Abstract
The periosteum contributes to bone repair and maintenance of cortical bone mass. In contrast to the understanding of bone development within the epiphyseal growth plate, factors that regulate periosteal osteogenesis have not been studied as intensively. Osteofibrous dysplasia (OFD) is a congenital disorder of osteogenesis and is typically sporadic and characterized by radiolucent lesions affecting the cortical bone immediately under the periosteum of the tibia and fibula. We identified germline mutations in MET, encoding a receptor tyrosine kinase, that segregate with an autosomal-dominant form of OFD in three families and a mutation in a fourth affected subject from a simplex family and with bilateral disease. Mutations identified in all families with dominant inheritance and in the one simplex subject with bilateral disease abolished the splice inclusion of exon 14 in MET transcripts, which resulted in a MET receptor (METΔ14) lacking a cytoplasmic juxtamembrane domain. Splice exclusion of this domain occurs during normal embryonic development, and forced induction of this exon-exclusion event retarded osteoblastic differentiation in vitro and inhibited bone-matrix mineralization. In an additional subject with unilateral OFD, we identified a somatic MET mutation, also affecting exon 14, that substituted a tyrosine residue critical for MET receptor turnover and, as in the case of the METΔ14 mutations, had a stabilizing effect on the mature protein. Taken together, these data show that aberrant MET regulation via the juxtamembrane domain subverts core MET receptor functions that regulate osteogenesis within cortical diaphyseal bone.
Introduction
The periosteum is a thin membranous structure that covers the external surfaces of all bones in mammals. It is composed of an outer fibrous layer and an inner layer that contains blood vessels and cells that have the potential to sponsor the maintenance of the underlying cortical bone in addition to contributing substantially to fracture healing.1, 2 The regulation of the periosteal cellular compartment is not well understood, although it is clear that a subfraction of cells from this population possess properties of osteochondroprogenitors in that they have the ability to differentiate into osteoblasts and chondrocytes.
Osteofibrous dysplasia (OFD [MIM: 607278]) is a developmental skeletal disorder characterized by radiolucent lesions located at the periosteal surface of the diaphyseal cortex, almost exclusively of the tibia and fibula (Figure 1A), although lesions in the radius and ulna have been occasionally described.3 These lesions are congenital and typically unilateral, and they spontaneously resolve during skeletal maturation;4 the residuum is most commonly mild bowing at the affected site. Prior to their resolution, secondary complications such as fractures and non-union and pseudoarthrosis formation can complicate the condition. Histologically, OFD lesions exhibit “zonal architecture” characterized by spindle-shaped fibroblast-like cells in the center of the lesions that are progressively replaced with peripherally located, more differentiated cells from the osteoblastic lineage (Figure 1B).5 The cells lying at the center of the lesions stain for markers of undifferentiated mesenchymal cell states, whereas bridging zones of osteoid with surface osteoblasts and embedded osteocytic cells are interspersed between the lesions.5, 6 In OFD, the unossified zones eventually mineralize after replacement with normal osteoid and, finally, bone. This histological progression corresponds with the clinical and radiographic resolution of the lesions, although some affected bones exhibit residual bowing (Figure 1A). Although typically sporadic and unilateral, familial and bilateral forms of OFD have been described.3, 7, 8, 9
Figure 1.
The Clinical, Pathological, and Genetic Basis of Bilateral Osteofibrous Dysplasia
(A) Radiograph of both lower limbs of subject III:6 from family 1 demonstrating gradual resolution of mid-diaphyseal lesions of the tibia and fibula over 12 years (reproduced with permission from the publisher of Beals and Fraser3). AP, antero-posterior; lat, lateral.
(B) Histology and immunohistochemistry of lesional tissue excised at surgery from subject III:6 (aged 9 years) from family 1 demonstrating spindle-shaped cells at the center of lesions (arrow) surrounded by osteoid (o); staining for alkaline phosphatase (ALP) is maximal in the periphery of lesions and absent at the center (arrows); lesions are characterized by rimming osteoblasts staining positively for the early osteoblastic marker osterix (OSX) and osteoclastic marker tartrate resistant acid phosphatase (TRAP).
(C) Pedigrees of the four families with bilateral OFD and with identified mutations in MET.
Here, we have identified germline and somatic mutations in the gene encoding the receptor tyrosine kinase MET that specifically disrupt the differentially spliced exon 14 to cause three familial and two simplex cases of OFD. Alternative splicing of MET (MIM: 164860) regulates the stability of the receptor by determining the inclusion or exclusion of an ubiquitination target within its cytoplasmic domain.10 Our results indicate that the stabilization of MET while ligand-dependent activation is maintained retards osteoblastic differentiation and, as such, presents MET as a key regulator of cortical bone osteogenesis.
Material and Methods
Ethical Review, Consent, Subject Ascertainment, and Clinical Descriptions
All subjects were ascertained by physician-initiated referral and consented to participate under approved protocols MEC/08/08/094 and 13/STH/56 from the Health and Disability Ethics Committee, New Zealand and the institutional review boards of the University of Texas Southwestern Medical Center and the Hospital for Sick Children, University of Toronto. All index subjects presented with lesions that were located at the periosteal surface of cortical bone and resolved with time post-fracture (Figure 1A).3, 7, 8 All individuals were clinically examined by at least one of the co-authors, and antero-posterior and lateral radiographs of both forelegs were obtained for all individuals. Individuals were assigned as affected if (1) they had clinically or radiologically evident involvement of the tibia and fibula or (2) historical clinical or radiographic evidence of involvement. Radiographs of individuals whose clinical or radiographic status was uncertain were examined by a radiologist who was blinded to the clinical or familial relationship of those persons within their respective families. A histological diagnosis of OFD was assigned to lesions that contained rare cells staining for cytoskeletal keratin and exhibited rimming osteoblasts embedded within osteoid. The distribution of the disease in affected individuals was almost entirely confined to the tibia and fibula, although one subject in family 1 had severe scoliosis, and he and several other family members had variable degrees of pectus excavatum. The condition did not appear to confer enhanced risk of fracture or other orthopedic complications except for fractures at the site of radiologically evident lesions. There was no history of malignant neoplasms, prior to the age of 50 years, in any family member. More extensive descriptions of families 1, 2, and 4 and their clinical presentations have been previously published.3, 7, 9 Control DNA samples and cell lines were obtained from sex- and age-matched healthy individuals. The University of Otago Animal Ethics Committee approved the protocols for use of mice in these investigations (ET11/13).
Immunohistochemistry
Human OFD tissue samples were fixed in formalin, decalcified, and embedded in paraffin. Serial sections were de-paraffinized and rehydrated through an alcohol gradient to water. A microwave pressure cooker was used for heat-induced epitope retrieval (15 min, 10 mM citrate buffer [pH 6.0]) for the alkaline phosphatase (ab108337) and MET (ab74217) antibodies. No antigen retrieval was required for the Sp7/osterix antibody (ab22552; Abcam). Endogenous peroxide activity was blocked with 3% (v/v) hydrogen peroxide in methanol for 15 min at room temperature, and nonspecific binding was blocked with 2% (v/v) normal goat sera (Vectorlabs) in PBS with 2% (w/v) BSA for 30 min. Primary antibody was incubated overnight at 4°C and Polinker-1 HRP secondary antibody was used to detect binding of the primary antibody (GBI Labs). Normal rabbit serum or monoclonal rabbit immunoglobulin G was used in control sections.
