Abstract
Template-directed incorporation of nucleotides at the terminus of a growing complementary strand is the basis of replication. For RNA, this process can occur in the absence of enzymes, if the ribonucleotides are first converted to an active species with a leaving group. Thus far, the activation required a separate chemical step, complicating prebiotically plausible scenarios. Here we show that a combination of a carbodiimide and an organocatalyst induces near-quantitative incorporation of any of the four ribonucleotides. Upon in situ activation, adenosine monophosphate was found to also form oligomers in aqueous solution. So, both de novo strand formation and sequence-specific copying can occur without an artificial synthetic step.
Keywords: genetic copying, nucleotides, oligomerization, replication, RNA
Ribonucleic acid (RNA) is found in all cells. It can encode genetic information, but it is also involved in protein synthesis,1 catalysis,2 and the regulation of gene expression.3 Because RNA can fulfil several pivotal roles in biochemistry, it is possible that life started with a so-called “RNA world”.4 It is therefore important to ask how oligoribonucleotides may form in the absence of enzymes, and how the genetic information they contain may be copied into complementary strands without the catalytic action of a polymerase. Current-day metabolism generates nucleoside triphosphates for replication, transcription, and encoded protein synthesis, but nucleoside triphosphates are largely unreactive in the absence of enzymes.5
The most common way to induce enzyme-free oligomerization of a ribonucleotide is to activate it in a separate chemical reaction, producing a monomer with an organic leaving group, or an anhydronucleotide.6 The product is isolated and then used in a subsequent oligomerization step (Figure 1). Following this protocol, strands have been shown to form in the presence of mineral surfaces7 or when exposed to elevated temperatures and/or organic solvents.8, 9 Heterogeneous media favor the incorporation of all four nucleotides,10, 11 and long polymers were found in eutectic phases.11 Pre-activated nucleotides were also used to demonstrate that copying of a given template sequence into a complementary strand can occur without enzymes, mostly in the form of enzyme-free primer extension. Pre-activated nucleotides typically used for copying include imidazolides,12 methylimidazolides,13 and oxyazabenzotriazolides.14 We recently showed that when the latter react with immobilized template–primer duplexes, near-quantitative incorporation of any of the four nucleotides (A/C/G/U) is found.15
Figure 1.
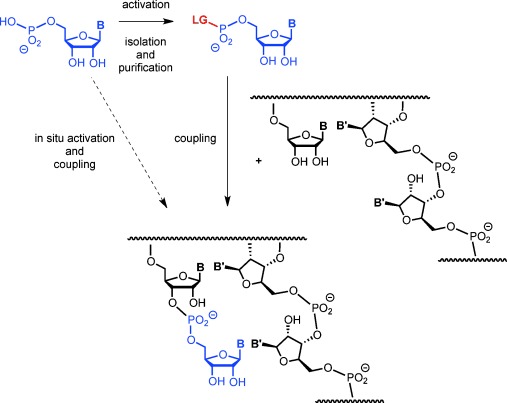
Copying of an RNA sequence via enzyme-free primer extension, with or without pre-activation of the ribonucleotide monomer. LG=Leaving group.
