Abstract
Somatic mutations in TEK, the gene encoding endothelial cell tyrosine kinase receptor TIE2, cause more than half of sporadically occurring unifocal venous malformations (VMs). Here, we report that somatic mutations in PIK3CA, the gene encoding the catalytic p110α subunit of PI3K, cause 54% (27 out of 50) of VMs with no detected TEK mutation. The hotspot mutations c.1624G>A, c.1633G>A, and c.3140A>G (p.Glu542Lys, p.Glu545Lys, and p.His1047Arg), frequent in PIK3CA-associated cancers, overgrowth syndromes, and lymphatic malformation (LM), account for >92% of individuals who carry mutations. Like VM-causative mutations in TEK, the PIK3CA mutations cause chronic activation of AKT, dysregulation of certain important angiogenic factors, and abnormal endothelial cell morphology when expressed in human umbilical vein endothelial cells (HUVECs). The p110α-specific inhibitor BYL719 restores all abnormal phenotypes tested, in PIK3CA- as well as TEK-mutant HUVECs, demonstrating that they operate via the same pathogenic pathways. Nevertheless, significant genotype-phenotype correlations in lesion localization and histology are observed between individuals with mutations in PIK3CA versus TEK, pointing to gene-specific effects.
Main Text
Venous malformations (VMs) are localized developmental defects composed of ectatic venous channels with a thin endothelial cell (EC) lining, surrounded by sparse, erratically distributed vascular smooth muscle cells (vSMCs) and a disorganized extracellular matrix (ECM).1, 2 VMs cause aesthetic problems, chronic and significant pain, and obstruction of organ function as a result of their size or localization. Currently, they are treated with surgery, sclerotherapy, or both; however, these approaches have significant limitations, including inaccessibility of lesions and regrowth after incomplete resection.1
We discovered that activating mutations in TEK, the gene encoding the EC tyrosine kinase receptor TIE2 (MIM 600221), cause familial3, 4 (MIM 600195) as well as sporadically occurring VMs,5, 6 and downstream PI3K/AKT signaling represents a major effector mechanism in lesion formation.2, 7, 8 This led to the first evidence-based application of molecular therapy to VMs: inhibition of the PI3K/AKT pathway by the mTOR inhibitor rapamycin (sirolimus) proved effective in ameliorating disease in a mouse model as well as a pilot cohort of individuals with difficult-to-treat VMs.8
PI3K signaling is initiated by binding of the p85 regulatory subunit to phosphorylated tyrosine residues of ligand-activated receptors, allowing for the activation of the p110 catalytic subunit.9 Chronic PI3K/AKT activation can instead occur as a result of somatic mutations in PIK3CA (the gene encoding p110α, a class IA PI3K catalytic protein; MIM 171834). Such mutations have been identified in a wide variety of cancers10, 11 (Catalogue of Somatic Mutations in Cancer; COSMIC); in overgrowth syndromes,12, 13, 14, 15, 16 some of which (congenital lipomatous overgrowth with vascular, epidermal, and skeletal anomalies [CLOVES]17 (MIM 612918); Klippel-Trenaunay syndrome [KTS]18 (MIM 149000); Fibro-adipose vascular anomaly [FAVA]18) have a vascular component;19 and in lymphatic malformation (LM18, 20, 21). Given the pivotal role of PI3K activation in VM causation, we hypothesized that PIK3CA mutations might cause VMs that lack detectable TIE2 alterations.5, 6
We screened surgically excised VMs from 87 unrelated individuals with unifocal VM. Informed consent was obtained from all participants, as approved by the ethics committee of the Faculty of Medicine at Université Catholique de Louvain. Tissues were collected in liquid nitrogen immediately after surgical excision, performed for reasons of medical necessity only. Frozen tissue homogenization was followed by overnight digestion with 100 μg/ml Proteinase K at 56° and DNA extraction (Wizard Genomic DNA extraction kit, Promega). Targeted deep sequencing of the 23 TEK and 20 PIK3CA coding exons (with ten nucleotides of flanking introns each) was carried out with an Ion AmpliSeq Custom DNA Panel (Life Technologies), which consists of primers for multiplexed PCR amplification of regions of interest (100% coverage of TEK; 98.75% coverage, missing 11 nucleotides in and flanking exon 5 and 34 nucleotides in exon 20, of PIK3CA). Ion Ampliseq libraries (with unique molecular bar codes and universal priming sequences ligated to each PCR-amplified sample according to manufacturers’ instructions) were pooled, loaded onto an Ion 316 Chip V2, and sequenced on an Ion PGM (all from Life Technologies) according to manufacturers’ instructions. Raw sequences (FASTQ files) were separated by bar code and aligned to the human genome reference sequence (assembly GRCh37/hg19) by the Torrent Server (Life Technologies). Aligned sequence (BAM) files were submitted to the Torrent Variant Caller for variant calling (Life Technologies). Called variants were subsequently annotated, filtered, and visualized with Highlander, an in-house bioinformatics framework.