Linkage
HumanLinkage-12 arrays (Illumina) were used to genotype individuals from family 1. Genotypes were imported into Alohomora11 for pedigree checking and removal of Mendelian errors prior to performance of parametric linkage analysis with Allegro v.2.12, 13
Custom Capture Chip Construction and Massively Parallel DNA Sequencing for Family 1
For family 1, customized NimbleGen 1.2M SeqCap capture arrays were designed according to the manufacturer’s instructions (NimbleGen). Targeted capture and sequencing was performed on the 454 Sequencing platform (Roche), and subsequent sequence alignment and calling were performed with proprietary analytic scripts (Roche).
Exome Sequencing in Family 2
The exomes of four family members from family 2—three affected (IV-4, IV-10, IV-12) and one unaffected (III-7)—were captured with the Agilent SureSelect Human All-Exon 50 Mb kit and sequenced with the Applied Biosystems (ABI) SOLiD 4 system. Sequences were aligned to the UCSC Genome Browser hg19 reference sequence with ABI LifeScope software; variants were annotated with SeattleSeq v.6.21 software. Variants predicting nonsynonymous or frameshift changes were further filtered for variants that were novel (absent in public databases dbSNP, 1000 Genomes, and the Exome Aggregation Consortium) and shared in exomes of affected, but not unaffected, family members.
Cell Culture and Exome Sequencing of an Individual with Sporadic Unilateral OFD
Cells from resected bone tissue were cultured in MEM-α 10% fetal bovine serum and penicillin/streptomycin (PS; 100 units of potassium penicillin and 100 μg of streptomycin sulfate per 1 ml of culture media) at 37°C and 5% CO2 as previously described.14 DNA was extracted from cultured cells after a single passage. Whole-exome capture was performed with the TruSeq Exome Kit (Illumina). Sequencing was performed with the 2× 150 bp paired-end protocol on an Illumina HiSeq 2500 instrument. Reads were mapped with the Burrows-Wheeler Aligner15, and quality-control was performed with Picard and the Genome Analysis Toolkit (GATK)16. Variants were detected with GATK, and high-quality novel nonsynonymous variants were identified after exclusion of those present in dbSNP, the NHLBI Exome Sequencing Project Exome Variant Server, and an in-house database of control exomes. For clonal cell isolation, cells were harvested and sorted into five 96-well plates with an Aria II cell sorter. The 96-well plates had a ratio of 1:1 fresh and conditioned media. Single cells were grown for three to four weeks at 37°C, and cDNA was prepared from surviving clonal lines. To detect the variant in lesional tissue, RNA was extracted with the RNeasy Mini Kit (QIAGEN), and RT-PCR was performed with the SuperScript III One-Step RT-PCR System (Invitrogen).
Haplotype Analysis
Microsatellites were genotyped on an Applied Biosystems 3730xl DNA Analyzer, and genotype files were analyzed in GeneMapper v.4.0 (Applied Biosystems) to assign alleles. Haplotypes were constructed manually. The locations of the microsatellites are presented in Table S5.
Cell Transfections and Western Blotting
Human MET WT (wild-type) and METΔ14 cDNAs were amplified from HEK293 and H596 cell lines, respectively, and cloned into commercially available pCMV6-AC-GFP plasmid (Origene) by restriction digestion. Previous studies have demonstrated that the GFP-tagged MET constructs express appropriately at the plasma membrane and are functionally unimpaired by this modification.17 The construct encoding METTyr1003Ser was generated from the MET WT GFP-tagged clone with a QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent). Using Lipofectamine 2000 and following the manufacturer’s recommendations, we transiently transfected HEK293T cells with WT or mutant constructs of GFP-tagged full-length human MET with or without CBL or SIAH1 expression constructs (an unrelated E3 ubiquitin ligase). After 24 hr, cells were washed with cold 1× PBS and lysed by the addition of RIPA buffer directly to the cell culture wells. Whole cell lysates were resolved by reducing SDS-PAGE on a 6% gel and were subjected to immunoblotting with rabbit anti-MET (1:500) (catalog no. sc-10; Santa Cruz) followed by rabbit anti-β–actin (1:1,000) (catalog no. 4970; Cell Signaling) antibodies in 5% non-fat milk.
In Situ Hybridization
In situ hybridization was performed on 12 μm cryosections. After 4% paraformaldehyde fixation, sections were acetylated and prehybridized for 2 hr at 60°C with hybridization solution (50% formamide, 5× saline sodium citrate, 500 μg/ml tRNA, 50 μg/ml heparin, 0.1% Tween 20, 1M citric acid). Hybridization (hybridization solution + 1 μg/ml probe) was performed overnight at 60°C in a humidified chamber. Slides were washed and processed for staining as previously described.18 To generate antisense and sense RNA probes, a mouse Met cDNA fragment was subcloned into pGEMt-easy with forward primer 5′-GCTTGTAAGTGCCCGAAGTG-3′ and reverse primer 5′-ACATGTCTCTGGCAAGACCG-3′. Antisense and sense RNA probes were prepared with Not1 and T7 RNA polymerase and Nco1 and SP6 RNA polymerase combinations, respectively. Probes were made with an mMESSAGE mMACHINE kit (Ambion) and purified with mini Quick Spin Columns (Roche).
RNA Extraction from Mouse Skeletal Tissue and Whole Transcriptome Sequencing
Total RNA from microdissected whole limb buds from time-mated C57BL/6 embryonic day (E) 12.5 mice was prepared with the Illumina TruSeq Total RNA LT kit to deplete rRNA. For tibial tissue RNA preparation from E15, E17, and adult Swiss Webster mice, hindlimbs were removed and tibiae were dissected from associated muscular tissue. For periosteal cell isolation from juvenile mice, periosteum was excised by a lengthwise scalpel cut along the dissected tibia and careful removal of the periosteum with watchman forceps. Tissue from this procedure, as well as from microdissected whole limb buds from time-mated C57BL/6 E10.5 mice, was collected in Trizol, homogenized, and RNA extracted as per the manufacturer’s recommendations. RNA was subsequently concentrated with a NucleoSpin RNA column (Macherey Nagel). For transcriptome sequencing, RNA was normalized and sequenced on an Illumina HiSeq 2500 and the paired-end protocol was used. Sequence reads were filtered for quality and mapped to the mouse reference sequence (mm9) with TopHat19 and the iGenomes annotation. Duplicate reads were marked with Picard. Alignments were visualized and plotted with the Integrative Genomics Viewer.