Discontinuous, two-step syntheses require complicated prebiotic scenarios. Conditions that induce activation and chain extension simultaneously make presumed prebiotic processes more likely. It is therefore important to ask whether such conditions exist and what activating chemistry supports them. Uronium salts are known to activate nucleotides16, 17 for subsequent coupling, but they are usually employed in organic solvents, and it is unclear whether they are prebiotically relevant. A combination of a phosphine and pyridyldisulfide has also been used to activate nucleotides,18, 19 but this approach is not suitable for in situ activation. Simple inorganic activation agents like COS have been shown to induce the formation of aminoacylnucleotides,20 but not RNA oligomers. Simple reagents are also problematic because the potential for side reactions in complex reaction mixtures involving highly reactive reagents or elevated temperatures is very high. Complex one-pot reactions often lead to intractable mixtures or tar.21
One class of activation reagents that is of prebiotic relevance is carbodiimides. Carbodiimide is a tautomer of cyanamide, a compound formed under presumed prebiotic conditions.22–24 Ligations between strands terminating in amino groups and phosphates have been induced by carbodiimides, including replication reactions.25–27 It is known that pre-activation can be induced with a conventional condensation agent, such as N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide (EDC) at pH 5.5,28 but no genetic copying occurs under these conditions. Untemplated oligomerization up to tetramers was recently reported in homogeneous solution at pH 6.5, accompanied by massive side reactions,29 but not genetic copying. In the 1960s, template-directed oligomerizations, not genetic copying, had been studied using in situ activation, without an organocatalyst, but the yields were low and the oligomers obtained were too short for duplex formation.30, 31 These results are understandable because free ribonucleotides were shown to act as inhibitors of enzyme-free primer extension.15, 32
Here we show that a combination of a carbodiimide and an N-alkyl heterocycle as catalyst induces efficient copying reactions on RNA templates using unactivated, free ribonucleotides. While additives like free imidazole give fairly unreactive imidazolides, alkylated imidazole as the organocatalyst can give a highly reactive imidazolium species. Both primer extension on preformed RNA strands and the untemplated de novo oligomerization of ribonucleotides to RNA strands were observed. This shows for the first time that genetic copying can occur without the synthetic step of pre-activation of the ribonucleotide. Both strand formation and copying of genetic information occur without the intervention of a synthetic chemist. Further, it was found that the conditions described here unleash other intrinsic reactivities of nucleotides and amino acids, leading to the spontaneous formation of peptidyl RNAs and cofactors, as reported in Ref. 33.
We started by studying primer extension with guanosine 5′-monophosphate (GMP) as the monomer and cytosine as the templating base in the RNA template (Figure 2 a). This is the most favorable case among the four copying reactions, as GMP pairs more strongly than the other three ribonucleotides (AMP, CMP, and UMP).32 Still, copying is demanding, as it requires both intrinsic reactivity and delicate molecular recognition in order to be sequence selective.34 Initially the challenge of achieving activation and coupling in one solution seemed all but insurmountable. The unactivated ribonucleotide was known to act as a competitive inhibitor to primer extension, blocking the extension site.15 Since NMR monitoring indicated that the vast majority of ribonucleotide molecules in solution remain unactivated upon treatment with EDC, reactions were expected to be inefficient. Further, the temperature optimum is different for the two steps, with coupling best performed at low temperatures,14 but activation at room temperature or above. Also, while activation is favored by organic solvents and acidic pH,28 coupling requires aqueous buffer to achieve the template effect and neutral or basic pH to ensure nucleophilic reactivity at the primer terminus.
Figure 2.
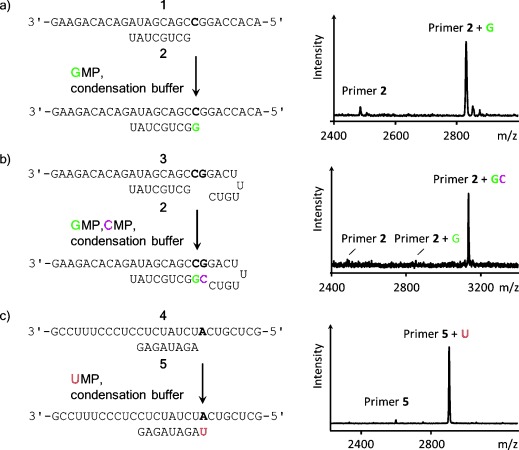
High-yielding incorporation of three of the four ribonucleotides opposite their complementary bases in aqueous condensation buffer with in situ activation. a) Extension of primer by GMP after 2 d at 20 °C; b) consecutive extension of primer by GMP and CMP after 6 d at 20 °C; c) extension of primer by UMP after 5 d at 0 °C. In each case, the reaction scheme is shown next to a MALDI mass spectrum; conditions: 0.8 m EDC, 0.1–0.15 m ethylimidazole, 0.4 m NaCl, 0.2–0.5 m HEPES buffer, and/or 0.08–0.16 m MgCl2. See the Supporting Information for details.
