding a 21-26-nt long miRNA:miRNA* duplex structure15,16,18. This miRNA duplex is loaded onto the RNA-induced silencing complex or RISC, where the guide miRNA strand is preferentially incorporated into the complex. The complementary passenger miRNA* strand is typically released and degraded, except for cases in which the miRNA* strand also mediates silencing14-16,19. The key effector at the core of RISC is a member of Argonaut (Ago) family of proteins, which directly interacts with single-stranded mature miRNAs and uses them as guides to recognize mRNAs bearing sequences complementary to the bound miRNA20-22. Ago binding to target mRNAs results in the recruitment of silencing effector proteins such as GW18223, ultimately leading to mRNA deadenylation and decay and/or translational repression13,14. Although the majority of miRNA regulatory sequences reside in the 3’-untranslated regions of transcripts, interactions of miRNAs with coding sequences have also been mapped24-26. The use of computational analyses, gene expression profiling, heterologous reporter assays, and biochemically coupled transcriptomic deep-sequencing approaches27,28 have provided a toolbox for systematic discovery of miRNA target genes.
Deregulated miRNA expression in human cancer
The link between deregulated miRNA expression and cancer had been well established29,30 prior to the identification of these small RNAs as regulators of metastasis Calin, Croce, and colleagues reported deregulated miRNA expression in cancer in a study showing that two clustered miRNAs, miR-15a and miR-16, were deleted or downregulated in the majority (> 60%) of B-cell chronic lymphocytic leukaemias tested31. Subsequent work demonstrated deregulated miRNA expression also in solid cancers: miR-145 exhibited reduced expression in colon32 and breast33 carcinomas, let-7 was found to be silenced in lung cancer34,35, while miR-155, miR-21, and the miR-17-92 cluster were upregulated in breast and lung cancers33,35-38. Interestingly, miR-21, miR-155, and miR-17-92 were also found to be overexpressed in B-cell and Hodgkin's lymphomas39-43, establishing them as potential oncogenic miRNAs in diverse cancers. Subsequent miRNA-profiling studies revealed distinct expression profiles in various solid and liquid cancers that could inform the diagnosis of these malignancies29,44-49. Although a small subset of specific oncogenic miRNAs have been found to be upregulated in cancer, global downregulation of miRNA expression and processing has emerged as a feature of these regulators during tumorigenesis50-54. Consistent with this, a miRNA family (miR-34a-c) was found to be transcriptionally induced by the key tumour suppressor p53, which is frequently mutated and inactivated in several types of cancers55-58.
Metastasis-suppressive miRNA networks
This early work raised the question of whether silencing of specific miRNAs could modulate metastasis by driving pro-metastatic gene expression programs. The search for miRNAs that repress the expression of pro-metastatic genes and consequently suppress metastasis originally focused on breast cancer6. Small RNA profiling of highly metastatic sub-lines derived from the in vivo selection of a human breast cancer population revealed a subset of miRNAs that were silenced in cells with high metastatic capacity. Functional testing demonstrated that overexpression of miR-335, miR-206, or miR-126 robustly suppressed metastatic colonization by human breast cancer lines. Conversely, antagomir-mediated inhibition of miR-335 enhanced metastasis, providing crucial loss-of-function in vivo evidence for endogenous miRNA activity in the regulation of metastasis. Importantly, the expression levels of these miRNAs in a collection of women's breast cancers correlated with these patients’ likelihood of metastatic relapse6. Several studies have since validated the silenced expression and prognostic capacity of these miRNAs in independent and larger clinical cohorts for diverse cancer types59-67, as well as their tumor-suppressive and/or metastasis-suppressive functions in other breast cancer models59,68 and cancer types67,69-72.
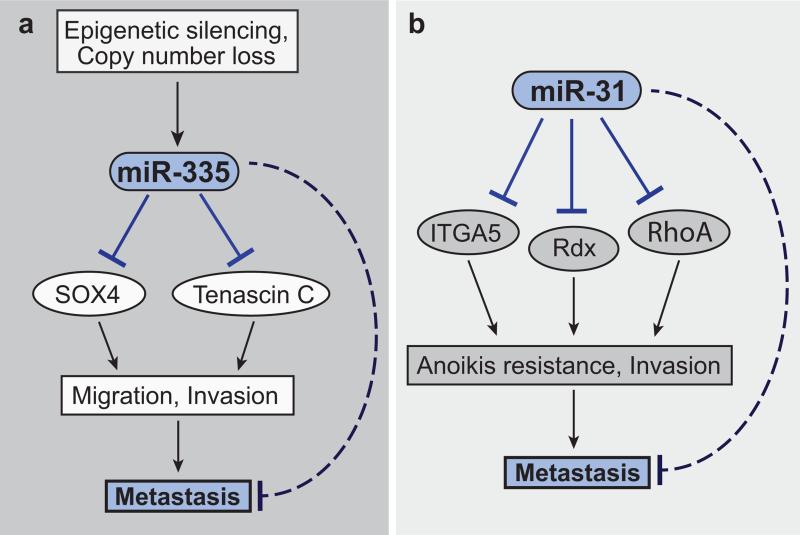
These miRNAs where shown to inhibit metastasis through several mechanisms. Whereas miR-335 and miR-206 suppressed cell invasion6, endogenous miR-126 inhibited tumour growth, endothelial recruitment, and metastatic initiation by breast cancer cells73. Consistent with a role for miRNAs in establishing a metastatic gene expression program, miR-335 silenced a set of pro-metastatic genes, the expression of which was found to correlate with metastatic outcomes in breast cancer patients. Two of these genes, SOX4 and Tenascin-C, were found to drive the miR-335-suppressed phenotypes—cell invasion, migration, and metastasis6 (Fig. 1a). SOX4 and Tenascin-C have been subsequently validated as promoters of cancer progression in additional models74-76.
