Abstract
How the biochemical machinery evolved from simple precursors is an open question. Here we show that ribonucleotides and amino acids condense to peptidyl RNAs in the absence of enzymes under conditions established for genetic copying. Untemplated formation of RNA strands that can encode genetic information, formation of peptidyl chains linked to RNA, and formation of the cofactors NAD+, FAD, and ATP all occur under the same conditions. In the peptidyl RNAs, the peptide chains are phosphoramidate-linked to a ribonucleotide. Peptidyl RNAs with long peptide chains were selected from an initial pool when a lipophilic phase simulating the interior of membranes was offered, and free peptides were released upon acidification. Our results show that key molecules of genetics, catalysis, and metabolism can emerge under the same conditions, without a mineral surface, without an enzyme, and without the need for chemical pre-activation.
Keywords: cofactors, genetic code, peptides, RNA, RNA world
Both amino acids and nucleobases have been shown to form from simple precursors under potentially prebiotic conditions.1, 2 Pathways for the abiotic formation of nucleotides have also been described.3 The emergence of the earliest forms of life from inanimate material is likely to have involved a phase in which biopolymers formed from monomers. The ability of the resulting chemical systems to survive then depended on replication.4 Further, during the emergence of the protein–RNA world,5, 6 a molecular process must have existed that linked specific RNA sequences to specific peptide sequences so that one could encode the other and the latter could support the former through catalysis.7 This process may have led to the genetic code.8 But, to the best of our knowledge, no experimental conditions have been described that induce the simultaneous formation of the key molecules of genetics, metabolism, and protein synthesis.
Cells rely heavily on molecules containing nucleoside phosphates.9 So, it is reasonable to focus on nucleotides as reactants when searching for reactions producing life-like chemical systems. The most conspicuous nucleotide in the cell is adenosine monophosphate (AMP). For example, the energy currency of the cell is ATP.10 The central hub of metabolism is acetyl coenzyme A, which also contains AMP. The universal redox cofactors NAD+, NADP+, and FAD all contain adenosine monophosphate.11 Among the second messengers of signal transduction, cAMP is the best established one.12 Further, AMP is one of the principal components of RNA. Encoded protein synthesis relies on RNA, in terms of the ribosomal machinery13 as well as in terms of the adaptor molecules (tRNAs) and for transcripts (mRNAs). Here, tRNA and mRNA both terminate in AMP residues (Figure 1). So, among the ribonucleotides, AMP either has a privileged structure or it had an important role when today’s biochemistry first arose. This begs the question whether such a role can be demonstrated experimentally.
Figure 1.
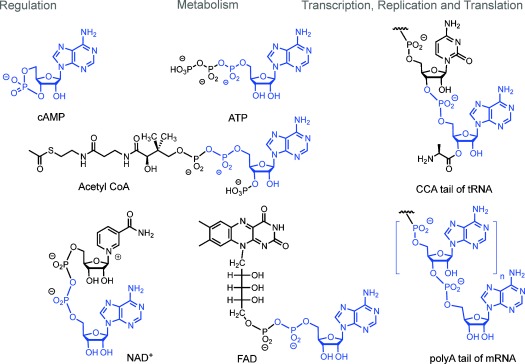
Selected functional biomolecules containing AMP residues (shown in blue).
Oligomerization of peptides and oligonucleotides requires activation to drive endergonic chain-growth reactions. As reported in Ref. 14, we have found that a combination of the water-soluble carbodiimide EDC and alkylated heterocycles, such as 1-ethylimidazole, induces copying processes in RNA via in situ activation of ribonucleotides, without the need for chemical pre-activation. Carbodiimide is a tautomer of cyanamide, a compound formed under presumed prebiotic conditions.15 We noticed that both template-directed chain extension and oligomerization of ribonucleotides occurs under condensation conditions, prompting us to investigate this chemistry more broadly.