We monitored primer extension with MALDI-TOF mass spectrometry under conditions that allow for quantitative detection.35 Initially, no extension of primer 2 by GMP was detectable, even in high-salt buffer containing 0.8 m EDC, and our experimental work aimed at high-yielding reactions was unsuccessful, in agreement with the low yields reported in the early literature.30, 31 Only when an organocatalyst was used did detectable copying set in.
Adding imidazole or 2-methylimidazole led to 2 % conversion after 24 h. Presumably, imidazolides of the nucleotide, known monomers for copying,29, 36 were formed in the solution, but steady-state levels of these species were not high enough for efficient extension. With 1-methyladenine as catalyst,37 12 % primer extension was observed after 1 d at 20 °C. When 1-ethylimidazole was used, the yield of extended primer was 32 % after 24 h (Table 1). A subsequent screen of pH values gave an optimum of 7.5.
Table 1.
Results of primer extension assays in solution.[a]
| Template, primer[b] | NMP[c] | Cat.[d] | Buffer[e] | pH | T [C°] | t [h] | Exten- sion [%] |
|---|---|---|---|---|---|---|---|
| 1, 2 | G | – | A | 6.7 | 20 | 24 | <1 |
| 1, 2 | G | Im | A | 6.7 | 20 | 24 | 2 |
| 1, 2 | G | 2-MeIm | A | 6.7 | 20 | 24 | 8 |
| 1, 2 | G | MeAde | A | 7.7 | 20 | 24 | 12 |
| 1, 2 | G | 1-EtIm | A | 6.7 | 20 | 24 | 32 |
| 1, 2 | G | 1-EtIm | A | 5.5 | 20 | 24 | 44 |
| 1, 2 | G | 1-EtIm | A | 6.5 | 20 | 24 | 62 |
| 1, 2 | G | 1-EtIm | A | 7.0 | 20 | 24 | 63 |
| 1, 2 | G | 1-EtIm | A | 7.5 | 20 | 24 | 74 |
| 1, 2 | G | 1-EtIm | A | 7.9 | 20 | 24 | 73 |
| 1, 2 | G | 1-EtIm | A | 7.9 | 20 | 48 | 90 |
| 3, 2 | G+C | 1-EtIm | A | 7.5 | 20 | 140 | 95 |
| 4, 5 | U | 1-EtIm | A | 7.5 | 20 | 50 | 75 |
| 4, 5 | U | 1-EtIm | B | 7.5 | 0 | 120 | 92 |
[a] For conditions, see General Protocol 1 in the Supporting Information.
[b] 60 μm Template, 50 μm primer.
[c] 20 mm GMP; or 150 mm UMP; or 50 mm GMP and 50 mm CMP. [d] Catalysts: Im, imidazole; 2-MeIm, 2-methylimidazole; MeAde, 1-methyladenine; 1-EtIm, 1-ethylimidazole (0.1 m of either for Buffer A; 0.15 m 1-ethylimidazole for Buffer B).
[e] Buffer A=0.2 m HEPES, 0.4 m NaCl, 0.16 m MgCl2, 0.8 m EDC; Buffer B=0.5 m HEPES, 0.08 m MgCl2, 0.8 m EDC.
Next we varied the salt conditions. In the absence of a divalent metal ion, no primer extension was observed. In the presence of 80 mm Mn2+, 10 % extended primer was detected after 24 h at pH 8. With 80 mm Ca2+, 25 % conversion was found after 24 h. The fastest reaction was observed with Mg2+, with 70 % extension after 1 d and 90 % conversion after 2 d (mass spectrum in Figure 2 a). With the same buffer and a mixture of GMP and CMP, two consecutive high-yielding copying steps occurred on a hairpin template that prevents uncontrolled further extension (Figure 2 b). When uridine 5′-monophosphate (UMP), the most weakly pairing nucleotide,32 was offered to a primer on a template with adenine as the first templating base, 75 % incorporation occurred after 2 d. Lowering the temperature to 0 °C, conditions close to those of eutectic ice phases, led to near-quantitative incorporation of UMP after 5 d in 0.5 m HEPES buffer (Figure 2 c, last entry in Table 1). High concentrations are characteristic of eutectic ice phases, and the beneficial effect of low temperatures for copying was known.17 The buffer thus optimized was dubbed “general condensation buffer” (0.8 m EDC, 0.15 m 1-ethylimidazole, 0.08 m MgCl2, 0.5 m HEPES, pH 7.5) and was used in all subsequent assays.