Figure 1.
Metastasis suppression by divergent miRNA regulatory networks. (a) miR-335 suppresses breast cancer metastasis by targeting SOX4 and Tenascin-C, which in turn promote cancer cell migration, invasion, and ultimately metastasis6. miR-335 is silenced in metastatic breast cancer cells through genetic copy number loss and epigenetic promoter hypermethylation59. (b) miR-31 inhibits breast cancer cell invasion, anoikis resistance, and metastasis by coordinately targeting ITGA5, RDX, and RhoA77-79.
Another well-characterized example of a metastasis-suppressive miRNA network is that of miR-3177-79 (Fig. 1b). Overexpression of miR-31 in human cancer cells strongly suppressed metastasis, whereas stable miRNA inhibition in otherwise non-metastatic cells led to a robust 10-fold increase in metastatic capacity, consistent with metastasis suppression by endogenous miR-31. Importantly, the expression of miR-31 in patients’ primary breast tumors was prognostic of metastatic relapse77. Silencing of miR-31 has also been observed in additional cancer types80-83. Mechanistically, miR-31 was found to inhibit metastatic progression through coordinated regulation of three genes—integrin α5 (ITGA5), radixin (RDX), and RhoA, each of which was shown to promote anoikis resistance and cell invasion77-79 (Fig. 1b). Collectively, these findings illustrate that silencing of robust metastasis-suppressor miRNAs (Table 1) in highly metastatic cells results in the lifting of key post-transcriptional regulatory barriers imposed on pro-invasive genes.
Table 1.
Examples of metastasis-regulatory miRNAs
| miRNA | Cancer type | Targets | Phenotype | Refs |
|---|---|---|---|---|
| Metastasis suppressor miRNAs | ||||
| miR-335 | Breast cancer | SOX4, Tenascin C | Invasion, Migration | 6 |
| miR-126 | Breast cancer | IGFBP2, PITPNC1 MERTK | Endothelial recruitment | 73 |
| miR-200s | Breast cancer | ZEB1, ZEB2 | EMT, Migration | 88-92 |
| miR-31 | Breast cancer | RhoA, RDX, ITGA5 | Invasion, Anoikis resistance | 77-79 |
| let-7 | Breast cancer | BACH1, HMGA2 | Invasion | 103-105 |
| miR-34a | Prostate cancer, Breast cancer, Colorectal cancer | CD44, Fra-1, SNAIL | Invasion, Migration EMT | 117, 128-129 |
| miR-29b | Breast cancer, Prostate cancer, HCC | ANGPTL4, LOX, MMP2, MMP9, VEGFA, PDGF | EMT, Invasion | 110-112 |
| miR-139 | HCC | ROCK2 | Invasion, Migration | 130 |
|
Metastasis promoter miRNAs | ||||
| miR-10b | Breast cancer | HOXD10 | Invasion, Migration | 84,86 |
| miR-373/520c | Breast cancer | CD44 | Invasion, Migration | 87 |
| miR-200s | Breast cancer | Sec23a | Colonization | 93 |
| miR-9 | Breast cancer | E-cadherin, LIFR | Invasion, Migration | 94-95 |
| miR-21 | Breast cancer, Colorectal cancer, Squamous carcinoma | PDCD4, TPM1, Maspin | Invasion, Tumor growth, Tumor cell survial | 96-99, 134-136 |
| miR-103/107 | Breast cancer | Dicer | Migration, EMT | 119 |
| miR-182 | Melanoma | FOXO3, MITF | Invasion, Apoptosis | 100 |
| miR-30b/30d | Melanoma | GALNT7, GALNT1 | Invasion | 101 |
| miR-199a-3p, miR-199a-5p, miR-1908 | Melanoma | DNAJA4, ApoE | Invasion, Endothelial recruitment | 102 |
| miR-214 | Melanoma | TFAP2C, ITGA3 | Migration, Extravasation, Anoikis resistance | 131 |
| miR-223 | Gastric cancer | EPB41L3 | Migration, Invasion | 132 |
| miR-143 | HCC | FNDC3B | Invasion | 133 |
HCC = hepatocellular carcinoma
Metastasis-promoting miRNA networks
Could a miRNA conversely promote metastasis by targeting a metastasis-suppressive gene? In a bold series of experiments, Ma and Weinberg demonstrated that miR-10b was a driver of metastasis84. Overexpression studies in human breast cancer cells demonstrated sufficiency of miR-10b in promoting metastasis. This miRNA was found to enhance cell migration and invasion through targeting of homeobox D10 (HOXD10), which in turn inhibited the metastasis-promoting gene, RhoC84,85 (Fig. 2a). Subsequent studies revealed a role for endogenous miR-10b in driving metastasis as well86.
Figure 2.