In the late 1950s, assays involving pre-activation of amino acids with DCC in organic solvents had been reported to yield peptides when performed in the presence of AMP.16 These findings led to studies on possible origins of the genetic code.8, 17 Cyanamide-fueled reactions of amino acids and ATP also yielded peptides, albeit in modest yield.18 The carbodiimide-induced formation of peptides was assumed to occur via mixed anhydrides (adenylates), but as Ponnamperuma and Banda wrote in 1971: “Attempts at purification of the adenylates only led to their extensive degradation.”17 Lacey et al. wrote in 1992 “so many properties of the ribonucleotides seem to conspire to make peptide synthesis possible and yet we are having trouble forming peptides.”19 A detailed analysis by Pascal and co-workers then concluded that mixed anhydrides are problematic intermediates.20 But, analyses of biochemical pathways suggested species with covalently linked oligopeptides and nucleotides as intermediates in the evolution of today’s translational machinery.21, 22 Since, experimentally, peptidyl RNA remained elusive, it was unclear how even a primitive form of RNA-induced protein synthesis emerged in the absence of translational machinery. Pathways that led to the simultaneous formation of genetic material and cofactors remained unclear as well.
Here we show that peptidyl RNAs form spontaneously when amino acids and ribonucleotides are exposed to a mixture of a condensing agent and a heterocyclic catalyst, that is, conditions inducing genetic copying.14 Much like the intermediates of translation, the peptidyl RNAs are species in which the biomolecules that underlie encoded protein synthesis are covalently linked to one another. Further, under the same condensation conditions, cofactors emerge through pyrophosphate-forming reactions. Adenosine monophosphate is particularly reactive in the processes named above, helping to explain why it is found in so many pivotal biomolecules today.
As mentioned above, signals for oligoadenylates had been observed in copying assays with in situ activation,14 indicating that de novo strand formation was occurring in the absence of mineral surfaces as templates for oligomerization.23–25 All ribonucleotides had shown some reactivity, but only AMP showed full conversion to oligomers after 7 d at 0 °C.14 This led to our current study with mixtures of AMP and other nucleotides. We used solutions of free ribonucleotides in the “general condensation buffer” (0.8 m EDC, 0.15 m 1-ethylimidazole, 0.08 m MgCl2, 0.5 m HEPES, pH 7.5),14 and a combination of ion-exchange HPLC and MALDI MS to analyze products. Figure 2 shows the result of an assay with UMP and AMP. Adenosine residues are more prominent in the product distribution, but the incorporation of uridine residues is very significant. The mass spectra do not reveal sequences, but even for a very simple mixed oligomer, (A5U), six different sequences are possible. This confirms that genetic material can form spontaneously from ribonucleotide mixtures in homogeneous aqueous solution.
Figure 2.
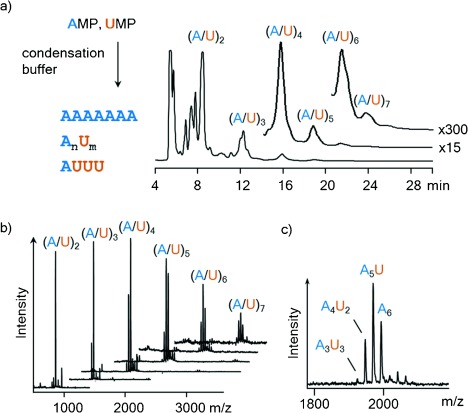
Oligomerization of AMP and UMP gives mixed sequences. a) Reaction scheme and ion-exchange HPLC chromatogram after 30 d at 0 °C, λdet=260 nm. b) Staggered plot of MALDI-TOF mass spectra of HPLC fractions. c) Expansion of peaks of the hexanucleotide fraction, showing the extent of incorporation of UMP. Further details can be found in Figure S1 in the Supporting information.
