Abstract
Carnitine is essential for fatty acid metabolism, but is associated with both health benefits and risks, especially heart diseases. We report the identification of potent, selective and cell active inhibitors of γ-butyrobetaine hydroxylase (BBOX), which catalyses the final step of carnitine biosynthesis in animals. A crystal structure of BBOX in complex with a lead inhibitor reveals that it binds in two modes, one of which adopts an unusual ‘U-shape’ conformation stabilised by inter- and intra-molecular π-stacking interactions. Conformational changes observed on binding of the inhibitor to BBOX likely reflect those occurring in catalysis; they also rationalise the inhibition of BBOX by high levels of its substrate γ-butyrobetaine (GBB), as observed both with isolated BBOX protein and in cellular studies.
Introduction
Carnitine plays a central role in the metabolism of long chain fatty acids, by enabling their transport into mitochondria where they undergo β-oxidation1-3. Carnitine is produced endogenously by all animals and is commonly used as a human food supplement. It is proposed that high levels of carnitine in red meat cause heart disease via metabolism of carnitine leading to production of trimethylamine N-oxide (TMAO), which promotes arteriosclerosis4. In animals carnitine biosynthesis starts from Nε-trimethyllysine, which is produced from proteins containing N-methylated lysine residues, and comprises four enzymatically catalysed steps5,6 (Fig. S1). The final step in carnitine biosynthesis is catalysed by γ-butyrobetaine hydroxylase (BBOX). BBOX and trimethyllysine hydroxylase (TMLH), which catalyses the first step of carnitine biosynthesis, belong to the superfamily of Fe(II) and 2OG dependent dioxygenases7 (Fig. 1A). BBOX catalyses the stereospecific8 C-3 hydroxylation of γ-butyrobetaine (GBB) to give l-carnitine (Fig. 1A, S1).
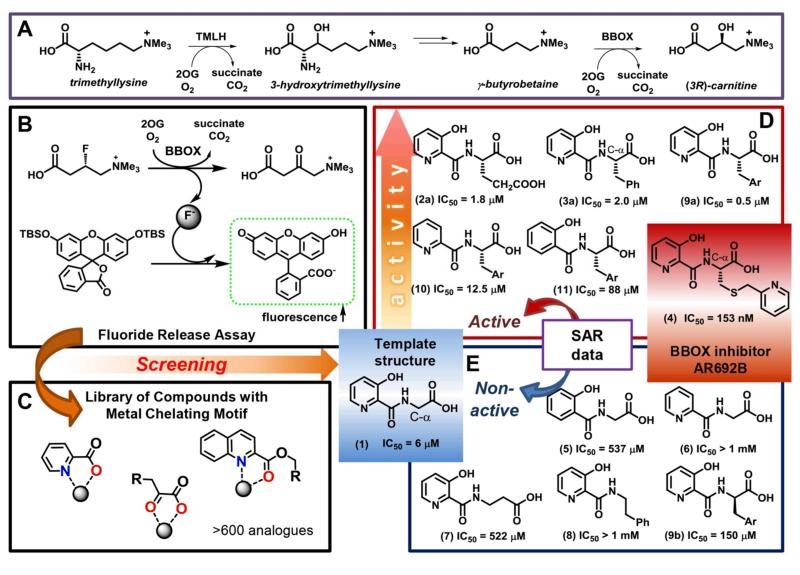
Figure 1. Identification of selective inhibitors of carnitine biosynthesis.
AR692B inhibits BBOX – the final enzyme of carnitine biosynthesis and an Fe(II)/ 2-oxoglutarate dependent oxygenase(A). A ‘fluoride release’ assay (B) enables efficient screening of potential inhibitors (C) and the identification of AR692B as a potent BBOX inhibitor. Structure–activity relationship results (D, E) imply that the pyridine nitrogen, the C-3 phenolic hydroxyl, hydrophobic side chain and carboxylate contribute to inhibition.
Cardiac ischemia and related conditions are major causes of mortality in many countries9. In normal heart glucose and fatty acid metabolism is tightly regulated with fatty acid acting as a main source of energy; however in failing heart this homeostasis is perturbed10,11. A shift towards glucose oxidation under ischemic conditions has been reported to aid in cardiac efficiency and improve heart function in both the long and short term12. The manipulation of the preferred cellular energy source is therefore of interest with respect to pharmacological intervention in cardiovascular disease. Such manipulation can potentially be achieved by modulating carnitine biosynthesis.