The following quality-control criteria were used in calling a sample positive for a variant: (1) total coverage depth of ≥100 reads across the position; (2) ≥5 mutant reads, including both forward and reverse orientation reads; and (3) ≥1% mutant read frequency. Variants that passed these criteria were evaluated as follows: (1) synonymous variants were discarded; (2) variants present at >1% frequency in databases of known polymorphisms (1000 Genomes, Genome of the Netherlands [GoNL], NHLBI Exome Sequencing Project (ESP 2500 and 6500), and Exome Aggregation Consortium [ExAC]) were discarded. Novel missense mutations were submitted to six programs (Polyphen2; SIFT; Mutation Taster; Mutation Assessor; Functional Analysis though Hidden Markov Models, or FATHMM; and Likelihood Ratio Test, or LRT; all implemented in Highlander) for their predicted effect on protein function.
We identified PIK3CA mutations in affected tissues from 27 out of 87 screened individuals (31%), and we identified TEK mutations in 37 individuals (Table 1). When combined with previously published data,5, 6 mutations in TEK account for 80 out of 130 (61.5%) individuals with unifocal VM (Table 1); c.2740C>T (NM_000459.4; p.Leu914Phe) remains the most frequent mutation (74%; Table 1). PIK3CA mutations account overall for 27 out of 130 individuals with unifocal VM (20.8%; Table 1).
Table 1.
Somatic Mutations Identified in Individuals with Unifocal VM
| Study |
TEK Mutation Detected |
PIK3CA Mutation Detected | TEK, PIK3CA Mutation Not Detected | Total Screened | ||
|---|---|---|---|---|---|---|
| c.2740C>T (p.Leu914Phe) | Other | Total | ||||
| Limaye et al., 20095 | 24 | 2 | 26 | - | - | - |
| Soblet et al., 20136 | 9 | 8 | 17 | - | - | - |
| Current study | 26 | 11 | 37 | 27 | 23a | - |
| Total (percent of total screened) | 59 (73.8%b) | 21 (26.2%b) | 80 (61.5%) | 27 (20.8%) | 23 (17.7%) | 130 |
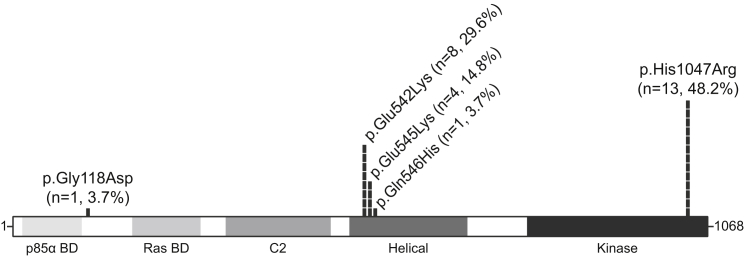
Five PIK3CA mutations were detected, at allele frequencies ranging from 1% to 17.5% of total reads (Figure 1, Table S1). “Hotspot” mutations (the most recurrent mutations in PIK3CA-associated cancers and developmental disorders) were identified in 25 out of the 27 individuals with PIK3CA mutations (Figure 1): The p110 α-helical-domain substitutions p.Glu542Lys (c.1624G>A, NM_006218.2) and p.Glu545Lys (c.1633G>A) were identified in eight and four individuals, respectively (together, 44.4%). The kinase domain substitution p.His1047Arg (c.3140A>G) was identified in 13 individuals (48.1%). p.Gly118Asp (c.353G>A) and p.Gln546His (c.1638G>T), both present in the COSMIC database, were identified in one individual each (Figure 1, Table S1). p.Gly118Asp is predicted to be damaging to protein function by six out of six in silico prediction programs, and p.Gln546His is predicted to be damaging by 3 out of 6 programs.
Figure 1.
Distribution of Amino Acid Changes and Frequency of PIK3CA Somatic Mutations in Unifocal VM-Affected Individuals
PIK3CA protein domains with mutations identified and their frequency (percentage) among individuals with somatic PIK3CA mutations. Abbreviations are as follows: n, number of individuals; BD, binding domain.