We then asked whether the condensation conditions we had established for ribonucleotides also induce peptide formation. We first studied individual couplings, producing one new amide bond. When an N-acetylated amino acid was allowed to react with an amino acid ester, a peptide formed within days at 0 °C (see Figures S2 and S3 in the Supporting Information for a representative example). For acetylglycine and phenylalanine ethyl ester, control assays performed in the presence of both AMP and 0.125 m dipalmitoylphosphatidylcholine (DPPC) also showed that neither the nucleotide nor a membrane-forming phospholipid prevent peptide formation.26 When mass spectra of the peptide-forming assays were analyzed, concomitant formation of a phosphoramidate of the phenylalanine ester was observed (Figure S4).
We then studied mixtures of amino acid building blocks, all four ribonucleotides, and immobilized primer–template complexes14 in one reaction volume. The presence of the other ribonucleotides improved the solubility of GMP in the buffer. Also, neither of the components prevented chain growth in the other reaction channels of this complex reaction mixture. Organic extraction showed formation of the dipeptide. Chromatographic analysis of the aqueous phase showed RNA strands containing all four different ribonucleotides (see Figure 3 and Figure S5). Further, despite the competition for the condensing reagent, a sequence-selective extension of the primer occurred in the same mixture (Figure 3), with a rate only modestly slower than those measured in the absence of the other components.14
Figure 3.
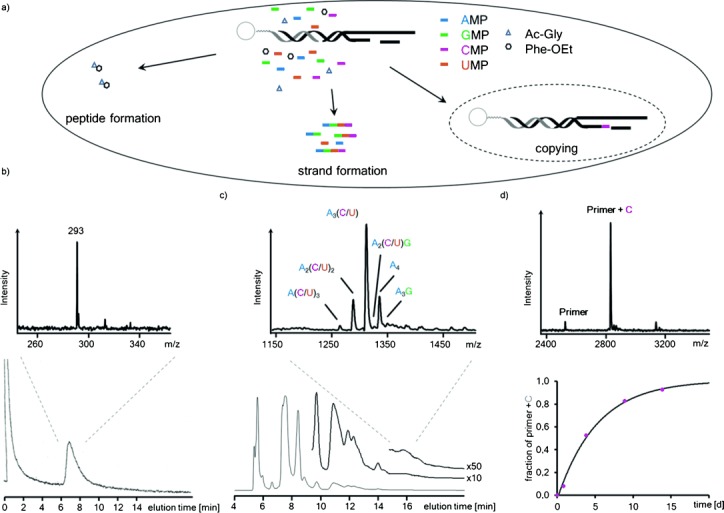
Simultaneous formation of a peptide, oligoribonucleotides, and a sequence-specifically extended primer in the same reaction volume. a) Reaction mixture and processes. b) Analysis of an organic extract containing dipeptide after 5 d, with HPLC trace at λdet 260 nm and MALDI mass spectrum. c) Newly formed RNA strands, as determined by ion-exchange HPLC after 5 d, together with a mass spectrum of one chromatographic fraction. d) Sequence-specific extension of primer by CMP, as detected by MALDI mass spectrometry after release from beads; bottom, right-hand side: reaction kinetics of the copying on the immobilized template.14 Conditions: 0.15 m riboncleotides (each) and 0.125 m amino acid building block (each), condensation buffer, 0 °C.
Next, we studied a mixture of AMP and a free amino acid in condensation buffer at 0 °C. We chose glycine, the most abundant amino acid in the product mixture of Miller.1, 27 An unexpected combination of oligomerization reactions occurred. Figure 4 a shows a chromatogram of the reaction mixture after 10 d. At this time point, the most prominent peak was that of oligoglycines linked to an AMP residue. Late-eluting peaks gave mass spectra of oligoglycyl oligoribonucleotides. Among them, masses for chains of seven amino acid residues and three adenylates were readily discernible. Control experiments with glycine in condensation buffer without AMP showed incomplete conversion of glycine and peaks for side products, as monitored by 13C NMR spectroscopy (Figure S6). So, AMP favors the efficient formation of peptidyl chains. Mechanistically, engaging the amino terminus may suppress cyclizations and other side reactions and may modulate the reactivity of the carboxy group via inductive effects. The phosphoramidate link to the nucleotide may also offer reaction channels via cyclic intermediates similar to those described for N-acyl amino acids.28
Figure 4.
