Summary
The malaria parasite develops sexually in the mosquito midgut upon entry with the ingested blood meal before it can invade the midgut epithelium and embark on sporogony. Recent data have identified a number of distinct transcriptional programmes operating during this critical phase of the parasite life cycle. We aimed at characterizing the parental contribution to these transcriptional programmes and establish the genetic framework that would guide further studies of P lasmodium zygotic development and ookinete‐to‐oocyst transition. To achieve this we used in vitro and in vivo cross‐fertilization experiments of various parasite lines expressing fluorescent reporters under the control of constitutive and stage‐specific promoters. The results revealed that the zygote/ookinete stage exhibits a maternal phenotype with respect to constitutively expressed reporters, which is derived from either maternal mRNA inheritance or transcription of the maternal allele. The respective paternal alleles are silenced in the zygote/ookinete but reactivated after midgut invasion and transformation to oocyst. Transcripts specifically produced in the zygote/ookinete are synthesized de novo by both parental alleles. These findings highlight a putative role of epigenetic regulation of P lasmodium zygotic development and add substantially to the emerging picture of the molecular mechanisms regulating this important stage of malaria transmission.
Introduction
Malaria is a parasitic disease affecting almost half of the world's population. WHO reported 627 000 deaths from malaria in 2012, mostly children below the age of 5 in sub‐Saharan Africa. The disease is caused by apicomplexan Plasmodium parasites transmitted to humans through bites of Anopheles mosquitoes. After an infective mosquito bite, haploid sporozoites are injected into the human body and travel to the liver where they invade and replicate within hepatocytes. After some days, the infected cells rupture and merozoites enter the blood stream to infect red blood cells and embark on a continuous asexual replication–release invasion cycle responsible for the disease symptoms. Eventually, some parasites escape this asexual cycle and transform to sexually dimorphic, non‐dividing gametocytes that are infective to mosquitoes upon a blood meal. Once inside the mosquito midgut, gametocytes exit the host cells and produce male and female haploid gametes that fuse to form zygotes. The diploid zygotes embark on meiosis and soon become tetraploid. Within hours, they transform into motile ookinetes that traverse the midgut cell wall and, upon arrival to the basal side, complete meiosis and transform to oocysts. Inside an oocyst, thousands of haploid sporozoites are produced over a period of 10–15 days. They are released into the hemolymph, invade the salivary glands and infect a human host during another mosquito bite.
The time required between parasite entry in the mosquito midgut and transformation to oocyst is about 22–32 h depending on the species and environmental conditions. This is one of the most decisive phases in the entire malaria transmission cycle; the vast majority of parasites are lost during this process, mostly due to robust immune reactions mounted by the mosquito host, and indeed only few parasites survive to continue the transmission cycle. Therefore, advanced understanding of the mechanisms regulating parasite development during this time could inform the design of new methods to block disease transmission.
The Plasmodium erythrocytic cycle in the human host is controlled by a tightly regulated cascade of transcriptional activation, whereby different subsets of genes are switched on and off as parasites transition from one stage to another (Bozdech et al., 2003). A similar pattern of distinct transcriptional repertoires has been recently reported for the development of the rodent malaria parasite Plasmodium berghei in the Anopheles gambiae mosquito (Akinosoglou et al., 2015). At least two transcriptional programmes involving differentially regulated transcripts have been shown to support the zygote/ookinete development. The first programme (henceforth TPR1) includes transcripts that are abundant during the first 24 h post‐infection (hpi) and diminish by 48 hpi. The second programme (henceforth TPR2) includes transcripts that are de novo produced in the zygote/ookinete.
Many of the TPR1 transcripts are produced in the female gametocyte but remain translationally repressed by the RNA helicase DOZI; they are then supplied to the zygote as maternal mRNAs where they are translated (Mair et al., 2006; 2010). This process is widely observed across life kingdoms whereby quiescent female germ cells store mRNAs that are translated later in the zygote (Bettegowda and Smith, 2006; Sheth et al., 2010). In Plasmodium, products of these genes are required for zygote and ookinete development and include the ookinete surface antigens P25 and P28 as well as the Apetela 2 (AP2) transcription factor AP2‐O (Mair et al., 2006; 2010; Yuda et al., 2009). Interestingly, the TPR2 programme includes several transcripts regulated by AP2‐O itself, linked to midgut invasion and the developmental transition to oocyst. Finally, a third transcriptional programme (henceforth TPR0) that also supports the developing zygote/ookinete involves genes that are constitutively expressed throughout the parasite's sexual and sporogonic development (Akinosoglou et al., 2015). It includes transcripts encoding proteins with housekeeping functions such as metabolism, translation and transport.
It has increasingly become evident that epigenetic regulation is an important hallmark of the parasite's development and survival strategy within the host (Voss et al., 2014). The identification of the aforementioned regulatory mechanisms (Mair et al., 2006) and transcriptional programmes (Akinosoglou et al., 2015), in conjunction with the general understanding that epigenetic genomic imprinting is common in zygotic development across life kingdoms, prompted us to further investigate the parental origin of the developing Plasmodium zygote. Our first aim was to determine the origin of proteins and phenotypes associated with the TPR0 programme, which support housekeeping function in the developing zygote/ookinete and facilitate the production of proteins from maternally inherited transcripts of the TPR1 programme. The second aim was to characterize the origin of proteins and phenotypes associated with the TPR2 programme in order to shed light into whether both the paternal and maternal alleles are involved in the transcriptional activation of genes in the TPR2 programme or whether allelic exclusion takes place. To address these questions we used both in vitro and in vivo cross‐fertilization assays of parasite lines expressing fluorescent reporters under the control of promoters of genes belonging to the two programmes. The results of this study can help uncover the foundations for the genetic and epigenetic dissection of the Plasmodium zygotic development.
Results
Transient allelic exclusion or silencing post‐fertilization
We used two transgenic P. berghei lines stably expressing mCHERRY and green fluorescent protein (GFP) under the control of the constitutive elongation factor 1 subunit alpha gene promoter (ef1αp), designated as wt_red230p and wt_green230p respectively. Transcripts of the ef1α gene are abundant throughout P. berghei development in the vector, falling within the TPR0 transcriptional programme (Akinosoglou et al., 2015). Constitutive GFP expression in P. berghei mosquito stages when GFP is placed under the control of the ef1αp has been previously reported (Franke‐Fayard et al., 2004). The wt_red230p line was newly generated as described in the Experimental procedures section (Fig. S1A,D), while generation of the wt_green230p line is reported in Janse et al. (2006). In both lines, the transgenic cassettes were integrated into the 230p genomic locus. The morphology and growth of asexual and sexual stages of the wt_red230p line including blood stages, round and retort form zygotes, ookinetes, oocysts and sporozoites, as well as transmission to mice, were comparable to those of wt_green230p parasites (data not shown).