In Vitro Mineralization Assay
Early passage MC3T3E1 (American Tissue Culture Collection; clone 4) cells were seeded in 6- or 24-well culture plates at 5× 104−105 cells/cm2. After 24 hr, we transfected cells with phosphorothioate oligonucleotides (TriLink Biotechnologies) by using RNAiMAX (Life Technologies) according to the manufacturer’s protocol. Matrix mineralization was initiated 24 hr after transfection by the addition of ascorbic acid and beta-glycerophosphate (final concentrations of 50 μg/ml and 10 mM, respectively). Complete media changes were performed every three days. Mineralization was assessed at 12 days with Alizarin red (Sigma-Aldrich). The Met- skipping oligo sequence was 5′-2′O-Me-AGU GUG UAC UCU UGC GUC AU-3′. The control-scrambled oligo sequence was 5′-2′O-Me-AUG ACG CAA GAG UAC ACA CU-3′.
RNA Extraction and qRT-PCR for Markers of Ossification
We extracted RNA from transiently transfected MC3T3E1 (clone 4) cells on day four after transfection by using a NucleoSpin RNA kit (Mackerey Nagel). RNA was treated with a TURBO DNA-free Kit (AM1907, Life Technologies) and cDNA was prepared with SuperScript III reverse transcriptase (Life Technologies). qRT-PCR was performed according to the manufacturer’s specifications with the LightCycler 480 SYBR Green I Master kit (Roche) and run on a LightCycler 480 instrument. Results were analyzed with Qbase.20
Results
OFD Lesions Exhibit Active Marginal Osteogenesis
We sought to extend our understanding concerning the zonal architecture of the lesions in OFD, including staining for markers of osteoblasts and osteoclasts in addition to cell proliferation. Analysis of lesional tissue from affected individual III:6 from family 1 displayed the typical zonal architecture of OFD (Figure 1B), as previously reported for family 27. Staining for proliferating cell nuclear antigen (PCNA) demonstrated that the centrally located spindle-shaped cells were not mitotically active but that the cells rimming these lesions stained for markers of osteoblastic differentiation (osterix, OSX; alkaline phosphatase, ALP). Osteoclasts staining for tartrate resistant acid phosphatase (TRAP) were also prominent at the periphery of the lesions (Figure S1). On the basis of these and previous observations5 demonstrating that the centrally located cells stained positively for markers that are associated with undifferentiated mesenchymal cells, we hypothesized that the pathogenesis of familial OFD was related to a delay in osteogenesis, with lesions resolving at the margins through the action of rimming osteoblasts and osteoclasts.
Mutations in MET Cause Familial OFD
The majority of previous clinical and histological studies on OFD concentrated on the unilateral, presumptive sporadic forms of the condition. To extend understanding of the etiology and pathogenesis of OFD, we initiated a study of four families with bilateral disease, three of them affected by OFD that segregated as an autosomal-dominant trait (Figure 1C). The ascertainment of rare familial instances of OFD presented the opportunity to locate a genetic regulator of osteogenesis that, considering the location of the lesions, was likely to relate to the periosteal contribution to this process. To identify the causative locus for familial OFD, we genotyped family 1 (Figure 1C) for linkage analysis by using HumanLinkage-12 arrays (Illumina) under a model of autosomal-dominant inheritance with incomplete penetrance. The two most significant regions, but with submaximal LOD scores, were identified on chromosomes 7 and 11 (Figure S2). Targeted capture of all coding and phylogenetically conserved elements (4.76 Mb of a combined 44 Mb region) within these two candidate regions was followed by massively parallel sequencing from three affected individuals (III:8, IV:2, and IV:8) with the Roche 454 system. Only one non-synonymous variant not represented in dbSNP build 131 or the Exome Variant Server was shared by all three individuals —a 26 bp deletion (c.3010_3028+8del ∼∼[p.Leu964_Asp1010del]) that included the donor splice site at the exon 14 and intron 14 junction of MET (GenBank: NM_000245.2; Figure 1C; Table 1; Tables S1 and S2), the gene that encodes the receptor tyrosine kinase MET. Family 2 was analyzed by a similar mapping and sequencing strategy. In this family, exome sequencing of four individuals revealed a second MET variant (c.3028+1G>T) at the phylogenetically invariant +1 position of the 3′ splice donor site of the same exon (Table 1; Figure 1C; Table S3). The same mutation was found in an affected mother and one son in family 3. To test whether the two identical mutations in families 2 and 3 had arisen as independent events, we constructed haplotypes by using flanking polymorphic microsatellite markers (Table S5). Results indicated that the c.3028+1G>T mutations had arisen on different haplotypes in families 2 and 3 (Figure S3). The c.3028+1G>T mutation was also observed in DNA extracted from normal tissue adjacent to an excised lesion from the affected individual in family 4. The presentation in this instance was that of bilateral OFD as previously reported,9 but insufficient DNA prevented complete verification of the haplotype on which this variant arose. Parental DNA samples were also unavailable, and therefore we were unable to confirm whether the mutation arose de novo in the germline, was an early post-zygotic mutation, or had been inherited from a parent. These MET variants segregated throughout families 1–3 in all affected and in four non-penetrant individuals (Table 1; Figure S4).
Table 1.
Mutations in MET and Penetrance in Bilateral Osteofibrous Dysplasia
|
Mutation |
Penetrancea |
Bones Affected | |||
|---|---|---|---|---|---|
| DNA | Protein | Penetrant | Non-penetrant | ||
| Family 1, familial | c.3010_3028+8del | p.Leu964_Asp1010del | 10 | 1 | T, F, U |
| Family 2, familial | c.3028+1G>T | p.Leu964_Asp1010del | 6 | 4 | T, F |
| Family 3, familial | c.3028+1G>T | p.Leu964_Asp1010del | 2 | 0 | T, F |
| Family 4, simplex | c.3028+1G>T | p.Leu964_Asp1010del | 1 | 0 | T, F; bilateral |
Reference transcript, GenBank: NM_000245.2. Abbreviations are as follows: F, fibula; T, tibia; U, ulna.
Numbers indicate the number of mutation-carrying family members who are affected (penetrant) versus unaffected (non-penetrant).
The variants identified in all four families affect the canonical splice-donor consensus sequence for exon 14 of MET. To assess the consequences of these mutations, we performed RT-PCR on cDNA prepared from cultured dermal fibroblasts (family 1), lymphoblastoid cell lines (family 2), and excised lesional tissue from bone (family 3) by using primers located in exons flanking exon 14 (Figure 2A). Both the 26 bp deletion and the c.3028+1G>T variants in MET were shown to result in the exclusion of exon 14 of MET from the mature transcript (METΔ14; Figure 2A). In all instances, compared to the WT allele, this splice form was not associated with instability, as judged by the presence of both transcripts in the respective sequence chromatograms.
Figure 2.