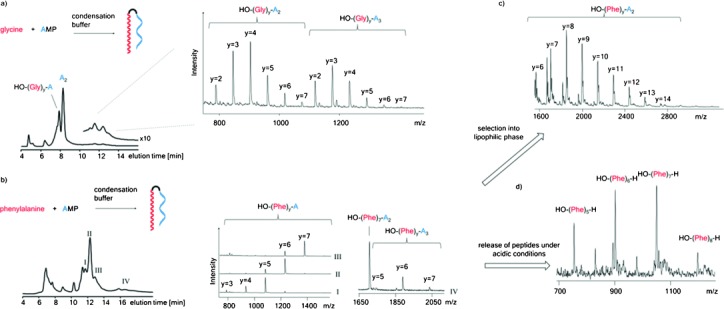
Spontaneous condensation of amino acids and AMP produces peptidyl RNAs. a) Condensation of glycine and AMP: reaction scheme, IE-HPLC trace (260 nm) after 10 d at 0 °C and MALDI MS of fractions with peptidyl RNAs. b) Reaction scheme for the condensation of phenylalanine and AMP, chromatogram of precipitated products after 21 d at 0 °C and mass spectra of selected fractions. c) Selection of long-chain peptidyl RNAs upon exposure to n-octanol: mass spectrum of peptidyl RNAs with up to 14 phenylalanine residues linked to dinucleotide A2. d) Release of peptides from peptidyl RNAs: mass spectrum (positive mode) of a sample of the product mixture exposed to aqueous acetic acid (4:1) for 5 d at 20 °C, followed by exposure to n-octanol.
Because in peptidyl RNA the peptide chain is covalently linked to a nucleic acid, a stable, “selectable” complex results, in which RNA and peptide cannot be separated by diffusion. When we allowed phenylalanine and AMP to oligomerize, oligophenylate RNAs formed (Figure 4 b) and slowly precipitated. Extraction of the product mixture with n-octanol to simulate the lipophilic part of a membrane bilayer gave peptidyl RNAs with long peptide chains (Figure 4 c). These chains begin to approach the lengths required for folding into a defined three-dimensional structure. The combination of long phenylalanines and adenosine-containing nucleic acids is also interesting because these are lipophilic nucleotides and very lipophilic amino acid residues. Such combinations are expected to emerge based on physico-chemical relationships found in the genetic code.19 Anticodonic relationships could be the result of attractive interactions between anticodons and the residues they code for.
When the original product mixture of the condensation of Phe and AMP was exposed to aqueous acetic acid, free peptides were found (Figure 4 d). So, much like in present-day translation, where tRNAs with the nascent peptide chain are intermediates, the peptidyl RNAs can release peptides, in this case upon a spontaneous drop in pH. When a mixture containing glycine, phenylalanine, and AMP was exposed to condensation conditions, a distribution of mixed-sequence peptidyl RNAs with species containing up to seven amino acids and up to at least five adenosine residues formed (Figure S7). When the oligomerization mixture contained UMP, AMP, phenylalanine, and glycine, mixed-sequence peptidyl RNAs with up to four amino acids and up to five nucleotide residues were readily detected (Figure S8). All four building blocks were represented in the products, suggesting that the condensation to peptidyl RNAs creates diverse sets of molecular species. This is advantageous for Darwinian evolution.29
We then asked whether the peptidyl RNAs detected are amino acid adenylates.16 Structural analysis by one- and two-dimensional NMR spectroscopy showed that this is not the case. First, control assays with AMP alone were analyzed by 31P NMR spectroscopy. In the absence of amines, a peak emerged in the chemical shift range typical for a pyrophosphate, together with a series of smaller peaks in the range typical for oligomers with phosphodiester linkages, as expected for oligomerization. In the presence of an amino acid, peaks in the ppm range typical for phosphoramidates appeared, indicating that the peptidyl RNAs formed are N-linked. In contrast, mixed anhydrides give 31P signals shifted approximately 10 ppm to higher field.30 The structure of the phosphoramidate-linked glycyl RNAs was confirmed by 31P,1H HMBC NMR spectroscopy (Figure S9). We note that phosphoramidates are known to be alternative substrates for present-day viral polymerases,31, 32 adding to the interconnectivity of this chemistry.