We examined the expression of fluorescent reporters in in vitro cross‐fertilization assays after mixing wt_red230p and wt_green230p gametocytes. Throughout the zygote and ookinete development, we observed parasites expressing only GFP or mCHERRY; GFP/mCHERRY double‐positive parasites were not detected (Fig. 1A). This observation suggested either that the (diploid and tetraploid) zygotes/ookinetes were derived exclusively from self‐fertilization of the haploid gametes of the two lines, i.e. no cross‐fertilization, or that allelic reporter gene exclusion and/or silencing occurred resulting in expression of only a single‐fluorescent protein. To confirm this observation we carried out in vivo cross‐fertilization assays in A. gambiae mosquitoes directly fed on mice previously infected with equal numbers of the two transgenic parasite lines. Microscopic observations of mosquito midguts at 24 hpi showed comparable numbers of GFP and mCHERRY expressing ookinetes invading the midgut epithelium and a total absence of GFP/mCHERRY double‐positive ookinetes (Fig. 1B). Three independent biological replicates of the in vivo cross‐fertilization assays were performed with highly consistent results, each using a different mouse and mosquito batch.
Figure 1.
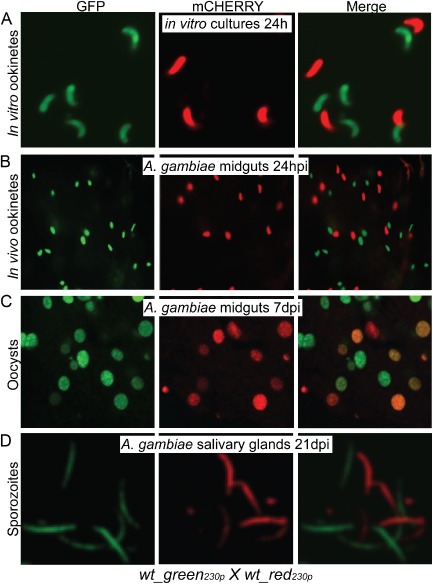
Imaging of wt_green230p and wt_red230p parasite lines derived from cross‐fertilization assays.
A. Purified ookinetes from in vitro cross‐fertilization assays.
B. Ookinetes crossing the mosquito midgut epithelium 24 hpi.
C. Confocal microscopy sections of 7‐day‐old oocysts on the basal side of A . gambiae midgut epithelium.
D. Confocal microscopy sections of sporozoites from homogenized A . gambiae salivary glands 21 dpi. Images were taken at ×63 magnification for the purified in vitro ookinetes and at ×40 magnification for mosquito tissues.
Intriguingly, when we continued monitoring the developmental progression of these parasites throughout their sporogonic development, a large number of GFP/mCHERRY double‐positive oocysts were detected in infected midguts 7 days post‐infection (dpi) in addition to the GFP and mCHERRY single‐positive oocysts (Fig. 1C). These observations confirmed that cross‐fertilization had occurred between the wt_red230p and wt_gfp230p gametes and that the absence of GFP/mCHERRY double‐positive zygotes/ookinetes is due to allelic exclusion or gene silencing. Only single‐positive GFP and mCHERRY sporozoites were detected in salivary glands dissected at 21 dpi, as expected due to the haploid nature of this parasite stage (Fig. 1D).
We investigated the allelic exclusion/silencing phenotype in in vivo cross‐fertilization assays by tightly timing the mosquito midgut dissections and quantifying the number of single‐ and double‐positive parasites in each midgut. The GFP/mCHERRY double‐positive phenotype was detected in young oocysts starting at 32 hpi (Fig. 2A; Table S1) but not in any of the preceding stages including round and retort form zygotes, mature ookinetes in the blood bolus and mature ookinetes traversing the mosquito midgut. These assays were repeated three times with similar results. These data led us to hypothesize that one of the parental allelic transgenes is silenced after fertilization in the zygote/ookinete and reactivated in the young oocyst, perhaps soon after the ookinete‐to‐oocyst transition.
Figure 2.
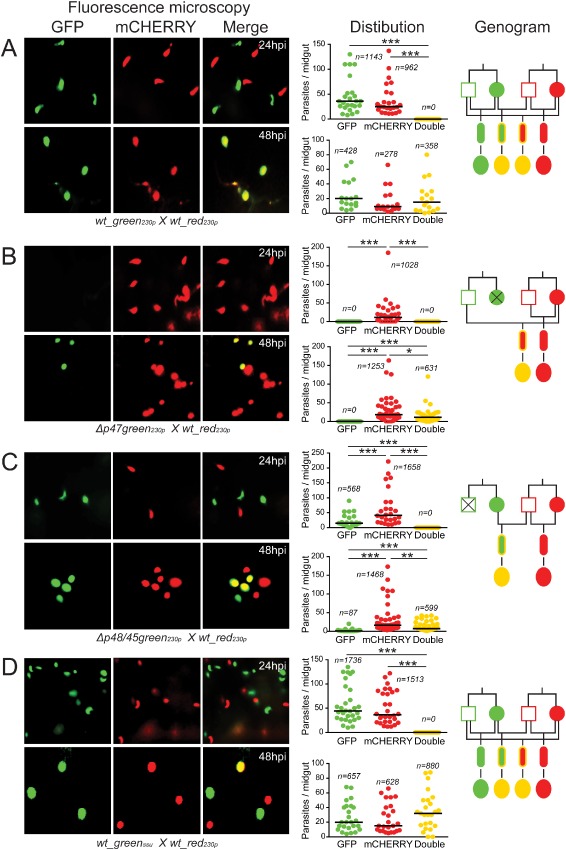
Allelic expression of fluorescent reporters placed under the control of a constitutive gene promoter.
A. wt_green230p x wt_red230p.
B. Δp47green230p x wt_red230p.
C. Δp48/45green230p x wt_red230p.
D. wt_greenssu x wt_red230p. Each part consists of three panels. The first composite panel is a collection of six representative fluorescence microscopy pictures of A . gambiae midguts fed on mice co‐infected with two transgenic parasite lines as indicated at the bottom of each part, taken at 24 and 48 hpi respectively. The GFP, mCHERRY and a combination of the two channels (merge) are shown. The second panel is a graph showing the distribution and median number of GFP‐positive, mCHERRY‐positive and GFP/mCHERRY double‐positive parasites per midgut at 24 and 48 hpi. The collective results from three biological replicates are shown, where n is the total number of parasites counted. The third panel shows a genogram summarizing the results of these cross‐fertilization experiments. Squares correspond to male gametocytes, circles correspond to female gametocytes, small ellipses correspond to ookinetes and large ellipses correspond to early stage oocysts. The colour of the outline indicates the genotype, while the fill‐in colour indicates the phenotype. Yellow outline indicates gfp/m C herry heterozygotes, while yellow fill‐in colour indicates GFP/mCHERRY double‐positive parasites. Black lines indicate the crosses and the resulting progeny. Horizontal black lines indicate the median parasite number. Stars indicate statistical significance determined with the Mann–Whitney U‐test (***P < 0.001; **P < 0.01; *P < 0.05).
