Abstract
Circumferential skin creases Kunze type (CSC-KT) is a specific congenital entity with an unknown genetic cause. The disease phenotype comprises characteristic circumferential skin creases accompanied by intellectual disability, a cleft palate, short stature, and dysmorphic features. Here, we report that mutations in either MAPRE2 or TUBB underlie the genetic origin of this syndrome. MAPRE2 encodes a member of the microtubule end-binding family of proteins that bind to the guanosine triphosphate cap at growing microtubule plus ends, and TUBB encodes a β-tubulin isotype that is expressed abundantly in the developing brain. Functional analyses of the TUBB mutants show multiple defects in the chaperone-dependent tubulin heterodimer folding and assembly pathway that leads to a compromised yield of native heterodimers. The TUBB mutations also have an impact on microtubule dynamics. For MAPRE2, we show that the mutations result in enhanced MAPRE2 binding to microtubules, implying an increased dwell time at microtubule plus ends. Further, in vivo analysis of MAPRE2 mutations in a zebrafish model of craniofacial development shows that the variants most likely perturb the patterning of branchial arches, either through excessive activity (under a recessive paradigm) or through haploinsufficiency (dominant de novo paradigm). Taken together, our data add CSC-KT to the growing list of tubulinopathies and highlight how multiple inheritance paradigms can affect dosage-sensitive biological systems so as to result in the same clinical defect.
Keywords: exome sequencing, Michelin tire baby, circumferential skin creases, tubulinopathy, TUBB, MAPRE2
Introduction
Congenital symmetrical circumferential skin creases are rare disorders, characterized by the folding of excess skin, which leads to ringed creases, mostly of the limbs. This feature was first described in 1969 by Ross, who introduced the unfortunate term “Michelin tire baby.”1 Subsequent reports described variable additional features of the Michelin-tire-baby syndrome (MIM: 156610), such as intellectual disability (ID), facial dysmorphism, and cardiac and genital anomalies.2, 3, 4, 5, 6, 7, 8, 9, 10, 11 Previously, we described two unrelated young individuals with an identical phenotype consisting of circumferential skin creases, cleft palate, facial dysmorphism, growth retardation, and ID and proposed the term “circumferential skin creases Kunze type” (CSC-KT), based on the phenotype’s resemblance to the original cases reported by Kunze and Riehm, to distinguish this specific syndrome from other affected individuals presenting with the same skin phenotype.6, 7 Given the distinctive phenotype, we were able to recruit five additional unrelated individuals presenting with this rare syndrome.
Here, we report that mutations in TUBB or in MAPRE2 underlie this genetic condition. TUBB is one of nine β-tubulin-encoding genes present in the human genome and is expressed widely among mammalian tissues; it has a particularly pronounced abundance in the developing CNS.12 Tubulins constitute the structural units of microtubules, which are essential for a number of cellular processes including intracellular trafficking, chromosome separation, and cell migration.13 MAPRE2 encodes a member of the microtubule end-binding family of proteins that bind to the GTP cap at growing microtubule plus ends and either contribute to the regulation of microtubule dynamics or to microtubule reorganization during cell differentiation.14 We show that mutations in MAPRE2 or TUBB result in either an altered affinity of MAPRE2 for microtubules or defects in the assembly of TUBB into tubulin heterodimers. In addition, in vivo functional studies in zebrafish gave us insight into the pathophysiological effect of the different MAPRE2 mutations during craniofacial development.
Subjects and Methods
Subjects
Through previously published case reports and collaboration, DNA from seven unrelated individuals with CSC-KT had been collected, as well as parental DNA when available. Written informed consent was obtained from all parents on behalf of the affected individual. This study was approved by the KU Leuven ethical board commission.
Clinical details are summarized in Table 1.
Table 1.
Overview of MAPRE2 and TUBB Mutations and Clinical Features Present in Individuals Included in This Study
| M2 | M8 | M9 | M1 | M3 | M11 | M15 | |
|---|---|---|---|---|---|---|---|
| Country of origin | Spain | Tunisia | Belgium | Belgium | Canada | Norway | Turkey |
| Gene (NCBI Genome browser GRCh38) | MAPRE2 (GenBank: NM_014268.3) | TUBB (GenBank: NM_178014.3) | |||||
| Mutation | c.203A>G (p.Asn68Ser) | c.260A>G (p.Tyr87Cys) | c.427C>T (p.Arg143Cys) | c.454C>T (p.Gln152∗) | c.43C>A (p.Gln15Lys) | c.43C>A (p.Gln15Lys) | c.665A>T (p.Tyr222Phe) |
| Inheritance | homozygous, parents are heterozygous carriers | homozygous, parents’ DNA not available | heterozygous, de novo | heterozygous, maternally inherited | de novo | de novo | de novo |
| Parents affected? | father might have had minor folds as a baby | no | no | mother has mild cognitive impairment, similar facial phenotype | no | no | no |
| Parents consanguineous? |
yes |
yes |
no |
no |
no |
no |
yes |
| Clinical Features | |||||||
| Age at assessment | 15 months | 19 years | 8 years and 9 months | 6 years | 15 years | 5 years and 6 months | 18 months |
| Length, at birth | 52.5 cm (0 SD) | 47 cm (−2.3 SD) | 47 cm (−2 SD) | 48 cm (−1.5 SD) | 48 cm (−2 SD) | 53 cm (+0.5 SD) | 48 cm (−1.5 SD) |
| Length, current | 77 cm (-1 SD) | 120 cm (<−3 SD) | 126 cm (−1.5 SD) | 108 cm (−2 SD) | 168 cm (−1 SD) | 103.5 cm (−2.8 SD) | nt |
| OFC, at birth | 36 cm (0 SD) | 32.5 cm (−2.5 SD) | 32.8 cm (−2 SD) | 33 cm (−2 SD) | 31.5 cm (<−2.5 SD) | 36.5 cm (+0.5 SD) | nt |