Targeted deep sequencing was performed on all available blood DNA from individuals with PIK3CA mutations identified in lesions. Blood DNA from an individual was considered positive for the variant detected in the corresponding tissue if (1) the variant was present in ≥5 reads, (2) the total read depth was ≥100 reads, and (3) the variant was detected at ≥3 SDs above the mean background frequency, calculated separately for each variant position, as described in Table S1.
PIK3CA mutations were not detected in affected individuals’ blood DNA, despite coverage depths ranging from 383–11,384 reads (mean: 4,940) across the five mutation positions (Table S1). Added to the relatively low frequency of mutant alleles detected in tissues (Table S1), this suggests that VM-causative PIK3CA mutations are indeed somatic.
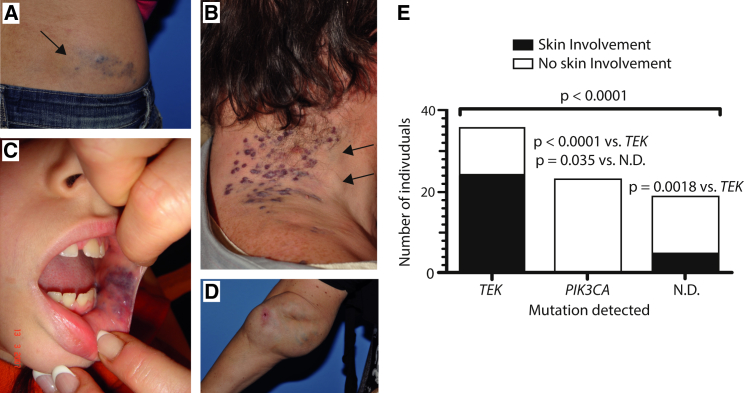
VMs are often cutaneous (Figures 2A and 2B), subcutaneous (Figures 2A and 2B), or mucosal (Figure 2C) and can extend deep into various tissues and organs, such as muscles, joints (Figure 2D), and intestines. We observed that in subjects with TEK mutations (n = 37 identified in this study; Table 1), VMs often affected, but were not restricted to, the skin (24 out of 36 individuals for whom information on lesion localization was available; 66.7%). By contrast, of the 23 (out of 27) PIK3CA-associated VMs with available localization information, none showed skin involvement (Figure 2C). This suggests that, unlike TEK-associated VMs, PIK3CA-associated VMs tend not to extend to the skin surface, perhaps as result of differences in where and when these mutations were acquired.
Figure 2.
VM Localization in Individuals with Mutations Detected in TEK, PIK3CA, or Neither
Single, extensive slow-flow malformations, present at birth and growing proportionately with individual. Lesions are bluish and compressible. They can be cutaneous (A and B), sub-cutaneous (A and B, arrows; D), mucosal (C), or extended deep into muscles (intra-muscular), joints (intra-articular, (D) or other tissues and organs. (A and B) Skin involvement; (C, D) no skin involvement. (E) Number of VMs with a mutation detected in TEK, PIK3CA, or neither (N.D.: none detected) that do or do not show skin involvement (p < 0.0001 overall, chi-square test). Inter-group p values: post-hoc Fisher’s exact tests.
VMs, uniquely among vascular anomalies, are associated with significant coagulopathy in the form of high D-dimer levels, sometimes accompanied by low fibrinogen levels.1, 22, 23 We found that D-dimer levels were significantly lower in individuals with no detected TEK or PIK3CA mutations than in individuals with mutations in either gene (Figure S1). D-dimer levels have been correlated with total lesion size;22 this might therefore simply reflect differences in the surface area, depth, or volume of VMs caused by TEK and PIK3CA mutations, as compared to those of as-yet-unknown cause. Alternatively, it could be due to dysregulation of coagulation cascade components in the endothelium; we demonstrate that such effects can be engendered by TEK2 and PIK3CA mutations (current study) in human umbilical vein endothelial cells (HUVECs).