Male alleles are silent in zygotes/ookinetes and reactivated in oocysts
We further investigated the putative allelic exclusion/silencing by carrying out in vivo cross‐fertilization assays in A. gambiae mosquitoes involving crossings of the wt_red230p line with the Δp47green230p line that produces only male fertile gametes (Van Dijk et al., 2010) and the wt_red230p line with the Δp48/45green230p (RMgm346) line that produces only female fertile gametes. Both the Δp47green230p and the Δp48/45green230p lines contain the same ef1αp:gfp transgenic cassette integrated into the 230p genomic locus.
The wt_red230p line crossing with the Δp47green230p line resulted in only mCHERRY single‐positive ookinetes in the mosquito midgut epithelium at 24 hpi (Fig. 2B; Table S1). At 48 hpi, a comparable number of mCHERRY single‐positive and GFP/mCHERRY double‐positive oocysts were observed on the basal side of the midgut wall. These observations confirmed that the male Δp47green230p gametes cross‐fertilized with the wt_red230p female gametes and indicated that the ef1αp:gfp allele, contributed by the male genome, began to be expressed after 32 hpi in the young hybrid oocysts.
The wt_red230p line crossing with the Δp48/45green230p line resulted in ookinetes exhibiting GFP or mCHERRY single‐positive phenotypes 24 hpi (Fig. 2C; Table S1). At the oocyst stage, parasites were mCHERRY single‐positive or GFP/mCHERRY double‐positive. A small number of GFP single‐positive oocysts are thought to have had derived from a known limited self‐fertilization between gametes of the Δp48/45green230p line (Fig. S2; Table S2). These results confirmed that fertile Δp48/45green230p female gametes fertilized with wt_red230p males, but the male ef1αp:mcherry allele was not expressed before 32 hpi. Together the results from the two crossing assays indicated that the non‐active allele in the developing zygote/ookinete is contributed by the male gamete; this allele is activated again following ookinete transformation to oocyst.
To examine whether these phenotypes were specific for the 230p genomic locus in which the fluorescent reporters had been integrated, we carried out in vivo cross‐fertilization assays of the wt_red230p line with the wt_greenssu line in which the ef1αp:gfp cassette was integrated into the silent ssu‐rRNA gene locus (529c12; Franke‐Fayard et al., 2004). The results revealed that the fluorescence patterns were similar to those described earlier, i.e. midgut invading ookinetes 24 hpi were either single GFP positive or mCHERRY positive whereas a large number of double‐positive oocysts were observed 48 hpi (Fig. 2D; Table S1). These data corroborated our findings that the male ef1αp:gfp allele is expressed only after 32 hpi and that our observations are independent of the genomic integration locus.
We also investigated whether our findings are specific to the P. berghei ef1a promoter or the dhfr 3′ UTR sequences, which are used in both the ef1αp:gfp and ef1αp:mcherry expression cassettes. For this, we used a transgenic line in which mCHERRY was expressed under the control of the constitutive hsp70 gene promoter and the hsp70 3′ UTR (Annoura et al., 2014). Like the ef1αp transgenic expression cassettes, this transgene is also integrated into the 230p genomic locus. In vivo cross‐fertilization assays in A. gambiae mosquitoes between this line, designated as wt_hsp70pred230p, and the wt_green230p line resulted in only single GFP‐positive or mCHERRY‐positive parasites at 24 hpi, while double‐positive oocysts began to appear only after 32 hpi (Table S1; Fig. S3). These data revealed that our findings are not limited to the ef1a promoter and 3′ UTR regulatory sequences.
D e novo gene expression post‐fertilization occurs by both parental alleles
In the experiments described earlier, we used promoters of the ef1α and hsp70 genes that are constitutively expressed throughout the Plasmodium life cycle. To investigate whether genes specifically expressed in developing zygotes are also subject to allelic exclusion/silencing, we generated two P. berghei transgenic lines expressing the GFP and mCHERRY fluorescent reporters under the control of the chitinase gene (cht1; PBANKA_080050) promoter (chtp). Cht1 is expressed specifically in mature ookinetes and contributes to the penetration of the chitinaceous peritrophic matrix surrounding the blood bolus (Dessens et al., 2001). The two lines were designated as wt_chtpgreen230p and wt_chtpred230p respectively. Integration of these expression cassettes into the 230p genomic locus was achieved by double crossover homologous recombination (Fig. S1B–C). Diagnostic polymerase chain reaction (PCR) and southern analysis on pulse field gel electrophoresis separated chromosomes (Fig. S1D) of transgenic clonal lines were used to confirm successful integration. In both lines, transcription of the transgenes began 2 h post‐fertilization (hpf) and the fluorescent reporters were detected 12 hpf onwards (Figs S4 and S5). The development of both transgenic lines was comparable with that of wt parasites (data not shown).
Phenotypic analysis of parasite stages in in vitro cross‐fertilization assays of the wt_chtpgreen230p and wt_chtpred230p lines revealed that all possible phenotypes are obtained at the ookinete stage: GFP and mCHERRY single‐positive and GFP/mCHERRY double‐positive ookinetes (Fig. 3A; Table S3). This fluorescent pattern could be detected starting at 12 hpf. No fluorescence could be detected in activated gametocytes, gametes and zygotes (data not shown). The same pattern of fluorescent reporter expression was observed in in vivo cross‐fertilization assays in A. gambiae (Fig. 3A; Table S4). These observations demonstrated that, when placed under the control of the cht1 promoter, the gfp and mCherry transgenes are de novo transcribed in developing zygotes/ookinetes from both parental alleles.
Figure 3.