| OFC, current | 47 cm (-0.5 SD) | 49 cm (<−3 SD) | 52 cm (0 SD) | 49.5 cm (−1 SD) | 51 cm (<−2.5 SD) | 41 cm (<−3 SD) | 43 cm (<−2.5 SD) |
| Cleft palate | cleft palate | cleft palate | no | cleft palate | cleft palate | cleft palate | high palate, no cleft |
| Creases | limbs and neck | limbs; improvement, but visible | limbs; disappeared around age 4 years | limbs; spontaneous improvement | limbs, fingers, neck, penis; improved at follow-up, but visible | limbs and neck; disappeared at age 4 years; still creases on wrists | limbs |
| Other dysmorphisms | flat face, microphthalmia, short palpebral fissures, epicanthal folds, low broad nasal bridge, low-set, small dysplastic ears, hypoplastic scrotum, coronal hypospadias | elongated face, hypertelorism, bilateral epicanthic folds, upslanting palpebral fissures, microphthalmia, strabismus, wide nasal bridge, aberrant teeth, low-set posteriorly rotated ears with overfolded thick helices, short neck, widely spaced nipples, hypospadias, undescended testes, second and third toe syndactyly | flat face, low set anterior hairline, microphthalmia, bilateral epicanthic folds, small downslanting palpebral fissures, ptosis, synophris, broad nasal bridge, dysplastic small, low-set and posteriorly rotated ears with overfolded helices, upturned ear lobes; microstomia with thin upper lip, small chin, short hands, tapering fingers | microphthalmia with small upslanting palpebral fissures, epicanthal folds, broad nasal bridge, flat midface, small mouth, small chin, clinodactyly of fifth fingers | microphthalmia, short palpebral fissures, epicanthal folds, flat supraorbital ridge, low nasal bridge, long philtrum, small mouth, small low-set posteriorly rotated malformed ears with thick overfolded helices, brachycephaly, wide-spaced nipples | short palpebral fissures, blepharophimosis, broad nasal bridge with epicanthal folds, flat face, small mouth, mild asymmetry in face and abdomen, low-set dysmorphic and posteriorly rotated ears, short neck, long fingers | elongated flat face, hypertelorism, upslanting short palpebral fissures, epicanthus, periorbital fullness, long eyelashes, blepharophimosis, broad and depressed nasal bridge, malformed low-set ears, microstomia, down-turned corners of the mouth, wide-spaced nipples, second and third toe syndactyly |
| Intellectual disability | moderate-severe | profound (unable to walk or speak) | mild to normal functioning | mild-moderate | mild | mild-moderate | mild developmental delay, particularly speech delay |
| Brain imaging | mildly dilated lateral ventricles, corpus callosum hypoplasia | hypoplastic vermis, hypoplastic corpus callosum, mild dilatation of ventricles | nt | nt | normal | normal | hypoplasia of corpus callosum, Dandy-Walker malformation |
| Other phenotypic remarks | seizures from age 3 years | deafness, seizures and ureterocele with vesical reflux without impairment of the renal function | – | – | strabismus, narrow ear canals (multiple infections), hearing aids, myopia and hyperopia | infantile hypotonia | – |
Individuals M1 and M2 have been described previously by Wouters et al.7 Re-assessment of these individuals was performed at the ages of 6 and 3 years, respectively. Tinsa et al. has previously reported individual M8.11 He was re-evaluated at the age of 18 years. Individual M3 has been previously reported by Leonard et al. in 2002 and individual M15 by Ulucan et al.4, 9 Abbreviation is as follows: nt, not tested.
Exome Sequencing and Data Analysis
Genomic DNA was extracted from peripheral blood via standard methods. DNA library preparation and exome capturing were performed for two subject-parent trios and two isolated subjects. Samples were sequenced on an Illumina HiSeq2000 platform, and the acquired reads were aligned to the reference human genome (UCSC Genome Browser hg19). Data processing was performed with the Genome Analysis Toolkit, and variants were annotated with Annovar and an in-house-developed web interface called Annotate-it.15, 16, 17 Variants were restricted to rare and novel variants and filtered according to a de novo or recessive hypothesis. Interesting candidate variants were validated with Sanger sequencing.
Sanger Sequencing
PCR amplification and Sanger sequencing of the complete coding regions of MAPRE2 and TUBB, including exon-intron boundaries, and 5′ UTRs was performed. Primers are available on request.
Binding of MAPRE2 Proteins to Microtubules
Wild-type and mutant forms of MAPRE2 were generated by coupled transcription and translation in rabbit reticulocyte lysate (TnT Quick Coupled Transcription/Translation System; Promega) containing 35S-methionine (specific radioactivity, 10 mCi/mMole), according to the manufacturer’s recommendations. The reactions were cleared of particulate material by centrifugation at 200,000× g at 4°C in a Beckman Optima ultracentrifuge. Bovine brain tubulin depolymerized by incubation on ice in 50 mM PIPES buffer (pH 6.8) and centrifugation at 100,000× g to remove aggregated material was added to the cleared translation cocktail, and the mixture was adjusted to approximately physiological ionic strength by addition of NaCl to a final concentration of 0.2 M. Polymerization was induced by the addition of GTP to 1 mM and glycerol (containing 0.2M NaCl) to 40% and incubation at 37°C for 30 min. The reactions were diluted 5-fold with 50 mM PIPES buffer containing 0.2 M NaCl, 1 mM GTP, and 10 μM taxol (the latter was present to stabilize polymerized microtubules) and loaded onto cushions (1.0 ml) of PIPES-buffered 1 M sucrose also containing 0.2 M NaCl and taxol. Microtubules were recovered by centrifugation for 10 min at 200,000 × g; microtubule pellets were analyzed by 8% SDS-PAGE. Gels were stained with Coomassie blue, dried, and subjected to autoradiography.