Examples of miRNA regulatory networks that promote metastasis. (a) miR-10b drives metastatic breast cancer progression by direct targeting and suppression of HOXD10, which in turn inhibits RhoC—a promoter of migration, invasion, and metastasis. TWIST1 transcriptionally induces miR-10b in breast cancer cells84. (b) miR-373 and miR-520c promote breast cancer metastasis by directly targeting CD44, which leads to increased cancer cell invasion and migratory capacities87. (c) miR-200s, which are negatively regulated by TGFβ127, display a pleiotropic role in metastatic progression. miR-200-mediated targeting of ZEB1/2 results in de-repression of E-cadherin expression, which maintains the epithelial tumor phenotype and suppresses migration and metastasis88-92 (left blocking arrow), while miR-200-mediated silencing of Sec23a promotes metastatic colonization by inhibiting secretion of TINAGL1 and IGFBP493 (right blocking arrow). (d) miR-9 enhances breast cancer metastasis by two independent pathways. Direct repression of E-cadherin by miR-9 leads to enhanced cell invasion and migration94 (left blocking arrow), whereas miR-9 targeting of LIFR results in activation of the Hippo signalling component YAP95 (right blocking arrow). MYC/MYCN drives transcription of miR-9 in breast cancer cells94. (e) miR-21 promotes cancer cell invasion and metastasis through coordinate suppression of TPM1, PDCD4, and Maspin96-98. The miR-21 target genes have also been shown to suppress cancer cell growth and survival96,134-136. Active mTOR and STAT3 signaling has been shown to correlate with miR-21 upregulation in cancer cells99. (f) miR-30b/30d increases melanoma invasion and metastasis by targeting GALNT1 and GALNT7. GALNT7 reduces the levels of IL-10—a putative immunosuppressive factor101.
miR-373 and miR-520c, which belong to the same miRNA family, were also identified as metastasis-promoting miRNAs87. In an innovative forward genetic screen using human breast cancer cells transduced with a miRNA-expression library and assessed for their migration ability, miR-373 and miR-520c were found to be enriched in cells with enhanced migratory capacity. These miRNAs were shown to promote cell migration and invasion by targeting CD44 (Fig. 2b). Importantly, introduction of miR-373 or miR-520c was sufficient to confer metastatic capacity to otherwise non-metastatic human breast cancer cells87.
The miR-200 family, which includes miR-200a, miR-200b, miR-200c, miR-141, and miR-429, is another prominent example of a metastasis-regulatory network driven by miRNAs. Early work by several independent groups demonstrated that this miRNA family is important in maintaining the tumor epithelial phenotype and inhibiting the epithelial-to-mesenchymal transition (EMT)88-91. This was supported by the strong correlation observed between the levels of miR-200s and the epithelial marker E-cadherin in a collection of cancer lines as well as in clinical samples88,90. Mechanistically, miR-200s were found to inhibit cell migration by directly targeting the transcription factors ZEB1 and ZEB2, which suppress E-cadherin (Fig. 2c). Overexpression of miR-200s was shown to decrease metastatic dissemination from primary tumors in a syngenic breast cancer mouse model92. In addition to the reports indicating a metastasis-suppressive role for the miR-200 family, recent findings have also demonstrated a role for these miRNA in promoting metastasis93. High levels of miR-200s correlated with shorter metastasis-free survival times in breast cancer patients, whereas overexpression of miR-200s enhanced lung colonization by murine breast cancer cells. Interestingly, the metastasis-promoting effects of miR-200s were mediated through downstream targets distinct from the ones implicated in EMT. Targeting of Sec23a, a COPII vesicle component, resulted in decreased secretion of TINAGL1 and IGFBP4—factors shown to suppress metastasis by mouse breast cancer cells (Fig. 2c)93.
These findings implicate miR-200s as potential pleiotropic regulators of metastatic progression. Whereas silencing of miR-200s may be beneficial at the early steps of metastasis by triggering EMT through ZEB1/2-dependent repression of E-cadherin, upregulation of miR-200s at distal metastatic sites may actually promote metastatic colonization through Sec23 targeting. These seemingly opposing roles of miR-200s in metastasis could be reconciled if one considers the multi-faceted nature of metastasis. It is conceivable that context-dependent miRNA modulation at different steps of the metastatic cascade may drive both promoting and suppressive roles in cancer progression through divergent miRNA targeting of distinct molecular and cellular pathways. Loss-of-function experiments on miR-200s should provide important insights into their complex roles during metastatic progression.
Whereas miR-200s indirectly promote E-cadherin expression to maintain an epithelial phenotype, miR-9, was found to directly target E-cadherin to drive cancer cell motility, invasion, and metastasis94 (Fig. 2d). Overexpression of miR-9 enhanced micrometastasis formation by human breast cancer cells, whereas its stable silencing inhibited murine breast cancer metastasis. Subsequent work demonstrated that overexpression of miR-9 in E-cadherin-negative human breast cancer cells enhanced metastasis, suggesting that miR-9 represses additional metastasis-suppressive target genes95. This study identified leukemia inhibitory factor receptor (LIFR) as a direct target of miR-9 that mediates its pro-metastatic effects in the absence of E-cadherin. Depletion of LIFR in non-metastatic cells was shown to increase invasion and migration, whereas its overexpression abrogated the metastatic capacity conferred by miR-9. The metastasis-suppressive role of LIFR was found to be mediated through inactivation of YAP, a previously known oncoprotein whose activity is regulated by the Hippo pathway95 (Fig. 2d).