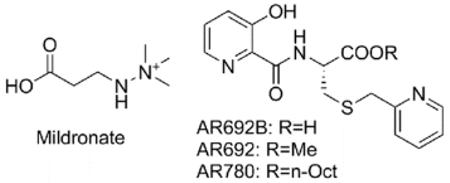
The clinically used compound Mildronate (THP, Met-88) is given to patients after myocardial infarction with the aim of reducing fatty acid oxidation and associated reactive oxidative species13-15. Mildronate is both a relatively weak BBOX inhibitor in vitro (IC50 value of 60 μM16) and a competitive substrate17,18, producing multiple products. Moreover, the mode of action of Mildronate may be non-selective19,20; Mildronate is a structural mimic of both GBB and carnitine (Fig. S2) and likely interacts with other GBB/ carnitine binding proteins/ import channels21.
Human 2-oxoglutarate (2OG) dependent dioxygenases catalyse multiple hydroxylations and N-demethylations via methyl group hydroxylations7,22. Several classes of human 2OG oxygenases are current medicinal targets23, including enzymes involved in chromatine regulation, histone demethylases, nucleic acid oxidising enzymes and the hypoxia inducible factor hydroxylases. However, other than the use of likely non-selective GBB-analogues24-27 there has been little reported on the inhibition of carnitine biosynthesis. Thus, there is a need for the development of selective and cell-active inhibitors of carnitine biosynthesis to investigate the pathophysiological roles of carnitine. We report the identification of selective BBOX inhibitors, active against both isolated enzyme and in cells. Crystallographic analyses reveal the lead inhibitor compound adopts an unusual binding mode and induces conformational changes that reflect regulation of carnitine biosynthesis by a substrate-mediated inhibition of BBOX.
Results and discussion
Inhibitor development
We focused on targeting the 2OG binding pocket of BBOX23, in part because this approach should enable specificity over other carnitine interacting enzymes. We applied an efficient assay, based on release of fluoride ions by BBOX catalysed hydroxylation of (3S)-fluoro-γ-butyrobetaine (GBBF), followed by fluoride mediated activation of a fluorescent reporter molecule (Fig. 1, S3)16. The assay was used to screen a set of > 600 compounds that are potential chelators of the essential ferrous ion28 in the active site of 2OG dependent oxygnases such as BBOX. This screen led to the identification of (3-hydroxypicolinoyl)glycine (1) as a potential scaffold for the development of selective BBOX inhibitors (Fig. 1).
Analysis of crystal structures for BBOX17,29 reveals the presence of an unoccupied hydrophobic pocket directly adjacent to the 2OG methylene binding region and located on the interior of the active site (Fig. 2). Docking studies implied that the glycine moiety (CH2COOH) of (1) likely binds analogously to the (CH2)2COOH group of 2OG. We therefore prepared analogues of (1) derivatised at the C-α position of the modified amino acid. We found that the derivatisation of glycine at its pro-(S) but not pro-(R) position led to increased potency, with bulky side chains, such as phenyl (3a), 3-indole (9a) or pyridinyl (4) groups, being preferred (Fig. 1, S4, S5). On the basis of structural work on 2OG oxygenase inhibitor complexes30,31, we predicted that the identified ‘template’ BBOX inhibitor would bind to the active site iron via its carbonyl group and either its pyridine-nitrogen or C-3 hydroxyl group (Fig. S6)30. We established that the pyridine-nitrogen, C-3 hydroxyl group and side chain carboxylate are essential for binding, since structures lacking any of these elements (compounds 5-6, 8) were not active (Fig. 1, S4). When the bulky side chain is present, analogues lacking either the pyridine-nitrogen or C-3 hydroxyl group retain some activity, whilst analogues lacking a side chain carboxylate are inactive (Fig. S4).
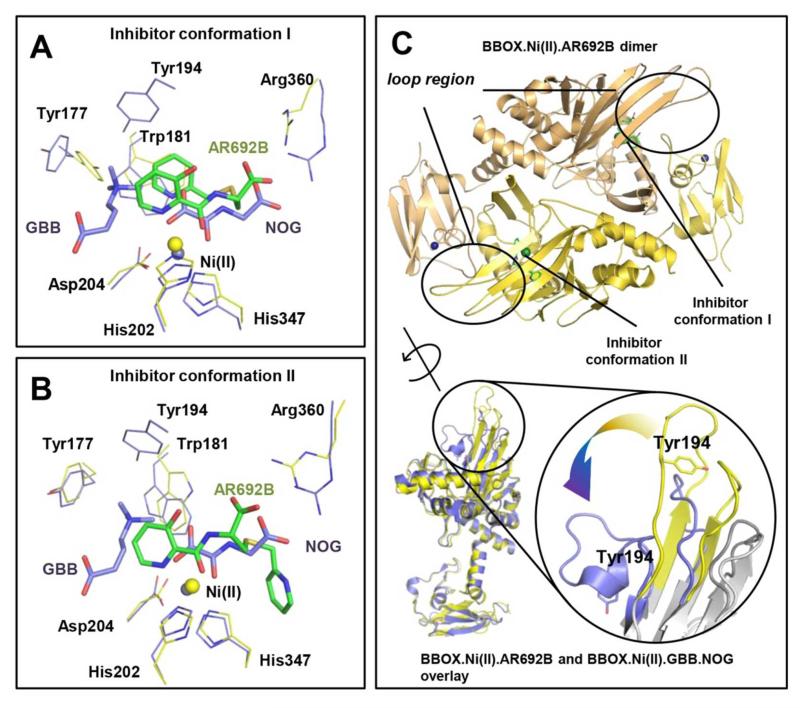
Figure 2. AR692B inhibits BBOX via a structurally unusual mechanism.