We then probed whether the condensation processes observed critically depend on an organic carbodiimide as the activating agent. For this, the condensation of AMP and glycine was initiated by an organic carbodiimide, the “inorganic” activating agent cyanamide, or the oxyazabenzotriazolide of AMP as the only activating species. Product distributions were determined by 31P NMR analysis (Figure 5 and Figure S10). The rate of condensation was slower with cyanamide, so that reaction temperatures of 20 °C or 50 °C were used, but all three activation modes gave a mixture of phosphoramidates and pyrophosphate, with small peaks in the phosphodiester range. This confirmed that all three reactions in Figure 5 a occur, independent of what condensation agent induces the formation of the reactive species.
Figure 5.
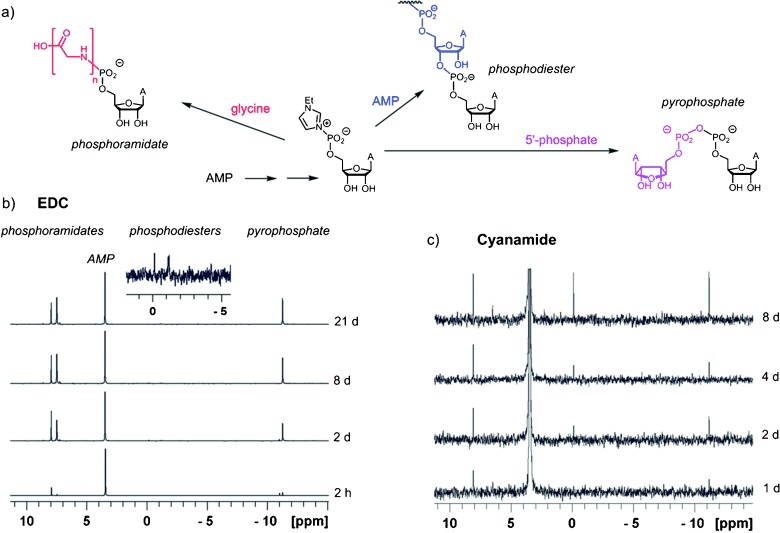
The reactivity pattern of AMP in the presence of glycine is similar for two different activating agents, as monitored by 31P NMR spectroscopy (125 MHz). a) Reaction scheme and mechanistic proposal. b) Spectra from an assay with EDC as the condensing agent. c) Spectra from an assay with cyanamide as the activating agent. Reaction times are given on the right of spectra. With EDC, reactivity is greater than with cyanamide, so that monoglycyl (8 ppm) and oligoglycyl species (7.5 ppm) form. But in either case, peaks for phosphoramidates phosphodiesters and pyrophosphate appear simultaneously. Conditions: condensation buffer in H2O/D2O (4:1) with 0.3 m AMP and 0.3 m glycine at 0 °C or 50 °C for (c). The expansion in (b) shows signals in the phosphodiester region.