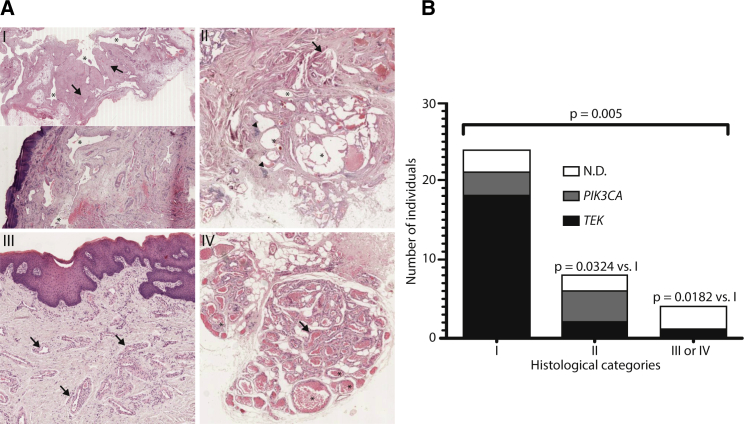
Blinded histological evaluation of hematoxylin and eosin-stained formalin-fixed and paraffin-embedded tissue sections from 36 unifocal VMs revealed four distinct categories (Figure 3A): the majority (n = 24), placed in category I, consisted of isolated or interconnected veins with large, highly irregular lumens and thick fibrotic walls containing irregularly distributed clumps of vSMCs running parallel to the ECs; their characteristics corresponding to the “classic” histological description of the malformation. Eight VMs, placed in category II, were similar to VMs observed in PTEN hamartoma tumor syndrome (PHTS; MIM 601728) and were characterized by interconnected veins with fibrotic walls containing foci of lymphoid cells and vSMCs radiating outward in irregular bundles perpendicular to the lumen. The minority of VMs were placed in categories III (n = 2) and IV (n = 2), both characterized by small, dilated veins with thick walls composed of regular layers of vSMCs. Veins in category III were isolated, whereas those in category IV were smaller and clustered together; individual lumens were separated by fibrous tissue, and a thick vSMC layer bordered the entire cluster. 18 out of 21 tissues with TEK mutations (75%) were in category I, whereas only two were in category II and only 1 was in category III. In contrast, tissues with PIK3CA mutations were equally distributed between categories I and II (three and four out of seven, respectively), and none were in categories III or IV. Intriguingly, categories III and IV were almost exclusively made up of samples with no detected TEK and PIK3CA mutation. Samples without a detected mutation also included three tissues in category I and two in category II (Figure 3E).
Figure 3.
Histological Categorization of Unifocal VM Tissues
(A) Hematoxylin and eosin-stained sections demonstrating the major features of four histological categories identified. (I) Interconnected (top) or isolated (bottom) veins with large, irregular lumens (asterisks) and thick fibrotic walls (arrows) containing irregular vascular smooth-muscle-cell aggregates (not visible at this magnification) oriented in parallel with ECs; (II) interconnected veins (asterisks) with fibrotic walls containing radial bundles of vascular smooth-muscle cells (arrow) and foci of lymphoid cells (arrowheads); (III, IV) small-to-medium sized dilated veins (asterisks) with thick walls composed of organized layers of vascular smooth-muscle cells (arrows), either isolated (III) or clustered with fibrous tissue separating lumens (IV).
(B) Number of samples with a mutation detected in TEK, PIK3CA, or neither (N.D.: none detected) in each histological category (p = 0.005 overall, chi-square test). Inter-group p values: post-hoc chi-square tests.