Expression of MET in Familial OFD
(A) MET splicing and expression in germline and OFD lesional tissue. (i) RT-PCR of cDNA from lymphoblastoid cell lines obtained from a normal control (lane 2), a lung cancer cell line (H596)10 with a mutation conferring constitutive skipping of exon 14 (lane 3), and lymphoblastoid cell lines from two affected individuals (IV:10, IV:12; Figure 1C) from family 2 (lanes 4 and 5). Primers designed to exons 13 and 15 amplified a 507 bp product in exon-14-inclusive templates and a 367 bp fragment in mutant-exon-14-excluded transcripts. (ii) Sequence verification from gel-excised RT-PCR products confirming exon 14 exclusion in transcripts from two affected individuals from families 1 and 2. (iii) RT-PCR with primers complementary to exons 13 and 15 and cDNA prepared from cells liberated from an OFD lesion. Control cells were grown from a femoral metaphyseal biopsy from an unrelated age-matched subject. FL, full-length MET+14 product; Δ, METΔ14 product.
(B) MET protein is detectable in OFD lesions and normal bone. (i) Western blot of cell lysates from lesional OFD tissue demonstrating MET in lesional tissue (OFD) and in metaphyseal bone from a healthy age-matched control subject. (ii) Immunohistochemistry of lesional tissue from subject III:6 (family 1) demonstrating MET immunoreactivity in rimming osteoblasts (red arrows) and osteoclasts (black arrows). o, osteoid.
(C) Schematic diagram of MET showing position of mutations and consequences to receptor structure conferred by the mutations affecting the splice donor site leading to exclusion of the cytoplasmic JMD (yellow).
Consistent with mutations in MET being causative of OFD, immunohistochemical staining in lesion tissue revealed the presence of MET in both osteoblasts and osteoclasts (Figure 2B). Additionally, western blot analysis of lesional tissue obtained from an affected individual from family 3 revealed detectable levels of MET, as did an assay on metaphyseal bone obtained from an age-matched healthy individual (Figure 2B). At the protein level, exclusion of exon 14 from the spliced transcript predicts the in-frame omission of a 47-amino-acid juxtamembrane domain (JMD) from the MET receptor (Figure 2C), although the resolution of western blot analysis is insufficient to differentiate this METΔJMD isoform from a receptor protein that retains the JMD.
Both the 26 bp deletion and the c.3028+1G>T variant have the same consequence—the in-frame deletion of exon 14 that encodes the phylogenetically conserved JMD of the MET receptor. The JMD regulates MET receptor stability and subcellular localization. Engagement of MET with its sole known ligand, hepatocyte growth factor (HGF), leads to homodimerization of the receptor and phosphorylation of at least five tyrosine residues located in its cytoplasmic domain.21 Whereas four of these phosphotyrosine residues are known to initiate the assembly of protein complexes that mediate signal transduction, phosphorylation of Tyr1003 within the JMD leads to its engagement with the ubiquitin ligase, CBL.22 Receptor ubiquitination within the JMD regulates MET endocytosis, downregulation of plasma membrane receptor abundance, and endosomal degradation and/or recycling of internalized receptors.23, 24 Consistent with Tyr1003 being located within the exon-14-encoded JMD, previous work has demonstrated that exclusion of this protein domain from MET attenuates receptor internalization and recycling and upregulates the duration of ligand-dependent signaling.10, 25 Furthermore, somatically acquired mutations in lung10, 26, 27, 28 and gastric cancer29 that lead to splice exclusion of exon 14, and hence aberrant levels of METΔ14, have been shown to prolong ligand-induced mitogen-activated protein kinase (MAPK) signaling as a result of stabilization of MET and enhanced retention of MET receptors at the plasma membrane.10, 29 The observation that similar, if not identical, germline mutations lead to OFD directly implicates MET as a developmental regulator of osteogenesis through a similar gain-of-function mechanism.
MET Exon 14 Skipping Occurs during Development in Mice
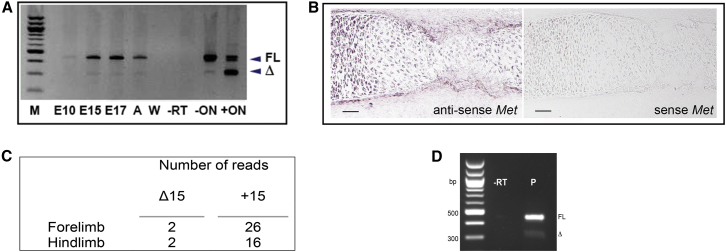
Splice exclusion of MET exon 14 (or orthologous sequences in other species) might be a developmentally regulated event. Splice forms orthologous to METΔ14 have been detected in embryonic and postnatal rats and mice,25, 30, 31 although data are lacking on the spatial and temporal regulation of these splice forms. Given the strong immunostaining for MET in osteoclasts and osteoblasts rimming OFD lesional tissue (Figure 2B), we sought evidence for expression of Met transcripts lacking the orthologous exon to human exon 14 (mouse exon 15; MetΔ15) in normal developing mouse skeletons. Preparations from either whole dissected mouse embryonic limb buds collected at E10.5 or tibiae collected at E15, E17, and adult time points showed consistent but low-level expression of MetΔ15 (Figure 3A). In situ hybridization of a riboprobe designed to detect both MetΔ15 and Met+15 in mouse E15 tibiae revealed prominent expression of Met in chondrocytes and, of relevance to the site of lesion formation in OFD, in the periosteum of long bones (Figure 3B). Furthermore, RNA sequencing (RNA-seq) analysis of RNA extracted from E12.5 mouse limb buds revealed the expression of both full-length and skipped isoforms in forelimbs and hindlimbs (Figure 3C; Figure S5).
Figure 3.
Met Expression and Exon 15 Splice Exclusion during Mouse Skeletal Development
(A) RT-PCR of exons 14–16 of Met using cDNA prepared from developing mouse limbs at E10 (whole limb), E15, and E17 and from adult (A) mouse limbs (all from dissected tibia). C2C12 cells alone (–ON) and cells transiently transfected with 2-O-methyl oligoribonucleotide (+ON) to induce exon 15 exclusion are shown. FL, full-length exon 15-inclusive product; Δ, exon 15-excluded product; W, no cDNA control; −RT, no reverse transcriptase control.
(B) In situ riboprobe staining of mouse E15.5 tibial diaphysis for Met. Strong expression of Met is demonstrated in the periosteum. The scale bar represents 200 μm.
(C) RNA-seq split reads spanning multiple exons for both MetΔ15 and Met15 isoforms obtained from RNA isolated from E12.5 mouse forelimb and hindlimb buds.
(D) RT-PCR with RNA from cells cultured from periosteum dissected from the tibial diaphysis of a 3-week-old mouse. Δ, MetΔ15; FL, Met15+; −RT, minus reverse transcriptase control; P, periosteal cDNA.
Because Met exon 15 skipping occurred physiologically and Met expression was prominent in embryonic periosteum, we sought evidence that Met exon 15 skipping occurred specifically in this tissue. Because the in situ hybridization study (Figure 3B) did not discriminate between the full-length exon-inclusive (Met15+) and exon-exclusive (MetΔ15) isoforms, we isolated a proliferating cell population from in vitro culture of explanted dissected periosteum from a 3-week-old mouse and showed evidence for production of both MetΔ15 and Met15+ transcripts by RT-PCR (Figure 3D). These results suggested that exon 15 splice exclusion was active in the periosteum during normal development. The germline mutations that we have shown to underlie OFD and subvert exon 14 skipping therefore have the potential to influence periosteal function and lead to the osseous lesions observed in OFD through the aberrant production of a MET receptor lacking the JMD domain (METΔJMD).
