Abstract
Background
The barnacle Balanus amphitrite is widely distributed in marine shallow and tidal waters, and has significant economic and ecological importance. Nauplii, the first larval stage of most crustaceans, are extremely abundant in the marine zooplankton. However, a lack of genome information has hindered elucidation of the molecular mechanisms of development, settlement and survival strategies in extreme marine environments. We sequenced and constructed the genome dataset for nauplii to obtain comprehensive larval genetic information. We also investigated iTRAQ-based protein expression patterns to reveal the molecular basis of nauplii development, and to gain information on larval survival strategies in the Red Sea marine environment.
Results
A nauplii larval transcript dataset, containing 92,117 predicted open reading frames (ORFs), was constructed and used as a reference for the proteome analysis. Genes related to translation, oxidative phosphorylation and cytoskeletal development were highly abundant. We observed remarkable plasticity in the proteome of Red Sea larvae. The proteins associated with development, stress responses and osmoregulation showed the most significant differences between the two larval populations studied. The synergistic overexpression of heat shock and osmoregulatory proteins may facilitate larval survival in intertidal habitats or in extreme environments.
Conclusions
We presented, for the first time, comprehensive transcriptome and proteome datasets for Red Sea nauplii. The datasets provide a foundation for future investigations focused on the survival mechanisms of other crustaceans in extreme marine environments.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-2262-1) contains supplementary material, which is available to authorized users.
Keywords: B. amphitrite, Red sea, Transcriptomics, Proteomics, Adaptation
Background
Barnacles are marine dominant fouling organisms accumulate on ship hulls, piers and under water structures; biofouling by these invertebrates results in an increase in power and fuel consumption [1–3] associated with marine transport. Removing biofouling barnacles is a major expense involving significant economic losses in shipping and other marine industries. However, some barnacles are of great commercial importance; for example, the gooseneck barnacle achieves an average market price of 30–60 USD per kilogram, depending on the season [4]. Such species are highly exploited, involving harvesting of approximately 500 kg per season in Spain and Portugal, and hence there is an urgent need for strategies to protect the stocks. The barnacle Balanus amphitrite is a widely distributed dominant fouling organism and a major component of rocky shore benthic communities [5–7]. Therefore, B. amphitrite is an excellent model organism for research on settlement biology, adaptation and biofouling. Nauplii are the earliest free-swimming stage in the development of most crustaceans, including barnacles [8], and are extremely abundant in the marine ecosystems. Barnacle embryos are brooded by the parents, and the nauplii are released into the water column as swimming larvae [9]. The nauplius I stage has an elongated and pear-shaped body, and the abdominal and caudal spines are partially differentiated. The eye is located in a front medial position and occurs in all developmental stages. The presence of posteriorly folded frontal horns and simple setae on the limbs are characteristics of the nauplius I stage. The newly released larvae swim actively and molt to the next developmental stage (nauplius II) within 30 min [10]. In the nauplius II stage the carapace extends in all directions and the entire body develops a bell shape that has numerous small spines and a pair of prominent spines. The nauplius II molts to the nauplius III stage in 38 h [11]. The nauplius III stage undergoes several moults before it transforms into a cyprid stage [12]. Molecular studies defining the role of gene or protein expression signatures in development and adaptive strategies of marine larvae in the natural marine environment are limited. Previous studies have used a broad range of proteomics and genomic tools to identify proteins and genes in larval stages of barnacles. However, these studies have mainly focused on exploring settlement and attachment processes. For example, Chen et al. (2011) identified 7954 putative differentially-expressed genes in larvae and adults of B. amphitrite using 454 pyrosequencing [13], and Yan et al. (2012) carried out in silico transcriptome data mining to study neuropeptides and their possible functional role in barnacle larval settlement [14]. Similarly, De Gregoris et al. (2012) generated deep sequencing EST libraries for nauplii and the cyprid and adult stages of B. amphitrite and identified unique contigs for each developmental stage [15]. More recently, Lin et al. (2014) performed Illumina sequencing of a membranous-based barnacle and identified cement- and adhesion-related genes [16]. Thiyagarajan and Qian (2008) reported the first study of global proteome expression patterns in cyprids and metamorphosed larvae of B. amphitrite [17], and Zhang et al. (2010) identified proteins from cyprid larvae using prefractionation methods to reduce sample complexity [18]. Chandramouli et al. (2012) used multiplex proteomics to describe a distinct protein glycosylation pattern during barnacle larval metamorphosis [19], and Chen et al. (2014) recently used label-free quantitative proteomics to identify 700 proteins from nauplii II and VI, cyprids, and juvenile stages of B. amphitrite [20]. In addition, several studies have demonstrated the feasibility of using proteomics tools to investigate the mode of action of antifoulant chemicals in preventing barnacle larval settlement [21–23]. These studies have provided a wealth of molecular information, but comprehensive genome and proteome datasets for nauplius larvae are still lacking. Thus, in the present study we revealed sequence information and investigated the dynamics of protein expression patterns of nauplius larvae. Patterns of differential protein expression between two larval populations from contrasting marine environments were screened to investigate the survival strategies of larvae. Extreme marine environments pose great challenges for the survival of marine larvae. For example, the Red Sea (RS) is one of the warmest, most saline bodies of water; it receives little rainfall and has reduced oxygen solubility and high evaporation rates [24, 25]. The ranges of salinity (36–41psu) and temperature (30–33 °C) in the RS are considered extreme. In contrast, in Hong Kong (HK) marine water the salinity and temperature ranges are 28–31psu and 26–28 °C, respectively [26, 27]. The RS has a low concentration of dissolved oxygen (4.3–5.6 mg/L) compared with that of HK marine water (6.1–7.3 mg/L). Therefore, we hypothesized that changes in protein expression patterns would reflect the adaptation of barnacle larvae to the RS marine environment. To test this hypothesis we adopted a population proteomics approach to compare protein expression in larvae from HK marine waters and the RS. The specific objectives were: (i) to generate a comprehensive transcriptome dataset for the nauplius larvae of B. amphitrite; (ii) to compare the proteomes of barnacle larvae populations from the RS and HK; and (iii) to explore survival strategies of larvae in the RS environment.
Results
Transcriptome of RS barnacle nauplii
Figure 1 shows the experimental design used for the transcriptome and proteome analyses. Sequencing data statistics, assembly and annotation information for newly released nauplii of RS B. amphitrite are shown in Table 1. The transcriptome data were deposited in the NCBI BioProject ID: PRJNA256251 (http://www.ncbi.nlm.nih.gov/bioproject/256251). Approximately 170 million paired-end reads were obtained. Following trimming of low quality reads (Q-score < 20) and adapters, the number of reads was reduced to 151 million, with an average read length of 87 bp. Assembly of the reads using ABySS v1.3.4 and Trans-ABySS v1.4.4 yielded 479,922 contigs with an average contig length (N50) of 471 bp. Using GETORF we predicted 2,485,979 open reading frames (ORFs), of which 92,117 appeared in at least one blast hit. Blastp against a non-redundant (nr) database resulted in 89,151 contigs having at least one sequence homologous with other species, while no homolog was found for 390,771 contigs. An Interproscan v5.4 search resulted in 54,082 contigs having at least one annotated PFAM domain, while no domain was found for 425,840 contigs. In total, 76,263 contigs showed at least one gene ontology (GO) annotation while 403,659 contigs remained hypothetical, and 33,992 contigs showed at least one Reactome annotation, whereas no annotation was found for 445,930 contigs. A total of 1,613,892 were assigned to ‘biological process’ (BP), and 508,916 and 664,282 contigs were assigned to ‘molecular function’ (MP) and ‘cellular component’ (CC), respectively (Additional file 1: Table S1-S4). Figure 2 shows the top 10 categories of assembled contigs belonging to the BP, MF and CC categories. The GOs having the top 10 hits and contigs under the BP category were ‘embryo development’ (14091), ‘transport’ (11260), ‘reproduction’ (11047), ‘transcription regulation’ (10440), ‘larval development’ (10417), ‘signal transduction’ (10150), ‘metabolic process’ (9767), ‘small molecule metabolic process’ (9316), ‘transcription’ (8757) and ‘locomotion’ (8743).
Fig. 1.
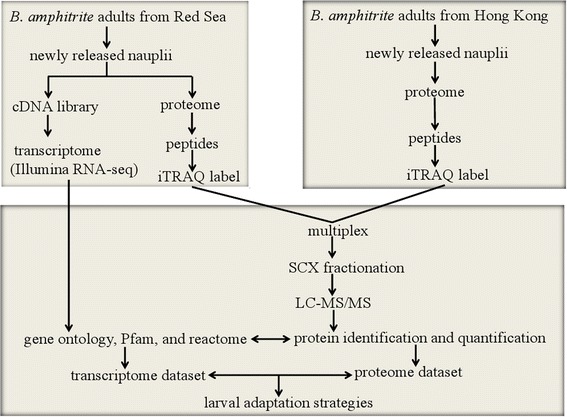
Schematic representation of the experimental design for the transcriptome and proteome analyses
Table 1.
Sequencing data, assembly, and annotation statistics of RS newly released nauplii larvae of B.amphitrite
| Raw read data | Before preprocessing | After preprocessing |
|---|---|---|
| Reads | ||
| No. of reads | 170,319,087 | 151,051,086 |
| Average read length | 101 | 87.17 |
| Assembly | ||
| No. of contigs | 479,922 | |
| Ave contig length (N50) | 471 | |
| Annotation | ||
| Predicted open reading frames (ORFs) | 2,485,979 | |
| ORFs with at least one blast result | 92,117 | |
| Contigs with at least one homolog | 89,151 | |
| Contigs with at least one annotated protein domain | 54,082 | |
| Contigs with at least one GO annontation | 76,263 | |
| Contigs with at least one REACTOME annontation | 33,992 | |
Fig. 2.