In mass spectra of copying assays with AMP as the monomer, strong peaks for oligoadenylates were observed. This was unexpected because similar assays with pre-activated monomers had not shown oligomers.14, 15, 17 We immobilized template–primer duplexes on magnetic beads via hybridization (Figure 3).15, 38 Now, magnetic separation, followed by washing the beads and denaturation allowed an unobstructed view on copying reactions, even for A (Figure 3 a). Kinetics showed that incorporation of AMP opposite U occurred with a half-life time of approximately 3 d. This is less than one order of magnitude slower than similar reactions involving pre-activated forms of AMP.15, 39
Figure 3.
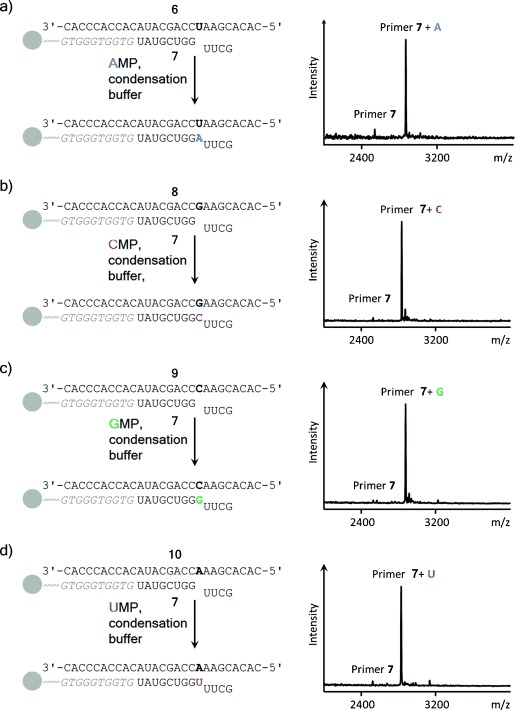
High-yielding incorporation of any of the four ribonucleotides opposite their complementary bases in condensation buffer. Reaction schemes are shown next to MALDI mass spectra. The oligodeoxynucleotide that immobilizes the primer–template duplex on beads is shown in italics; the tetramer downstream of the extension site limits the chain growth to a single step, facilitating analysis.15 a) Extension by AMP, spectrum after 21 d at 0 °C; b) extension by CMP, spectrum after 8 d at 0 °C; c) extension by GMP, spectrum after 8 d at 0 °C; d) extension by UMP after 21 d at 0 °C. Conditions: 0.8 m EDC, 0.15 m 1-ethylimidazole, 0.5 m HEPES buffer, and 0.08 m MgCl2. See the Supporting Information for details.
To confirm that the copying conditions found are general, primer 7 was hybridized to three other templates, each offering a different template base (C, G, or U). The resulting primer–template duplexes were allowed to react with the corresponding ribonucleotide (AMP, CMP, GMP or UMP). In each case near-quantitative primer extension was observed (Figure 3 b–d). Despite the high concentrations, which are reminiscent of what is found in eutectic ice phases,11 the reactions were surprisingly well behaved, with little to no side products detectable by MALDI MS. This confirmed that enzyme-free copying of RNA does not require pre-activation.