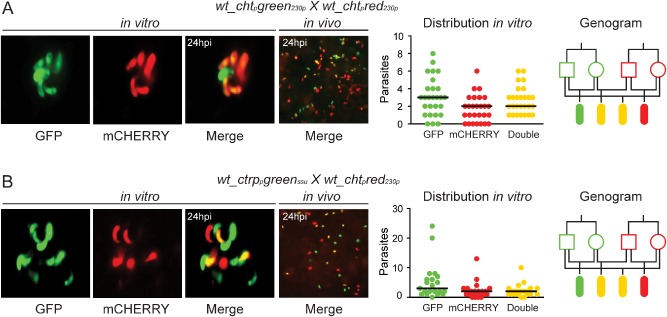
Allelic expression of fluorescent reporters placed under the control of zygote/ookinete specific promoters.
A. wt_chtpgreen230p x wt_chtpred230p.
B. wt_ctrppgreenssu x wt_chtpred230p. Each part consists of four panels. The first composite panel is a collection of three representative fluorescence microscopy pictures of in vitro cultured ookinetes initiated with blood from mice co‐infected with two transgenic parasite lines as indicated at the top of each part. The GFP, mCHERRY and a combination of the two channels (merge) are shown. The second panel shows a representative fluorescence microscopy picture of an A . gambiae midgut fed on mice co‐infected with the two transgenic parasite lines taken at 24 hpi. The third panel is a graph showing the distribution and median number of GFP‐positive, mCHERRY‐positive and GFP/mCHERRY double‐positive ookinetes in the in vitro culture. The fourth panel shows a genogram summarizing the results of these crosses. Squares correspond to male gametocytes, circles correspond to female gametocytes and ellipses correspond to ookinetes. The colour of the outline indicates the genotype, and the fill‐in colour indicates the phenotype. Yellow outline indicates gfp/m C herry heterozygotes, while yellow fill‐in indicates to GFP/mCHERRY double‐positive parasites. Black lines show the crosses and the resulting progeny.
To confirm that the earlier results are not limited to the chtp, we used the wt_ctrppgreenssu parasite line that expresses GFP under the control of the circumsporozoite and thrombospondin‐related adhesive protein gene promoter (ctrpp; PBANKA_041290). In this line, GFP is detected in zygotes as soon as 4 hpf (Vlachou et al., 2004). Phenotypic analysis of ookinetes from in vitro cross‐fertilization assays of wt_ctrppgreenssu and wt_chtpred230p parasites revealed an all‐encompassing fluorescent pattern 24 hpf similar to that of the chtp assays: GFP and mCHERRY single‐positive as well as GFP/mCHERRY double‐positive ookinetes (Fig. 3B; Table S3). Similar results were obtained in in vivo cross‐fertilization assays using the wt_ctrppgreenssu and wt_chtpred230p lines. At 24 hpi, both single‐ and double‐fluorescent ookinetes were detected in the invaded mosquito midgut epithelium (Fig. 3B; Table S4). These observations indicated that genes specifically expressed in the developing zygote/ookinete are de novo transcribed by both parental alleles.
Discussion
We present data suggesting that, during P. berghei zygotic (including ookinete) development in the A. gambiae midgut, transcripts produced by constitutive promoters and encoding proteins involved in housekeeping functions are provided exclusively by maternal alleles. We reveal that the respective paternal alleles are silenced during this time but reactivated for transcription and translation after invasion of the mosquito midgut, following the ookinete transformation to oocyst. We also reveal that, at the same time, transcripts specifically found in the zygote/ookinete, encoding proteins that function in midgut invasion and the developmental transition to oocyst, are synthesized de novo by both parental alleles. A final group of transcripts involved in zygotic development are synthesized in the female gametocyte and supplied to the zygote during fertilization as maternal mRNAs (Mair et al., 2006; 2010). These data add to an emerging picture of complex and tightly regulated gene expression repertoires and deduced phenotypes in the developing Plasmodium zygote (Akinosoglou et al., 2015) and expose a sophisticated genetic framework within which further studies of this important stage of malaria transmission can be conducted.
We present three hypotheses with regard to the origin of zygotic transcripts of constitutively expressed genes (TPR0 transcripts). The first hypothesis is that both the paternal and maternal alleles are silenced during zygotic development, and that transcripts are inherited to the zygote by the female gametocyte as maternal mRNAs (Fig. 4A). Indeed, a defining feature of sexual development in metazoans is the detainment of a subset of transcripts in quiescent messenger ribonucleoprotein particles in the oocytes for later translation in the zygote (Bettegowda and Smith, 2006; Sheth et al., 2010). These particles involve DDX6 class DEAD box RNA helicases that directly bind maternal mRNAs and are often located in structures designated as P granules (Pitt et al., 2000; Schisa et al., 2001). This hypothesis is strongly supported by the discovery of a translational repression mechanism in P. berghei involving the RNA helicase DOZI and additional proteins homologous to those found in P granules (Mair et al., 2006; 2010). This mechanism controls the expression of a large number of genes involved in zygotic development corresponding to almost half of the parasite transcriptome (Guerreiro et al., 2014) and including the surface proteins P25 and P28 (TPR1 programme). In conclusion, according to this first hypothesis, the maternal TPR0 phenotype of the zygote/ookinete gradually diminishes as maternal mRNAs are exhausted and is replaced by an integrated parental phenotype derived from transcriptional reactivation of both parental alleles in the young oocyst.
Figure 4.
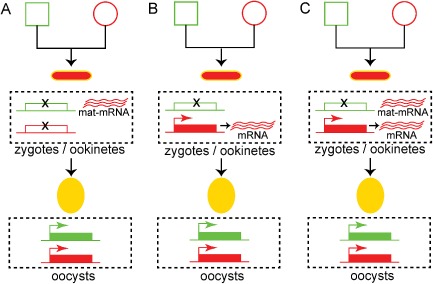
Models explaining the maternal phenotype related to the constitutively expressed reporters.
A. Maternal mRNA (mat‐mRNA) contribution. Both parental alleles are silenced during zygotic development. Maternal mRNA is supplied by the female gametocyte and translated.
B. Paternal allele silencing. The parental allele is silenced during zygotic development. The maternal allele is de novo transcribed and translated.
C. Paternal allele silencing and maternal mRNA contribution. A combined model, whereby the paternal allele is silenced, while maternal mRNA is supplied by the female gametocyte and the maternal allele is de novo transcribed and translated. Squares correspond to male gametocytes, circles correspond to female gametocytes, small ellipses correspond to ookinetes and large ellipses correspond to early stage oocysts. The colour of the outline indicates the genotype, and the fill‐in colour indicates the phenotype. Yellow outline indicates gfp/mCherry heterozygotes, while yellow fill‐in indicates GFP/mCHERRY double‐positive parasites.