Mechanistically, after the initial activation, for example, via formation of a carbodiimide adduct, it is probably the organocatalyst (ethylimidazole in the current case) that gives the highly reactive species, which then reacts with amines, phosphates, or alcohols to give phosphoramidates, pyrophosphates, or phosphodiesters, respectively. Phosphodiester formation appears to be the slowest of these processes, so that improvements in the rate and fidelity of RNA replication can lead to the survival of self-replicating systems.29
The condensation reactions were accompanied by pyrophosphate formation. Several key coenzymes of today’s primary metabolism contain pyrophosphate-linked AMP residues. When a mixture of AMP and nicotinamide mononucleotide (NMN+) was allowed to react in the condensation buffer at 0 °C, the formation of NAD+ was observed, together with that of symmetrical pyrophosphates (Figure 6 and Figures S11 and S12, Supporting Information). A mixture of flavin mononucleotide (FMN) and AMP gave FAD with partial suppression of the usual cyclization of FMN.33 Further, AMP and the tetrasodium salt of inorganic pyrophosphate gave significant amounts of ATP, demonstrating that the reaction conditions inducing copying and oligomerization also lead to the spontaneous formation of pivotal compounds of primary metabolism.
Figure 6.
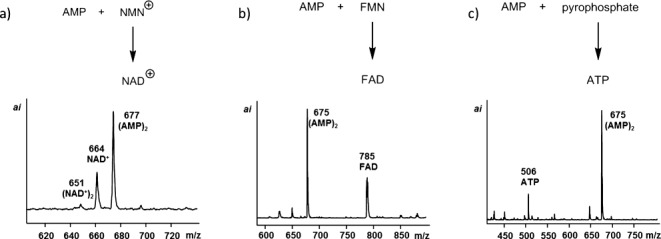
Formation of pyrophosphate-linked cofactors and ATP in condensation buffer at 0 °C, as detected by MALDI MS. a) Solution of AMP and NMN+ (0.15 m each) after 4 d; spectrum in positive mode. b) AMP and FMN after 4 d. c) AMP and sodium pyrophosphate (0.15 m) after 20 d.
In summary, assays under “general condensation conditions” reveal that AMP has favorable properties for the emergence of functional biomolecules. Of the four ribonucleotides, AMP oligomerizes most readily in the absence of mineral surfaces14 probably because it possesses proper self-assembly properties. The fast oligomerization of AMP is in contrast to the slow copying of A-rich sequences,34, 35 probably resulting in a kinetic compensation during replication. Secondly, AMP and other phosphates form cofactors under condensation conditions, thus facilitating the emergence of a simple metabolism. Thirdly, AMP forms peptidyl RNAs without a cellular machinery for protein synthesis. All processes occur under conditions and at concentrations typical for eutectic ice phases.36
Peptidyl substituents at the 5′-terminus of oligonucleotides can increase the stability of duplexes with RNA.37 Duplex RNA degrades more slowly than single-stranded RNA. Strands complementary to the RNA portion of the peptidyl RNA may have stabilized it against degradation, and may have eventually become precursors of today’s mRNAs. Further, lipophilic peptidyl chains may have anchored peptidyl RNAs on a membrane, retaining it in a protocell.38 Further, specific RNA sequences of peptidyl RNAs may have templated peptide growth or vice versa, and intramolecular interactions during chain growth (that are entropically favored over intermolecular interactions) may have led to the emergence of a first genetic code.19 If so, our findings support Eigen’s proposal of primitive “tRNAs” as the earliest components of the translational machinery.39 We conclude that ribonucleotide-dependent condensation reactions produce more functional biomolecules than previously thought.
Experimental Section
Representative condensation assay: An aqueous solution of HEPES (0.5 m), MgCl2 (0.08 m), 1-ethylimidazole (0.15 m), and the appropriate amount of one or more of the substrates [NMP (0.02–0.15 m), amino acid (0.1–0.3 m), and/or other phosphate (0.15 m)] was adjusted to pH 7.5 with NaOH solution. An aliquot (65 μL) of this solution was taken to dissolve EDC hydrochloride (10 mg, 52 μmol) to give an EDC concentration of approximately 0.8 m. For primer extension with immobilized strands, 5 μL of the solution was added to 25 μg of magnetic beads, loaded with the primer–template complex14 and then used to dissolve the downstream-binding strand (5 nmol). The mixture was allowed to react at 0 °C, with in situ monitoring by NMR spectroscopy or sampling and analysis by HPLC and MS. See the Supporting Information for protocols and additional data.