Folding and Limited Proteolysis Experiments
In the case of TUBB, folding and assembly of wild-type and mutant forms was followed kinetically in coupled transcription and translation reactions as described previously.12 In the case of MAPRE2, coupled transcription and translation reactions (10 μl) driven by plasmids encoding wild-type and mutant forms were done in the presence of 35S-methionine (specific radioactivity, 10 mCi/mMole), under the conditions recommended by the manufacturer (Promega). Reaction products were diluted to 0.1 ml by the addition of buffer (20 mM Tris-HCl [pH 7.5], 0.2 M NaCl, 1 mM DTT) and were centrifuged at 200,000× g for 10 min to remove all particulate material, and the supernatants were applied to a Superdex 200 HR 10/20 gel filtration column (GE Healthcare) equilibrated and run in the same buffer. Aliquots (0.1 ml) of fractions (0.5 ml) emerging from this column were analyzed for their radioactivity content by scintillation counting. Material contained in the major radioactive peak was detected by autoradiography after SDS-PAGE. In experiments to compare the susceptibility of wild-type and mutant forms of MAPRE2 to proteolysis, labeled wild-type and mutant forms generated by coupled transcription and translation as described above were subjected to limited digestion by proteinase K, and the reaction products were analyzed by SDS-PAGE as described previously.18
Neuro-2a Cell Transfection, Immunostaining, and Surface-Area Quantification
Neuro-2a cells were cultured and transfected as described previously.12 Mutant constructs were generated with a site-directed mutagenesis kit (Quickchange Lightning; Agilent Technologies). Immunostaining employing an anti-α-tubulin antibody (ab7291, 1:1,000; Abcam,) and an anti-FLAG antibody (ab1162, 1:1,000; Abcam) was performed as previously described.12 For detection of the primary antibodies Alexa-488 and Alexa-568, labeled secondary antibodies (Molecular Probes) were employed. For the determination of surface areas, cells (n ≥ 18 cells for each condition) were imaged and their area was measured with ImageJ. Statistical analysis (one-way ANOVA with Bonferroni post hoc test for multiple comparisons) was performed with GraphPad Prism. Images were acquired on a Zeiss LSM 710.
Live Cell Imaging and Analysis of EB3 Comets
Neuro-2a cells were seeded in 35 mm glass-bottom culture dishes (MatTek Corporation) that were coated with Poly-L-Lysine. Cell culture medium was exchanged with imaging medium (DMEM with no phenol red [GIBCO Life Sciences], 10% fetal bovine serum [Sigma], and 1% L-Glutamine [Sigma]), and dishes were placed in the microscope stage incubator (37°C, 5% CO2) for 30 min prior to image acquisition. Images were acquired with a spinning disc confocal microscope UltraVIEW VoX (PerkinElmer) mounted on an AXIO Observer Z1 (Zeiss) by scanning bidirectionally at three Z-positions with 0.3 μm spacing every second for 2 min. Recorded stacks were analyzed with ImarisTrack 7.4 (Bitplane) by application of an automatic tracking algorithm. The analyzed region of interest was selected manually. Statistical analyses were done with GraphPad Prism 6, applying a one-way ANOVA and a Bonferroni post hoc test after pooling the data of at least 16 cells from four different imaging days (see Table S1).
Plasmid Preparation and In Vitro Transcription
We obtained a commercial MAPRE2 open reading frame (ORF) clone corresponding to the full-length human transcript GenBank: NM_014268.3 (NCBI Genome browser GRCh38) (IOH5850, Ultimate ORF Clones; Invitrogen). We conducted site-directed mutagenesis to generate mutant constructs according to the manufacturer’s instructions (QuikChange Site-Directed Mutagenesis Kit, Agilent) and sequence confirmed the resulting vectors. Wild-type and mutant ORF constructs were subsequently cloned into the pCS2+ vector, linearized with NotI, and in vitro transcribed with the SP6 mMessage mMachine kit (Ambion).
Zebrafish Embryo Injections
We obtained two splice-blocking antisense morpholino oligonucleotides (MOs) targeting mapre2 exon 2 (e2i2; 5′-GAGCTTCACATACCTGACGACAGCT-3′) and exon 3 (e3i3; 5′-TGATGTCGGCTCACCTTATCAACAT-3′) splice-donor sites, respectively. To test MO efficiency, wild-type (AB background) zebrafish embryos were injected with 1 nl of increasing doses (4 ng, 6 ng, 8 ng) of MO at the one-to-four cell stage (n = 20 embryos per injection batch). At 1 day post-fertilization (dpf), embryos were harvested in Trizol (Invitrogen) and total RNA was extracted according to the manufacturer’s instructions. We synthesized cDNA by using the QuantiTect Reverse Transcription Kit (QIAGEN); we used the resulting cDNA as a template for PCR to monitor the MO effect on mRNA splicing. PCR products were separated by agarose gel electrophoresis, gel-purified, and sequenced directly via Sanger methodology. For craniofacial phenotyping studies, we collected embryos from natural matings of heterozygous -1.4col1a1:egfp transgenic adults (AB) outcrossed with wild-type (AB) adults.19 A 1 nl cocktail of either MO (6 ng e2i2 or 6 ng e3i3) and/or 100 pg capped human MAPRE2 mRNA was injected into embryo batches at the one-to-four cell stage (n = 50–100 embryos per injection batch) and maintained at 28°C in embryo media (0.3 g/L NaCl, 75 mg/L CaSO4, 37.5 mg/L NaHC03, 0.003% methylene blue) and was screened for the transgene at 1 dpf.
Automated Zebrafish Imaging
Larvae were positioned and imaged live with the Vertebrate Automated Screening Technology (VAST; software version 1.2.2.8) platform (Union Biometrica) in a manner similar to previously described methods.20 Larvae from each experimental condition were anesthetized with 0.2 mg/mL Tricaine prior to being loaded into the sample reservoir. Dorsal and lateral image templates of wild-type and morphant larvae were created for each experimental time point (2, 3, and 4 dpf) and compared to each larva in the capillary; images were acquired at a >70% minimum similarity for the pattern-recognition algorithms. All VAST operational mode settings were set to “auto,” including rotational position, high-resolution imaging, output, and bubbles and debris. Once recognized inside the 600 μm borosilicate capillary of the VAST module on the microscope stage (AxioScope A1, Zeiss), the larvae were rotated 180° to capture a ventral image via a 5× fluar objective and fluorescent excitation at 470 nm to detect GFP (Axiocam 503 monochromatic camera, Zen Pro software; Zeiss). After imaging, the larvae were transferred to a collection beaker with fresh embryo media then stored at 28°C until subsequent imaging time points.
Zebrafish Phenotypic Analysis
We assessed craniofacial patterning by either measuring the angle of the ceratohyal cartilage (2, 3, and 4 dpf) or by counting the number of ceratobranchial arch pairs visible at 3 dpf. Pairwise comparisons to determine statistical significance were made via a Student’s t test (ceratohyal measurements) or a χ-squared test (ceratobranchial-arch-pair counts). Experiments were repeated at least twice.