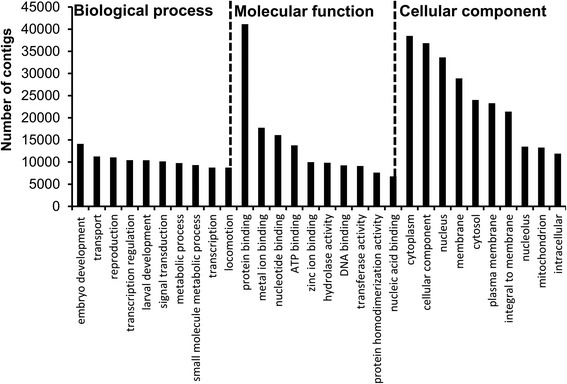
Gene ontology (GO) of assembled contigs of newly-released nauplii larvae of B. amphitrite. The top 10 categories for biological processes, molecular function and cellular components are shown
Abundant transcripts, protein domains and Reactome pathways
Functional analysis of contigs revealed that transcripts related to translation, oxidative phosphorylation and cytoskeletal development were highly abundant. The top 40 abundant transcripts and the number of reads are shown in Additional file 2: Table S5. Elongation factor-1 alpha was ranked first (1,698,408 reads), followed by NADH dehydrogenase subunit 4 (1,100,166 reads). Notably, four transcripts encoding cytochrome oxidase subunit I were also well represented. Interestingly, 10 of the 40 most abundant transcripts encoded hypothetical or predicted proteins. Additional file 3: Table S6 lists the top 30 Pfam domains predicted in the transcriptome. We were able to map 26 pathways present in the Reactome database (Additional file 4: Table S7). These pathways were highly overlapping in terms of the GO BP (Fig. 2, Additional file 1: Table S2) and differentially-expressed protein categories (Table 2).
Table 2.
Up-regulated proteins involved in stress response and osmoregulation in Red Sea newly released nauplii larvae of B.amphitrite
| Accession no. | Contig no.a) | Protein description | Unique peptides | Sequence coverage (%) | Fold changeb) (RS/HK) | STDVE |
|---|---|---|---|---|---|---|
| Stress response/defense/energy | ||||||
| AAN74984.1 | 348386_5 | 70 kDa heat shock protein 70 kDa (HSP70) | 3 | 28 | 2.07 | 0.078 |
| XP_001951792.1 | 284615_1 | heat shock 70 kDa protein 4 L ((HSP70-4 L) | 2 | 31 | 2.78 | 0.043 |
| XP_969171.1 | 221326_2 | chaperonin (CPN) | 1 | 34 | 1.71 | 0.283 |
| AER95784.1 | 112034_2 (+1) | chaperonin containing TCP1subunit 4(CPN-TCP1-4) | 2 | 30 | 1.6 | 0.07 |
| BAG72108.1 | 333804_5 | chaperonin eta subunit (CPN-eta) | 3 | 29 | 1.6 | 0.028 |
| XP_004206182.1 | 216349_6 (+2) | major egg antigen (MGN) | 8 | 28 | 2.31 | 0.057 |
| XP_001606473.2 | 235958_8 | T-complex protein 1 subunit zeta- isoform 1 (TCP1) | 2 | 20 | 2 | 0.043 |
| BAA92814.1 | 379389_7 | annexin IX-A (AnxIXA) | 2 | 30 | 3 | 0.043 |
| BAG41813.1 | 149743_6 | cyclophilin A (CyPA) | 4 | 44 | 2.21 | 0.088 |
| XP_001521816.2 | 331314_1 (+2) | collagen alpha-3(IV) chain (Col4) | 1 | 13 | 2.7 | 0.068 |
| ABZ91537.1 | 11340_21 | vitellogenin (VTG) | 3 | 9 | 5.53 | 0.821 |
| XP_002741413.1 | 9957_2 | vitellogenin structural gene-1 (VTG1) | 3 | 12 | 1.96 | 0.076 |
| Osmoregulation | ||||||
| AEX07319.1 | 136693_6 | Na+/K + −ATPase alpha subunit (H + K + αATPase) | 6 | 21 | 1.85 | 0.168 |
| AAA35578 | 356633_2 | ATPase | 2 | 50 | 2.03 | 0.017 |
| XP_003817086.1 | 110200_6 | sodium-dependent phosphate transport protein 2C(SLC2C) | 3 | 60 | 4.35 | 0.2 |
| XP_003244453.1 | 333233_4 | v-type proton ATPase 116 kDa subunit a isoform 1(ATP6V0A1) | 2 | 12 | 2.91 | 0.432 |
| P31401.1 | 358029_1 | v-type proton ATPase subunit B (ATPV0B) | 2 | 64 | 3.05 | 0.367 |
| ADI56517.1 | 263296_2 | voltage-dependent anion channel 2 (VDAC2). | 2 | 62 | 1.75 | 0.017 |
| XP_004073305.1 | 259151_2 | neuronal acetylcholine receptor subunit non-alpha-3 (α7-nAChR) | 1 | 17 | 4.09 | 0.076 |
| ADO27772.1 | 30169_8 (+1) | carbonic anhydrase (CA) | 1 | 5 | 4 | 0.13 |
| AFS60116.1 | 324487_3 | selenoprotein M (SelM) | 1 | 28 | 4.4 | 0.119 |
| Muscle contraction | ||||||
| EFX86510.1 | 142581_8 | myosin essential light chain (MLC) | 1 | 16 | 2.65 | 0.249 |
| XP_002425291.1 | 233725_2 | paramyosin(PM) | 1 | 9 | 2.71 | 0.057 |
| XP_002432355.1 | 385389_2 (+1) | paramyosin, long form (PM-L) | 1 | 32 | 1.9 | 0.139 |
| EGI57423.1 | 295447_2 | titin(TTN) | 1 | 8 | 2.91 | 0.07 |
| AAT77811.1 | 33648_2 (+1) | calpain B(CalpB) | 1 | 35 | 1.84 | 0.035 |
| Development/morphogenesis | ||||||
| ADV40137.1 | 350939_3 | actin 5C (ACT5C) | 2 | 60 | 3.6 | 0.07 |
| XP_003696514.1 | 286277_1 | actin, indirect flight muscle-like isoform 2(ACTIFM2) | 2 | 96 | 2.29 | 0.135 |
| XP_003696514.1 | 103842_3 | actin, indirect flight muscle-like isoform 2 (ACTIFM2) | 3 | 62 | 2.17 | 0.043 |
| ACO11823.1 | 368856_3 | actin, muscle (ACTM) | 3 | 26 | 1.72 | 0.143 |
| NP_001029376.1 | 358955_6 | tubulin alpha-1C chain (TUBA1C) | 2 | 15 | 3.01 | 0.204 |
| XP_003220514.1 | 322186_3 | tubulin alpha-1C chain (TUBA1C) | 2 | 67 | 1.63 | 0.064 |
| XP_001928233.1 | 258099_4 (+1) | tubulin alpha-1D chain (TUBA1D) | 1 | 22 | 4.17 | 0 |
| XP_624843.2 | 161549_1 (+1) | filamin-B (FLNB) | 2 | 26 | 2.31 | 0.028 |
| Cellular metabolism | ||||||
| ACX53990.1 | 291866_1 (+3) | ubiquinol-cytochrome c reductase complex (UQCCR) | 1 | 13 | 1.83 | 0.057 |
| EEZ99338.1 | 333062_1 | cytochrome P450 6BQ13 (CY-P450 6BQ13) | 1 | 8 | 4.13 | 0.224 |
| XP_970215.1 | 359961_2 | cytochrome P450 CYP6BK17 (CY-P450 6BQ13) | 1 | 2 | 3.15 | 0.028 |
| XP_004079962.1 | 302118_1 (+1) | dehydrogenase/reductase SDR family member 4(DHRS4) | 2 | 44 | 3.21 | 0.132 |
| ZP_09403919.1 | 328617_1 | short chain oxidoreductase (SCOR) | 2 | 25 | 2.14 | 0.043 |
| XP_003204363.1 | 272396_1 | reticulon-4-interacting protein 1, mitochondrial (RTN4IP1) | 1 | 26 | 3.64 | 0.081 |
| ZP_01905843.1 | 388126_2 | oxidoreductase, short chain dehydrogenase/reductase family protein(SDR) | 1 | 13 | 2.38 | 0.078 |
| XP_003701373.1 | 142763_2 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase(DHDH) | 1 | 7 | 1.83 | 0.028 |
| Signaling pathways | ||||||
| XP_966512.1 | 40595_3 | GTP-binding nuclear protein Ran isoform 1 (RanGTPBP1) | 6 | 37 | 1.93 | 0.067 |
| XP_002432661.1 | 92938_6 | GTP-binding protein SAR1B (GTP-BP-SAR1b) | 1 | 75 | 4.8 | 0.247 |
| XP_001945117.1 | 198588_2 | guanine nucleotide-binding protein G(i) subunit alpha(GNBPα) | 1 | 6 | 1.93 | 0.067 |
| NP_001040272.1 | 194501_4 | receptor for activated protein kinase C isoform 2 (RACK2) | 3 | 41 | 1.63 | 0.092 |
| NP_001037203.1 | 238662_1 (+1) | innexin 2 (INX2) | 3 | 23 | 1.96 | 0.073 |
| XP_003488474.1 | 362217_1 | epidermal growth factor receptor substrate 15 (EGFR15) | 1 | 19 | 2 | 0.159 |
a) Refer to contig no. in transcriptome database
b) Averaged expression values derived from independent replicates of Red Sea (RS)/Hong Kong (HK) samples
A large number of genes related to larval development were found to be expressed (Fig. 3a), among which those for brain (GO:0007420; 3884 contigs), muscle (GO:0007517; 2826 contigs), eye (GO:0048749; 1842 contigs), digestive track (GO:0048565; 722 contigs) and cuticle (GO:0040003; 652 contigs) were dominant. Notably, 491 genes related to metamorphosis (GO:0007552) were also identified. A substantial number of genes assigned to signaling pathways involved in larvae development were also detected (Fig. 3b). We found that a large number of genes involved in stress and osmoregulation were over-represented. Genes expressed in response to high salinity, high temperature, cellular stress and ultraviolet (UV) light exposure were identified (Fig. 4a). We also identified several genes involved in homeostasis (Fig. 4b).