The inhibitor adopts 2 conformations (A and B) in a complexed with BBOX. In 3 of the monomers in the asymmetric unit inhibitor ‘folds-back’ on itself to form a sandwich type structure stabilised by inter- and intra-molecular π-stacking interactions involving both of its pyridine rings and Trp181, Tyr 177 of BBOX. In the alternative binding mode, observed in the 3 other monomers of the asymmetric unit, the thioether linked pyridine side chain occupies a hydrophobic pocket adjacent to the 2OG binding site. Overlays of BBOX.AR692B complex (yellow) and BBOX.GBB.NOG complex (blue) reveals a steric clash of inhibitor with GBB. NOG (N-oxalylglycine) is a close structural mimic of 2OG. C – BBOX functions as a dimer. Overlay of BBOX.Ni(II).NOG.GBB (blue, PDB: 3O2G) and BBOX.Ni(II).AR692B (yellow, PDB: 4C8R) complexes reveals substantial movement of the ‘βI/βII loop’ (observed in both conformers). Note, the position of Tyr194 changes from an ‘open’ (yellow) to a ‘closed’ (blue) conformation upon binding of GBB and formation of the –NMe3+ binding pocket.
We also examined scaffolds with bicyclic aromatic rings, i.e. with quinoline and isoquinoline derivatives substituting for the (hydroxyl)pyridine ring. Quinolines were found to have rather weak potency (Table S1B), but isoquinolines were relatively good inhibitors with the IC50 values in the low micromolar range, providing they were functionalised with a hydroxyl group in the meta- position to the isoquinoline nitrogen (Fig. S7, Table S1C). As in the case of the pyridine derivatives, removal of the nitrogen or hydroxyl group from the N-acyl aromatic ring led to loss of potency (Fig. S7). In the isoquinoline series, C-α side chains with the (S)-stereochemistry were also preferred over those with the (R)-stereochemistry. However, in contrast to the pyridine series small side chains were preferred with the hydroxy-isoquinolines, with the methyl group having the best inhibitory properties (Table S1C).
Collectively the results imply that the different series all bind the active site iron in a bidentate manner, but either the C-α side chain can adopt different conformations or different coordination modes are possible, i.e. exploiting hydroxyl and carbonyl groups (Fig. S6)31-33. They also suggested that sufficent potency might be achieved with the 3-hydroxy pyridine series, so we carried out the synthesis of further analogues in this series including with the extended link between the C-α centre and the aromatic group on the side chain. These studies led to the identification of the thioether linked pyridine derivative AR692B (4) as a candidate for further investigation.
Kinetic evaluation of BBOX inhibition by AR692B using 1H NMR17, employing GBB – the natural BBOX substrate - reveals AR692B to be a potent inhibitor (IC50 153 nM, Fig. S8), and that inhibition is both 2OG and GBB concentration dependent (Fig. S9). We have also examined binding of inhibitors containing 3-hydroxypyridine scaffold to BBOX. A binding assay was developed based on the observation that the potent BBOX inhibitors caused quenching of the intrinsic fluorescence of BBOX (Fig. S10). This method enabled determination of binding curves (Fig. S11) and KD values of 0.5 μM and 10.8 μM for AR692B and (1), respectively. The results further demonstrate how high potency and selective oxygenase inhibition can be achieved by simultaneously targeting both substrate and 2OG binding pockets34,35. We tested AR692B against a panel of seven other human 2OG oxygenases including histone demethylases (KDMs) and hypoxia inducible factor (HIF) hydroxylases33,36,37. AR692B was inactive against the tested KDMs at 100 μM, and had only weak affinity towards the catalytic domain of the HIF prolyl hydroxylase 2 isoform (PHD2) (Table S2), with an IC50 of 28 μM (Fig. S12). The reduced activity with PHD2 likely reflects the relatively tight 2OG binding pocket of PHD238 compared to that of BBOX, which hinders binding of the bulky side chain of AR692B.