Diminished Osteoblastic and Osteoclastic Function Resulting from MET Exon Skipping
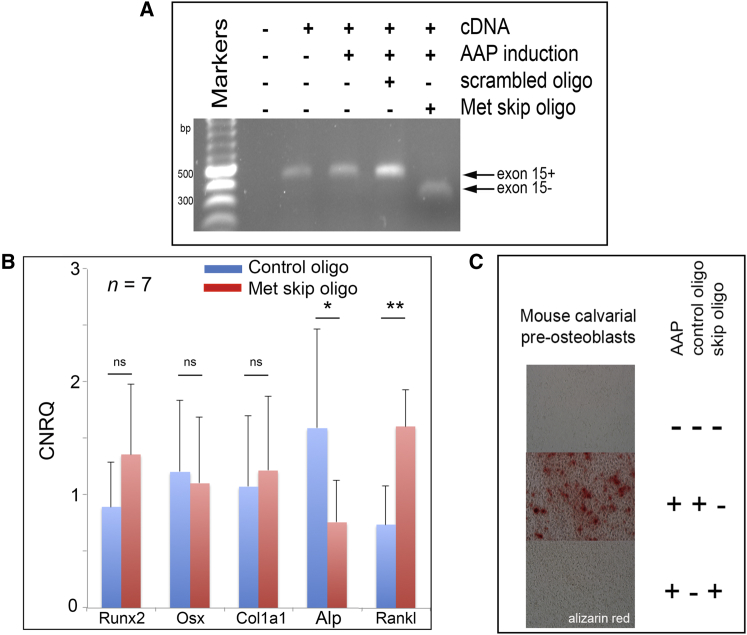
MET levels are prominent in both osteoblasts and osteoclasts in OFD lesional tissue (Figure 2B; Figure S1) and in normal bone.32 To address the possibility that OFD is primarily due to a defect in osteoblast function, we assessed the effect of the MetΔ15 splice form in an in vitro assay of osteoblastic differentiation. We designed and validated 2′-O-methyl-oligoribonucleotides that can induce exon 15 splice exclusion when transfected into mouse cell lines (Figures 3A and 4A). Using the mouse MC-3T3 pre-osteoblastic cell line,33 we measured the effect of induced MetΔ15 exon skipping at 4 days by using RT-PCR for multiple markers of osteogenic differentiation and at 12 days by using alizarin red to assess matrix mineralization. At day 4 after the induction of exon exclusion, the osteoblastic marker Alp showed significantly reduced expression (p < 0.05), whereas Rankl, an osteoblast-derived osteoclastogenic factor, showed elevated expression (p < 0.01, Figure 4B). Early markers of osteoblastic differentiation (Osx, Runx2) and Col1a1 were not significantly altered by induction of MetΔ15 expression, indicating that either the level of exon skipping achieved by the oligoribonucleotide was insufficient to have an effect on these functions or that MetΔ15 does not impact the earliest phases of osteoblastic differentiation. At day 12, matrix mineralization was markedly repressed in these cells compared to mineralization in cells transfected with a scrambled control oligoribonucleotide (Figure 4C). These results support the hypothesis that a forced overexpression of MetΔ15 relative to Met+15 conferred a block in progression to late stages of osteoblast differentiation. Furthermore, these data suggest that physiological deployment of this event might regulate the speed at which osteoblasts transition to more differentiated states during skeletogenesis.
Figure 4.
Skipping of Met Exon 15 in Mice Impairs Both Osteoclast and Osteoblast Function
(A) Induced MC-3T3 cells with and without treatment with exon-skipping or scrambled control 2-O-methyl oligoribonucleotides assayed for the presence or absence of skipped and unskipped Met transcripts by RT-PCR.
(B) qRT-PCR for osteoblastic markers 4 days after induced osteoblastic differentiation of MC-3T3 cells with AAP (ascorbic acid, glycerol phosphate) and transfection with exon-skipping or scrambled control oligoribonucleotides. Transcripts assayed were Runx2b, Osx, Col1a1, Alp, and Rankl. n = 7; CNRQ, calibrated normalized relative quantities; scale bars represent SEM, ∗p < 0.05; ∗∗p < 0.01, paired two-tailed Student’s t test.
(C) Endpoint matrix mineralization assay (alizarin red) 12 days after AAP induction. Oligoribonucleotides were re-applied to cultures every 3 days.
Somatic Mutation of Tyr1003 in Unilateral OFD
To test whether the mutational basis of familial OFD was the same in sporadic, unilateral cases of the disorder, DNA extracted from 20 formalin-fixed, paraffin-embedded tissue samples from individuals with sporadically occurring, unilateral OFD were Sanger-sequenced across exon 14 of MET, including the flanking intron-exon boundaries. No mutations predicted to lead to disturbance of exon 14 splicing were found in these samples, although low levels of mosaicism could not be excluded by this technique. An additional lesional sample was expanded in culture and then subjected to exome sequencing, revealing a missense mutation in exon 14, c.3008A>C (p.Tyr1003Ser). This substitution affects the phosphorylation site (Tyr1003) known to regulate JMD-mediated receptor ubiquitination and recycling34 through binding of the ubiquitin ligase CBL.22 Eight clonal cell lines six of which were shown not to have the c.3008A>C variant, were isolated from this lesion, indicating that it was somatically acquired (Figure 5A). RT-PCR followed by Sanger sequencing of lesional tissue demonstrated the presence of both WT and mutant alleles, confirming that the mutation had not arisen as an artifact in vitro.
Figure 5.
Mutations in MET Render the Receptor Insensitive to CBL-Mediated Degradation
(A) Of eight clonal lines isolated from primary lesional tissue, Sanger sequencing confirmed the presence of the c.3008A>C mutation (mutant) predicting the p.Tyr1003Ser substitution in two, and six clonal cell lines were WT (wild-type). Genomic DNA was extracted from a fragment of uncultured lesional tissue (tissue), and sequencing showed the presence of both alleles, demonstrating that the mutation was somatically acquired in this individual.
(B) OFD mutations stabilize MET in the presence of the ubiquitin ligase CBL. HEK293F cells were transfected with equal amounts of WT MET-GFP (lanes 1–3), METp.Tyr1003Ser-GFP (lanes 4–6), and METΔ14-GFP (lanes 7–9). Cells were co-transfected with equal amounts of a construct encoding SIAH1, an unrelated ubiquitin ligase (lanes 2, 5, and 8) or c-CBL-DDK (lanes 3, 6, and 9). The level of WT MET-GFP β-chain is reduced (relative to MET precursor) in the presence of CBL but not of SIAH1. The destabilizing effect of CBL is not evident in the presence of MET constructs with the p.Tyr1003Ser or Δ14 variants. This blot is an illustrative example of three independent experiments.