Fig. 3.
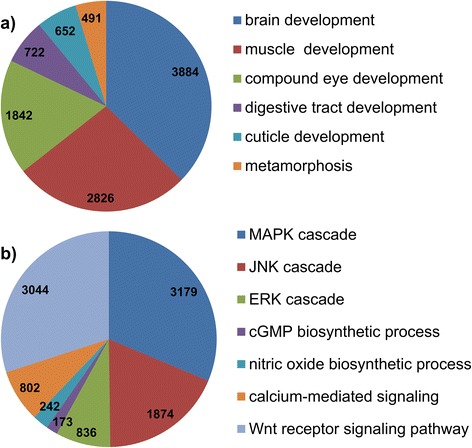
Selected genes involved in (a) larval development and (b) signaling mechanisms. The contig numbers inside the pie chart were obtained from the GO biological functions listed in Supplementary Table S1
Fig. 4.
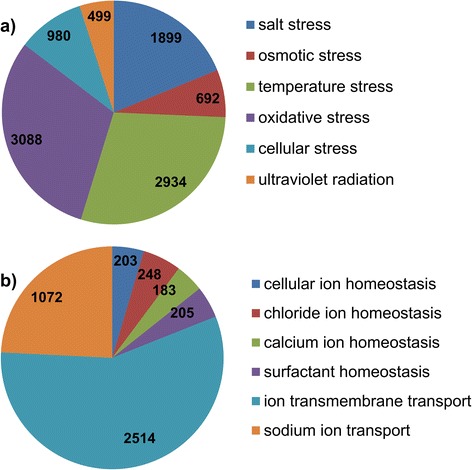
Selected genes involved in (a) response to environment stressors and (b) osmoregulation. The contig numbers inside the pie chart were obtained from the GO biological functions listed in Additional file 11: Figure S1
The larvae proteome
A total of 1264 proteins from HK and RS nauplii were identified based on 4434 unique peptides and 5286 unique spectra (Additional file 5: Table S8). Proteins were quantified using HK larvae as a reference (reporter ion intensities were adjusted to 1; fold change > 1.5). Among these proteins, 170 showed differential expression in RS larvae, based on an average of 1.9 unique peptides, 2.22 spectra and 22.76 % sequence coverage. Of these proteins, 89 were up-regulated and 81 were down-regulated in RS larvae (Tables 2 and 3). Additional file 6: Table S9, Additional file 7: Table S10 and Additional file 8: Table S11 shows the number of peptides and the spectra, sequence coverage, peptide sequences and fold changes for each differential protein.
Table 3.
Down-regulated proteins involved in homeostasis and energy metabolism in Red Sea newly released nauplii larvae of B.amphitrite
| Accession no. | Contig no. a) | Protein description | Unique peptides | Sequence coverage (%) | Fold changeb) | STDVE |
|---|---|---|---|---|---|---|
| Detoxification | ||||||
| AGA83299.1 | 143917_1 | glutamine synthetase (GS) | 1 | 6 | 2.1 | 0.046 |
| EKC40774.1 | 381680_1 | glutaredoxin(GRX) | 1 | 15 | 2.34 | 0.021 |
| EFX81633.1 | 352718_6 | glutathione S-transferase (GST) | 2 | 18 | 2.24 | 0.053 |
| NP_001108460.1 | 176458_3 | glutathione S-transferase epsilon 4 (GST4) | 4 | 44 | 2.24 | 0.014 |
| XP_537038.2 | 68758_2 | glutathione S-transferase Mu 1 isoform 1 (GST Mu1) | 2 | 9 | 1.73 | 0.098 |
| XP_003693286.1 | 382037_1 | aldehyde oxidase 2 (AO2) | 1 | 6 | 2.01 | 0.129 |
| EHJ73729.1 | 382498_5 | alcohol dehydrogenase (ADH) | 1 | 4 | 1.77 | 0.028 |
| AEE36486.1 | 326926_1 (+1) | protein disulfide isomerase 2 (PDI2) | 6 | 47 | 1.6 | 0.017 |
| AFL70280.1 | 281344_8 (+1) | protein disulfide-isomerase A6 (PDIA6) | 1 | 8 | 1.83 | 0.09 |
| Transport | ||||||
| XP_002431951.1 | 15248_12 | dynein heavy chain, cytosolic (DHC) | 1 | 1 | 5.16 | 0.228 |
| XP_002426223.1 | 129454_5 | mitochondrial import inner membrane translocase subunit Tim23 (TIMM23) | 1 | 9 | 1.75 | 0.043 |
| EFX90424.1 | 283903_1 | mitochondrial inner membrane protein (MIM) | 2 | 14 | 1.84 | 0.128 |
| XP_967948.2 | 299347_2 | bumetanide sensitive NaK2Cl cotransporter isoform 1 (NKCC2) | 1 | 9 | 1.75 | 0.095 |
| ADM64456.1 | 378620_1 (+1) | fatty acid binding protein (FABP) | 5 | 44 | 1.93 | 0.028 |
| XP_393519.1 | 237152_4 (+3) | fatty acid-binding protein, adipocyte (FABPA) | 1 | 11 | 1.92 | 0.177 |
| XP_001601236.1 | 34337_3 (+1) | nuclear transport factor 2 (NTF2) | 1 | 9 | 2.06 | 0.017 |
| XP_973673.1 | 301328_8 | vesicle associated protein (VAP) | 3 | 5 | 1.78 | 0.067 |
| XP_002737711.1 | 360183_6 (+2) | coated vesicle membrane protein- isoform 2 (VAMP-2) | 1 | 11 | 1.91 | 0.014 |
| EGF79988.1 | 198964_4 | peptidyl-prolyl cis-trans isomerase (PPI) | 2 | 94 | 2.81 | 0.123 |
| ACJ05313.1 | 307444_1 (+1) | leucine aminopeptidase(LAPs) | 1 | 9 | 1.74 | 0.064 |
| Glycolysis/gluconeogenesis | ||||||
| ACD75898.1 | 360305_3 | enolase (EN) | 1 | 43 | 2.21 | 0.159 |
| AAS02310.1 | 286350_2 (+1) | glyceraldehyde 3-phosphate dehydrogenase (GPDH) | 1 | 77 | 5.75 | 0.043 |
| AGC97435.1 | 281394_3 | glycogen phosphorylase (GPs) | 1 | 12 | 1.66 | 0.057 |
| XP_002400796.1 | 278577_6 | GDP-mannose pyrophosphorylase (GDP-MP) | 1 | 3 | 2.1 | 0.043 |
| XP_003393705.1 | 130725_2 (+2) | transketolase 2 isoform 1 (TKT2) | 2 | 14 | 1.7 | 0.014 |
| Q91XV4.1 | 312847_2 | L-xylulose reductase (XRs) | 1 | 11 | 1.71 | 0.028 |
| BAJ23881.1 | 360259_3 | fructose 1,6-bisphosphatase (FDPase) | 2 | 21 | 3.54 | 0.035 |
| ACH81781.1 | 242946_2 | fructose 1,6-bisphosphate aldolase (FBPA) | 7 | 32 | 2.05 | 0.014 |
| XP_001601687.1 | 383275_9 | phosphoglucomutase-2- isoform 1 (PGM 2) | 1 | 4 | 1.66 | 0.057 |
| XP_003240970.1 | 383186_3 | pyruvate carboxylase, mitochondrial- isoform 3(PCm3) | 2 | 15 | 1.91 | 0.095 |
| EKC25154.1 | 284278_2 (+1) | UTP--glucose-1-phosphate uridylyltransferase (GalU) | 1 | 7 | 2.28 | 0.035 |
| XP_001847482.1 | 255952_3 | UTP-glucose-1-phosphate uridylyltransferase 2 (GalU2) | 2 | 11 | 2.53 | 0.176 |
| Actin-binding proteins | ||||||
| XP_001944078.1 | 370072_2 | vinculin (VCL) | 1 | 19 | 1.62 | 0.035 |
| CCI71880.1 | 97015_4 | gelsolin (GSN) | 1 | 15 | 1.55 | 0.039 |
| XP_001657431.1 | 377569_1 (+1) | gelsolin precursor(GSNP) | 2 | 18 | 1.71 | 0.043 |
| XP_003691411.1 | 216_3 | plastin-3(PLS3) | 2 | 10 | 1.71 | 0 |
| JAA59814.1 | 237239_4 | villin-1 (VIL1) | 3 | 27 | 1.81 | 0.028 |
a) Refer to contig no. in transcriptome database
b) Averaged expression values derived from independent replicates of Red Sea (RS) /Hong Kong (HK) samples
Differences in proteome expression between RS and HK larvae
Principle component analysis (PCA) of the proteome datasets showed clear separation of the HK and RS samples (Fig. 5), and hierarchical clustering analysis revealed two clusters of down-regulated (green) and up-regulated (red) proteins in the RS larvae that were distinct from those in the HK larvae (Fig. 6). A slight variation in expression pattern between the replicates (R1–R3 and R4–R6) of HK larvae was observed. It is important to note that the adult barnacles were collected from wild natural conditions. Habitat conditions could differ substantially because of factors including origin, age, gender and physiological state. It is difficult to account for uncontrolled sources of variation among specimens collected from natural habitats at the same time in the same area. However, such variability may not have a significant impact as the average individual protein expression was only slightly different among replicates. Furthermore, the average coefficient of variation (CV) among replicate samples from HK was lower (within the standard range for iTRAQ experiments), and not enough to mask the overall difference in the expression patterns of HK samples. Table 2 and Additional file 6: Table S9 lists the up-regulated proteins and their fold change values. Nine stress-activated proteins were up-regulated in the RS larvae relative to the HK larvae (Fig. 7a). The expression of osmoregulatory proteins was also significantly increased in the RS larvae (Fig. 7b). Additional file 7: Table S10 lists some of the down-regulated proteins and their fold change values. In RS larvae 9 proteins involved in cellular detoxification (Fig. 7c) and 11 involved in glycolysis and gluconeogenesis were down-regulated (Fig. 7d). Notably, all 5 actin-binding proteins, including gelsolins, vinculin, plastin-3 and villin-1 were also down-regulated.