Cell based studies
To evaluate the activity of AR692B in cells, we developed an LC-MS based method for detection of carnitine biosynthesis metabolites (Fig. S13). Because AR692B displayed lower activity in cells compared to the in vitro assays with isolated protein; prodrug forms of AR692B were also synthesized for use in cellular studies, i.e. methyl (AR692) and octyl ester (AR780) derivatives (Table 1). The octyl ester was chosen to improve liphophilicity of the AR692B in order to increase cell permeability. Studies on HEK 293T cells demonstrated both AR692B and AR692 were not cytotoxic up to concentrations of 0.5 mM; AR780 induced reduction in cell growth and survival at > 0.06 mM. Both AR692B and AR692 induced > 40% reduction in carnitine levels at 100 μM. The octyl ester AR780 was more potent with ~60% reduction of free carnitine levels when used at 10 μM and > 75% reduction at 50 μM (Fig. S14). The improved cellular potency of the octyl ester (AR780) is likely due to its increased cell-membrane permeability. Mildronate was less active than AR780 giving 63% reduction of carnitine levels at 50 μM and 30% reduction at 20 μM (Table 1). Mildronate was not active below 5 μM. Both methyl and octyl ester derivatives were inactive against BBOX in an in vitro assay, implying that they are hydrolysed to AR692B in cells.
Table 1.
Inhibition of carnitine biosynthesis in HEK 293T cells.
| ||
|---|---|---|
| Compound | Concentration of compound [μM] | Carnitine levels* |
| AR692 | 100 | 52 ± 4 |
| AR692 | 300 | 33 ± 3 |
| AR692B | 100 | 57 ± 9 |
| AR780 | 50 | 21 ± 7 |
| AR780 | 10 | 40 ± 4 |
| Mildronate | 50 | 37 ±10 |
| Mildronate | 20 | 70 ± 6 |
Measured carnitine levels are given as percentage of free carnitine levels obtained for cells dosed with DMSO (control). Control was set to 100% (SEM ± 13%).
Crystallographic analyses
To investigate the mode of AR692B inhibition, we solved a crystal structure of hBBOX in complex with AR692B and Ni (an Fe(II) surrogate), P212121 space group, 2.8 Å resolution. The overall fold of BBOX in complex with AR692B is similar to the fold observed previously for BBOX17,29, i.e. comprised of eight β-strands (βI-VIII), surrounded by structural elements common to other 2OG oxygenases and some specific to the BBOX subfamily (DSBH: the core double stranded β-helix of the 2OG oxygenases, which has eight β-strands, I→VIII39,40). Attempts to crystallize BBOX with GBB in the presence of AR692B were unsuccessful, likely reflecting a competitive mode of BBOX inhibition with GBB as well as 2OG. The structure contains 6 BBOX molecules per asymmetric unit corresponding to 3 homodimers. In each of the 6 chains, additional difference density (Fo-Fc), assigned as AR692B, was observed at the active site (Fig. S15). Consistent with the SAR results, in all 6 chains the active site metal is chelated in a bidentate fashion by the AR692B pyridine nitrogen (trans to His347) and the side chain carbonyl oxygen (trans to Asp204) of AR692B, in addition to the conserved HXD ..H motif and a water molecule. Like the C5-carboxylate of 2OG17, the AR692B carboxylate side chain is positioned to form electrostatic interactions with the guanidinium group of Arg360 (Fig. 2A and 2B). The pyridine ring of the C4 side chain appears to have 2 different binding modes (from the thioether bond) (Fig. 2A and 2B). Based on the OMIT Fo-Fc electron density maps we modelled the binding mode with >70% occupancy (Fig. S16). In conformer I (modelled in 3 monomers), the AR692B aromatic side chain is ‘sandwiched’ between the metal-coordinating pyridine ring of AR692B and a hydrophobic region of BBOX formed by the side chains of Tyr177, Trp181 and Val183 (Fig. 2A, S17). Such a ‘U-shaped’ inhibitor binding mode is unprecedented, in available 2OG oxygenases inhibitor complexes. In conformer II the aromatic side chain is positioned to interact in the hydrophobic cleft (adjacent to the 2OG methylene group binding site) formed by the Leu217 (DSBH β-III), Ile338 and Phe340 (DSBH β-III) side chains (Fig. 2B, S18). The observation of two binding modes for the inhibitor is notable, because it has been proposed that the ability of an inhibitor to adopt multiple conformations may help to enable a ‘biologically robust’ inhibition mechanism41,42.