Results
Identification of Mutations in TUBB and MAPRE2
We performed whole-exome sequencing in four unrelated individuals with CSC-KT (Figure 1, Table 1). One heterozygous nonsense mutation (c.454C>T [p.Gln152∗]) and two homozygous missense mutations (c.203A>G [p.Asn68Ser] and c.260A>G [p.Tyr87Cys]) in MAPRE2 (MIM: 605789), encoding a microtubule-associated protein, member 2, from the rp/eb family, were identified in three of the four individuals (Table 1). None of the mutations were present in the 1000 Genomes, Exome Variant Server, or Exome Aggregation Consortium (ExAC) databases. All affected residues were highly conserved and predicted to be deleterious by various in silico methods.21, 22, 23 Sequencing of the coding sequence and intron-exon boundaries of MAPRE2 in three additional individuals with CSC-KT led to the identification of one additional de novo missense mutation, c.427C>T (p.Arg143Cys), in individual M9 (Table 1). Next, we re-analyzed the exome data of individual M3, in whom we failed to identify a MAPRE2 mutation, for mutations in other genes that encode proteins associated with microtubule biology. This revealed a de novo mutation, c.43C>A (p.Glu15Lys), in the β-tubulin-encoding gene TUBB (MIM: 191130). Sequencing of TUBB in the remaining two CSC-KT-affected individuals also identified de novo missense mutations (c.665A>T [p.Tyr222Phe] and c.43C>A [p.Glu15Lys]), one of which was identical to the mutation in individual M3 (Table 1).
Figure 1.
Clinical Features of Affected Individuals with a MAPRE2 or TUBB Mutation
(A and B) Facial phenotype of individual M9 with MAPRE2 p.Arg143Cys substitution.
(C) Individual M9 at the age of 6 years.
(D–F) Individual M11 with a TUBB p.Gln15Lys substitution.
(G and H) Individual M3 with a TUBB p.Gln15Lys substitution and at the age of 15 years.
(I and J) Individual M11 at the age of 5.5 years.
Note the circumferential skin creases on arms and legs, most pronounced at a young age, and the similar facial features including hypertelorism, small palpebral fissures, and low-set ears with overfolded helices and prominent lobes. For a detailed description, see Table 1.
MAPRE2 Mutations Affect Microtubule Binding
The dynamic behavior of microtubules is subject to regulation by several factors, including the local concentration of heterodimers available for incorporation, post-translational modifications, the binding of associated proteins to the microtubule polymer, and transient interactions of the GTP cap with members of a sizable family of proteins termed +TIPs.24, 25, 26 Among these, the EB family of proteins, to which MAPRE2 belongs, is among the best characterized; they bind to microtubule plus ends and act as a link to a network of other +TIPs that regulate interactions of microtubules with a spectrum of cell structures and organelles.27
Unlike MAPRE1 and MAPRE3, MAPRE2 does not promote microtubule growth or suppress catastrophe; rather, MAPRE2 is critical for microtubule reorganization during early stages of apico-basal epithelial differentiation.14 We explored the mechanism of defective function conferred by the mutations we identified. In the case of MAPRE2, we first examined the ability of the mutant proteins to fold to the native state as well as their structural integrity. We found no detectable differences in the behavior of wild-type and mutant proteins when newly translated sequences were analyzed by gel filtration (Figure S1A). Similarly, kinetic analysis of reactions in which these proteins were incubated with the non-specific protease proteinase K revealed no difference between wild-type and mutant proteins in terms of vulnerability to degradation (Figure S1B). We conclude that none of the MAPRE2 mutations significantly compromise the secondary or tertiary structure of the protein.
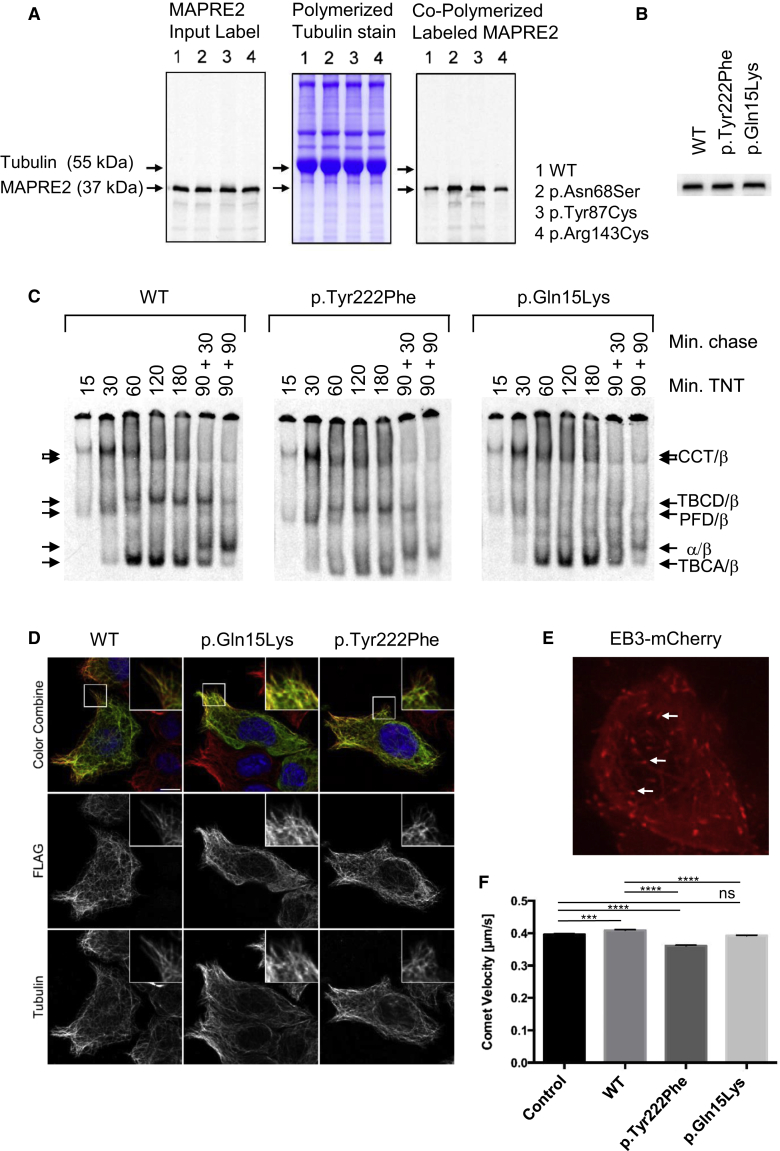
We next considered the possibility that the MAPRE2 mutations might affect microtubule binding, given that all four MAPRE2 mutations we identified reside in the calponin-homology (CH) domain of the protein, previously shown to be responsible for interaction with microtubules.28 To test this hypothesis, 35S-methionine labeled wild-type and mutant MAPRE2 proteins were compared for their ability to co-polymerize with equal aliquots of unfractionated depolymerized bovine brain microtubules. Because the stoichiometric ratio of input labeled MAPRE2 to tubulin is identical in each reaction, this experiment showed that, under physiological conditions of ionic strength, there was significantly enhanced binding of mutant proteins (4-fold in the case of p.Asn68Ser and p.Tyr87Cys and about 2-fold in the case of p.Arg143Cys) to microtubules in comparison to binding of the wild-type control proteins to microtubules (Figure 2A). These data imply an increased dwell time at microtubule plus ends, which could influence the initial MAPRE2-dependent microtubule reorganization that occurs during apico-basal epithelial differentiation.14
Figure 2.