Fig. 5.

2-D view of the principal component analysis of 1264 identified proteins in two natural larval populations. Log2-transformed fold change ratios from six replicates were used for the analysis. HKR: replicates of Hong Kong larvae; RSR: replicates of RS larvae
Fig. 6.
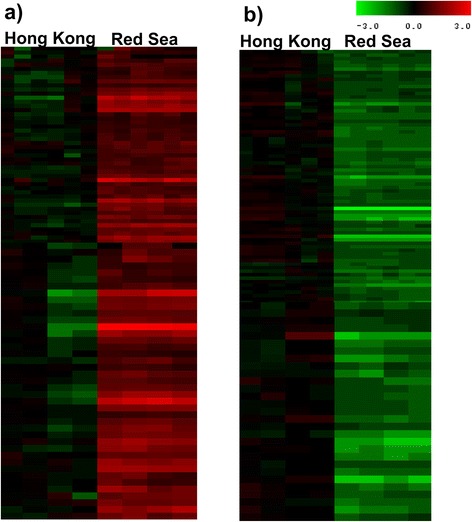
Hierarchical clustering of differentially-expressed proteins in Hong Kong and Red Sea larvae of B. amphitrite. a) Hierarchical cluster 1, b) Hierarchical cluster 2. Red: up-regulation; green: down-regulation. The horizontal axis indicates the replicates in the order R1 to R6
Fig. 7.
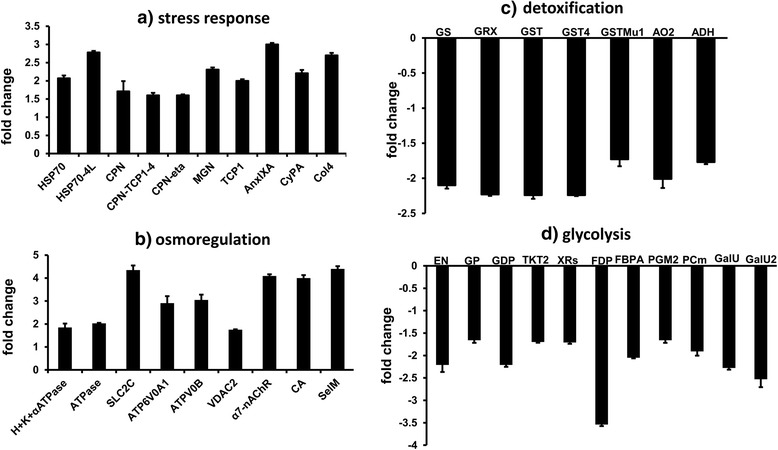
Proteins involved in the stress response (a) and osmoregulation (b) were up-regulated in Red Sea B. amphitrite larvae, whereas detoxification (c) and glycolysis (d) proteins were down-regulated. The full protein names are shown in Tables 2 and 3
Correlation between the proteome and transcriptome of RS larvae
The number of genes in RS larvae of B. amphitrite that were identified in each functional category, and their corresponding protein expression patterns, are shown in Table 4 and Additional file 9: Table S12. The profiles of the two datasets revealed overlap in the majority of abundant transcripts and differentially expressed proteins. These categories, which included stress response, and development and signaling pathways, showed similar trends of expression at both the transcript and protein levels. We identified 30 stress response proteins from the proteome analysis and 16,367 contigs in the transcriptome; these belonged to various categories of stress. Similarly, the ‘developmental’ category showed 34 proteins and 24,597 transcripts, while ‘signaling pathways’ showed 18 proteins and 18,171 transcripts in the respective datasets. Further, the proteins of binding and catalytic activity were also equally represented in the proteome and the transcriptome. Overall, abundant proteins showed high degree of correlation with the corresponding transcripts.
Table 4.
Parallel comparison of protein regulation to their corresponding transcript abundance in Red Sea larvae of B.amphitrite
| Protein category | No. of proteins identified | Protein regulation | Transcripts abundance(no. of contigs) |
|---|---|---|---|
| Stress | 12 | up-regulation | 10092 |
| Osmoregulation | 9 | up-regulation | 4425 |
| Detoxification | 9 | down-regulation | 1850 |
| Cytoskeleton | 8 | up-regulation | 11615 |
| Actin binding | 5 | down-regulation | 3916 |
| Muscle development | 5 | up-regulation | 323 |
| Cuticle development | 10 | n/a | 735 |
| Locomotion | 6 | dual regulation | 8743 |
| Apoptosis | 12 | dual regulation | 8561 |
| Signaling pathways | 6 | up-regulation | 10150 |
Discussion
Deep sequencing of the larval transcriptome
The transcriptome analysis yielded approximately 150 million reads and 479,922 contigs (N50, 471 bp) for the RS larvae, including 92,117 unique predicted ORFs, and constitutes the largest available sequence dataset for nauplii larvae. Chen et al. (2011) obtained 630,845 reads using 454 pyrosequencing and 23,451 contigs from pooled larvae and adult samples of B. amphitrite from HK seawater [13], while De Gregoris et al. (2011) generated 575,666 reads from nauplius, cyprid and adult stages of B. amphitrite [15]. More recently, Lin et al. (2014) obtained 77,528,326 reads and 104,610 contigs from the prosoma, and 59,244,468 reads and 123,534 contigs from the base of the barnacle Tetraclita japonica formosana [16]. The sequencing depth of the present study provides much greater read coverage than the previous studies. Using this in-house transcriptome database we successfully used mass spectrometry to identify 1264 proteins; of these, 170 showed distinct expression patterns between the HK and RS larval populations. We also identified a large number of transcripts related to development, locomotion and signal transduction.
Molecular basis of larval development
The abundant expression of cuticular proteins in both the transcriptome and proteome datasets may reflect the importance of exoskeleton dynamics during development. The barnacle body is covered with exoskeleton, which is composed of carbohydrates and proteins [28]. During molting the larvae partially resorb the cuticle to provide space for newly forming tissues. The larval proteome showed that several structural proteins were differentially expressed. For example, myosins, titin, actin, tubulins, talin-1 and fasciclin-1 were up-regulated in RS larvae (Table 2), whereas settlement pheromone, cement secretion protein 2, retinol dehydrogenase 13 and NMDA receptor protein 1 were down-regulated (Table 3). This is consistent with our previous reports showing differential expression of cytoskeletal proteins during development in polychaetes [29–32]. The myosins and titins contribute to muscle development and the contraction of striated muscle. In barnacles, striated muscles in antennules and thoracopods help in locomotion and sensory detection for substratum attachment [33]. Barnacle cyprid-specific protein 2 (BCS-2) is abundantly expressed in cyprids, but decreases following larval attachment [34]. The protein BCL-4 is less abundant in young cyprids, but gradually increases in older cyprids. Apoptosis and protein degradation occur spontaneously during larval development and metamorphosis [35], and are critical in the removal and subsequent degradation of early larval structures, and in cellular and tissue remodeling. In the present study we found several differentially expressed proteins related to apoptosis. Our previous proteomic analyses identified several apoptosis proteins in early stages of larval development in barnacles [19].
Signal transduction pathways
Genes that participate in the MAPK cascade, cGMP, nitric oxide (NO) biosynthesis, calcium and Wnt pathways were identified in the present study. Our previous studies showed that in barnacles these pathways are closely associated with larval settlement and metamorphosis. For example, p38-mediated MAPK signaling has been found to be crucial for larval attachment in barnacles [36, 37]. Zhang et al. (2012) found that NO regulates attachment of barnacle larvae by modulating cGMP signaling [38]. It appears that NO signaling contributes to stress-activated responses (in the present case, up-regulation of heat shock proteins), and thereby influences larval survival under unfavorable environmental conditions. Based on our findings, we hypothesize that MAPK, NO, cGMP and calcium signaling are important for larval development and stress tolerance in larvae of B. amphitrite. Furthermore, the proteome data showed up-regulation of six proteins related to G protein signaling or protein kinase. Among these, GTP-binding nuclear protein (RanGTPBP1), GTP-binding protein SAR1B (GTP-BP-SAR1b) and guanine nucleotide-binding protein (GNBPα) belong to G protein signaling, whereas receptor for protein kinase C (RACK2), innexin 2 (INX2) and epidermal growth factor receptor (EGFR15) belong to the protein kinase and calcium receptor-mediated signal transduction cascades. Chen et al. (2014) reported up-regulation of the G protein-coupled receptor in barnacle cyprid larvae [20], and G protein receptors bind to the metamorphic cue, while the protein kinase independent pathway participates in settlement signaling.