Acknowledgments
This work was supported by the DFG (grant no. RI 1063/8-2) and EU COST action CM1304. We thank Marilyne Sosson for sharing results, T. Sabirov for skilled technical assistance, Dr. Birgit Claasen for recording NMR spectra, and Dr. Eric Kervio and Svenja Kaspari for discussions.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
miscellaneous_information
References
- 1.Miller SL. Science. 1953;117:528–529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- 2.Oró J. Biochem. Biophys. Res. Commun. 1960;2:407–412. doi: 10.1016/j.bbrc.2011.07.075. [DOI] [PubMed] [Google Scholar]
- 3.Powner MW, Gerland B, Sutherland JD. Nature. 2009;459:239–242. doi: 10.1038/nature08013. [DOI] [PubMed] [Google Scholar]
- 4.Eigen M, Schuster P. Naturwissenschaften. 1977;64:541–565. doi: 10.1007/BF00450633. [DOI] [PubMed] [Google Scholar]
- 5.da Silva JAL. J. Theor. Biol. 2015;370:197–201. doi: 10.1016/j.jtbi.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Strazewski P. Isr. J. Chem. 2015;55:851–864. [Google Scholar]
- 7.Sutherland JD, Blackburn JM. Chem. Biol. 1997;4:481–488. doi: 10.1016/s1074-5521(97)90318-5. [DOI] [PubMed] [Google Scholar]
- 8.Crick FH. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 9.Westheimer F. Science. 1987;235:1173–1178. doi: 10.1126/science.2434996. [DOI] [PubMed] [Google Scholar]
- 10.Lipmann F. Werkman C. In: Advances in Enzymology and Related Areas of Molecular Biology. Nord FF, editor; Hoboken: Wiley; 1941. pp. 99–162. [Google Scholar]
- 11.Benner SA, Ellington AD, Tauer A. Proc. Natl. Acad. Sci. USA. 1989;86:7054–7058. doi: 10.1073/pnas.86.18.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beavo JA, Brunton LL. Nat. Rev. Mol. Cell Biol. 2002;3:710–718. doi: 10.1038/nrm911. [DOI] [PubMed] [Google Scholar]
- 13.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 14.Jauker M. Griesser H. Richert C. Angew. Chem. Int. Ed. 2015 Angew. Chem2015DOI: 10.1002/anie.201506592 , DOI: 10.1002/ange.201506592. [Google Scholar]
- 15.Schimpl A, Lemmon RM, Calvin M. Science. 1965;147:149–150. doi: 10.1126/science.147.3654.149. [DOI] [PubMed] [Google Scholar]
- 16.Berg P. J. Biol. Chem. 1958;233:608–611. [PubMed] [Google Scholar]
- 17.Banda PW, Ponnamperuma C. Space Life Sci. 1971;3:54–62. doi: 10.1007/BF00924215. [DOI] [PubMed] [Google Scholar]
- 18.Nooner DW, Sherwood E, More MA, Oró J. J. Mol. Evol. 1977;10:211–220. doi: 10.1007/BF01764596. [DOI] [PubMed] [Google Scholar]
- 19.Lacey JC, Wickramasinghe NSMD, Cook GW. Origins Life Evol. Biospheres. 1992;22:243–275. doi: 10.1007/BF01810856. [DOI] [PubMed] [Google Scholar]
- 20.Pascal R, Boiteau L, Commeyras A. In: Topics in Current Chemistry. Walde P, editor. Berlin/Heidelberg: Springer; 2005. [Google Scholar]