In addition to breast cancer, metastasis-regulatory miRNAs have been identified in multiple other cancer types (Table 1). At roughly the same time as several reports demonstrating pro-tumorigenic and pro-metastatic roles for miR-21 in breast cancer96-97, another group showed that miR-21 drives colorectal cancer invasion and metastasis by targeting PDCD498 (Fig. 2e). TPM1 and Maspin were two additional miR-21 targets uncovered in breast cancer.97 Recently, miR-21 was also shown to drive metastasis of squamous cell carcinoma tumors deficient for p5399. Metastasis-regulatory miRNAs have also been described in melanoma, a highly intractable malignancy. Overexpression of miR-182 was initially shown to increase metastasis by mouse melanoma cells100. Subsequently, miR-30b/30d were found to correlate with human melanoma progression outcomes and their overexpression in human melanoma cells enhanced micrometastasis formation by targeting the GALNT1 and GALNT7 suppressors of cell invasion101 (Fig. 2f). Finally, miR-199a-3p, miR-199a-5p, and miR-1908 were recently discovered as endogenous miRNA drivers of human melanoma metastasis. Inhibition of these clinico-pathologically correlated miRNAs in a series of human melanoma lines robustly suppressed metastasis102.
Cell-intrinsic versus cell-extrinsic effects of metastasis-regulatory miRNAs
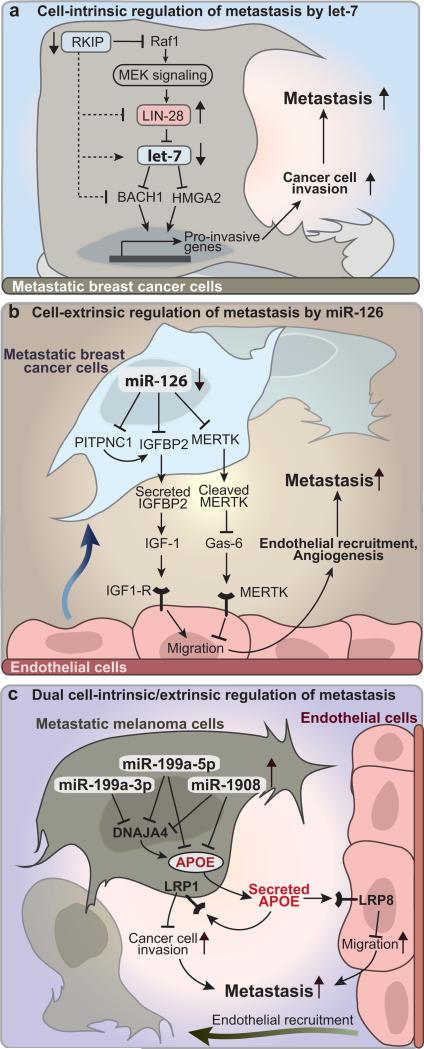
Early studies ascribed cell-autonomous phenotypes to miRNAs implicated in metastasis. For example, miR-3356 and miR-3177 were identified as suppressors of cell migration and invasion (Fig. 1), whereas miR-10b84, miR-373/miR-520c87, miR-994, and miR-2196-98 were shown to promote these phenotypes (Fig. 2). Another prominent example of a miRNA regulatory network controlling a cell-intrinsic phenotype is that of the let-7 family103-105 (Fig. 3a). let-7 overexpression in human breast cancer cells was initially shown to decrease the incidence of metastatic events to the liver and lung103. Subsequent work revealed that let-7-mediated repression of the chromatin-remodeling protein HMGA2 and the transcription factor BACH1, both of which promote the transcription of pro-invasive genes, suppressed cell invasion and metastasis to the bone104-105 . Thus, let-7 halts the cell-intrinsic phenotype of cell invasion, which is required for efficient metastasis, by coordinately targeting factors that regulate pro-invasive gene expression programs (Fig. 3a).
Figure 3.
Cell-intrinsic versus cell-extrinsic control of metastatic progression by miRNAs. (a) The let-7-regulatory network suppresses metastasis by inhibiting the cell-intrinsic phenotype of cancer cell invasion. Direct targeting of HMGA2 and BACH1 by let-7 leads to transcriptional suppression of a set of pro-invasive genes. Let-7 is regulated by LIN-28, a miRNA-binding protein that destabilizes the let-7 pre-miRNA and ultimately promotes metastasis. RKIP, an indirect repressor of LIN-28, suppresses metastasis through upregulation of let-7104-105. (b) miR-126 halts metastasis by impairing the cell-extrinsic ability of breast cancer cells to recruit endothelial cells into the metastatic niche. Cancer-expressed miR-126 coordinately targets PITPNC1, IGFBP2, and MERTK. Cancer cell-secreted IGFBP2 promotes endothelial cell migration by binding to endothelial IGF1-R, while PITPNC1 induces IGFBP2 expression. In parallel to this IGFBP2-driven pathway, cancer cell-cleaved MERTK ectodomain increases endothelial cell migration by binding and sequestering Gas-6—an extracellular factor that inhibits endothelial cell migration through its action on endothelial MERTK receptors73. (c) Three miRNAs—miR-199a-3p, miR-199a-5p, and miR-1908—promote melanoma metastasis by convergent targeting of DNAJA4 and ApoE. DNAJA4 suppresses metastasis by inducing ApoE expression. Melanoma cell-secreted ApoE halts metastatic progression by both cell-autonomous and non-cell-autonomous mechanisms. ApoE suppresses melanoma cell invasion by targeting melanoma LRP1 receptors, while its inhibition of endothelial cell migration results from its engagement of endothelial LRP8 receptors102.