Adaptive strategies under extreme marine conditions
Adult barnacles live in various habitats and are subject to differing selection pressures that could have a major influence on the survival strategies of their offspring (in this case nauplii) [39, 40]. Temperature and salinity influence larval recruitment and the survival of marine invertebrates [41, 42]. In addition, the intertidal zone is highly variable with respect to temperature conditions and among different marine environments [43]. For example, in summer the offshore RS and HK water temperatures average 30.8 °C and 28 °C, respectively [27, 44]. Nauplii may have developed remarkable adaptive mechanisms for survival in the pelagic stage and display a variety of survival strategies. The higher expression levels among stress response genes in RS larvae, and the up-regulation of stress-activated proteins including heat shock proteins (HSPs), chaperonins (CPNs), major egg antigen (MGN), T-complex protein (TCP1), annexin (AnxIXA), cyclophilin (CyPA) and collagen alpha (Col4) suggest the development of adaptive strategies to cope with extreme conditions. This observation is consistent with previous studies showing significant up-regulation of HSPs in barnacle larvae challenged with antifoulants [16, 23]. The cellular response of larvae to stress leads to abundant expression of genes encoding HSPs. Some HSPs act as molecular chaperons (CPNs, MGN, TCP1) by modulating the correct folding of other proteins under stress, and are involved in various developmental signaling pathways, and assembly and transport mechanisms [45, 46]. For instance, in ascidians and gastropods, HSP90 activity regulates the repression of larval metamorphosis by nitric oxide signaling [47]. AnxIXA expression patterns have been shown to change significantly when cells undergo proliferation or differentiation [20]. The differential expression of AnxIXA in RS nauplii and juveniles of B. amphitrite could contribute to anti-inflammatory responses. CyPA secreted by cells in response to inflammatory stimuli is known to play an important role in protein folding. We identified a large number of genes involved in osmotic stress tolerance. Furthermore, the RS larval proteome displayed dramatic up-regulation of osmoregulatory proteins including H + K + αATPase, v-type proton ATPase, phosphate transport protein (SLC2C), voltage-dependent anion channel (VDAC2), neuronal acetylcholine receptor (α7-nAChR), carbonic anhydrase (CA) and selenoprotein (SelM) (Table 2). During barnacle larval development, biomineralization and osmoregulation facilitate larval tolerance to a wide range of osmotic stresses via active ion transport [48]. CA is widely distributed in barnacle tissues, and is involved in the synthesis of calcium carbonate [20] and the molting cycle in crustaceans. In other marine species osmotic stress affects the expression patterns of the proteins ATP6V0A1 and ATPV0B, and Na + −K + −ATPase and V-H + −ATPase are required for ion regulation [49]. The sodium-phosphate symporter SLC2C plays a role in osmoregulation by absorbing phosphate from interstitial fluid. The Ca2+ transport regulator VDAC plays an important role in regulating the exchange of ions and solute molecules in mitochondria, and α7-nAChR facilitates the opening of ion channels across the plasma membrane. Consequently, we believe that under extreme environmental conditions larvae may be protected by abundant expression of osmoregulatory proteins that maintain cellular homeostasis, and that the large number of genes involved in various homeostasis mechanisms (including for chloride, calcium and sodium ions, and regulation of ion transmembrane and surfactant concentrations) are probably involved in osmoregulation (Fig. 4b).
Metabolic depression
Under extreme environmental conditions marine invertebrates typically show greater metabolic depression by minimizing energy loss [50, 51]. It has been estimated that protein translation alone may consume up to 50 % of cellular energy; by suppressing metabolism, energy can be diverted to synthesis of stress tolerance proteins [52]. Metabolic depression may lead to functional cellular modification, post-translational modification of proteins (phosphorylation), and maintenance of homeostasis by ion transport. Consistent with these observations, in RS larvae we found down-regulation of 81 proteins involved in various metabolic processes (Table 3). For instance, glutathione S-transferases (GSTs) are antioxidant isoenzymes that provide cellular defense against oxidative stress and harmful chemicals [53]. Han et al. (2013) reported increased expression of GSTs in barnacle cyprid larvae following exposure to the antifoulant meleagrin [23], suggesting a role in chemical detoxification. GST also regulates apoptosis by inhibiting kinases, thereby affecting downstream signaling pathways. It has been shown in barnacle cyprid larvae that proteins involved in glycolysis and gluconeogenesis are differentially expressed, suggesting roles in maintaining homeostasis [20, 23]. Fatty acid binding proteins (FABPs) and fructose 1,6-bisphosphatase (FDPase), which are involved in the synthesis of byproducts of glycolysis and gluconeogenesis, have been identified in barnacle larvae [20]. Membrane proteins play an important role in the transport of solutes and toxic materials across the cell membrane. In particular, mitochondrial inner membrane proteins (MIM and TMIM 23) transport metabolites across the membrane in a highly controlled manner [54]. FABPs act as carrier proteins for fatty acids and other lipophilic substances [55]. The regulation of energetically expensive transport systems appears to be an important strategy used by RS larvae to survive in extreme conditions. The actin binding proteins may regulate cytoskeletal dynamics in developing larvae. It is hypothesized that larval tolerance of stress is regulated by complex stress response mechanism and strategies, and metabolic depression is probably more diverse and complicated than previously assumed. We believe this study is of international significance in advancing marine biology, as most previous studies have focused on response to specific environmental factors in controlled environment; it also highlights the importance of studying the cumulative effects of environmental conditions in natural habitats.
Conclusion
The transcriptome and proteome data generated in this study provide a valuable resource that will enhance molecular studies of crustacean larvae, which are of ecological and economic importance. The large differences found in the protein expression patterns of larval populations from the two contrasting environments were striking. The genes, proteins and signaling pathways identified may play crucial roles in the development of nauplii in the RS environment. For example, the abundant expression of cuticular and muscle proteins in nauplii is critical in the molting cycle, and in muscle development that influences swimming and sensory detection. We also identified genes that participate in several biochemical pathways that are important for larval settlement, metamorphosis and the stress response in barnacles. Specifically, heat shock proteins, chaperonin, major egg antigen, annexin and NO signaling participate in stress-activated responses, and thereby contribute to larval strategies for survival in the RS environment. Proteins involved in osmoregulation, including ATPase, transport proteins, acetylcholine receptor and carbonic anhydrase facilitate larval tolerance to osmotic stress. Overall, the findings of this study contribute significantly to knowledge of larval development and adaptation in extreme conditions, and the settlement biology of crustaceans.
Methods
Environmental conditions
Marine environmental data were collected from each sampling location. Measurements were carried out using a YSI Professional Pro Plus Multiparameter meter (YSI, Ohio, USA), which was calibrated prior to use. The probe was immersed in the water at the designated locations, and measurements were recorded after 2–3 min; it was rinsed with fresh water between measurements and locations. The average values of salinity, temperature, pH and dissolved oxygen were 42.31 psu, 34.23 °C, 8.2 and 5.06 mg/L, respectively. The values for HK water for salinity, temperature, pH and dissolved oxygen were 31.3 psu, 29.16 °C, 8.13 and 6.53 mg/L, respectively (Additional file 10: Table S13).
Barnacle collection
Barnacles were collected from the intertidal zone between August 2012 and November 2012. Adult B. amphitrite barnacles attached to hard rocks were collected from the intertidal rocky shore of the King Abdullah University of Science and Technology, Thuwal, Kingdom of Saudi Arabia (27°18′168″N, 35°05′661″E). Approximately 100 collected adults were washed to remove attached debris and sediment, and were kept in a 10 L tank containing aged filtered sea water. The tank was covered with black plastic sheet, but had a small opening enabling light to penetrate, as a means of attracting newly released larvae. Newly hatched nauplii (hereafter, newly released RS larvae) were collected at 30-min intervals using a pipette until sufficient larvae were obtained for subsequent studies. Excess water was removed and approximately 3–4 × 103 larvae derived from independent replicates were separated into two groups, one of which was used for transcriptome analysis and the other for proteome analysis (Additional file 11: Figure S1).
Approximately 100 individuals of adult B. amphitrite barnacles were collected from concrete columns at Pak Sha Wan, HK (22°21′45″N, 114°15′35″E), and placed in a 10 L tank containing filtered seawater (FSW; 0.22 μm) to induce the release of larvae. Approximately 3–4 × 103 newly hatched nauplii (hereafter, newly released HK larvae) were collected as described above, and transferred overnight on dry ice to the King Abdullah University of Science and Technology. No significant size difference in the adults from the two populations was observed. On arrival the larvae were thawed, washed in seawater, and used for proteome analysis.
For the RS larval samples the total RNA was extracted from 600 to 700 larvae using TRIzol reagent (Invitrogen), following the manufacturer's instructions. TURBO DNA-free treatment was performed using RNeasy spin columns (Qiagen). The RNA was purified and eluted in 14 μl of MilliQ water. The purity was checked using a NanoDrop ND-1000 UV–vis spectrophotometer (NanoDrop Technologies). The integrity of the extracted RNA was then checked using an Agilent 2100 Bioanalyzer (Agilent Technologies), and confirmed on a 1 % agarose gel.