The second hypothesis is that, although paternal alleles are silent, maternal alleles remain transcriptionally active throughout zygotic development (Fig. 4B). This hypothesis is supported by the fact that another transcriptional programme, TPR2, is operational during the same time, starting soon after fertilization. This programme includes de novo synthesized transcripts encoding proteins that are exclusive to the zygote/ookinete and are involved in ookinete midgut invasion such as CHT1, WARP and CTRP. It appears to be tightly and specifically regulated by the transcription factor AP2‐O (Yuda et al., 2009). Therefore, it is probable that the basal transcriptional machinery remains constitutively active throughout zygotic development transcribing non‐silenced maternal alleles of the TPR0 programme, while the TPR2 programme is initiated upon availability of the AP2‐O factor, supplied as maternal mRNA by the TPR1 programme, and possibly additional transcriptional factors. Finally, a combined hypothesis is also possible, whereby both maternal and de novo produced mRNAs of maternal allele origin are involved in the maternal TPR0 phenotype of the developing zygote (Fig. 4C).
A conserved feature of all three hypotheses is the silencing of the paternal and perhaps the maternal alleles throughout zygotic development and their reactivation upon transformation to oocyst. This observation hints at epigenetic regulation mechanisms, pertaining to monoallelic or biallelic gene silencing respectively. In support of the paternal allele silencing concept is the finding that the paternal DNA is packaged differently in the male than the female gametocyte (Laurentino et al., 2011), which may explain its unavailability for transcription in the zygote/ookinete and until mitotic replication commences in the oocyst. In the last decade, it has become increasingly evident that epigenetic mechanisms are an important hallmark of the parasite's survival strategy within the host (Voss et al., 2014). Genes involved in host–parasite interactions, coding for virulence factors or ligands involved in red blood cell invasion, are epigenetically regulated (Cortés et al., 2007; Flueck et al., 2009), while genes involved in drug resistance are switched on or off epigenetically in an environment‐dependent manner (Sharma et al., 2013). Therefore, it is reasonable to hypothesize that processes involved in parasite survival in the hostile mosquito gut environment are also controlled at an epigenetic level, allowing parasites to adapt to their environment. This is supported by recent evidence showing that parasite virulence in mice is greatly affected by the passage through the mosquito gut (Spence et al., 2013), suggesting that epigenetic imprinting may occur during sexual development. Although the P. falciparum genome is largely euchromatic (active) and dominated by a code of histone modifications associated with euchromatic marks (Salcedo‐Amaya et al., 2009; Trelle et al., 2009; Gupta et al., 2013), clonally variant gene families found in heterochromatic loci are associated with the evolutionary conserved regulator of heterochromatin, HP1 (Flueck et al., 2009; Lopez‐Rubio et al., 2009). It remains to be explored whether such differential histone marking is also relevant during zygotic development.
The zygote/ookinete is the only stage in the entire life cycle at which parasites are non‐haploid. Therefore, infections of mosquitoes with non‐clonal parasite populations, e.g. parasite strains harbouring variations or carrying mutations in different genes, lead to allele heterozygosity. The resulting combinations of heterozygote alleles increase exponentially as the number of input strains increases. This hinders the design of genetic approaches to study the function of genes during these critical stages of malaria transmission when parasites are not clonal or when conducted in a high throughput manner. The data we present here offer an elegant genetic framework in which such approaches can be envisaged, whereby genes or genetic modifications can be placed on either side of the allelic exclusion ‘seesaw’ in order for them to be expressed or kept silent until a later time point respectively.
Experimental procedures
Ethics statement
This study was carried out in strict accordance with the United Kingdom Animals (Scientific Procedures) Act of 1986. The protocols for mosquito maintenance or infection with P. berghei through blood feeding on naïve or parasite‐infected mice, respectively, as well as for culturing parasites in mice were approved and carried out under the UK Home Office License PLL70/7185.
Parasite cultivation and mosquito infections
The P. berghei clones constructed previously and reused in this study were the gametocyte‐producer ANKA 15cy1A, Δp48/45green230p (764acl1; RMgm346), Δp47green230p (765acl1;RMgm347; Van Dijk et al., 2010), wt_ctrppgreenssu (Vlachou et al., 2004), wt_green230p (507m6cl1; Janse et al., 2006), wt_greenssu (259cl2; Franke‐Fayard et al., 2004) and wt_hsp70pred230p (1804cl1; RMgm928; Annoura et al., 2014). Parasite handling and purification of blood stages were performed as described (Janse and Waters, 1995). Ookinete in vitro culturing was carried out as described (Rodrıguez et al., 2002). A. gambiae mosquitoes of the N'gousso strain were infected with P. berghei by direct feeding on mice using standard methods (Sinden, 1997; Sinden et al., 2002).
Generation of P . berghei reporter lines
To introduce a constitutively expressed mCherry cassette into the P. berghei ANKA 15cy1A genome and generate the wt_red230p stable transgenic line, the pmCherrycon vector plasmid was constructed. The plasmid pL0018 (MRA‐787, MR4) served as a backbone for this vector. The plasmid pL0018 contains two expression cassettes, one for the expression of the selectable marker Toxoplasma gondii dhfr (tgdhfr) under the control of the P. berghei dhfr promoter (dhfrp) and a second under the control of the P. berghei ef1αp that allows constitutive expression of any inserted downstream transgene. The 230p cassette allows integration via double crossover homologous recombination. mCherry was amplified from pmCherry vector (Clonetech) as a BamHI fragment and cloned into the pCR®2.1‐TOPO® vector (Invitrogen). The BamHI fragment of the GFP mutant 3 gene of plasmid pL0018 was replaced with the BamHI mCherry gene fragment.
For the generation of the wt_chtpred230p and wt_chtpgreen230p lines, the pchtpmCherry and pchtpgfp vectors, respectively, were constructed. First, a 820 bp fragment directly upstream of the cht1 open‐reading frame was amplified from P. berghei genomic DNA as an AflII/BamHI fragment and cloned into the pCR 2.1‐TOPO vector (Life Technologies). To construct the pchtpmCherry vector, the plasmid pmCherrycon (see earlier) served as a backbone for this vector. The ef1αp promoter of pmCherrycon was replaced by the AflII/BamHI fragment of the chtp promoter. Similarly, to construct the pchtpgfp vector, the plasmid pchtpmCherry served as a backbone for this vector. The BamHI fragment of the mCherry gene of pchtpmCherry plasmid was replaced with the BamHI fragment of pL0018 containing the gfp mutant 3 gene.