Finally, we performed an exploratory study on strand formation occurring under our in situ activation conditions (vide supra). Using a combination of ion-exchange HPLC and MALDI MS, we tested for the formation of oligomers in solutions of AMP, UMP, or CMP (150 mm each) after one week at 0 °C. For GMP, a 20 mm solution was used to avoid precipitation and aggregation. Only for AMP >90 % of the monomer was converted after 7 d, whereas for CMP ≥25 % of the monomer remained, and ≥40 % of UMP was unreacted after 7 d. In the case of GMP, peaks were too broad for unambiguous assignment, but it appeared that significant concentrations of both monomers and short oligomers were present. Figure 4 shows results from an assay with AMP run for 30 d at 0 °C. Peaks for chains of up to at least nine AMP residues are discernable. The formation of mixed sequences that can encode genetic information from any of the four ribonucleotides (A/C/G/U) is presented in Ref. 33. Taken together, our results confirm that “general condensation conditions” induce both high-yielding copying reactions and the untemplated de novo formation of RNA strands.
Figure 4.
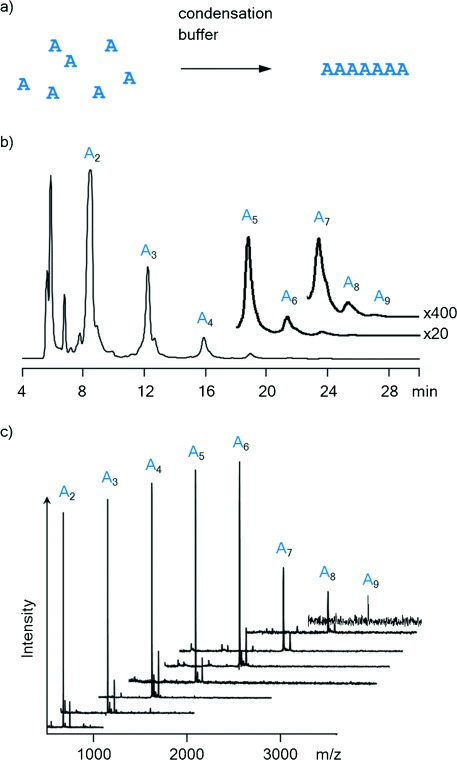
Oligomerization of AMP in condensation buffer at 0 °C after 30 d. a) Reaction scheme. b) Ion-exchange HPLC chromatogram at λdet=260 nm. c) Overlay of MALDI-TOF mass spectra of HPLC fractions, showing peaks of oligoribonucleotides with 2–9 residues.
In conclusion, we report that the combination of a carbodiimide and an N-alkylated heterocycle as the catalyst induces high-yielding genetic copying. The efficiency of the reaction strongly depends on the choice of the covalent catalyst. A positively charged imidazolium species, expected to form with 1-ethylimidazole, is apparently much more reactive than the well-established imidazolides. Chain growth is not limited to template-directed reactions. Oligomerization of ribonucleotides occurs in the absence of a mineral surface. It takes place at a fast enough rate to provide strands that may act as templates or primers, but slowly enough not to dominate the reaction landscape, so that is still allows for sequence-selective copying steps. The fast oligomerization of AMP is an interesting contrast to the slow primer extension on A-rich templates,40, 41 the second step of replication scenarios with strand formation and subsequent copying, suggesting a kinetic compensation. Based on our results, a more conclusive picture of the emergence of RNAs can be formulated, and much simpler experimental setups can be used to study their formation. A number of other processes leading to pivotal biomolecules occur under the same general condensation conditions, as reported in Ref. 33.
Experimental Section
Condensation buffer: The optimized reaction medium, referred to as “general condensation buffer” was an aqueous solution of HEPES (0.5 m), MgCl2 (0.08 m), 1-ethylimidazole (0.15 m), and the appropriate concentration of reactants, adjusted to pH 7.5. A fresh aliquot (65 μL) of this solution was added to EDC hydrochloride (10 mg, 52 μmol) to give an initial EDC concentration of approximately 0.8 m.