- 21.Di Giulio M. J. Mol. Evol. 1997;45:571–578. doi: 10.1007/pl00006261. [DOI] [PubMed] [Google Scholar]
- 22.Cavalier-Smith T. J. Mol. Evol. 2001;53:555–595. doi: 10.1007/s002390010245. [DOI] [PubMed] [Google Scholar]
- 23.Ferris JP, Ertem G, Agarwal V. Origins Life Evol. Biospheres. 1989;19:165–178. doi: 10.1007/BF01808150. [DOI] [PubMed] [Google Scholar]
- 24.Ferris JP, Hill AR, Liu RH, Orgel LE. Nature. 1996;381:59–61. doi: 10.1038/381059a0. [DOI] [PubMed] [Google Scholar]
- 25.Cafferty BJ, Hud NV. Curr. Opin. Chem. Biol. 2014;22:146–157. doi: 10.1016/j.cbpa.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 26.See also. Strazewski P. Colloids Surf. B. 2009;74:419–425. doi: 10.1016/j.colsurfb.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Johnson AP, Cleaves HJ, Dworkin JP, Glavin DP, Lazcano A, Bada JL. Science. 2008;322:404. doi: 10.1126/science.1161527. [DOI] [PubMed] [Google Scholar]
- 28.Danger G, Michaut A, Bucchi M, Boiteau L, Canal J, Plasson R, Pascal R. Angew. Chem. Int. Ed. 2013;52:611–614. doi: 10.1002/anie.201207730. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013;125 [Google Scholar]
- 29.de Duve C. Nature. 2005;433:581–582. doi: 10.1038/433581a. [DOI] [PubMed] [Google Scholar]
- 30.Moriguchi T, Yanagi T, Wada T, Sekine M. J. Chem. Soc. Perkin Trans. 1. 1999:1859–1866. [Google Scholar]
- 31.Adelfinskaya O, Terrazas M, Froeyen M, Marlière P, Nauwelaerts K, Herdewijn P. Nucleic Acids Res. 2007;35:5060–5072. doi: 10.1093/nar/gkm498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terrazas M, Marlière P, Herdewijn P. Chem. Biodiversity. 2008;5:31–39. doi: 10.1002/cbdv.200890013. [DOI] [PubMed] [Google Scholar]
- 33.Forrest HS, Mason HS, Todd AR. J. Chem. Soc. 1952:2530–2534. [Google Scholar]
- 34.Hill AR, Orgel LE, Wu T. Origins Life Evol. Biospheres. 1993;23:285–290. doi: 10.1007/BF01582078. [DOI] [PubMed] [Google Scholar]
- 35.Kervio E, Hochgesand A, Steiner UE, Richert C. Proc. Natl. Acad. Sci. USA. 2010;107:12074–12079. doi: 10.1073/pnas.0914872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monnard P-A, Kanavarioti A, Deamer DW. J. Am. Chem. Soc. 2003;125:13734–13740. doi: 10.1021/ja036465h. [DOI] [PubMed] [Google Scholar]
- 37.Sarracino DA, Steinberg JA, Vergo MT, Woodworth GF, Tetzlaff CN, Richert C. Bioorg. Med. Chem. Lett. 1998;8:2511–2516. doi: 10.1016/s0960-894x(98)00449-1. [DOI] [PubMed] [Google Scholar]
- 38.Isaad ALC, Carrara P, Stano P, Krishnakumar KS, Lafont D, Zamboulis A, Buchet R, Bouchu D, Albrieux F, Strazewski P. Org. Biomol. Chem. 2014;12:6363–6373. doi: 10.1039/c4ob00721b. [DOI] [PubMed] [Google Scholar]
- 39.Eigen M, Winkler-Oswatitsch R. Naturwissenschaften. 1981;68:282–292. doi: 10.1007/BF01047470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
miscellaneous_information