METΔJMD and METp.Tyr1003Ser Receptors Are Resistant to CBL-Mediated Destabilization
Previous work in cancer cell lines and transient transfection experiments have indicated that the JMD and phosphorylation of Tyr1003 regulate the ubiquitination of MET at the plasma membrane, hence its stability and signaling half-life.10, 25, 28 Therefore, we sought to investigate whether the familial METΔ14 mutations and the somatic p.Tyr1003Ser substitution affected CBL-mediated receptor degradation as previously indicated.10 Using a transient transfection approach in HEK293 cells, we introduced human constructs of MET incorporating either a deletion of exon 14 or the p.Tyr1003Ser variant into cells alongside either the ubiquitin ligase CBL or an unrelated ubiquitin ligase, SIAH1. CBL-dependent MET receptor stability was assessed by western blot analysis. The results showed that in the presence of CBL, but not of the unrelated ubiquitin ligase SIAH1, WT MET was destabilized, whereas the METΔ14 and METp.Tyr1003Ser mutations ablated the sensitivity of MET to the effects of CBL (Figure 5B). These results indicate that the germline and somatic mutations that underlie OFD stabilize the MET receptor and, as such, confer a gain of function to the protein.
Discussion
These genetic, cellular, and biochemical data indicate that OFD is caused by MET mutations that can be inherited or, occasionally, somatically acquired. In our series, all bilateral cases were caused by mutations that result in exclusion of exon 14 (METΔ14), whereas a single instance of sporadically arising unilateral OFD was associated with a somatic missense mutation within the JMD encoded by exon 14. Both mutational mechanisms exert the same effect—they stabilize MET, an effect that relates to the role that the JMD plays in mediating receptor recycling and degradation.10 Phosphorylation of residues within the JMD, including Tyr1003, results in receptor internalization to the endosomal compartment, where they can either continue to signal,35 be redirected back to the cell membrane, or undergo degradation.34
Limited evidence exists to implicate a developmental role for MET in skeletogenesis. Mouse models of Met overexpression or insufficiency are characterized by placental, muscle, and liver developmental anomalies and do not exhibit detectable skeletal developmental phenotypes.36, 37, 38, 39 These data are not necessarily inconsistent with our experiments linking mutations in MET with a skeletal disorder because these gain-of-function mutations act through a ligand-dependent mechanism, and therefore its effect is spatially restricted to tissues in which both MET and HGF are normally co-localized.
Mutations similar to those reported here that lead to splice exclusion of exon 14 have been reported as somatic events in gastric cancer,29, 40 lung cancer,26, 27, 41, 42 and other tumors.28 In lung cancer, these variants have been suggested to be probable driver mutations, acting in concert with other oncogenic mutations, to protract ligand-dependent signaling mediated by MET through prolongation of receptor occupancy at the plasma membrane.10, 28 Anecdotal evidence suggests that treatment of individuals with lung tumors and METΔ14 mutations with MET-antagonist drugs leads to a favorable clinical response, implying that the clinical resolution of OFD in individuals who have a mutation at this locus might also be hastened by treatment with these same agents.
The MET-HGF axis is well described as a mediator of the early developmental program known as the epithelial-mesenchymal transition.43, 44 In this scenario, HGF production is restricted to mesenchymal cells, where it is secreted in an paracrine fashion to activate MET receptors in epithelial cells, stimulating mesenchymal transition and cell dispersal. Uncoupling of this mechanism can lead to oncogenic activation in certain cell types.34 The mechanism by which exclusion of the functional JMD in MET leads to OFD differs from the activating germline missense mutations affecting the tyrosine kinase domain of MET, which are associated with a predisposition to develop renal papillary carcinoma. In the latter instance, ligand-independent signal activation leads to tumor formation,45 whereas in OFD, the gain of function is dependent on engagement of the receptor with its ligand, HGF.10 Consequently, the requirement for HGF could be one factor that explains the different clinical presentations of these two allelic disorders. The gain of function in OFD might therefore only manifest in tissues that possess significant levels of ligand, although it remains enigmatic as to why lesions manifest predominantly within the tibia and fibula and not in other skeletal components. In this respect, OFD is reminiscent of the tibial lesions observed in a minority of individuals with neurofibromatosis type 1. In this condition, functional haploinsufficiency for NF1, the product of which represses RAS-MAPK signaling, also predisposes individuals to pseudoarthrosis of the tibia. However, there are key differences between the two diagnoses. For example, the tibial lesions typically do not self-resolve in NF1, and evidence exists that a second mutational event is required to give rise to an NF1 tibial lesion.14, 46, 47, 48 The question still remains as to why it is the tibial diaphysis that has a diathesis to develop lesions when MAPK signaling is increased. Possibilities include vascular or mechanical factors or the presence of a hypoxic or vascular watershed that could influence MET expression and activity.49, 50
Data reported here show that exclusion of the JMD in MET alters osteoblastic function in vitro and that this same mechanism could be the predominant factor that produces OFD lesions. Osteoblasts and osteoclasts are known to co-regulate one another, however, and MET and HGF also participate in reciprocal signaling between these two cell types, leaving open the possibility that osteoclastic functions are also impaired in this condition.32 Given the potential for such complexity, modeling this condition in the mouse might improve our understanding of the cellular basis for the pathogenesis of OFD and the wider role that MET plays in osteogenesis.
The regulation of periosteal osteoprogenitor cell differentiation and function is presently poorly understood despite this cell pool being a major contributor to fracture repair51, 52 and cortical bone mass.25, 26 Our results suggest that MET-mediated signaling is a regulator of osteogenesis in the tibial and fibular diaphysis, and this insight offers new avenues to explore the possibility of therapeutically altering these processes under circumstances when bone repair and maintenance needs to be enhanced or sustained.
Acknowledgments
We thank the families and individuals with OFD for their participation in this study. This work was funded by the Wishbone Trust (New Zealand Orthopaedic Association) and Cure Kids (to S.P.R.); the Rare Diseases: Models & Mechanisms Network, the Rare Disease Foundation, and the University of Toronto McLaughlin Centre (to P.K.); G.O.O.D for Kids (to C.A.W.); and the Pediatric Orthopedic Society of North America (to J.J.R.). A.M.M. was supported by the National Institute for Health Research and University College London (UCL) Hospitals NHS Foundation Trust Biomedical Research Centre and the UCL Experimental Cancer Centre. Samples were obtained from the Stanmore Musculoskeletal Biobank. M.J.G. was supported by the Elman Poole and Sir Claude McCarthy fellowships. The authors thank the Next-Generation Sequencing and Bioinformatics core facilities at the University of Texas Southwestern Medical Center and staff at The Centre for Applied Genomics in Toronto. Research reported in this publication was supported in part by the National Center for Advancing Translational Sciences of the NIH under award number UL1TR001105. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Published: December 3, 2015
Footnotes
Supplemental Data include five figures and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.11.001.
Contributor Information
Peter Kannu, Email: peter.kannu@sickkids.ca.