Mutant MAPRE2 Proteins Bind to Microtubules with Enhanced Affinity, Whereas Substitutions in TUBB Compromise Heterodimer Assembly and Microtubule Dynamics
(A) 35S-labeled wild-type and mutant MAPRE2 proteins were mixed with depolymerized native bovine brain tubulin and polymerized, and the resulting microtubules were isolated by sedimentation. (Left) Analysis of equal aliquots of input labeled MAPRE2; note the indistinguishable translational efficiency among all MAPRE2 sequences. (Center) Coomassie stain of SDS-PAGE of pelleted microtubules showing identical recovery of tubulin in each case. (Right) Autoradiograph of the gel shown in the center panel. Upper and lower arrows show the migration positions of tubulin (at 55 kDa) and MAPRE2 (at 37 kDa), respectively.
(B) Analysis by SDS-PAGE of 35S-labeled wild-type and mutant TUBB proteins. Note the indistinguishable translational efficiency among all TUBB sequences.
(C) Kinetic analysis of TUBB heterodimer assembly reactions. Arrows show the migration positions of various intermediate species in the assembly pathway, each assigned on the basis of their characteristic electrophoretic mobility.18 Note various quantitative differences between the wild-type and mutant reactions, including a relative reduction in the yields of α/β heterodimer.
(D) Localization of FLAG-tagged wild-type and TUBB mutants in cultured Neuro-2a cells. Staining with anti-FLAG antibody is shown in green, with DM1alpha antibody for endogenous tubulin in red and Hoechst staining to visualize nuclei in blue. The individual channels for FLAG and DM1alpha staining in gray scale and magnifications of the boxed regions indicated in the color combine image are shown. Note that the wild-type FLAG-tagged TUBB, as well as both mutants, are incorporated into the microtubule lattice. Scale bar, 10 μm.
(E) Still image of a Neuro-2a cell transfected with EB3-mCherry. Note the comets located at the plus tips of growing microtubules (arrows, see also Movies S1, S2, and S3).
(F) Quantification of comet speeds in Neuro-2a cells transfected with EB3-mCherry alone (control), or EB3-mcherry with wild-type or mutant TUBB (p.Tyr222Phe and p.Gln15Lys). n ≥ 16 cells, n ≥ 1651 tracks per condition. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, nsp > 0.05. Error bars show SEM.
Mutations in TUBB Compromise Heterodimer Assembly and Microtubule Dynamics
Mutations that affect the C-terminal domain of TUBB have been previously reported to cause microcephaly with structural brain malformations (MIM: 615771).12 In contrast, the mutations identified in this study affect the N-terminal part of the protein. Like all α- and β-tubulins, newly synthesized TUBB polypeptides cannot form heterodimers without facilitation via interaction with a series of molecular chaperones, including prefoldin (PFD), the cytosolic chaperonin CCT, and five tubulin-specific chaperones termed TBCA-E that function collectively as a GTP-dependent heterodimer-assembly nanomachine.18 To establish the mechanistic basis of defects caused by the newly identified TUBB mutations, we investigated their influence on the chaperone-dependent assembly of the α/β-tubulin heterodimer. To do this, the folding and assembly of wild-type and mutant-bearing TUBB polypeptides were followed kinetically in coupled in vitro transcription and translation reactions and the various intermediates were resolved on non-denaturing polyacrylamide gels.12, 18 For wild-type TUBB, the kinetic analysis showed the characteristic flow of label from the PFD/β- and CCT/β-tubulin binary complexes to TBCA/β-tubulin and TBCD/β-tubulin, respectively, and ultimately to de novo assembled heterodimers (Figure 2C). In the case of the p.Tyr222Phe substitution, however, the yield of TBCA/β was greatly diminished, as was the yield of de novo assembled heterodimers. In the case of the p.Gln15Lys substitution, we found a profound reduction in the formation of the TBCD/β-tubulin intermediate and an even greater reduction (compared to the wild-type control and p.Tyr222Phe) in the yield of de novo assembled heterodimers (Figure 2C). We conclude that both the TUBB mutations we describe here result in defective interactions with the chaperones that participate in de novo heterodimer assembly. However, we found that those heterodimers that did form were capable of co-polymerization into the microtubule cytoskeleton upon expression in cultured Neuro-2a cells and NIH 3T3 fibroblasts (Figure 2D, Figure S2).
To assess the potential effect of the TUBB mutations on microtubule dynamics in vivo, we measured the speed of microtubule plus ends by tracking EB3-mCherry comets (Figure 2E). We first established microtubule co-localization of FLAG-tagged TUBB wild-type and mutant tubulins with EB3-mCherry in a co-transfection experiment in which fixed cells were stained with an anti-FLAG antibody. Co-localization was found in >98% of cases, and a morphological assessment of cells transfected with wild-type and mutant constructs showed no detectable difference (Figure S2). The plus-end tracking experiments showed that overexpression of wild-type TUBB resulted in a small but significant increase of microtubule plus end velocity (p < 0.001, relative to control, Figure 2F), consistent with our previous results.29 This increase in EB3 velocity was not observed in the p.Gln15Lys mutant (p < 0.0001, relative to wild-type, Figure 2F), whereas overexpression of the p.Tyr222Phe mutant significantly decreased microtubule plus end velocity, even below control EB3 levels (p < 0.0001, relative to EB3 control; Figure 2F, Table S1, and Movies S1, S2, and S3). These data suggest that both TUBB mutations compromise microtubule dynamics.