Comparison of the BBOX-AR692B structure with those with and without substrate/ inhibitor (PDB ID: 3O2G17/ 3N6W29) reveals substantial conformational differences (Fig. S19). In both AR692B binding modes, the metal-coordinating pyridine of AR692B causes a steric clash with a loop (residues 183-199, ‘βI/βII loop’) involved in GBB binding17. In the BBOX.GBB.NOG complex structure this loop conatins a short 310 helix linking DSBH strands βI and βII (residues 183-199, ‘βI/βII loop’ hereafter, NOG – N-oxalylglycine is a close, non-reactive 2OG analogue). Tyr194 of the ‘βI/βII loop’ forms part of an aromatic cage that binds the GBB trimethyl ammonium group via hydrophobic and π-cation interactions43 and acts as a ‘lid’ that encloses the GBB binding site. The clash between AR692B and the ‘βI/βII loop’ causes this part of the structure to unfold and form extended βI and βII strands (Fig. 2C).
The observation that the ‘βI/βII loop’ adopts an ‘open’ conformation with AR692B and is apparently unstructured in the BBOX apo form29 (PDB ID: 3N6W), but folds to isolate the active site once GBB is bound17 (PDB ID: 3O2G) reveals conformational flexibility as a feature of BBOX catalysis (Fig. 3). The ‘closed’ conformation is likely stabilised by π-cation interactions of the trimethylammonium group of GBB with the residues from the aromatic cage. As judged from the structure of BBOX in complex with AR692B (which is a 2OG mimetic), 2OG likely does not stabilise the βI/βII loop in a ‘closed’ conformation, so enabling binding of GBB substrate after that of 2OG.
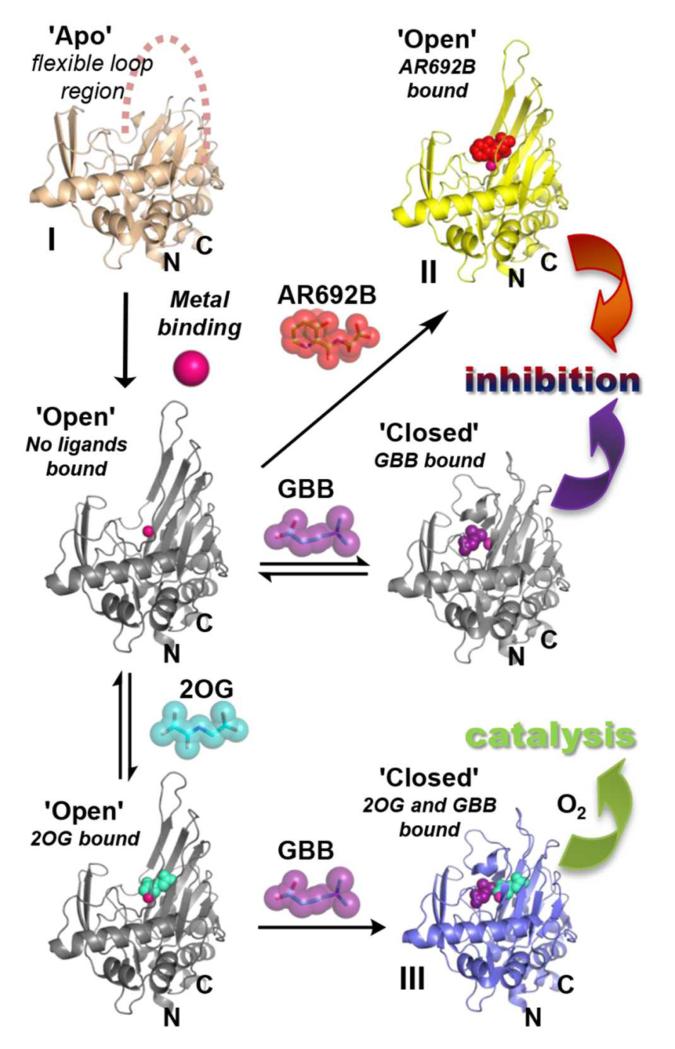
Figure 3. Conformational changes during BBOX inhibition/catalysis.
The combined BBOX structures imply a flexible loop (residues 183-199, ‘βI/βII loop’) can fold to enclose the active site. During catalysis 2OG binding is followed by GBB, then binding of O2. Binding of GBB stabilises the ‘closed’ conformation by π-cation interactions in an aromatic cage (Figure 2C). BBOX substrate inhibition may occur when GBB binds prior to 2OG and ‘closed’ loop conformation is stabilised hindering 2OG binding. Binding of AR692 competes with both 2OG and GBB and induces the open loop conformation. Only the catalytic domain is shown (residues 106 – 384); grey structures represent predicted conformations with modeled ligands. The catalytic domains in structures II and III havean active site metal bound (Ni(II)), while the structure of the apo-catalytic domain (I) does not have active site metal bound. Metals are shown as pink spheres. PDB codes of BBOX complexes: I – 3N6W (in this structure the loop was unresolved)29, II – 4C8R, III – 3O2G.