Library preparation and Illumina sequencing
cDNA library preparation was performed according to the Illumina TruSeq standard RNA Seq library preparation protocol, and sequencing was performed using an Illumina HiSeq 2000 genome sequencer. Briefly, the quantity of total RNA was measured using a Qubit® 2.0 fluorometer (Invitrogen). rRNA was removed using the Ribo-Zero Magnetic Gold Kit (Epicenter, Illumina). The RNA was subjected to thermal fragmentation, and first strand cDNA was synthesized using Super-Script III reverse transcriptase (Invitrogen). The cDNA was converted into double stranded DNA using the Second Strand Making Master Mix (Illumina). After dA-tailing, ligation of adaptors and purification, library amplification (15 cycles) was performed using a thermal cycler (Applied Biosystems). The libraries were purified using AMPure XP beads (Beckman Coulter, Inc.). Library dilution and pooling was carried out as described in the Illumina standard protocol. A quantity of 7.5 picomoles of DNA was used for cluster generation (CBOT Illumina) and sequencing.
Assembly and annotation
Low quality reads (Q-score < 20) and adapters were trimmed using java scripts designed in-house. Reads were assembled using ABySS v1.3.4 [56] with setting parameters of every odd k-mer from 45 to 85, and consolidated using Trans-ABySS v1.4.4 [57]. Contig redundancy was reduced using CD-HIT-EST with the threshold sequence identity set to 0.99, to discard contigs with < 10 bp mismatches per 1 kb [58]. To obtain coding amino acid sequences we used GETORF to translate contigs from start to stop codons for all six possible reading frames [59]. Two approaches were used for functional annotation of contigs. First, homologous proteins from other species were searched using blastp against a non-redundant (nr) database (minimum bit score threshold 50; match length 20 amino acids). Second, the PFAM domain of the proteins was searched using Interproscan v5.4 [60]. Gene ontology (GO) and Reactome pathway information from homologous genes was transferred to barnacle contigs using a transcript designed in-house.
Larval proteome extraction, digestion and iTRAQ labeling
The HK and RS larvae were suspended in 8 M urea and protease inhibitor (Roche Diagnostics, Germany) and homogenized (Wheaton homogenizer, USA). The homogenate was sonicated at 4 W using five × 1 s bursts (Q Sonica, LLC, USA), and the protein concentration was measured using a 2-D Quant kit (GE Healthcare, United Kingdom). Protein (100 μg) from each sample was reduced, alkylated, and diluted by a factor of 8 using 50 mM iTRAQ dissolution buffer as previously described [61]. The protein was digested using a trypsin–protein ratio of 1:50 for 14–16 h at 37 °C (Promega, USA). The digest was desalted using Sep-Pak C18 Vac cartridges (Water Corporation, USA), and labeled using the iTRAQ Reagents Multiplex (4plex) Kit (Applied Biosystems, USA), according to the manufacturer’s protocol. Briefly, dried peptides were reconstituted in 30 μL of resolubilization buffer, and 70 μL of ethanol was added to the iTRAQ reagents; the reagents 114 and 115 were labeled to HK larval peptides, and reagents 116 and 117 were labeled to the RS larval peptides. Labeling was carried out at room temperature for 60 min, and the labeled peptides were multiplexed and dried prior to fractionation.
Peptide fractionation coupled with mass spectrometry
The iTRAQ-labeled peptides were suspended in 85 μL strong cation exchange chromatography (SCX) buffer A, and centrifuged at 10,000 rpm for 5 min at room temperature. The supernatant containing peptides was transferred to liquid chromatography (LC) sample vials and fractionated using an Accela 1250 LC system (Thermo Scientific, USA). The SCX-fractionated peptides were desalted using Sep-Pak C18 Vac cartridges and resuspended in 20 μL of LC sample buffer (97 % H20, 3 % ACN, 0.1 % formic acid), centrifuged at 10,000 rpm for 5 min, and then transferred to LC-MS sample vials. A total of 15 SCX fractions were run in a LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific, Germany) coupled to a Proxean Easy-nLC liquid chromatography system (Bruker, Denmark), as described previously [61].
Protein identification
Raw MS data were processed following a previously described procedure [62]. Briefly, higher-energy collisional dissociation (HCD) and collision-induced dissociation (CID) spectra were extracted independently using Proteome Discoverer 1.2 software (Thermo Scientific), and processed using a script designed in-house. Mascot generic format (MGF) files were submitted to MASCOT v2.2 (Matrix Sciences Ltd, United Kingdom) for searching against a newly released RS barnacle larval protein dataset (92,117 target sequence entries that matched to 43,921 unique GI numbers), developed in-house from transcriptome data. The mass tolerance for the peptide was set to 10 ppm, and the MS/MS fragment ion tolerance was 0.5 Da. A maximum of one missed cleavage was allowed. Variable modifications were set to 4-plex iTRAQ and oxidation was set to methionine (M). The fixed modifications were set to methylethanethiosulfonate at cysteine and lysine, and 4-plex iTRAQ at N-terminal. The MASCOT files (.dat files) were analyzed using Scaffold v4.1.1 (Proteome Software Inc. USA) software for validation and quantitation of peptide and protein identifications. The peptide and protein identification threshold was set at 95 % using the Peptide and Protein Prophet algorithm with Scaffold delta-mass correction [63, 64]. Using these criteria the false positive rates for peptide and protein were 1 % and 0.1 %, respectively.
Protein quantitation
iTRAQ label-based quantitation of the identified proteins was performed using the Scaffold Q+ algorithm. The intensities of all labeled peptides were normalized across all runs. Individual quantitative data acquired in each run were normalized using the i-Tracker algorithm, as described previously [65]. Peptide intensity was normalized within the assigned protein. The reference channel (e.g. 114) was normalized to produce a 1:1 fold change, and the iTRAQ ratios were then transformed to a log scale. P values were calculated using a paired t-test. All normalization calculations were performed as described previously [61].
Statistical analysis
The sources of variation between biological and technical replicates of HK and RS samples were evaluated using unsupervised multivariate principal component analysis (PCA), using the Multiple Array Viewer (MeV) data analysis and visualization tool [66]. Log2-transformed fold changes from six replicates were used for the analysis. Sample selection was set to Cluster, and the centering mode was set to Median. A hierarchical cluster analysis for differentially-expressed proteins from replicates of HK and RS samples was performed using MeV. Tree selection was set to Gene Tree, sample was set to Tree, the Pearson correlation was set to distance matrix selection, average linkage clustering was set to linkage method selection, and centroid linkage was selected as the clustering method.
Supporting data
The sequence datasets obtained during this project have been deposited in the NCBI BioProject ID: PRJNA256251 (http://www.ncbi.nlm.nih.gov/bioproject/256251).
Acknowledgements
The authors thank Harris Mavromatis for technical assistance in RNA extraction and PCA analysis, and Gen Zhang for larval collection. This study was supported by the King Abdullah University of Science and Technology, and a grant [SA-C0040/UK-C0016] from the King Abdullah University of Science and Technology to P-Y Qian and Timothy Ravasi.
Abbreviations
- AnxIXA
Annexin
- BCS-2
Barnacle cyprid-specific protein 2
- BP
Biological process
- CA
Carbonic anhydrase
- CC
Cellular component
- CID
Collision-induced dissociation
- Col4
Collagen alpha 4
- CPNs
Chaperonins
- CV
Coefficient variation
- CyPA
Cyclophilin
- EGFR15
Epidermal growth factor receptor
- FABP
Fatty acid binding proteins
- FDPase
Fructose 1,6-bisphosphatase
- GNBPα
Guanine nucleotide-binding protein
- GO
Gene ontology
- GSTs
Glutathione S-transferases
- HCD
Higher-energy collisional dissociation
- HK
Hong Kong
- HSP
Heat shock proteins
- INX2
Innexin 2
- LC
Liquid chromatography
- MeV
Multiple array viewer
- MGF
Mascot generic format
- MGN
Major egg antigen
- MIM
Mitochondrial inner membrane proteins
- MP
Molecular function
- NO
Nitric oxide
- ORF
Open reading frame
- PCA
Principal component analysis
- RACK2
Receptor for protein kinase C
- RanGTPBP1
GTP-binding nuclear protein
- RS
Red Sea
- SCX
Strong cation exchange chromatography
- SelM
Selenoprotein
- SLC2C
Phosphate transport protein
- TCP1
T-complex protein
- VDAC2
Voltage-dependent anion channel
- α7-nAChR
Neuronal acetylcholine receptor
Additional files
Gene ontology (biological process) of assembled unigenes of newly released nauplii larvae of B. amphitrite. Biological process information was obtained by transferring homologous genes to barnacle contigs using a custom transcript. Table S2. Top 10 gene ontology terms for assembled unigenes of newly released nauplii larvae of B. amphitrite. Selected genes involved in a) larval development, b) signal transduction, c) in response to environment stressors, and d) osmoregulation. Table S3. Gene ontology (molecular function) for assembled unigenes of newly released nauplii larvae of B. amphitrite. Molecular process information was obtained by transferring homologous genes to barnacle contigs using a custom transcript. Table S4. Gene ontology (cellular component) for assembled contigs of newly released nauplii larvae of B. amphitrite. Cellular component information was obtained by transferring homologous genes to barnacle contigs using a custom transcript. (XLSX 57 kb)
Top 40 abundant genes identified from the transcriptome of newly released nauplii larvae of B. amphitrite. (XLSX 14 kb)
Top 30 Pfam domains predicted from the transcriptome of newly released nauplii of B. amphitrite. (XLSX 11 kb)
The number of contigs linked to the Reactome pathway in newly released nauplii larvae of B. amphitrite. (XLSX 10 kb)
Identified proteins and fold change ratios for Hong Kong and Red Sea larvae. (XLSX 168 kb)
Up-regulated proteins involved in the stress response and osmoregulation in newly released nauplii larvae of Red Sea B. amphitrite. Hits were based on the number of unique peptides identified, the spectrum count and sequence coverage. (XLSX 24 kb)
Down-regulated proteins involved in homeostasis and energy metabolism in newly released nauplii larvae of Red Sea B. amphitrite. Hits were based on the number of unique peptides identified, the spectrum count and sequence coverage. (XLSX 31 kb)
List of differentially expressed proteins involved in cellular metabolism in newly released nauplii larvae of Red Sea B. amphitrite. Hits were based on the number of unique peptides identified, the spectrum count and sequence coverage. (XLSX 59 kb)
Abundance of transcripts categorized to stress response, development, and signaling mechanisms, compared in parallel with protein regulation. (XLSX 9 kb)
Environmental differences between the Red Sea and Hong Kong marine environments. (PDF 99 kb)
Newly-released nauplii of the Red Sea barnacle B. amphitrite used for transcriptome and proteome analyses. (PDF 56 kb)
Footnotes
Competing interests
The authors declare no competing financial interests.