MAPRE2 Is Involved in Craniofacial Patterning in Zebrafish
Microtubules are known to be essential for proper migration of the neural crest into the branchial arches during facial and palatal development. The typical facial features observed in individuals with CSC-KT, including cleft palate and low-set and dysplastic ears, are reminiscent of defective neural-crest cell migration. To investigate this possibility, we coupled a zebrafish model of craniofacial development, in which GFP marks the developing cartilage in live embryos, with newly developed automated in vivo imaging technology.19, 20 Although reciprocal BLAST searches failed to identify a clear ortholog for TUBB (due to the complexity of the β-tubulin gene family), we identified a single reciprocal MAPRE2 ortholog in the D. rerio genome (78% identity, 85% similarity). Transient mapre2 suppression via two non-overlapping, efficient splice-blocking MOs resulted in quantifiable and reproducible defects in early craniofacial patterning (Figure 3, Figure S3). First, we noted aberrant formation of the angle between early bilateral cartilaginous structures at 2 dpf, which persisted to a broadened angle of the ceratohyal at 3 and 4 dpf as indicated by GFP-positive cells in -1.4col1a1:egfp larvae (p < 0.0001 for all comparisons between controls and e2i2 or e3i3 MOs; Figures 3A and 3B). Second, we detected a significant delay in rostro-caudal ceratobranchial (cb) arch patterning, most evident at 3 dpf. Whereas 94% of control larvae displayed at least three cb arch pairs at this stage, morphants showed a significant reduction in these structures: only 8% or 22% of morphant larval batches had at least three equivalent pairs of structures (e2i2 and e3i3 MOs, respectively, p < 0.0001; Figure 3C). Importantly, the observed phenotypic concordance between our MOs for two different readouts spanning three different time points suggested that their effects were specific to mapre2 suppression, and were unlikely to be a result of off-target effects.
Figure 3.
In Vivo Analyses of MAPRE2 Variants Indicate a Role in Craniofacial Patterning and Differing Functional Effects of Recessive versus De Novo Variants
(A) Suppression of mapre2 in zebrafish results in altered craniofacial patterning, including broadening of the ceratohyal (ch) and delay in the formation of the ceratobranchial (cb) arches. Representative ventral views of -1.4col1a1:egfp control and morphant larvae imaged live at 2, 3, and 4 days post-fertilization (dpf). Scale bar, 200 μm.
(B) Measurement of the ch angle indicates abnormal formation of craniofacial structures as early as 2 dpf, which persists until at least 4 dpf. Images were measured as shown in (A) (angle between dashed lines) and ch angle was increased significantly for both MOs in comparison to controls at the three time points assessed. n = 20–48 embryos per batch, repeated twice. Error bars, SEM.
(C) Distribution of cb arch pairs at 3 dpf shows a significant delay for both e2i2 and e3i3 MOs in comparison to controls at the same stage. n = 36–48 larvae per batch, repeated twice.
(D) In vivo complementation assay scoring cb arch pair counts at 3 dpf indicates that the recessive c.203A>G (p.Asn68Ser) is a hypermorph, and the de novo c.427C>T (p.Arg143Cys) change is a hypomorph; (+) indicates an ameliorating effect; (−) indicates an exacerbative effect. n = 29–53 larvae per batch, repeated.
∗p < 0.0001.
Motivated by our in vitro studies that suggested that the MAPRE2 variants induced enhanced end-binding, we used our in vivo craniofacial model to assess the functional consequences of p.Asn68Ser and p.Arg143Cys-encoding variants by using in vivo complementation.30 Focusing on the cb arch formation delay at 3 dpf as the most robust phenotypic readout, we co-injected the e2i2 MO with wild-type human MAPRE2 mRNA. We were able to significantly rescue the cb arch patterning defect, indicating phenotypic specificity (p < 0.0001, wild-type rescue versus e2i2 MO alone; Figure 3D). Next, we compared the efficiency of MO-induced phenotypic rescue between wild-type mRNA and mRNA harboring either of the two missense mutations. The c.427C>T [p.Arg143Cys] mutation, occurring de novo in individual M9, significantly improved the presence of cb arches at 3 dpf, but was still significantly worse than wild-type rescue, suggesting that this variant causes partial loss of function in this assay. In contrast, the recessively inherited c.203A>G [p.Asn68Ser] mutation produced cb arch counts that were significantly improved from the wild-type rescue larval batches (p < 0.0001 for mutant versus wild-type rescue batches; n = 29–53 larvae per batch, repeated with similar results; Figure 3D). These reproducible data suggest that the p.Asn68Ser substitution gives rise to a hyperactive protein, suggesting that this in vivo effect on craniofacial development might be correlated with the four-fold increase in microtubule binding for this change seen in our in vitro assay (Figure 2A). Importantly, expression of mutant mRNAs in the absence of e2i2 MO did not result in any appreciable craniofacial phenotypes (97% and 84% with three or more cb arch pairs for c.203A>G [p. Asn68Ser] and c.427C>T [p.Arg143Cys], respectively, compared to 90% for wild-type mRNA alone; Figure 3D), arguing against the possibility of these changes having dominant-negative effects.
Tubb Is Expressed in Mouse Skin
Multiple circumferential skin folds of the limbs are rare and should be differentiated from underlying nevus lipomatosis or smooth-muscle hamartoma.7 Clinical follow-up showed that in the majority of affected individuals, skin creases become less pronounced with age (Figure 1). Asymmetric cell division is known to drive the development and differentiation of the skin and the epidermis in particular, with a distinct role for microtubules in spindle orientation and cell polarity.31 It is conceivable that the circumferential skin creases observed are a consequence of altered progenitor output associated with defects in the plane of cell division.32 To investigate this possibility, we explored whether Tubb is expressed in the developing murine skin in 4-day-old mice. We exploited a BAC transgenic mouse model that drives GFP expression under the endogenous Tubb promoter because no specific antibodies are available for this protein. Immunostaining with the m-phase marker (pH 3) revealed co-localization with GFP in the proliferative layers of the epidermis and in the developing hair follicle (Figure S4).