Recently, miRNAs were shown to regulate metastasis also by cell-extrinsic modulation of the metastatic microenvironment. Combined loss-of-function, gain-of-function, and epistasis experiments demonstrated that miR-126 inhibits the recruitment of endothelial cells by multiple human breast cancer lines in vitro. Endogenous miR-126 was also found to suppress metastatic endothelial recruitment (MER), the ability of cancer cells to recruit endothelial cells to incipient metastases in vivo73. Human breast cancer cells displaying silenced expression of miR-126 were shown to upregulate a set of miR-126 target genes—IGFBP2, MERTK, and PITPNC1—molecules that individually and collectively correlate in expression with human metastatic progression (Fig. 3b)73. These previously uncharacterised angiogenesis genes were shown to modulate the cell-extrinsic recruitment of endothelial cells by forming two divergent signalling pathways that emanate from cancer cells and engage endothelial cells. Cancer cell-secreted IGFBP2 was shown to promote endothelial cell migration by driving IGF1-dependent activation of the endothelial IGF1-Receptor (IGF1-R). PITPNC1, a phospho-inositide binding protein106, was found to promote endothelial cell migration by enhancing extracellular IGFBP2 levels. Finally, the receptor tyrosine kinase MERTK was shown to be proteolytically cleaved from the surface of cancer cells and released into the extracellular space. This MERTK ectodomain promoted endothelial cell migration by acting as a decoy receptor and sequestering circulating GAS6—a molecule shown to inhibit endothelial cell migration through its action on endothelial MERTK receptors (Fig. 3b)73.
The metastasis-suppressive role of miR-126 and its clinical association with human metastatic relapse were recently validated also in a mouse xenograft model of breast cancer metastasis107. This report found that, in addition to its ability to suppress angiogenesis which was previously demonstrated for human breast cancer lines of triple-negative73, luminal73,108, and HER-2 positive73 backgrounds, as well as for a variety of additional cancer types such as lung72 and oral109 carcinomas, miR-126 overexpression can reduce mesenchymal stem cell (MSC) and monocyte content in primary xenograft tumors established by mouse 4T1 cells107. Interestingly, miR-126 silencing did not enhance monocyte or leukocyte recruitment by human breast cancer cells in vivo73, suggesting that the inflammatory cell recruitment observed by these investigators may be limited to the 4T1 mouse system or be the consequence of overexpression effects.
Additional metastasis-regulatory miRNAs likely govern other non-cell-autonomous phenotypes by altering the tumor microenvironment. For instance, a recent report suggested a cell-extrinsic role for miR-29b, a miRNA transcriptionally driven by GATA3 whose expression is reduced in breast cancer110. This miRNA, which was also shown to inhibit prostate111 and liver cancer metastasis112 in addition to breast cancer metastasis110, was found to target a set of genes that had been previously implicated in cell-extrinsic processes such as angiogenesis and collagen remodelling110 (Table 1). Some miRNAs may even directly impact neighbouring cell types, as is the case for miR-9, which was shown by Ferrara and colleagues to regulate endothelial cell migration through its secretion from cancer cells inside microvesicles113. Such microvesicles or exosomes have also been found to contain multiple proteins that could modulate the tumour microenvironment114-115.
Convergent control of metastatic progression by multiple miRNAs
Nearly all previously described metastasis-regulatory miRNAs act through divergent targeting of multiple genes. However, recent work has delineated a network of miRNAs that drive metastasis through convergent targeting of functionally coupled genes102 (Fig. 3c). Gain- and loss-of-function approaches demonstrated that miR-199a-3p, miR-199a-5p, and miR-1908 promote metastasis of multiple human melanoma lines of diverse mutational backgrounds. The combined expression of the three miRNAs exhibited stronger prognostic capacity in predicting metastatic outcomes than each individual miRNA. These findings, which are suggestive of miRNA cooperativity in human metastatic progression, were corroborated by in vivo loss-of-function studies. Inhibition of each miRNA suppressed metastasis by roughly 4-fold, while concurrent silencing of the three miRNAs decreased metastatic colonization by more than 70-fold, attesting to their remarkable functional cooperativity102.
miR-199a-3p, miR-199a-5p, and miR-1908 were shown to convergently target two phenotypically related genes, the heat-shock factor DNAJA4 and the metabolic protein apolipoprotein-E (ApoE). Consistent with these two genes acting as the key downstream effectors of the miRNAs, the metastasis-suppressive effects observed following miRNA inhibition were rescued by depletion of either DNAJA4 or ApoE. Interestingly, DNAJA4 was shown to suppress metastasis through positive regulation of ApoE expression, establishing ApoE as the central node in this convergent miRNA regulatory network. The metastasis-suppressive role of ApoE was corroborated by the correlation observed between ApoE expression levels and human melanoma progression outcomes; by the ability of ApoE to abrogate metastasis of multiple human melanoma lines; and by the robust enhancement of metastasis following genetic deletion of ApoE in immunocompent mice. ApoE, which is secreted by melanoma cells, was shown to suppress both melanoma cell-intrinsic and cell-extrinsic metastatic phenotypes. The cell-autonomous targeting of melanoma LRP1 receptors by ApoE decreased melanoma cell invasion, whereas its non-cell-autonomous engagement of endothelial LRP8 receptors inhibited MER102 (Fig. 3c). These findings provide an example of a convergent miRNA regulatory network, with miRNAs playing dual cell-intrinsic/cell-extrinsic roles in metastatic progression. Interestingly, MER has emerged as a common feature driving both breast cancer and melanoma metastasis73,102.