Genotypic analysis of transgenic parasites
P. berghei genomic DNA was prepared from transfected blood stage parasite populations. White blood cells were removed by filtration over CF‐11 column (Whatman) and red blood cells were lysed by incubation for 20 min on ice in 0.17 M ammonium chloride. Genomic DNA was extracted using DNeasy kit (Qiagen) and subjected to diagnostic PCR to assess successful integration. Southern blot analysis on pulse field gel electrophoresis separated chromosomes of purified blood stage parasites was also performed. The blot was hybridized against a probe encompassing the Pbdhfr 3′ UTR cassette obtained by the Hind III/EcoRV digest of the pBS‐TgDHFR vector.
Characterization of the chtp promoter
To characterize the timing of the expression of the fluorescent reporters under the control of the chtp promoter, a time course analysis was carried out. Briefly, blood from mice infected with either wt_chtpgreen230p or wt_chtpred230p parasite lines was used to set up an in vitro ookinete culture. The culture was split into several flasks and incubated at 21°C. At 2, 4, 8, 12, 16 and 24 h, a sample was taken and analysed by fluorescence microscopy. The rest of the culture was spun down, the red blood cells lysed in 0.17 M ammonium chloride and parasites pelleted for subsequent RNA extraction.
Transcriptional profiling using RT‐PCR
Using the Trizol® reagent (Invitrogen), total RNA was isolated from purified mixed blood stages (MBS) and gametocytes and in vitro ookinetes of the wt_chtpgreen230p or wt_chtpred230p parasite lines. Gene‐specific primers (Table S5) were designed using Primer3 (v. 0.4.0) and used in RT‐PCR.
In vivo and in vitro cross‐fertilization assays
We performed both in vitro and in vivo cross‐fertilization assays to assess the developmental profiles of transgenic lines and the expression of fluorescence reporters. Briefly, blood from mice infected with individual parasites was mixed in equal proportions and used to co‐infect new mice. For in vitro cross‐fertilization assays, infected blood at parasitaemia of 15–20% was acquired from these mice via heart puncture and used to setup in vitro ookinete cultures. At 24 hpf, mature ookinetes were analysed for fluorescence and also counted. Similarly, for in vivo cross‐fertilization assays, A. gambiae mosquitoes (N'gousso strain) were allowed to feed directly on the co‐infected mice with a parasitaemia 5–6%. Mosquito midgut tissues were dissected and parasite numbers and expression of fluorescent reporters were assessed by fluorescent microscopy at 24 and 32–48 hpi, 7 and 21 dpi.
Imaging and enumeration of parasites
Following mosquito infection and dissection, midguts were fixed in 4% formaldehyde (v/v) (16% methanol free, ultrapure stock diluted in PBS, Polysciences Inc.) for 20 min at room temperature and washed three times for 10 min each in PBS. Fixed midguts were mounted in Vectashield® (VectorLabs) on glass slides under sealed coverslips. Oocyst numbers were counted at 7 dpi using fluorescence microscopy under ×10 magnification. Salivary gland sporozoites were observed at day 21 dpi. Parasites were visualized using either a Leica DMT fluorescence microscope or a Leica SP5 MP inverted confocal microscope, and images were taken using a Zeiss AxioCam HRc camera coupled to Zeiss Axiovision40 version 4.6.1.0 software or Leica LAS AF software (Leica Microsystems) respectively.
Statistics
Statistical analysis of parasite loads in the mosquito midguts were performed using the Mann–Whitney U‐test.
Supporting information
Fig. S1. Generation of wt_red230p, wt_chtpred230p and wt_chtpgreen230p transgenic parasites. Schematic representation of the pmCherrycon (A), pchtpmCherry (B) and pchtpgfp (C) expression cassettes that are inserted into the 230p locus via double crossover homologous recombination resulting in the loss of a 1 kb region of the native locus.
D. Diagnostic PCR of clonal parasites corroborating successful integration of the aforementioned expression cassettes.
E. Southern blot analysis of clonal parasites corroborating successful integration of the expression cassettes. The positions of the PCR primers used for the diagnostic PCR reactions are shown (P5, P6 and P7).
Fig. S2. Oocyst load in A. gambiae mosquitoes infected with the Δp48/45green230p parasite. The incomplete defective phenotype of the male gamete defective Δp48/45green230p transgenic parasite results in the escape of very few male gametes that are able to fertilize the normal Δp48/45green230p female gametes to form few ookinetes and thus the few oocysts observed in A. gambiae mosquitoes.
Fig. S3. Allelic expression of fluorescent reporters placed under the control of constitutive gene promoters. In vivo cross‐fertilization assays in A. gambiae mosquitoes directly fed on mice infected with equal numbers of the transgenic parasite lines wt_green230p (ef1α gene promoter) and wt_hsp70pred230p. The first composite panel includes representative fluorescence microscopy pictures of A. gambiae midguts fed on mice co‐infected with two transgenic parasite lines as indicated at the bottom of each part, taken at 24 and 48 hpi respectively. The GFP, mCHERRY and a combination of the two channels (merge) are shown. The second panel is a graph showing the distribution and median number of GFP‐positive, mCHERRY‐positive and GFP/mCHERRY double‐positive parasites per midgut at 24 and 48 hpi. The collective results from three biological replicates are shown, where n is the total number of parasites counted. The third panel shows a genogram summarizing the results of these cross‐fertilization experiments. Squares correspond to male gametocytes, circles correspond to female gametocytes, small ellipses correspond to ookinetes and large ellipses correspond to early stage oocysts. The colour of the outline indicates the genotype, whereas the fill‐in colour indicates the phenotype. Yellow outline indicates gfp/mCherry heterozygotes, whereas yellow fill‐in colour indicates GFP/mCHERRY double‐positive parasites. Black lines indicate the crosses and the resulting progeny. Horizontal black lines indicate the median parasite number. Stars indicate statistical significance determined with the Mann–Whitney U‐test (***P < 0.001).
Fig. S4. GFP expression in the wt_chtpgreen230p parasite line.
A. RT‐PCR analysis of gfp transcripts from mixed blood stages, activated (A) and non‐activated (nA) gametocytes, and parasites purified from in vitro ookinete cultures at 2, 4, 8, 12, 14, 16 and 24 h post‐fertilization. Tubulin transcripts served as a loading control.
B. Fluorescence microscopy analysis of wt_chtpgreen230p parasites purified from in vitro ookinete cultures at 2, 4, 8, 12, 14, 16 and 24 hpa. Parasites were also stained with a Cy3 conjugated α‐P28 antibody. Images were taken at ×40 magnification.
Fig. S5. Expression of mCHERRY in the wt_chtpred230p parasite line.
A. RT‐PCR analysis of mCherry transcripts from mixed blood stages, activated (A) and non‐activated (nA) gametocytes, and parasites purified from in vitro ookinete cultures at 2, 4, 8, 12, 14, 16 and 24 h post‐fertilization. Tubulin transcripts served as a loading control.