Authors’ contributions
TIR, PYQ, KHC conceived and designed the experiments. KHC, SA participated in collection of larvae. KHC performed the experiments: HZ performed MS analysis. KHC, TAR, LS, YD analyzed the data. KHC interpreted the data and drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
Kondethimmanahalli H. Chandramouli, Email: Kondethimmanahalli.Chandramouli@kaust.edu.sa
Sarah Al-Aqeel, Email: sarah.aqeel@kaust.edu.sa.
Taewoo Ryu, Email: taewoo.ryu@kaust.edu.sa.
Huoming Zhang, Email: Huoming.Zhang@kaust.edu.sa.
Loqmane Seridi, Email: loqmane.seridi@kaust.edu.sa.
Yanal Ghosheh, Email: yanal.ghosheh@kaust.edu.sa.
Pei-Yuan Qian, Email: boqianpy@ust.hk.
Timothy Ravasi, Email: timothy.ravasi@kaust.edu.sa.
References
- 1.Holm ER. Barnacles and biofouling. Integr Comp Biol. 2012;52:348–55. doi: 10.1093/icb/ics042. [DOI] [PubMed] [Google Scholar]
- 2.Evans LV. Editor’s note. Biofouling. 2009;25:81. doi: 10.1080/08927010802568222. [DOI] [PubMed] [Google Scholar]
- 3.Schultz MP, Bendick JA, Holm ER, Hertel WM. Economic impact of biofouling on a naval surface ship. Biofouling. 2011;27:87–98. doi: 10.1080/08927014.2010.542809. [DOI] [PubMed] [Google Scholar]
- 4.Rivera A, Gelcich S, García-Florez L, Alcázar JL, Acuña JL. Co management in Europe: insights from the gooseneck barnacle fishery in Asturias, Spain. Mar Policy. 2014;50:300–8. doi: 10.1016/j.marpol.2014.07.011. [DOI] [Google Scholar]
- 5.Clare AS, Høeg JT. Balanus amphitrite or Amphibalanus amphitrite? A note on barnacle nomenclature. Biofouling. 2008;24:55–7. doi: 10.1080/08927010701830194. [DOI] [PubMed] [Google Scholar]
- 6.Maruzzo D, Aldred N, Clare AS, Høeg JT. Metamorphosis in the cirripede crustacean Balanus amphitrite. PLoS One. 2012;7:e37408. doi: 10.1371/journal.pone.0037408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman WA: Cirripedia. In: Light SF, Carlton JT, editors. The Light and Smith Manual: Intertidal Invertebrates from Central California to Oregon (4th ed.). University of California Press. 2007. p. 475–84.
- 8.Dahms HU, Fornshell JA, Fornshell BJ. Key for the identification of crustacean nauplii. Org Divers Evol. 2006;6:47–56. doi: 10.1016/j.ode.2005.04.002. [DOI] [Google Scholar]
- 9.Dahms HU. A pictorial key for the identification of crustacean nauplii from the marine meiobenthos. J Crust Biol. 1993;13:609–16. doi: 10.2307/1548802. [DOI] [Google Scholar]
- 10.Thiyagarajan V, Venugopalan VP, Subramoniam T, Nair VK. Description of the naupliar stages of Megabalanus tintinnabulum (Cirripedia: Balanidae) J Crust Biol. 1997;17:332–42. doi: 10.2307/1549282. [DOI] [Google Scholar]
- 11.Dionisio M, Rodrigues A, Cost A. A description of the larval development of Megabalanus azoricus (Pilsbry, 1916) reared in the laboratory. Helgol Mar Res. 2014;68:89–98. doi: 10.1007/s10152-013-0370-3. [DOI] [Google Scholar]
- 12.Dahms HU. Phylogenetic implications of the crustacean nauplius. Advances in copepod taxonomy. Hydrobiologia. 2000;417:91–9. doi: 10.1023/A:1003886818724. [DOI] [Google Scholar]
- 13.Chen ZF, Chen ZF, Matsumura K, Wang H, Arellano SM, Yan X, et al. Toward an understanding of the molecular mechanisms of barnacle larval settlement: a comparative transcriptomic approach. PLoS One. 2011;6:e22913. doi: 10.1371/journal.pone.0022913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan XC, Chen ZF, Sun J, Matsumura K, Wu RS, Qian P-Y. Transcriptomic analysis of neuropeptides and peptide hormones in the barnacle Balanus amphitrite: evidence of roles in larval settlement. PLoS One. 2012;7:e46513. doi: 10.1371/journal.pone.0046513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Gregoris TB, Rupp O, Klages S, Knaust F, Bekel T, Kube M, et al. Deep sequencing of naupliar-, cyprid- and adult-specific normalised Expressed Sequence Tag (EST) libraries of the acorn barnacle Balanus amphitrite. Biofouling. 2011;27:367–74. doi: 10.1080/08927014.2011.577211. [DOI] [PubMed] [Google Scholar]
- 16.Lin HC, Wong YH, Tsang LM, Chu KH, Qian PY, Chan BK. First study on gene expression of cement proteins and potential adhesion-related genes of a membranous-based barnacle as revealed from Next-Generation Sequencing technology. Biofouling. 2014;30:169–81. doi: 10.1080/08927014.2013.853051. [DOI] [PubMed] [Google Scholar]
- 17.Thiyagarajan V, Qian P-Y. Proteomic analysis of larvae during development, attachment, and metamorphosis in the fouling barnacle, Balanus amphitrite. Proteomics. 2008;8:3164–72. doi: 10.1002/pmic.200700904. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Xu Y, Arellano SM, Xiao K, Qian P-Y. Comparative proteome and phosphoproteome analyses during cyprid development of the barnacle Balanus (=Amphibalanus) amphitrite. J Proteome Res. 2010;9:3146–57. doi: 10.1021/pr1000384. [DOI] [PubMed] [Google Scholar]
- 19.Chandramouli KH, Zhang Y, Wong YH, Qian P-Y. Comparative glycoproteome analysis: dynamics of protein glycosylation during metamorphic transition from pelagic to benthic life stages in three invertebrates. J Proteome Res. 2012;11:1330–40. doi: 10.1021/pr200982k. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Zhang H, Wang H, Matsumura K, Wong YH, Ravasi T, et al. Quantitative proteomics study of larval settlement in the barnacle Balanus amphitrite. PLoS One. 2014;9:e88744. doi: 10.1371/journal.pone.0088744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiyagarajan V, Wong T, Qian P-Y. 2D gel-based proteome and phosphoproteome analysis during larval metamorphosis in two major marine biofouling invertebrates. J Proteome Res. 2009;8:2708–19. doi: 10.1021/pr800976u. [DOI] [PubMed] [Google Scholar]
- 22.Dash S, Chandramouli KH, Zhang Y, Qian P-Y. Effects of poly-ether B on proteome and phosphoproteome expression in biofouling Balanus amphitrite cyprids. Biofouling. 2012;28:405–15. doi: 10.1080/08927014.2012.679731. [DOI] [PubMed] [Google Scholar]
- 23.Han Z, Sun J, Zhang Y, He F, Xu Y, Matsumura K, et al. iTRAQ-based proteomic profiling of the barnacle Balanus amphitrite in response to the antifouling compound meleagrin. J Proteome Res. 2013;12:2090–2100. doi: 10.1021/pr301083e. [DOI] [PubMed] [Google Scholar]
- 24.Edwards AJ, Head SM. Key Environments: Red Sea. London: Pergamon; 1987. [Google Scholar]
- 25.Roder C, Berumen ML, Bouwmeester J, Papathanassiou E, Al-Suwailem A, Voolstra CR. First biological measurements of deep-sea corals from the Red Se. Sci Rep. 2013;3:2802. doi: 10.1038/srep02802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu JW, Qian P-Y. Tolerance of the barnacle Balanus amphitrite amphitrite to salinity and temperature stress: effects of previous experience. Mar Ecol Prog Ser. 1999;188:123–32. doi: 10.3354/meps188123. [DOI] [Google Scholar]
- 27.Thiyagarajan V, Harder T, Qian P-Y. Combined effects of temperature and salinity on larval development and attachment of the subtidal barnacle Balanus trigonus Darwin. J Exp Mar Biol. 2003;287:223–36. doi: 10.1016/S0022-0981(02)00570-1. [DOI] [Google Scholar]
- 28.Roer R, Dillman R. The structure and calcification of the crustacean cuticle. Am Zool. 1984;24:893–909. doi: 10.1093/icb/24.4.893. [DOI] [Google Scholar]
- 29.Chandramouli KH, Mok FS, Wang H, Qian P-Y. Phosphoproteome analysis during larval development and metamorphosis in the spionid polychaete Pseudopolydora vexillosa. BMC Dev Biol. 2011;11:31. doi: 10.1186/1471-213X-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandramouli KH, Reish D, Qian P-Y. Gel-based and gel-free identification of proteins and phosphopeptides during egg-to-larva transition in polychaete Neanthes arenaceodentata. PLoS One. 2012;7:e38814. doi: 10.1371/journal.pone.0038814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandramouli KH, Ravasi T, Reish D, Qian P-Y. Proteomic changes between male and female worms of the polychaetous annelid Neanthes arenaceodentata before and after spawning. PLoS One. 2013;8:e72990. doi: 10.1371/journal.pone.0072990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandramouli KH, Reish D, Zhang H, Qian P-Y, Ravasi T. Proteomic changes associated with successive reproductive periods in male polychaetous Neanthes arenaceodentata. Sci Rep. 2015;5:13561. doi: 10.1038/srep13561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagersson NC. The ultrastructure of two types of muscle fibre cells in the cyprid of Balanus amphitrite (Crustacea: Cirripedia) J Mar Biol Ass. 2002;82:573–78. doi: 10.1017/S0025315402005891. [DOI] [Google Scholar]