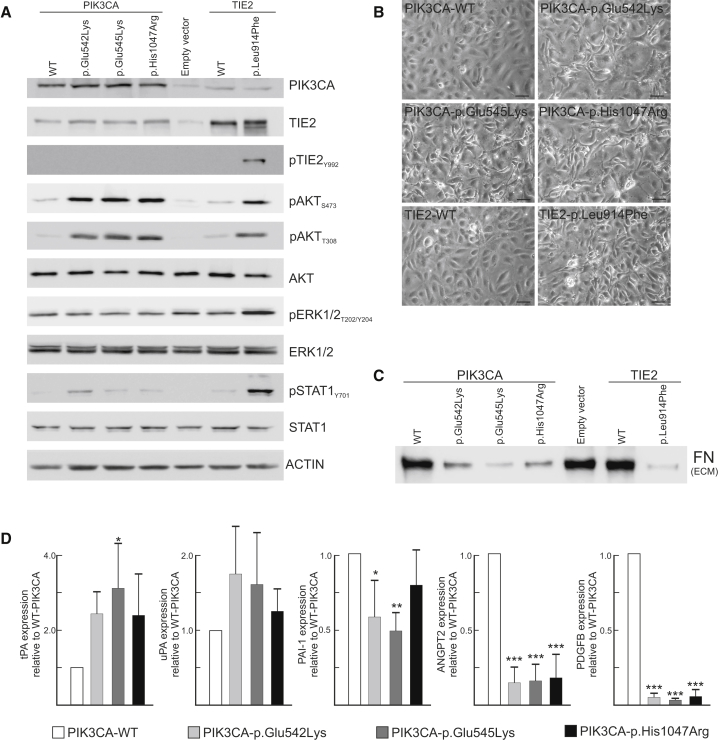
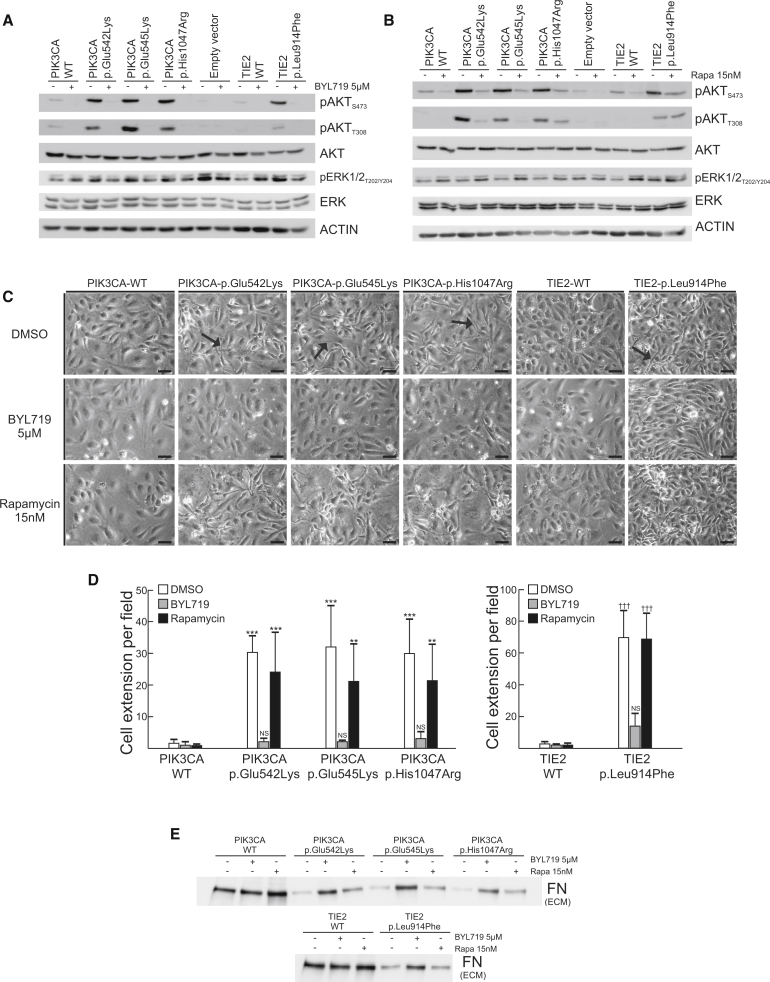
We previously showed that TEK mutations cause cell-intrinsic alterations when expressed in HUVECs2, 5, 8 and that activation of PI3K/AKT is a universal feature of HUVECs expressing mutated TEK.2, 7 We hypothesized that, because PIK3CA encodes the major isoform mediating growth-factor-induced PI3K activation in ECs,24 PIK3CA mutations have similar effects. HUVECs were retrovirally transfected with PIK3CA wild-type or recurrent VM-causative mutant forms (c.1624G>A, c.1633G>A, and c.3140A>G [p.Glu542Lys, p.Glu545Lys, and p.His1047Arg]) in the pMXs-puro vector.27 Similar to the most common VM-causative mutant, TEK c.2740C>T (TIE2 p.Leu914Phe2, 7, 8), PIK3CA variants activated AKT (Figure 4A), disrupted normal EC-characteristic monolayer morphology as visualized by phase-contrast microscopy (Figure 4B), resulted in loss of ECM fibronectin (Figure 4C), and strongly downregulated ANGPT2 and PDGF-B mRNA expression as measured by real-time quantitative PCR with normalization to GAPDH (Figure 4D). Plasminogen system-components were also somewhat dysregulated by PIK3CA variants (Figure 4D). Inhibition of PIK3CA with BYL7129, which selectively targets p110α,25 abolished both PIK3CA-variant and TIE2-p.Leu914Phe-induced AKT phosphorylation (Figure 5A), indicating that these proteins participate in the same signaling pathway. In addition, PIK3CA inhibition restored cell morphology (Figures 5C and 5D) and ECM fibronectin levels (Figure 5E) in both PIK3CA- and TIE2-p.Leu914Phe variants. Collectively, these data indicate that TIE2 and PIK3CA variants induce the same VM-causative signaling pathways.