Primer extension: A suspension of beads with capture oligonucleotide (5 μL, 5 mg mL−1) in HEPES buffer (0.5 m with 0.08 m MgCl2, pH 7.5) was treated by addition of solutions of template (0.6 μL, 100 μm, 60 pmol) and primer (0.5 μL, 100 μm, 50 pmol) and left at 0 °C for 15 min. The supernatant was aspirated, and 5 μL of a freshly prepared condensation buffer containing either AMP (0.15 m), GMP (0.02 m), CMP (0.15 m), or UMP (0.15 m) at pH 7.5 was added. The mixture was transferred to a vessel containing 5 nmol of the downstream-binding tetramer, vortexed for 5 s, and incubated at 0 °C. At 5 d intervals the supernatant was drawn and replaced with a fresh solution. The reaction was monitored with MALDI-MS.15
Acknowledgments
We thank Dr. C. Deck and Dr. E. Kervio for discussions. A grant from DFG (No. RI 1063/8-2 to C.R.) and the EU COST action CM1304 supported this work.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
miscellaneous_information
References
- 1.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 2.Cech TR. Angew. Chem. Int. Ed. Engl. 1990;29:759–768. [Google Scholar]; Angew. Chem. 1990;102 [Google Scholar]
- 3.Mello CC. Angew. Chem. Int. Ed. 2007;46:6985–6994. doi: 10.1002/anie.200701713. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2007;119 [Google Scholar]
- 4.Gilbert W. Nature. 1986;319:618. [Google Scholar]
- 5.Westheimer F. Science. 1987;235:1173–1178. doi: 10.1126/science.2434996. [DOI] [PubMed] [Google Scholar]
- 6.Verlander MS, Lohrmann R, Orgel LE. J. Mol. Evol. 1973;2:303–316. doi: 10.1007/BF01654098. [DOI] [PubMed] [Google Scholar]
- 7.Ferris JP, Hill AR, Liu RH, Orgel LE. Nature. 1996;381:59–61. doi: 10.1038/381059a0. [DOI] [PubMed] [Google Scholar]
- 8.Pino S, Ciciriello F, Costanzo G, Di Mauro E. J. Biol. Chem. 2008;283:36494–36503. doi: 10.1074/jbc.M805333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costanzo G, Saladino R, Botta G, Giorgi A, Scipioni A, Pino S, Di Mauro E. ChemBioChem. 2012;13:999–1008. doi: 10.1002/cbic.201200068. [DOI] [PubMed] [Google Scholar]
- 10.Monnard P-A, Szostak JW. J. Inorg. Biochem. 2008;102:1104–1111. doi: 10.1016/j.jinorgbio.2008.01.026. [DOI] [PubMed] [Google Scholar]
- 11.Monnard P-A, Kanavarioti A, Deamer DW. J. Am. Chem. Soc. 2003;125:13734–13740. doi: 10.1021/ja036465h. [DOI] [PubMed] [Google Scholar]
- 12.Weimann BJ, Lohrmann R, Orgel LE, Schneider-Bernloehr H, Sulston JE. Science. 1968;161:387. doi: 10.1126/science.161.3839.387. [DOI] [PubMed] [Google Scholar]
- 13.Joyce GF, Inoue T, Orgel LE. J. Mol. Biol. 1984;176:279–306. doi: 10.1016/0022-2836(84)90425-x. [DOI] [PubMed] [Google Scholar]
- 14.Vogel SR, Deck C, Richert C. Chem. Commun. 2005:4922. doi: 10.1039/b510775j. [DOI] [PubMed] [Google Scholar]
- 15.Deck C, Jauker M, Richert C. Nat. Chem. 2011;3:603–608. doi: 10.1038/nchem.1086. [DOI] [PubMed] [Google Scholar]
- 16.Hagenbuch P, Kervio E, Hochgesand A, Plutowski U, Richert C. Angew. Chem. Int. Ed. 2005;44:6588–6592. doi: 10.1002/anie.200501794. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2005;117 [Google Scholar]
- 17.Vogel SR, Richert C. Chem. Commun. 2007:1896. doi: 10.1039/b702768k. [DOI] [PubMed] [Google Scholar]
- 18.Mukaiyama T, Hashimoto M. J. Am. Chem. Soc. 1972;94:8528–8532. doi: 10.1021/ja00779a039. [DOI] [PubMed] [Google Scholar]