- 34.Okazaki Y, Shizuri Y. Structures of six cDNAs expressed specifically at cypris larvae of barnacles, Balanus amphitrite. Gene. 2000;250:127–135. doi: 10.1016/S0378-1119(00)00184-0. [DOI] [PubMed] [Google Scholar]
- 35.Chandramouli KH, Soo L, Qian P-Y. Differential expression of proteins and phosphoproteins during larval metamorphosis of the polychaete Capitella sp. I. Proteome Sci. 2011;9:51. doi: 10.1186/1477-5956-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He LS, Xu Y, Matsumura K, Zhang Y, Zhang G, Qi SH, et al. Evidence for the involvement of p38 MAPK activation in barnacle larval settlement. PLoS One. 2012;7:e47195. doi: 10.1371/journal.pone.0047195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang G, He LS, Wong YH, Qian P-Y. MKK3 was involved in larval settlement of the barnacle Amphibalanus amphitrite through activating the kinase activity of p38MAPK. PLoS One. 2013;8:e69510. doi: 10.1371/journal.pone.0069510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, He LS, Zhang G, Xu Y, Lee OO, Matsumura K, et al. The regulatory role of the NO/cGMP signal transduction cascade during larval attachment and metamorphosis of the barnacle Balanus (=Amphibalanus) amphitrite. J Exp Biol. 2012;215:3813–22. doi: 10.1242/jeb.070235. [DOI] [PubMed] [Google Scholar]
- 39.Anil AC, Chiba K, Okamoto K, Kurokura H. Influence of temperature and salinity on larval development of Balanus amphitrite: implications in fouling ecology. Mar Ecol Prog Ser. 1995;118:159–66. doi: 10.3354/meps118159. [DOI] [Google Scholar]
- 40.Burton T, Metcalfe NB. Can environmental conditions experienced in early life influence future generations? Proc R Soc B. 2014;281:20140311. doi: 10.1098/rspb.2014.0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pechenik JA. Environmental influences on larval survival and development. In: Giese AC, Pearse JS, Pearse VB, editors. Reproduction of Marine Invertebrates. Palo Alto: Blackwell; 1987. pp. 551–608. [Google Scholar]
- 42.Papakostas S, Vasemägi A, Vähä JP, Himberg M, Peil L, Primmer CR. A proteomics approach reveals divergent molecular responses to salinity in populations of European whitefish (Coregonus lavaretus) Mol Ecol. 2012;21:3516–30. doi: 10.1111/j.1365-294X.2012.05553.x. [DOI] [PubMed] [Google Scholar]
- 43.Davis KA, Lentz SJ, Pineda J, Farrar JT, Starczak VR, Churchill JH. Observations of the thermal environment on Red Sea platform reefs: a heat budget analysis. Coral Reefs. 2011;30:25–36. doi: 10.1007/s00338-011-0740-8. [DOI] [Google Scholar]
- 44.Ziegler M, Roder C, Büchel C, Voolstra CR. Niche acclimatization in Red Sea corals is dependent on flexibility of host-symbiont association. Mar Ecol Prog Ser. 2015;533:149–161. doi: 10.3354/meps11365. [DOI] [Google Scholar]
- 45.Heikkila JJ, Ohan N, Tam Y, Ali A. Heat shock protein gene expression during Xenopus development. Cell Mol Life Sci. 1997;53:114–21. doi: 10.1007/PL00000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chandramouli KH, Sun J, Mok FS, Liu L, Qiu JW, Ravasi T, et al. Transcriptome and quantitative proteome analysis reveals molecular processes associated with larval metamorphosis in the polychaete Pseudopolydora vexillosa. J Proteome Res. 2013;12:1344–58. doi: 10.1021/pr3010088. [DOI] [PubMed] [Google Scholar]
- 47.Bishop CD, Brandhorst BP. No/Cgmp signaling and Hsp90 activity represses metamorphosis in the sea urchin Lytechinus Pictus. Biol Bull. 2001;201:394–404. doi: 10.2307/1543617. [DOI] [PubMed] [Google Scholar]
- 48.Gohad NV, Dickinson GH, Orihuela B, Rittschof D, Mount AS. Visualization of putative ion-transporting epithelia in Amphibalanus amphitrite using correlative microscopy: Potential function in osmoregulation and biomineralization. J Exp Mar Biol Ecol. 2009;380:88–98. doi: 10.1016/j.jembe.2009.09.008. [DOI] [Google Scholar]
- 49.Papakostas S, Vasemägi A, Himberg M, Primmer CR. Proteome variance differences within populations of European whitefish (Coregonus lavaretus) originating from contrasting salinity environments. J Proteomics. 2014;105:144–50. doi: 10.1016/j.jprot.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 50.Wong KK, Lane AC, Leung PT, Thiyagarajan V. Response of larval barnacle proteome to CO(2)-driven seawater acidification. Comp Biochem Phys D. 2011;6:310–21. doi: 10.1016/j.cbd.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 51.Marshall DJ, McQuaid CD. Warming reduces metabolic rate in marine snails: adaptation to fluctuating high temperatures challenges the metabolic theory of ecology. Proc Biol Sci. 2011;278:281–88. doi: 10.1098/rspb.2010.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gracey AY, Chaney ML, Boomhower JP, Tyburczy WR, Connor K, Somero GN. Rhythms of gene expression in a fluctuating intertidal environment. Curr Biol. 2008;18:1501–07. doi: 10.1016/j.cub.2008.08.049. [DOI] [PubMed] [Google Scholar]
- 53.Zou FM, Lou DS, Zhu YH, Wang SP, Jin BR, Gui ZZ. Expression profiles of glutathione S-transferase genes in larval midgut of Bombyx mori exposed to insect hormones. Mol Biol Rep. 2011;38:639–47. doi: 10.1007/s11033-010-0150-y. [DOI] [PubMed] [Google Scholar]
- 54.Gieffers C, Korioth F, Heimann P, Ungermann C, Frey J. Mitofilin is a transmembrane protein of the inner mitochondrial membrane expressed as two isoforms. Exp Cell Res. 1997;232:395–99. doi: 10.1006/excr.1997.3539. [DOI] [PubMed] [Google Scholar]
- 55.Chmurzyńska A. The multigene family of fatty acid-binding proteins (FABPs): function, structure and polymorphism. J Appl Gene. 2006;47:39–48. doi: 10.1007/BF03194597. [DOI] [PubMed] [Google Scholar]
- 56.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–23. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, et al. De novo assembly and analysis of RNA-seq data. Nat Methods. 2010;7:909–12. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 58.Fu L, Niu B, Zhu Z, Wu SY, Li W. CD-HIT: accelerated for clustering the next generation sequencing data. Bioinformatics. 2012;28:3150–52. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–7. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 60.Mulder N, Apweiler R. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol Biol (Clifton, NJ) 2007;396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- 61.Zhang H, Zhao C, Li X, Zhu Y, Gan CS, Wang Y, et al. Study of monocyte membrane proteome perturbation during lipopolysaccharide-induced tolerance using iTRAQ-based quantitative proteomic approach. Proteomics. 2010;10:2780–89. doi: 10.1002/pmic.201000066. [DOI] [PubMed] [Google Scholar]
- 62.Zhu J, Zhang H, Guo T, Li W, Li H, Zhu Y, et al. Quantitative proteomics reveals differential biological processes in healthy neonatal cord neutrophils and adult neutrophils. Proteomics. 2014;14:1688–97. doi: 10.1002/pmic.201400009. [DOI] [PubMed] [Google Scholar]
- 63.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 64.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 65.Shadforth IP, Dunkley TP, Lilley KS, Bessant C. i-Tracker: for quantitative proteomics using iTRAQ. BMC Genomics. 2005;6:145. doi: 10.1186/1471-2164-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–78. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]