Discussion
Here, we have shown that mutations in either MAPRE2 or TUBB can cause CSC-KT. This syndrome is characterized by genetic heterogeneity but a highly similar and recognizable clinical phenotype. Within our cohort, the two individuals with a homozygous MAPRE2 mutation (M2 and M8) developed a more severe neurological involvement consisting of severe ID and seizures, absent in the two individuals with a heterozygous MAPRE2 mutation (M1 and M9) and the individuals with a de novo TUBB mutation (Table 1). We are reluctant to infer any possible genotype-phenotype correlations because a larger allelic series would be necessary in order to do so.
Our genetic studies highlight two emergent themes in rare genetic disorders. First, the mutations discovered here in TUBB significantly extend the phenotypic spectrum of β-tubulin beyond microcephaly and structural brain malformations.12 The CSC-KT individuals with a TUBB mutation in our study do not show gross brain malformations on imaging. On the contrary, the individuals reported by Breuss et al. carry a more C-terminal TUBB mutation and do not present the distinctive CSC-KT craniofacial and skin phenotype.12 This observation raises the possibility that the mutations discovered here affect other or additional functions of the molecule, thus inducing greater phenotypic pleiotropy.
Second, for MAPRE2, our studies revealed an initial genetic conundrum, wherein the same clinical phenotype can apparently be induced through either a recessive or a de novo presumed paradigm. Our in vivo functional studies potentially resolve this paradox by showing that the de novo events most likely induced haploinsufficiency, whereas the mutations inherited recessively impart increased activity of the protein, presumably requiring a threshold to be reached to trigger a pathological effect. In this regard, we speculate that MAPRE2 exhibits a “Goldilocks effect” whereby, at least for the maturation of the branchial arches, either excessive or insufficient protein can cause mispatterning and, ultimately, the same clinical pathology. Such dosage insufficiency has been reported previously; for example, hyperactive or hypoactive complement factor I (CFI) confers susceptibility to age-related macular degeneration, and deletion or duplication of a variety of copy-number variants can likewise give rise to the same phenotype.33, 34 Further studies will be required to understand whether the mechanism of pathology is the same for hypo- and hyperactive MAPRE2.
In summary, our data add CSC-KT to an expanding compendium of tubulinopathies and highlight the emergent phenomenon in which multiple inheritance paradigms can affect dosage-sensitive biological systems and cause the same clinical defect.
Acknowledgments
We thank the affected individuals and their families for their participation. We thank Shannon Fisher for the 1.4col1a1:egfp zebrafish line and acknowledge Igor Pediaditakis and Gaëlle Hayot (zebrafish studies) and Christelle Golzio and Mikalai Malinouski (VAST Bioimager) for their technical assistance. This work was supported by a grant from Concerted Research Actions KU Leuven (GOA/12/015) and funding from the Belgian Science Policy Office Interuniversity Attraction Poles program through the project IAP P7/43-BeMGI. H.V.E. is a clinical investigator of FWO-Vlaanderen and acknowledges receipt of a FWO grant (ZKC5737). N.J.C. acknowledges receipt of a grant (R01GM097376) from the NIH. D.A.K. is an EMBO Young Investigator and is supported by FWF grants I914 and P24367. E.K. is supported by the NHGRI/NHLBI grant (U54HG006542) to the Baylor-Hopkins Center for Mendelian Genomics. N.K. is supported by a grant from the NIH-NIDDK (P50DK096415) and is a distinguished George W. Brumley Professor.
Published: December 3, 2015
Footnotes
Supplemental Data include four figures, one table, and three movies and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.10.014.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
ExAC Browser (December 2014), http://exac.broadinstitute.org/
NCBI Gene, http://www.ncbi.nlm.nih.gov/gene
NHLBI Exome Sequencing Project (ESP) Exome Variant Server (December 2014), http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Ross C.M. Generalized folded skin with an underlying lipomatosus nevus: the Michelin Tire baby Arch. Dermatol. 1969;100:320–323. [PubMed] [Google Scholar]
- 2.Cohen M.M., Jr., Gorlin R.J., Clark R., Ewing S.G., Camfield P.R. Multiple circumferential skin folds and other anomalies: a problem in syndrome delineation. Clin. Dysmorphol. 1993;2:39–46. [PubMed] [Google Scholar]
- 3.Elliott A.M., Ludman M., Teebi A.S. New syndrome?: MCA/MR syndrome with multiple circumferential skin creases. Am. J. Med. Genet. 1996;62:23–25. doi: 10.1002/(SICI)1096-8628(19960301)62:1<23::AID-AJMG5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 4.Leonard N.J. A second patient with MCA/MR syndrome with multiple circumferential skin creases. Am. J. Med. Genet. 2002;112:91–94. doi: 10.1002/ajmg.10665. [DOI] [PubMed] [Google Scholar]
- 5.Basel-Vanagaite L., Sprecher E., Gat A., Merlob P., Albin-Kaplanski A., Konen O., Solomon B.D., Muenke M., Grzeschik K.H., Sirota L. New syndrome of congenital circumferential skin folds associated with multiple congenital anomalies. Pediatr. Dermatol. 2012;29:89–95. doi: 10.1111/j.1525-1470.2011.01403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunze J., Riehm H. A new genetic disorder: autosomal-dominant multiple benign ring-shaped skin creases. Eur. J. Pediatr. 1982;138:301–303. doi: 10.1007/BF00442501. [DOI] [PubMed] [Google Scholar]