Carol A. Wise, Email: carol.wise@tsrh.org.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
ExAC Browser, http://exac.broadinstitute.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
Picard, http://picard.sourceforge.net/
SnpEff, http://snpeff.sourceforge.net/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Colnot C., Zhang X., Knothe Tate M.L. Current insights on the regenerative potential of the periosteum: molecular, cellular, and endogenous engineering approaches. J. Orthop. Res. 2012;30:1869–1878. doi: 10.1002/jor.22181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murao H., Yamamoto K., Matsuda S., Akiyama H. Periosteal cells are a major source of soft callus in bone fracture. J. Bone Miner. Metab. 2013;31:390–398. doi: 10.1007/s00774-013-0429-x. [DOI] [PubMed] [Google Scholar]
- 3.Beals R.K., Fraser W. Familial congenital bowing of the tibia with pseudarthrosis and pectus excavatum: report of a kindred. J. Bone Joint Surg. Am. 1976;58:545–548. [PubMed] [Google Scholar]
- 4.Campanacci M., Laus M. Osteofibrous dysplasia of the tibia and fibula. J. Bone Joint Surg. Am. 1981;63:367–375. [PubMed] [Google Scholar]
- 5.Taylor R.M., Kashima T.G., Ferguson D.J., Szuhai K., Hogendoorn P.C., Athanasou N.A. Analysis of stromal cells in osteofibrous dysplasia and adamantinoma of long bones. Mod. Pathol. 2012;25:56–64. doi: 10.1038/modpathol.2011.141. [DOI] [PubMed] [Google Scholar]
- 6.Sakamoto A., Oda Y., Iwamoto Y., Tsuneyoshi M. A comparative study of fibrous dysplasia and osteofibrous dysplasia with regard to expressions of c-fos and c-jun products and bone matrix proteins: a clinicopathologic review and immunohistochemical study of c-fos, c-jun, type I collagen, osteonectin, osteopontin, and osteocalcin. Hum. Pathol. 1999;30:1418–1426. doi: 10.1016/s0046-8177(99)90162-4. [DOI] [PubMed] [Google Scholar]
- 7.Karol L.A., Brown D.S., Wise C.A., Waldron M. Familial osteofibrous dysplasia. A case series. J. Bone Joint Surg. Am. 2005;87:2297–2307. doi: 10.2106/JBJS.D.02575. [DOI] [PubMed] [Google Scholar]
- 8.Hunter A.G., Jarvis J. Osteofibrous dysplasia: two affected male sibs and an unrelated girl with bilateral involvement. Am. J. Med. Genet. 2002;112:79–85. doi: 10.1002/ajmg.10664. [DOI] [PubMed] [Google Scholar]
- 9.Sunkara U.K., Sponseller P.D., Hadley Miller N., McCarthy E.F. Bilateral osteofibrous dysplasia: a report of two cases and review of the literature. Iowa Orthop. J. 1997;17:47–52. [PMC free article] [PubMed] [Google Scholar]
- 10.Kong-Beltran M., Seshagiri S., Zha J., Zhu W., Bhawe K., Mendoza N., Holcomb T., Pujara K., Stinson J., Fu L. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 2006;66:283–289. doi: 10.1158/0008-5472.CAN-05-2749. [DOI] [PubMed] [Google Scholar]
- 11.Rüschendorf F., Nürnberg P. ALOHOMORA: a tool for linkage analysis using 10K SNP array data. Bioinformatics. 2005;21:2123–2125. doi: 10.1093/bioinformatics/bti264. [DOI] [PubMed] [Google Scholar]
- 12.Gudbjartsson D.F., Jonasson K., Frigge M.L., Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat. Genet. 2000;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- 13.Gudbjartsson D.F., Thorvaldsson T., Kong A., Gunnarsson G., Ingolfsdottir A. Allegro version 2. Nat. Genet. 2005;37:1015–1016. doi: 10.1038/ng1005-1015. [DOI] [PubMed] [Google Scholar]
- 14.Paria N., Cho T.J., Choi I.H., Kamiya N., Kayembe K., Mao R., Margraf R.L., Obermosser G., Oxendine I., Sant D.W. Neurofibromin deficiency-associated transcriptional dysregulation suggests a novel therapy for tibial pseudoarthrosis in NF1. J. Bone Miner. Res. 2014;29:2636–2642. doi: 10.1002/jbmr.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moshitch-Moshkovitz S., Tsarfaty G., Kaufman D.W., Stein G.Y., Shichrur K., Solomon E., Sigler R.H., Resau J.H., Vande Woude G.F., Tsarfaty I. In vivo direct molecular imaging of early tumorigenesis and malignant progression induced by transgenic expression of GFP-Met. Neoplasia. 2006;8:353–363. doi: 10.1593/neo.05634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilhelm D., Hiramatsu R., Mizusaki H., Widjaja L., Combes A.N., Kanai Y., Koopman P. SOX9 regulates prostaglandin D synthase gene transcription in vivo to ensure testis development. J. Biol. Chem. 2007;282:10553–10560. doi: 10.1074/jbc.M609578200. [DOI] [PubMed] [Google Scholar]
- 19.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trusolino L., Bertotti A., Comoglio P.M. MET signalling: principles and functions in development, organ regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2010;11:834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 22.Peschard P., Ishiyama N., Lin T., Lipkowitz S., Park M. A conserved DpYR motif in the juxtamembrane domain of the Met receptor family forms an atypical c-Cbl/Cbl-b tyrosine kinase binding domain binding site required for suppression of oncogenic activation. J. Biol. Chem. 2004;279:29565–29571. doi: 10.1074/jbc.M403954200. [DOI] [PubMed] [Google Scholar]
- 23.Kermorgant S., Parker P.J. c-Met signalling: spatio-temporal decisions. Cell Cycle. 2005;4:352–355. doi: 10.4161/cc.4.3.1519. [DOI] [PubMed] [Google Scholar]
- 24.Lefebvre J., Ancot F., Leroy C., Muharram G., Lemière A., Tulasne D. Met degradation: more than one stone to shoot a receptor down. FASEB J. 2012;26:1387–1399. doi: 10.1096/fj.11-197723. [DOI] [PubMed] [Google Scholar]
- 25.Lee J.H., Gao C.F., Lee C.C., Kim M.D., Vande Woude G.F. An alternatively spliced form of Met receptor is tumorigenic. Exp. Mol. Med. 2006;38:565–573. doi: 10.1038/emm.2006.66. [DOI] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seo J.S., Ju Y.S., Lee W.C., Shin J.Y., Lee J.K., Bleazard T., Lee J., Jung Y.J., Kim J.O., Shin J.Y. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22:2109–2119. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frampton G.M., Ali S.M., Rosenzweig M., Chmielecki J., Lu X., Bauer T.M., Akimov M., Bufill J.A., Lee C., Jentz D. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5:850–859. doi: 10.1158/2159-8290.CD-15-0285. [DOI] [PubMed] [Google Scholar]
- 29.Asaoka Y., Tada M., Ikenoue T., Seto M., Imai M., Miyabayashi K., Yamamoto K., Yamamoto S., Kudo Y., Mohri D. Gastric cancer cell line Hs746T harbors a splice site mutation of c-Met causing juxtamembrane domain deletion. Biochem. Biophys. Res. Commun. 2010;394:1042–1046. doi: 10.1016/j.bbrc.2010.03.120. [DOI] [PubMed] [Google Scholar]