Divergent versus convergent metastasis regulation by miRNAs
Initial findings on the connectivity of miRNA regulatory networks suggested that they control metastasis through their ability to coordinately target multiple genes. Divergent metastasis control was demonstrated for miR-3356 and subsequently shown to be the pervasive mode of regulation for many other miRNAs, including miR-3177, the miR-200 family88-93, miR-994-95, miR-2196-98, let-7104-105, and miR-12673. The advantage that such divergent gene targeting confers to metastasis is conceptually intuitive: a single miRNA can coordinately regulate the expression of multiple genes, all of which participate in a common metastatic phenotype. The concurrent silencing of a set of genes by a given miRNA leads to more profound modulation of the phenotype than independent silencing of individual genes.
In contrast, the pervasiveness of the convergent target regulation by multiple unique miRNAs in additional metastasis models remains to be explored. The advantage of convergent miRNA regulation stems from the maximum silencing of a single gene, such as ApoE, that is the central determinant of the metastatic phenotype102. Cooperative targeting by multiple miRNAs ensures more robust gene expression control than that afforded by a single miRNA and may have an added advantage in cases where genetic inactivation of a key metastasis-suppressive factor may not be tolerated by cells. The divergent and convergent models of connectivity that characterize miRNA regulatory networks in metastasis are unlikely to be coincidental. Instead, they could have arisen from evolutionarily conserved connectivity that likely serves important roles during development and physiology, making them vulnerable to co-option during pathogenesis.
Mechanisms underlying aberrant miRNA expression in metastasis
Altered miRNA expression results from diverse mechanisms operating during metastatic progression. For instance, the metastasis-suppressor miR-335 (Fig. 1a) was found to be inactivated in human breast cancer through both genetic deletion of the miR-335 locus and through epigenetically-driven transcriptional silencing of the locus by promoter hypermethylation59. Moreover, several metastasis-promoting miRNAs have been shown to be direct targets of well-known oncogenic transcription factors. miR-10b and miR-9 were found to be transcriptionally activated by TWIST184 and MYC/MYCN94, respectively. Conversely, miR-200s have been reported to be transcriptionally repressed by ZEB1/2, allowing for negative feedback regulation of miR-200s by their metastasis-promoting target genes91,116. The metastasis-suppressor miR-34 was also shown to form a double-negative feedback loop with one of its targets, the oncogenic transcription factor SNAIL117.
The components of the miRNA processing machinery have also been implicated in cancer progression. Consistent with expression analyses indicating downregulation of Dicer and Drosha in breast cancer progression54,118, direct repression of Dicer by miR-103/107 enhanced breast cancer metastasis119, whereas transcriptional induction of Dicer by Tap63 suppressed metastasis120. Additionally, inactivating mutations in Exportin-5 have been identified in a small subset of human colorectal tumors, leading to defective miRNA nuclear export121. Finally, small RNA-binding proteins that regulate the processing of a specific miRNA have also been implicated in metastasis. For instance, LIN-28, which binds and destabilizes the let-7 pre-miRNA, was shown to promote breast cancer metastasis104. Taken together, these findings reveal a diversity of inactivating and activating molecular mechanisms governing the expression of metastasis-regulatory miRNAs, which underscores their importance amongst the regulatory molecules expressed by cancer cells.
Translational potential for basic discoveries
The ability of small RNAs to regulate gene expression has generated much hope for exploiting them as potential therapeutic molecules or therapeutic targets of human diseases. The two main problems that have hindered the clinical translation of miRNAs are their limited half-lives in serum and their limited delivery into target tissues. Although there is no broadly accepted or applied methodological approach for systemic in vivo delivery of therapeutic miRNAs, two recent reports raise renewed hope by using an alternative approach to “naked” nucleic acid delivery, which involves viral-based delivery methods122-123. Adeno-associated viral delivery of miR-26a, for instance, was found to significantly suppress tumorigenesis in a murine model of hepatocellular carcinoma—a cancer known to display reduced miR-26a expression122.
In vivo inhibition of miRNAs through the systemic administration of locked nucleic acids (LNAs) has progressed to a much greater degree in the pre-clinical and clinical arenas than miRNA delivery124. For instance, systemic delivery of an LNA targeting miR-122—a hepatic miRNA that regulates cholesterol—led to significant decreases in plasma cholesterol in non-human primates125. Therapeutic targeting of this miRNA, which is also required for hepatitis C virus (HCV) replication, was additionally shown to successfully suppress HCV viremia126. These successes have motivated studies involving therapeutic LNA delivery in pre-clinical cancer models. To this end, systemic delivery of an LNA targeting the breast cancer metastasis-promoter miR-10b was shown to suppress metastasis86. Recently, silencing of miR-199a-3p, miR-199a-5p, and miR-1908 using a cocktail of LNAs was found to dramatically suppress melanoma metastasis102, a condition that currently lacks effective therapeutic options.
Summary and future directions
miRNAs have been established as key metastasis regulators by numerous investigators working on diverse cancer types, with concepts such as divergent and convergent control by miRNA regulatory networks emerging more recently. Metastasis-regulatory miRNAs have been found to govern both cell-intrinsic processes and cell-extrinsic features of the tumour microenvironment. The prognostic capacity of these non-coding RNAs in predicting human metastatic outcomes suggests an active role for these molecules in human cancer progression. Most importantly, miRNAs have accelerated the discovery of genes and signalling pathways mediating metastasis. These basic explorations will expedite translational efforts aimed at reducing cancer mortality.