It is known that BBOX is inhibited by high concentrations of its GBB substrate in assays with isolated enzyme (Fig. S20)17,44. Analysis of the combined structures reveals that neither GBB nor 2OG (at least efficiently) binds to BBOX once the ‘βI/βII loop’ is in the closed conformation, because of inaccessibility of the active site. The structural studies suggest that the observed substrate inhibition is due to GBB binding prior to 2OG (Fig. 3) and disrupting of the ordered sequential mechanism of the 2OG oxygenases7, i.e. initial GBB binding prevents that of 2OG, by supporting a ‘closed’ loop conformation. In support of a biological role for the regulation of carnitine biosynthesis via GBB-mediated inhibition of BBOX, we found that carnitine levels in cells are reduced by ~50% after the addition of GBB at 20 μM (Fig. S20), supporting the proposal that GBB substrate inhibition may be involved in a negative feedback mechanism.
Conclusions
In conclusion, we have identified AR692B as a potent and selective inhibitor for human BBOX. AR692B was demonstrated to reduce carnitine levels in cells and will act as a tool for understanding the roles of carnitine biosynthesis in health and disease. A structure of BBOX complexed with AR692B reveals dual binding modes including a protein assisted formation of the inhibitor ‘sandwich’ type complex. Observation of the conformational differences, i.e. active site loop movement, likely reflect BBOX dynamics during the catalytic cycle in solution and prompted work leading to the finding that BBOX activity in cells is regulated by GBB-mediated substrate inhibition. BBOX is also kinetically unusual among 2OG oxygenases in that it has relatively high KM for 2OG (160 μM versus more typical values of 1-10 μM)33, leading to the possibility that its activity in cells may be linked to 2OG availability. Thus, modulation of BBOX activity may be of interest from the perspective of the range of diseases linked to abnormal fatty acid and TCA cycle metabolism45.
Supplementary Material
Acknowledgments
We thank the Biotechnology and Biological Sciences Research Council (BBSRC), the Wellcome Trust, European Union, BHF Centre of Research Excellence, Oxford (A.K.) and Dulverton Trust (A.R.) for funding. We thank the following for the preparation of the compounds used in the initial screening set for BBOX: M. Demetriades, Dr. C. Lejeune, C. C. Thinnes, O. Chang, K. S. England, Dr. E. C. Y. Woon, Dr. K. K. Yeoh, Dr. J. I. Candela-Lena, V. Gomez-Perez, Dr. L. Henry, and Dr. B. Banerji. Details will be published elsewhere.
Footnotes
Electronic Supplementary Information (ESI) available: Experimental details including full synthetic procedures, in vitro and cell based assay conditions and crystallography data. See DOI: 10.1039/b000000x/
References
- 1.Rebouche CJ, Seim H. Annu. Rev. Nutr. 1998;18:39. doi: 10.1146/annurev.nutr.18.1.39. [DOI] [PubMed] [Google Scholar]
- 2.Bremer J. J. Clin. Chem. Clin. Biochem. 1990;28:297. [PubMed] [Google Scholar]
- 3.Hoppel C. Am. J. Kidney Dis. 2003;41:S4. doi: 10.1016/s0272-6386(03)00112-4. [DOI] [PubMed] [Google Scholar]
- 4.Koeth RA, Wang ZE, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu XM, Wu YP, Li L, Smith JD, DiDonato JA, Chen J, Li HZ, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WHW, Bushman FD, Lusis AJ, Hazen SL. Nat. Med. 2013;19:576. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strijbis K, Vaz FM, Distel B. IUBMB Life. 2010;62:357. doi: 10.1002/iub.323. [DOI] [PubMed] [Google Scholar]