- 30.Lee C.C., Yamada K.M. Identification of a novel type of alternative splicing of a tyrosine kinase receptor. Juxtamembrane deletion of the c-met protein kinase C serine phosphorylation regulatory site. J. Biol. Chem. 1994;269:19457–19461. [PubMed] [Google Scholar]
- 31.Lee C.C., Yamada K.M. Alternatively spliced juxtamembrane domain of a tyrosine kinase receptor is a multifunctional regulatory site. Deletion alters cellular tyrosine phosphorylation pattern and facilitates binding of phosphatidylinositol-3-OH kinase to the hepatocyte growth factor receptor. J. Biol. Chem. 1995;270:507–510. doi: 10.1074/jbc.270.2.507. [DOI] [PubMed] [Google Scholar]
- 32.Grano M., Galimi F., Zambonin G., Colucci S., Cottone E., Zallone A.Z., Comoglio P.M. Hepatocyte growth factor is a coupling factor for osteoclasts and osteoblasts in vitro. Proc. Natl. Acad. Sci. USA. 1996;93:7644–7648. doi: 10.1073/pnas.93.15.7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sudo H., Kodama H.A., Amagai Y., Yamamoto S., Kasai S. In vitro differentiation and calcification in a new clonal osteogenic cell line derived from newborn mouse calvaria. J. Cell Biol. 1983;96:191–198. doi: 10.1083/jcb.96.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peschard P., Fournier T.M., Lamorte L., Naujokas M.A., Band H., Langdon W.Y., Park M. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol. Cell. 2001;8:995–1004. doi: 10.1016/s1097-2765(01)00378-1. [DOI] [PubMed] [Google Scholar]
- 35.Joffre C., Barrow R., Ménard L., Calleja V., Hart I.R., Kermorgant S. A direct role for Met endocytosis in tumorigenesis. Nat. Cell Biol. 2011;13:827–837. doi: 10.1038/ncb2257. [DOI] [PubMed] [Google Scholar]
- 36.Bladt F., Riethmacher D., Isenmann S., Aguzzi A., Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature. 1995;376:768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- 37.Haines L., Neyt C., Gautier P., Keenan D.G., Bryson-Richardson R.J., Hollway G.E., Cole N.J., Currie P.D. Met and Hgf signaling controls hypaxial muscle and lateral line development in the zebrafish. Development. 2004;131:4857–4869. doi: 10.1242/dev.01374. [DOI] [PubMed] [Google Scholar]
- 38.Jeffers M., Fiscella M., Webb C.P., Anver M., Koochekpour S., Vande Woude G.F. The mutationally activated Met receptor mediates motility and metastasis. Proc. Natl. Acad. Sci. USA. 1998;95:14417–14422. doi: 10.1073/pnas.95.24.14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang T.J., Reid A.E., Xavier R., Cardiff R.D., Wang T.C. Transgenic expression of tpr-met oncogene leads to development of mammary hyperplasia and tumors. J. Clin. Invest. 1996;97:2872–2877. doi: 10.1172/JCI118744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J.H., Han S.U., Cho H., Jennings B., Gerrard B., Dean M., Schmidt L., Zbar B., Vande Woude G.F. A novel germ line juxtamembrane Met mutation in human gastric cancer. Oncogene. 2000;19:4947–4953. doi: 10.1038/sj.onc.1203874. [DOI] [PubMed] [Google Scholar]
- 41.Ma P.C., Jagadeeswaran R., Jagadeesh S., Tretiakova M.S., Nallasura V., Fox E.A., Hansen M., Schaefer E., Naoki K., Lader A. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res. 2005;65:1479–1488. doi: 10.1158/0008-5472.CAN-04-2650. [DOI] [PubMed] [Google Scholar]
- 42.Ma P.C., Kijima T., Maulik G., Fox E.A., Sattler M., Griffin J.D., Johnson B.E., Salgia R. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res. 2003;63:6272–6281. [PubMed] [Google Scholar]
- 43.Birchmeier C., Gherardi E. Developmental roles of HGF/SF and its receptor, the c-Met tyrosine kinase. Trends Cell Biol. 1998;8:404–410. doi: 10.1016/s0962-8924(98)01359-2. [DOI] [PubMed] [Google Scholar]
- 44.Ponzetto C., Panté G., Prunotto’ C., Ieraci A., Maina F. Met signaling mutants as tools for developmental studies. Int. J. Dev. Biol. 2000;44:645–653. [PubMed] [Google Scholar]
- 45.Schmidt L., Duh F.M., Chen F., Kishida T., Glenn G., Choyke P., Scherer S.W., Zhuang Z., Lubensky I., Dean M. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat. Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 46.Sant D.W., Margraf R.L., Stevenson D.A., Grossmann A.H., Viskochil D.H., Hanson H., Everitt M.D., Rios J.J., Elefteriou F., Hennessey T., Mao R. Evaluation of somatic mutations in tibial pseudarthrosis samples in neurofibromatosis type 1. J. Med. Genet. 2015;52:256–261. doi: 10.1136/jmedgenet-2014-102815. [DOI] [PubMed] [Google Scholar]
- 47.Lee S.M., Choi I.H., Lee D.Y., Lee H.R., Park M.S., Yoo W.J., Chung C.Y., Cho T.J. Is double inactivation of the Nf1 gene responsible for the development of congenital pseudarthrosis of the tibia associated with NF1? J. Orthop. Res. 2012;30:1535–1540. doi: 10.1002/jor.22121. [DOI] [PubMed] [Google Scholar]
- 48.Stevenson D.A., Zhou H., Ashrafi S., Messiaen L.M., Carey J.C., D’Astous J.L., Santora S.D., Viskochil D.H. Double inactivation of NF1 in tibial pseudarthrosis. Am. J. Hum. Genet. 2006;79:143–148. doi: 10.1086/504441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pennacchietti S., Michieli P., Galluzzo M., Mazzone M., Giordano S., Comoglio P.M. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell. 2003;3:347–361. doi: 10.1016/s1535-6108(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 50.Hermanns-Sachweh B., Senderek J., Alfer J., Klosterhalfen B., Büttner R., Füzesi L., Weber M. Vascular changes in the periosteum of congenital pseudarthrosis of the tibia. Pathol. Res. Pract. 2005;201:305–312. doi: 10.1016/j.prp.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 51.Zhang X., Xie C., Lin A.S., Ito H., Awad H., Lieberman J.R., Rubery P.T., Schwarz E.M., O’Keefe R.J., Guldberg R.E. Periosteal progenitor cell fate in segmental cortical bone graft transplantations: implications for functional tissue engineering. J. Bone Miner. Res. 2005;20:2124–2137. doi: 10.1359/JBMR.050806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seeman E. Periosteal bone formation--a neglected determinant of bone strength. N. Engl. J. Med. 2003;349:320–323. doi: 10.1056/NEJMp038101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.