Acknowledgements
We thank members of our laboratory for insightful discussions. We thank Colin Buss for artistic assistance in figure design. N.P. is an Anderson Cancer Center graduate fellow. S.F.T. was supported by a DOD Era of Hope Scholar Award, an NIH New Innovator Award, the Rita Allen Foundation, CTSA award 5KL2 RR024142-04 , and additional grant support. We apologize to many colleagues, whose important work was not cited due to space limitations.
References
- 1.Talmadge JE, Fidler IJ. AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Res. 2010;70:5649–5669. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang Y, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 6.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 10.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 12.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 14.Ul Hussain M. Micro-RNAs (miRNAs): genomic organisation, biogenesis and mode of action. Cell Tissue Res. 2012;349:405–413. doi: 10.1007/s00441-012-1438-0. [DOI] [PubMed] [Google Scholar]
- 15.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 16.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 19.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 21.Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding L, Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 2007;17:411–416. doi: 10.1016/j.tcb.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forman JJ, Coller HA. The code within the code: microRNAs target coding regions. Cell Cycle. 2010;9:1533–1541. doi: 10.4161/cc.9.8.11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 30.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 31.Calin GA, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michael MZ, O' Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 33.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 34.Takamizawa J, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 35.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 36.Farazi TA, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71:4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayashita Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 38.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eis PS, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kluiver J, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 41.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 42.Tagawa H, Seto M. A microRNA cluster as a target of genomic amplification in malignant lymphoma. Leukemia. 2005;19:2013–2016. doi: 10.1038/sj.leu.2403942. [DOI] [PubMed] [Google Scholar]
- 43.Mu P, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Calin GA, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ciafrè SA, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 47.Murakami Y, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 48.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 49.Roldo C, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 50.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 51.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 52.Thomson JM, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 54.Yan M, et al. Dysregulated expression of dicer and drosha in breast cancer. Pathol Oncol Res. 2012;18:343–348. doi: 10.1007/s12253-011-9450-3. [DOI] [PubMed] [Google Scholar]
- 55.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 58.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 59.Png KJ, et al. MicroRNA-335 inhibits tumor reinitiation and is silenced through genetic and epigenetic mechanisms in human breast cancer. Genes Dev. 2011;25:226–231. doi: 10.1101/gad.1974211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang F, Zheng Z, Guo J, Ding X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol. 2010;119:586–593. doi: 10.1016/j.ygyno.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 61.Hafez MM, et al. MicroRNAs and metastasis-related gene expression in Egyptian breast cancer patients. Asian Pac J Cancer Prev. 2012;13:591–598. doi: 10.7314/apjcp.2012.13.2.591. [DOI] [PubMed] [Google Scholar]
- 62.Schmitz KJ, et al. Differential expression of microRNA-675, microRNA-139-3p and microRNA-335 in benign and malignant adrenocortical tumours. J Clin Pathol. 2011;64:529–535. doi: 10.1136/jcp.2010.085621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dohi O, et al. Epigenetic silencing of miR-335 and its host gene MEST in hepatocellular carcinoma. Int J Oncol. 2013;42:411–418. doi: 10.3892/ijo.2012.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White NM, et al. miRNA profiling for clear cell renal cell carcinoma: biomarker discovery and identification of potential controls and consequences of miRNA dysregulation. J Urol. 2011;186:1077–1083. doi: 10.1016/j.juro.2011.04.110. [DOI] [PubMed] [Google Scholar]
- 65.Missiaglia E, et al. MicroRNA-206 expression levels correlate with clinical behaviour of rhabdomyosarcomas. Br J Cancer. 2010;102:1769–1777. doi: 10.1038/sj.bjc.6605684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Donnem T, et al. Independent and tissue-specific prognostic impact of miR-126 in nonsmall cell lung cancer: coexpression with vascular endothelial growth factor-A predicts poor survival. Cancer. 2011;117:3193–3200. doi: 10.1002/cncr.25907. [DOI] [PubMed] [Google Scholar]
- 67.Xu Y, et al. MicroRNA-335 acts as a metastasis suppressor in gastric cancer by targeting Bcl-w and specificity protein 1. Oncogene. 2012;31:1398–1407. doi: 10.1038/onc.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heyn H, et al. MicroRNA miR-335 is crucial for the BRCA1 regulatory cascade in breast cancer development. Int J Cancer. 2011;129:2797–2806. doi: 10.1002/ijc.25962. [DOI] [PubMed] [Google Scholar]
- 69.Taulli R, et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–2378. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hamada S, et al. MiR-126 acts as a tumor suppressor in pancreatic cancer cells via the regulation of ADAM9. Mol Cancer Res. 2012;10:3–10. doi: 10.1158/1541-7786.MCR-11-0272. [DOI] [PubMed] [Google Scholar]
- 71.Feng R, et al. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298:50–63. doi: 10.1016/j.canlet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 72.Liu B, Peng XC, Zheng XL, Wang J, Qin YW. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009;66:169–175. doi: 10.1016/j.lungcan.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 73.Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 74.Oskarsson T, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O'Connell JT, et al. VEGF-A and Tenascin-C produced by S100A4+ stromal cells are important for metastatic colonization. Proc Natl Acad Sci U S A. 2011;108:16002–16007. doi: 10.1073/pnas.1109493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J, et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72:4597–4608. doi: 10.1158/0008-5472.CAN-12-1045. [DOI] [PubMed] [Google Scholar]