- 6.Vaz FM, Wanders RJA. Biochem. J. 2002;361:417. doi: 10.1042/0264-6021:3610417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hausinger RP. Crit. Rev. Biochem. Mol. Biol. 2004;39:21. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 8.Englard S, Blanchard JS, Midelfort CF. Biochemistry. 1985;24:1110. doi: 10.1021/bi00326a007. [DOI] [PubMed] [Google Scholar]
- 9.McMurray JJV, Pfeffer MA. Lancet. 2005;365:1877. doi: 10.1016/S0140-6736(05)66621-4. [DOI] [PubMed] [Google Scholar]
- 10.Wanders RJ, Vreken P, den Boer ME, Wijburg FA, van Gennip AH, Ijlst L. J. Inherit. Metab. Dis. 1999;22:442. doi: 10.1023/a:1005504223140. [DOI] [PubMed] [Google Scholar]
- 11.Jaswal JS, Keung W, Wang W, Ussher JR, Lopaschuk GD. Biochim. Biophys. Acta-Mol. Cell Res. 2011;1813:1333. doi: 10.1016/j.bbamcr.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 12.Lopaschuk GD. Semin. Cardiothorac. Vasc.Anesth. 2006;10:228. doi: 10.1177/1089253206291150. [DOI] [PubMed] [Google Scholar]
- 13.Dambrova M, Liepinsh E, Kalvinsh I. Trends Cardiovasc. Med. 2002;12:275. doi: 10.1016/s1050-1738(02)00175-5. [DOI] [PubMed] [Google Scholar]
- 14.Liepinsh E, Vilskersts R, Loca D, Kirjanova O, Pugovichs O, Kalvinsh I, Dambrova M. J. Cardiovasc. Pharmacol. 2006;48:314. doi: 10.1097/01.fjc.0000250077.07702.23. [DOI] [PubMed] [Google Scholar]
- 15.Vilskersts R, Liepinsh E, Mateuszuk L, Grinberga S, Kalvinsh I, Chlopicki S, Dambrova M. Pharmacology. 2009;83:287. doi: 10.1159/000210015. [DOI] [PubMed] [Google Scholar]
- 16.Rydzik AM, Leung IKH, Kochan GT, Thalhammer A, Oppermann U, Claridge TDW, Schofield CJ. ChemBioChem. 2012;13:1559. doi: 10.1002/cbic.201200256. [DOI] [PubMed] [Google Scholar]
- 17.Leung IKH, Krojer TJ, Kochan GT, Henry L, von Delft F, Claridge TDW, Oppermann U, McDonough MA, Schofield CJ. Chem. Biol. 2010;17:1316. doi: 10.1016/j.chembiol.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 18.Henry L, Leung IKH, Claridge TDW, Schofield CJ. Bioorg. Med. Chem. Lett. 2012;22:4975. doi: 10.1016/j.bmcl.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 19.Zvejniece L, Svalbe B, Makrecka M, Liepinsh E, Kalvinsh I, Dambrova M. Behav. Pharmacol. 2010;21:548. doi: 10.1097/FBP.0b013e32833d5a59. [DOI] [PubMed] [Google Scholar]
- 20.Sjakste N, Gutcaits A, Kalvinsh I. CNS Drug Rev. 2005;11:151. doi: 10.1111/j.1527-3458.2005.tb00267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaudzems K, Kuka J, Gutsaits A, Zinovjevs K, Kalvinsh I, Liepinsh E, Liepinsh E, Dambrova M. J. Enzym. Inhib. Med. Chem. 2009;24:1269. doi: 10.3109/14756360902829527. [DOI] [PubMed] [Google Scholar]
- 22.Walport LJ, Hopkinson RJ, Schofield CJ. Curr. Opin. Chem. Biol. 2012;16:525. doi: 10.1016/j.cbpa.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Rose NR, McDonough MA, King ON, Kawamura A, Schofield CJ. Chem. Soc. Rev. 2011;40:4364. doi: 10.1039/c0cs00203h. [DOI] [PubMed] [Google Scholar]
- 24.Petter RC, Banerjee S, Englard S. J. Org. Chem. 1990;55:3088. [Google Scholar]
- 25.Ziering DL, Pascal RA. J. Am. Chem. Soc. 1990;112:834. [Google Scholar]
- 26.Rebouche CJ. Faseb J. 1996;10:2918. [Google Scholar]
- 27.Blanchard JS, Englard S. Biochemistry. 1983;22:5922. doi: 10.1021/bi00294a036. [DOI] [PubMed] [Google Scholar]
- 28.Schofield CJ, Zhang Z. Curr. Opin. Struct. Biol. 1999;9:722. doi: 10.1016/s0959-440x(99)00036-6. [DOI] [PubMed] [Google Scholar]
- 29.Tars K, Rumnieks J, Zeltins A, Kazaks A, Kotelovica S, Leonciks A, Sharipo J, Viksna A, Kuka J, Liepinsh E, Dambrova M. Biochem. Biophys. Res. Commun. 2010;398:634. doi: 10.1016/j.bbrc.2010.06.121. [DOI] [PubMed] [Google Scholar]