- 7.Wouters L., Rodriguez Rodriguez C.M., Dapena E.P., Poorten V.V., Devriendt K., Van Esch H. Circumferential skin creases, cleft palate, typical face, intellectual disability and growth delay: “circumferential skin creases Kunze type”. Eur. J. Med. Genet. 2011;54:236–240. doi: 10.1016/j.ejmg.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Schnur R.E., Zackai E.H. Circumferential ringed creases (“Michelin tire babies”) with specific histologic findings and/or karyotype abnormalities: clues to molecular pathogenesis? Am. J. Med. Genet. 1997;69:221. doi: 10.1002/(sici)1096-8628(19970317)69:2<221::aid-ajmg22>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 9.Ulucan H., Koparir E., Koparir A., Karaca E., Emre R., Gezdirici A., Yosunkaya E., Seven M., Ozen M., Yuksel A. Circumferential skin folds and multiple anomalies: confirmation of a distinct autosomal recessive Michelin tire baby syndrome. Clin. Dysmorphol. 2013;22:87–90. doi: 10.1097/MCD.0b013e32835cd5df. [DOI] [PubMed] [Google Scholar]
- 10.Kondoh T., Eguchi J., Hamasaki Y., Doi T., Kinoshita E., Matsumoto T., Abe K., Ohtani Y., Moriuchi H. Hearing impairment, undescended testis, circumferential skin creases, and mental handicap (HITCH) syndrome: a case report. Am. J. Med. Genet. A. 2004;125A:290–292. doi: 10.1002/ajmg.a.20461. [DOI] [PubMed] [Google Scholar]
- 11.Tinsa F., Aissa K., Meddeb M., Bousnina D., Boussetta K., Bousnina S. Multiple congenital anomalies/mental retardation syndrome with multiple circumferential skin creases: a new syndrome? J. Child Neurol. 2009;24:224–227. doi: 10.1177/0883073808322673. [DOI] [PubMed] [Google Scholar]
- 12.Breuss M., Heng J.I.T., Poirier K., Tian G., Jaglin X.H., Qu Z., Braun A., Gstrein T., Ngo L., Haas M. Mutations in the β-tubulin gene TUBB5 cause microcephaly with structural brain abnormalities. Cell Rep. 2012;2:1554–1562. doi: 10.1016/j.celrep.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breuss M., Keays D.A. Microtubules and neurodevelopmental disease: the movers and the makers. Adv. Exp. Med. Biol. 2014;800:75–96. doi: 10.1007/978-94-007-7687-6_5. [DOI] [PubMed] [Google Scholar]
- 14.Goldspink D.A., Gadsby J.R., Bellett G., Keynton J., Tyrrell B.J., Lund E.K., Powell P.P., Thomas P., Mogensen M.M. The microtubule end-binding protein EB2 is a central regulator of microtubule reorganisation in apico-basal epithelial differentiation. J. Cell Sci. 2013;126:4000–4014. doi: 10.1242/jcs.129759. [DOI] [PubMed] [Google Scholar]
- 15.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sifrim A., Van Houdt J.K., Tranchevent L.-C., Nowakowska B., Sakai R., Pavlopoulos G.A., Devriendt K., Vermeesch J.R., Moreau Y., Aerts J. Annotate-it: a Swiss-knife approach to annotation, analysis and interpretation of single nucleotide variation in human disease. Genome Med. 2012;4:73. doi: 10.1186/gm374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian G., Kong X.-P., Jaglin X.H., Chelly J., Keays D., Cowan N.J. A pachygyria-causing alpha-tubulin mutation results in inefficient cycling with CCT and a deficient interaction with TBCB. Mol. Biol. Cell. 2008;19:1152–1161. doi: 10.1091/mbc.E07-09-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kague E., Gallagher M., Burke S., Parsons M., Franz-Odendaal T., Fisher S. Skeletogenic fate of zebrafish cranial and trunk neural crest. PLoS ONE. 2012;7:e47394. doi: 10.1371/journal.pone.0047394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pardo-Martin C., Allalou A., Medina J., Eimon P.M., Wählby C., Fatih Yanik M. High-throughput hyperdimensional vertebrate phenotyping. Nat. Commun. 2013;4:1467. doi: 10.1038/ncomms2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 22.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 24.Cowan N.J., Lewis S.A. Type II chaperonins, prefoldin, and the tubulin-specific chaperones. Adv. Protein Chem. 2001;59:73–104. doi: 10.1016/s0065-3233(01)59003-8. [DOI] [PubMed] [Google Scholar]
- 25.Nogales E., Wang H.-W. Structural intermediates in microtubule assembly and disassembly: how and why? Curr. Opin. Cell Biol. 2006;18:179–184. doi: 10.1016/j.ceb.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Brouhard G.J. Dynamic instability 30 years later: complexities in microtubule growth and catastrophe. Mol. Biol. Cell. 2015;26:1207–1210. doi: 10.1091/mbc.E13-10-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galjart N. Plus-end-tracking proteins and their interactions at microtubule ends. Curr. Biol. 2010;20:R528–R537. doi: 10.1016/j.cub.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 28.Juwana J.P., Henderikx P., Mischo A., Wadle A., Fadle N., Gerlach K., Arends J.W., Hoogenboom H., Pfreundschuh M., Renner C. EB/RP gene family encodes tubulin binding proteins. Int. J. Cancer. 1999;81:275–284. doi: 10.1002/(sici)1097-0215(19990412)81:2<275::aid-ijc18>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 29.Ngo L., Haas M., Qu Z., Li S.S., Zenker J., Teng K.S.L., Gunnersen J.M., Breuss M., Habgood M., Keays D.A., Heng J.I. TUBB5 and its disease-associated mutations influence the terminal differentiation and dendritic spine densities of cerebral cortical neurons. Hum. Mol. Genet. 2014;23:5147–5158. doi: 10.1093/hmg/ddu238. [DOI] [PubMed] [Google Scholar]
- 30.Niederriter A.R., Davis E.E., Golzio C., Oh E.C., Tsai I.C., Katsanis N. In vivo modeling of the morbid human genome using Danio rerio. J. Vis. Exp. 2013;78:e50338. doi: 10.3791/50338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lechler T., Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437:275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams S.E., Beronja S., Pasolli H.A., Fuchs E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 2011;470:353–358. doi: 10.1038/nature09793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van de Ven J.P., Nilsson S.C., Tan P.L., Buitendijk G.H., Ristau T., Mohlin F.C., Nabuurs S.B., Schoenmaker-Koller F.E., Smailhodzic D., Campochiaro P.A. A functional variant in the CFI gene confers a high risk of age-related macular degeneration. Nat. Genet. 2013;45:813–817. doi: 10.1038/ng.2640. [DOI] [PubMed] [Google Scholar]
- 34.Golzio C., Katsanis N. Genetic architecture of reciprocal CNVs. Curr. Opin. Genet. Dev. 2013;23:240–248. doi: 10.1016/j.gde.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.