- 77.Valastyan S, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.Valastyan S, Benaich N, Chang A, Reinhardt F, Weinberg RA. Concomitant suppression of three target genes can explain the impact of a microRNA on metastasis. Genes Dev. 2009;23:2592–2597. doi: 10.1101/gad.1832709. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 79.Valastyan S, Chang A, Benaich N, Reinhardt F, Weinberg RA. Concurrent suppression of integrin alpha5, radixin, and RhoA phenocopies the effects of miR-31 on metastasis. Cancer Res. 2010;70:5147–5154. doi: 10.1158/0008-5472.CAN-10-0410. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 80.Wszolek MF, et al. A MicroRNA expression profile defining the invasive bladder tumor phenotype. Urol Oncol. 2011;29:794–801. e791. doi: 10.1016/j.urolonc.2009.08.024. [DOI] [PubMed] [Google Scholar]
- 81.Schaefer A, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 82.Creighton CJ, et al. Molecular profiling uncovers a p53-associated role for microRNA-31 in inhibiting the proliferation of serous ovarian carcinomas and other cancers. Cancer Res. 2010;70:1906–1915. doi: 10.1158/0008-5472.CAN-09-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y, et al. Down-regulation of miR-31 expression in gastric cancer tissues and its clinical significance. Med Oncol. 2010;27:685–689. doi: 10.1007/s12032-009-9269-x. [DOI] [PubMed] [Google Scholar]
- 84.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 85.Hakem A, et al. RhoC is dispensable for embryogenesis and tumor initiation but essential for metastasis. Genes Dev. 2005;19:1974–1979. doi: 10.1101/gad.1310805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma L, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341–347. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang Q, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 88.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 89.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burk U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gibbons DL, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Korpal M, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ma L, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen D, et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat Med. 2012;18:1511–1517. doi: 10.1038/nm.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 97.Zhu S, et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 98.Asangani IA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 99.Bornachea O, et al. EMT and induction of miR-21 mediate metastasis development in Trp53-deficient tumours. Sci Rep. 2012;2:434. doi: 10.1038/srep00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Segura MF, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gaziel-Sovran A, et al. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell. 2011;20:104–118. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pencheva N, et al. Convergent Multi-miRNA Targeting of ApoE Drives LRP1/LRP8-Dependent Melanoma Metastasis and Angiogenesis. Cell. 2012;151:1068–1082. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yu F, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 104.Dangi-Garimella S, et al. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009;28:347–358. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yun J, et al. Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. EMBO J. 2011;30:4500–4514. doi: 10.1038/emboj.2011.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garner K, et al. Phosphatidylinositol transfer protein, cytoplasmic 1 (PITPNC1) binds and transfers phosphatidic acid. J Biol Chem. 2012;287:32263–32276. doi: 10.1074/jbc.M112.375840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang Y, et al. miR-126 and miR-126(*) repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol. 2013;15:284–294. doi: 10.1038/ncb2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhu N, et al. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem. 2011;351:157–164. doi: 10.1007/s11010-011-0723-7. [DOI] [PubMed] [Google Scholar]
- 109.Sasahira T, et al. Downregulation of miR-126 induces angiogenesis and lymphangiogenesis by activation of VEGF-A in oral cancer. Br J Cancer. 2012;107:700–706. doi: 10.1038/bjc.2012.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chou J, et al. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat Cell Biol. 2012;15:201–213. doi: 10.1038/ncb2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ru P, et al. miRNA-29b suppresses prostate cancer metastasis by regulating epithelial mesenchymal transition signaling. Mol Cancer Ther. 2012;11:1166–1173. doi: 10.1158/1535-7163.MCT-12-0100. [DOI] [PubMed] [Google Scholar]
- 112.Fang JH, et al. MicroRNA-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology. 2011;54:1729–1740. doi: 10.1002/hep.24577. [DOI] [PubMed] [Google Scholar]
- 113.Zhuang G, et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012;31:3513–3523. doi: 10.1038/emboj.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wolfers J, et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7:297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- 115.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bracken CP, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 117.Siemens H, et al. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 118.Grelier G, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. 2009;101:673–683. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Martello G, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 120.Su X, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Melo SA, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 122.Kota J, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Miyazaki Y, et al. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat Med. 2012;18:1136–1141. doi: 10.1038/nm.2791. [DOI] [PubMed] [Google Scholar]
- 124.Obad S, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Elmén J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 126.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gregory PA, et al. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol Biol Cell. 2011;22:1686–1698. doi: 10.1091/mbc.E11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu C, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yang S, et al. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 2012 doi: 10.1038/onc.2012.432. [DOI] [PubMed] [Google Scholar]
- 130.Wong CC, et al. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology. 2011;140:322–331. doi: 10.1053/j.gastro.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 131.Penna E, et al. microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J. 2011;30:1990–2007. doi: 10.1038/emboj.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li X, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9:824–833. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- 133.Zhang X, Liu S, Hu T, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490–499. doi: 10.1002/hep.23008. [DOI] [PubMed] [Google Scholar]
- 134.Zou Z, et al. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994;263:526–529. doi: 10.1126/science.8290962. [DOI] [PubMed] [Google Scholar]
- 135.Dorrello NV, et al. S6K1-and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 136.Latha K, Zhang W, Cella N, Shi HY, Zhang M. Maspin mediates increased tumor cell apoptosis upon induction of the mitochondrial permeability transition. Mol Cell Biol. 2005;25:1737–1748. doi: 10.1128/MCB.25.5.1737-1748.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]