- 19.Chu BC, Orgel LE. Nucleic Acids Res. 1988;16:3671–3691. doi: 10.1093/nar/16.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leman LJ, Orgel LE, Ghadiri MR. J. Am. Chem. Soc. 2006;128:20–21. doi: 10.1021/ja056036e. [DOI] [PubMed] [Google Scholar]
- 21.Benner SA, Kim H-J, Carrigan MA. Acc. Chem. Res. 2012;45:2025–2034. doi: 10.1021/ar200332w. [DOI] [PubMed] [Google Scholar]
- 22.Schimpl A, Lemmon RM, Calvin M. Science. 1965;147:149–150. doi: 10.1126/science.147.3654.149. [DOI] [PubMed] [Google Scholar]
- 23.Duvernay F, Chiavassa T, Borget F, Aycard J-P. J. Am. Chem. Soc. 2004;126:7772–7773. doi: 10.1021/ja048721b. [DOI] [PubMed] [Google Scholar]
- 24.Kilpatrick ML. J. Am. Chem. Soc. 1947;69:40–46. [Google Scholar]
- 25.von Kiedrowski G. Angew. Chem. Int. Ed. Engl. 1986;25:932–935. [Google Scholar]; Angew. Chem. 1986;98 [Google Scholar]
- 26.Zielinski WS, Orgel LE. Nucleic Acids Res. 1987;15:1699–1715. doi: 10.1093/nar/15.4.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zielinski WS, Orgel LE. Nature. 1987;327:346–347. doi: 10.1038/327346a0. [DOI] [PubMed] [Google Scholar]
- 28.Rojas Stütz JA, Kervio E, Deck C, Richert C. Chem. Biodiversity. 2007;4:784–802. doi: 10.1002/cbdv.200790064. [DOI] [PubMed] [Google Scholar]
- 29.Burcar BT, Jawed M, Shah H, McGown LB. Origins Life Evol. Biospheres. 2015;45:31–40. doi: 10.1007/s11084-015-9412-y. [DOI] [PubMed] [Google Scholar]
- 30.Sulston JE, Lohrmann R, Orgel LE, Miles HT. Proc. Natl. Acad. Sci. USA. 1968;59:726–733. doi: 10.1073/pnas.59.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sulston J, Lohrmann R, Orgel LE, Miles HT. Proc. Natl. Acad. Sci. USA. 1968;60:409–415. doi: 10.1073/pnas.60.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kervio E, Claasen B, Steiner UE, Richert C. Nucleic Acids Res. 2014;42:7409–7420. doi: 10.1093/nar/gku314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jauker M Griesser H Richert C Angew. Chem. Int. Ed 2015. Angew. Chem 2015 , DOI: 10.1002/anie.201506593 , DOI: [DOI]
- 34.Kaiser A, Richert C. J. Org. Chem. 2013;78:793–799. doi: 10.1021/jo3025779. [DOI] [PubMed] [Google Scholar]
- 35.Sarracino D, Richert C. Bioorg. Med. Chem. Lett. 1996;6:2543–2548. doi: 10.1016/s0960-894x(01)00284-0. [DOI] [PubMed] [Google Scholar]
- 36.Lohrmann R, Orgel LE. J. Mol. Biol. 1980;142:555–567. doi: 10.1016/0022-2836(80)90263-6. [DOI] [PubMed] [Google Scholar]
- 37.Prabahar KJ, Ferris JP. J. Am. Chem. Soc. 1997;119:4330–4337. doi: 10.1021/ja9700764. [DOI] [PubMed] [Google Scholar]
- 38.Kaiser A, Spies S, Lommel T, Richert C. Angew. Chem. Int. Ed. 2012;51:8299–8303. doi: 10.1002/anie.201203859. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2012;124 [Google Scholar]
- 39.Adamala K, Szostak JW. Science. 2013;342:1098–1100. doi: 10.1126/science.1241888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hill AR, Orgel LE, Wu T. Origins Life Evol. Biospheres. 1993;23:285–290. doi: 10.1007/BF01582078. [DOI] [PubMed] [Google Scholar]
- 41.Kervio E, Hochgesand A, Steiner UE, Richert C. Proc. Natl. Acad. Sci. USA. 2010;107:12074–12079. doi: 10.1073/pnas.0914872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
miscellaneous_information