- 30.Chowdhury R, Candela-Lena JI, Chan MC, Greenald DJ, Yeoh KK, Tian Y-M, McDonough MA, Tumber A, Rose NR, Conejo-Garcia A, Demetriades M, Mathavan S, Kawamura A, Lee MK, van Eeden F, Pugh CW, Ratcliffe PJ, Schofield CJ. ACS Chem. Biol. 2013;8:1488. doi: 10.1021/cb400088q. [DOI] [PubMed] [Google Scholar]
- 31.Poppe L, Tegley CM, Li V, Lewis J, Zondlo J, Yang E, Kurzeja RJ, Syed R. J. Am. Chem. Soc. 2009;131:16654. doi: 10.1021/ja907933p. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, Ren J, Harlos K, McKinnon CH, Clifton IJ, Schofield CJ. FEBS Lett. 2002;517:7. doi: 10.1016/s0014-5793(02)02520-6. [DOI] [PubMed] [Google Scholar]
- 33.Rose NR, Woon ECY, Tumber A, Walport LJ, Chowdhury R, Li XS, King ONF, Lejeune C, Ng SS, Krojer T, Chan MC, Rydzik AM, Hopkinson RJ, Che KH, Daniel M, Strain-Damerell C, Gileadi C, Kochan G, Leung IKH, Dunford J, Yeoh KK, Ratcliffe PJ, Burgess-Brown N, von Delft F, Muller S, Marsden B, Brennan PE, McDonough MA, Oppermann U, Klose RJ, Schofield CJ, Kawamura A, A. J. Med. Chem. 2012;55:6639. doi: 10.1021/jm300677j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo X, Liu Y, Kubicek S, Myllyharju J, Tumber A, Ng S, Che KH, Podoll J, Heightman TD, Oppermann U, Schreiber SL, Wang X. J. Am. Chem. Soc. 2011;133:9451. doi: 10.1021/ja201597b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woon ECY, Tumber A, Kawamura A, Hillringhaus L, Ge W, Rose NR, Ma JHY, Chan MC, Walport LJ, Che KH, Ng SS, Marsden BD, Oppermann U, McDonough MA, Schofield CJ. Angew. Chem.-Int. Edit. 2012;51:1631. doi: 10.1002/anie.201107833. [DOI] [PubMed] [Google Scholar]
- 36.King ON, Li XS, Sakurai M, Kawamura A, Rose NR, Ng SS, Quinn AM, Rai G, Mott BT, Beswick P, Klose RJ, Oppermann U, Jadhav A, Heightman TD, Maloney DJ, Schofield CJ, Simeonov A. PloS one. 2010;5:e15535. doi: 10.1371/journal.pone.0015535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hopkinson RJ, Tumber A, Yapp C, Chowdhury R, Aik W, Che KH, Li XS, Kristensen JBL, King ONF, Chan MC, Yeoh KK, Choi H, Walport LJ, Thinnes CC, Bush JT, Lejeune C, Rydzik AM, Rose NR, Bagg EA, McDonough MA, Krojer TJ, Yue WW, Ng SS, Olsen L, Brennan PE, Oppermann U, Muller S, Klose RJ, Ratcliffe PJ, Schofield CJ, Kawamura A. Chem. Sci. 2013;4:3110. doi: 10.1039/C3SC51122G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonough MA, Li V, Flashman E, Chowdhury R, Mohr C, Liénard BMR, Zondlo J, Oldham NJ, Clifton IJ, Lewis J, McNeill LA, Kurzeja RJM, Hewitson KS, Yang E, Jordan S, Syed RS, Schofield CJ. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9814. doi: 10.1073/pnas.0601283103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clifton IJ, McDonough MA, Ehrismann D, Kershaw NJ, Granatino N, Schofield CJ. J. Inorg. Biochem. 2006;100:644. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 40.Aik W, McDonough MA, Thalhammer A, Chowdhury R, Schofield CJ. Curr. Opin. Struct. Biol. 2012;22:691. doi: 10.1016/j.sbi.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Wilkinson A-S, Ward S, Kania M, Page MGP, Wharton CW. Biochemistry. 1999;38:3851. doi: 10.1021/bi990030i. [DOI] [PubMed] [Google Scholar]
- 42.Kerry PS, Mohan S, Russell RJM, Bance N, Niikura M, Pinto BM. Sci. Rep. 2013;3:2871. doi: 10.1038/srep02871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dougherty DA. J. Nutr. 2007;137:1504S. doi: 10.1093/jn/137.6.1504S. [DOI] [PubMed] [Google Scholar]
- 44.Lindstedt G, Lindstedt S, Nordin I. Scand. J. Clin. Lab. Invest. 1982;42:477. [PubMed] [Google Scholar]
- 45.Yang M, Soga T, Pollard PJ. J. Clin. Invest. 2013;123:3652. doi: 10.1172/JCI67228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.