B. Fluorescence microscopy analysis of wt_chtpred230p parasites purified from in vitro ookinete cultures at 4, 8, 12, 14, 16 and 24 hpa. Bright field images are also shown. Images were taken at ×40 magnification.
Table S1. Parasite development in A. gambiae mosquitoes co‐infected with parasites constitutively expressing GFP and mCHERRY.
Table S2. Oocyst numbers in A. gambiae infections.
Table S3. In vitro cross‐fertilization assays of GFP and mCHERRY expressing parasites.
Table S4. Parasite development in A. gambiae mosquitoes co‐infected with parasites expressing GFP and mCHERRY during the zygote/ookinete stages.
Table S5. Primers for the generation of transgenic parasites and RT‐PCR.
Acknowledgements
We thank Fotic C. Kafatos for supporting this research, Chris J. Janse for valuable feedback during drafting of the manuscript and Carolina Barillas‐Mury for critical review and suggestions. We also thank Kathrin Witmer for discussions on epigenetics. The work was funded by the Wellcome Trust as part of the project WT093587, and the EVIMalaR Network of Excellence grant of the European Commission Framework Programme (FP7/2007–2013) under grant agreement N°242095.
References
- Akinosoglou, K.A. , Bushell, E.S. , Ukegbu, C.V. , Schlegelmilch, T. , Cho, J.S. , Redmond, S. , et al (2015) Characterization of plasmodium developmental transcriptomes in Anopheles gambiae midgut reveals novel regulators of malaria transmission. Cell Microbiol 17: 254–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annoura, T. , van Schaijk, B.C. , Ploemen, I.H. , Sajid, M. , Lin, J.W. , Vos, M.W. , et al (2014) Two plasmodium 6‐Cys family‐related proteins have distinct and critical roles in liver‐stage development. FASEB J 28: 2158–2170. [DOI] [PubMed] [Google Scholar]
- Bettegowda, A. , and Smith, G.W. (2006) Mechanisms of maternal mRNA regulation: implications for mammalian early embryonic development. Front Biosci 12: 3713–3726. [DOI] [PubMed] [Google Scholar]
- Bozdech, Z. , Llinás, M. , Pulliam, B.L. , Wong, E.D. , Zhu, J. , and DeRisi, J.L. (2003) The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum . PLoS Biol 1: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés, A. , Carret, C. , Kaneko, O. , Lim, B.Y.Y. , Ivens, A. , and Holder, A.A. (2007) Epigenetic silencing of Plasmodium falciparum genes linked to erythrocyte invasion. PLoS Pathog 3: e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessens, J.T. , Mendoza, J. , Claudianos, C. , Vinetz, J.M. , Khater, E. , Hassard, S. , et al (2001) Knockout of the rodent malaria parasite chitinase pbCHT1 reduces infectivity to mosquitoes. Infect Immun 69: 4041–4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flueck, C. , Bartfai, R. , Volz, J. , Niederwieser, I. , Salcedo‐Amaya, A.M. , Alako, B.T. , et al (2009) Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog 5: e1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke‐Fayard, B. , Trueman, H. , Ramesar, J. , Mendoza, J. , van der Keur, M. , van der Linden, R. , et al (2004) A Plasmodium berghei reference line that constitutively expresses GFP at a high level throughout the complete life cycle. Mol Biochem Parasitol 137: 23–33. [DOI] [PubMed] [Google Scholar]
- Guerreiro, A. , Deligianni, E. , Santos, J.M. , Silva, P.A. , Louis, C. , Pain, A. , et al (2014) Genome‐wide RIP‐Chip analysis of translational repressor‐bound mRNAs in the Plasmodium gametocyte. Genome Biol 15: 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, A.P. , Chin, W.H. , Zhu, L. , Mok, S. , Luah, Y.‐H. , Lim, E.‐H. , and Bozdech, Z. (2013) Dynamic epigenetic regulation of gene expression during the life cycle of malaria parasite Plasmodium falciparum. PLoS Pathog 9: e1003170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse, C. , and Waters, A. (1995) Plasmodium berghei: the application of cultivation and purification techniques to molecular studies of malaria parasites. Parasitol Today 11: 138–143. [DOI] [PubMed] [Google Scholar]
- Janse, C.J. , Franke‐Fayard, B. , Mair, G.R. , Ramesar, J. , Thiel, C. , Engelmann, S. , et al (2006) High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol Biochem Parasitol 145: 60–70. [DOI] [PubMed] [Google Scholar]
- Laurentino, E.C. , Taylor, S. , Mair, G.R. , Lasonder, E. , Bartfai, R. , Stunnenberg, H.G. , et al (2011) Experimentally controlled downregulation of the histone chaperone FACT in Plasmodium berghei reveals that it is critical to male gamete fertility. Cell Microbiol 13: 1956–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Rubio, J.‐J. , Mancio‐Silva, L. , and Scherf, A. (2009) Genome‐wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host & Microbe 5: 179–190. [DOI] [PubMed] [Google Scholar]
- Mair, G.R. , Braks, J.A. , Garver, L.S. , Wiegant, J.C. , Hall, N. , Dirks, R.W. , et al (2006) Regulation of sexual development of Plasmodium by translational repression. Science 313: 667–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair, G.R. , Lasonder, E. , Garver, L.S. , Franke‐Fayard, B.M. , Carret, C.K. , Wiegant, J.C. , et al (2010) Universal features of post‐transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog 6: e1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt, J.N. , Schisa, J.A. , and Priess, J.R. (2000) P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev Biol 219: 315–333. [DOI] [PubMed] [Google Scholar]
- Rodrıguez, M. , Margos, G. , Compton, H. , Ku, M. , Lanz, H. , Rodrıguez, M. , and Sinden, R. (2002) Plasmodium berghei: routine production of pure gametocytes, extracellular gametes, zygotes, and ookinetes. Exp Parasitol 101: 73–76. [DOI] [PubMed] [Google Scholar]
- Salcedo‐Amaya, A.M. , van Driel, M.A. , Alako, B.T. , Trelle, M.B. , van den Elzen, A.M. , Cohen, A.M. , et al (2009) Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum . PNAS 106: 9655–9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schisa, J.A. , Pitt, J.N. , and Priess, J.R. (2001) Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development 128: 1287–1298. [DOI] [PubMed] [Google Scholar]
- Sharma, P. , Wollenberg, K. , Sellers, M. , Zainabadi, K. , Galinsky, K. , Moss, E. , et al (2013) An epigenetic antimalarial resistance mechanism involving parasite genes linked to nutrient uptake. Journal of Biological Chemistry 288: 19429–19440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, U. , Pitt, J. , Dennis, S. , and Priess, J.R. (2010) Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development 137: 1305–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden, R.E. (1997) Infection of mosquitoes with rodent malaria In The molecular biology of insect disease vectors. Crampton J.M., Beard C.B., and Christos L. (eds). Netherlands: Springer, pp. 67–91. [Google Scholar]
- Sinden, R.E. , Butcher, G.A. , and Beetsma, A. (2002) Maintenance of the Plasmodium berghei life cycle In Malaria methods and protocols. Doolan D.L. (ed). New York City: Springer, pp. 25–40. [DOI] [PubMed] [Google Scholar]
- Spence, P.J. , Jarra, W. , Lévy, P. , Reid, A.J. , Chappell, L. , Brugat, T. , et al (2013) Vector transmission regulates immune control of Plasmodium virulence. Nature 498: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelle, M.B. , Salcedo‐Amaya, A.M. , Cohen, A.M. , Stunnenberg, H.G. , and Jensen, O.N. (2009) Global histone analysis by mass spectrometry reveals a high content of acetylated lysine residues in the malaria parasite Plasmodium falciparum . J Proteome Res 8: 3439–3450. [DOI] [PubMed] [Google Scholar]
- Van Dijk, M.R. , Van Schaijk, B.C. , Khan, S.M. , Van Dooren, M.W. , Ramesar, J. , Kaczanowski, S. , et al (2010) Three members of the 6‐cys protein family of Plasmodium play a role in gamete fertility. PLoS Pathog 6: e1000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachou, D. , Zimmermann, T. , Cantera, R. , Janse, C.J. , Waters, A.P. , and Kafatos, F.C. (2004) Real‐time, in vivo analysis of malaria ookinete locomotion and mosquito midgut invasion. Cell Microbiol 6: 671–685. [DOI] [PubMed] [Google Scholar]
- Voss, T.S. , Bozdech, Z. , and Bártfai, R. (2014) Epigenetic memory takes center stage in the survival strategy of malaria parasites. Curr Opin Microbiol 20: 88–95. [DOI] [PubMed] [Google Scholar]
- Yuda, M. , Iwanaga, S. , Shigenobu, S. , Mair, G.R. , Janse, C.J. , Waters, A.P. , et al (2009) Identification of a transcription factor in the mosquito‐invasive stage of malaria parasites. Mol Microbiol 71: 1402–1414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Generation of wt_red230p, wt_chtpred230p and wt_chtpgreen230p transgenic parasites. Schematic representation of the pmCherrycon (A), pchtpmCherry (B) and pchtpgfp (C) expression cassettes that are inserted into the 230p locus via double crossover homologous recombination resulting in the loss of a 1 kb region of the native locus.
D. Diagnostic PCR of clonal parasites corroborating successful integration of the aforementioned expression cassettes.
E. Southern blot analysis of clonal parasites corroborating successful integration of the expression cassettes. The positions of the PCR primers used for the diagnostic PCR reactions are shown (P5, P6 and P7).
Fig. S2. Oocyst load in A. gambiae mosquitoes infected with the Δp48/45green230p parasite. The incomplete defective phenotype of the male gamete defective Δp48/45green230p transgenic parasite results in the escape of very few male gametes that are able to fertilize the normal Δp48/45green230p female gametes to form few ookinetes and thus the few oocysts observed in A. gambiae mosquitoes.
Fig. S3. Allelic expression of fluorescent reporters placed under the control of constitutive gene promoters. In vivo cross‐fertilization assays in A. gambiae mosquitoes directly fed on mice infected with equal numbers of the transgenic parasite lines wt_green230p (ef1α gene promoter) and wt_hsp70pred230p. The first composite panel includes representative fluorescence microscopy pictures of A. gambiae midguts fed on mice co‐infected with two transgenic parasite lines as indicated at the bottom of each part, taken at 24 and 48 hpi respectively. The GFP, mCHERRY and a combination of the two channels (merge) are shown. The second panel is a graph showing the distribution and median number of GFP‐positive, mCHERRY‐positive and GFP/mCHERRY double‐positive parasites per midgut at 24 and 48 hpi. The collective results from three biological replicates are shown, where n is the total number of parasites counted. The third panel shows a genogram summarizing the results of these cross‐fertilization experiments. Squares correspond to male gametocytes, circles correspond to female gametocytes, small ellipses correspond to ookinetes and large ellipses correspond to early stage oocysts. The colour of the outline indicates the genotype, whereas the fill‐in colour indicates the phenotype. Yellow outline indicates gfp/mCherry heterozygotes, whereas yellow fill‐in colour indicates GFP/mCHERRY double‐positive parasites. Black lines indicate the crosses and the resulting progeny. Horizontal black lines indicate the median parasite number. Stars indicate statistical significance determined with the Mann–Whitney U‐test (***P < 0.001).
Fig. S4. GFP expression in the wt_chtpgreen230p parasite line.
A. RT‐PCR analysis of gfp transcripts from mixed blood stages, activated (A) and non‐activated (nA) gametocytes, and parasites purified from in vitro ookinete cultures at 2, 4, 8, 12, 14, 16 and 24 h post‐fertilization. Tubulin transcripts served as a loading control.
B. Fluorescence microscopy analysis of wt_chtpgreen230p parasites purified from in vitro ookinete cultures at 2, 4, 8, 12, 14, 16 and 24 hpa. Parasites were also stained with a Cy3 conjugated α‐P28 antibody. Images were taken at ×40 magnification.
Fig. S5. Expression of mCHERRY in the wt_chtpred230p parasite line.
A. RT‐PCR analysis of mCherry transcripts from mixed blood stages, activated (A) and non‐activated (nA) gametocytes, and parasites purified from in vitro ookinete cultures at 2, 4, 8, 12, 14, 16 and 24 h post‐fertilization. Tubulin transcripts served as a loading control.
B. Fluorescence microscopy analysis of wt_chtpred230p parasites purified from in vitro ookinete cultures at 4, 8, 12, 14, 16 and 24 hpa. Bright field images are also shown. Images were taken at ×40 magnification.
Table S1. Parasite development in A. gambiae mosquitoes co‐infected with parasites constitutively expressing GFP and mCHERRY.
Table S2. Oocyst numbers in A. gambiae infections.
Table S3. In vitro cross‐fertilization assays of GFP and mCHERRY expressing parasites.
Table S4. Parasite development in A. gambiae mosquitoes co‐infected with parasites expressing GFP and mCHERRY during the zygote/ookinete stages.
Table S5. Primers for the generation of transgenic parasites and RT‐PCR.