Figure 4.
VM-Causative PIK3CA and TIE2 Variants Cause Similar Phenotypes in Transduced HUVECs
Full-length cDNAs encoding TIE2-WT and TIE2-Leu914Phe cloned into pMXs vector, full-length cDNAs for PIK3CA-WT cloned into pMXs-puro retroviral vector,27 and mutations generated with the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies) according to the manufacturer’s protocol. 293-GPG-VSV-G viral packaging cells28 were transfected with Fugene HD transfection reagent (Promega) to produce viruses. HUVECs were transduced with pMXs-puro-PIK3CA constructs or empty vector selected with 1 μg/ml puromycin (Sigma) for 5 days. (A) Activation of signaling molecules by HUVECs retrovirally transduced to express variant and wild-type proteins. A representative immunoblot from three independent experiments is shown.
(B) Elongated cell morphology with loss of cobblestone pattern in PIK3CA- and TIE2-variant HUVECs (the scale bar represents 50 μm).
(C) Loss of ECM fibronectin: representative immunoblot from three independent experiments. HUVECs were removed by washing with 0.05% Triton X-100 and 50 mM NH4OH in PBS, followed by one wash with 50 mM NH4OH and three washes with PBS. Lysis was carried out with ECM lysis buffer (6.5M Urea, 9.1 mM Na2HPO4, 1.7 mM NaH2PO4, 1% NP-40, 0.25% Sodium Deoxycholate, 150 mM NaCl, 0.1% SDS, and 1 mM EDTA).
(D) Normalized n-fold change in gene expression measured by real-time quantitative PCR on HUVEC cDNA (means + SD, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus PIK3CA-WT, ANOVA with Tukey’s post-hoc test). WT indicates wild-type.
Figure 5.
Normalization of PIK3CA- and TIE2-Variant HUVEC Phenotypes by PIK3CA Inhibition
(A) Effect of PIK3CA (p110α) inhibitor BYL719 versus DMSO control on AKT and ERK1/2 phosphorylation in confluent PIK3CA- and TIE2-variant cells at 24 hr in full-serum media; a representative immunoblot from two independent experiments is shown.
(B) Effect of mTOR inhibitor rapamycin versus DMSO control on AKT and ERK1/2 phosphorylation in confluent PIK3CA- and TIE2-variant cells at 24 hr in full-serum media; a representative immunoblot from two independent experiments is shown.
(C and D) Effects of DMSO (control), BYL719, or rapamycin on cell morphology at 24 hr. Cell extensions (arrows) were quantitated from digital images; representative images from four independent experiments are shown. The scale bar represents 50 μm. Three images per dish were randomly captured with a phase-contrast microscope, and the number of thin cell extensions overlapping neighboring cells was manually counted from digitalized images. The average number of extensions from three images was considered a single observation. (Mean+ SD, ∗∗∗p < 0.001, ∗∗p < 0.01 versus PIK3CA-WT DMSO, †††p < 0.001, †p < 0.05 versus TIE2-WT DMSO, ANOVA followed by Tukey’s test).
(E) Effect of BYL719 and rapamycin on ECM fibronectin; a representative immunoblot from two independent experiments is shown. WT indicates wild-type.
Interestingly, unlike TIE2-p.Leu914Phe,2, 7 PIK3CA variants induced low levels of pSTAT1 when they were compared to the wild-type, and they induced no pERK1/2 (Figure 4A). Added to the phenotypic differences observed between individuals with TEK versus PIK3CA mutations, this suggests certain gene-specific features that are yet to be understood.
Rapamycin, probably because of its inhibitory effect on AKT phosphorylation, was shown to control VM growth in a murine transplantation model, and it had therapeutic benefits in a clinical pilot study of six individuals (five with VM and one with KTS).8 Three individuals with VM had a TEK mutation,8 and the remaining three (one individual with KTS and individuals 10 and 57 with VMs; Table S1) were found to have a PIK3CA mutation. We compared the effects of BYL719 and rapamycin on abnormal HUVEC phenotypes. Rapamycin diminished AKT phosphorylation but did not restore ECM fibronectin to wild-type levels in PIK3CA-variant or TIE2-p.Leu914Phe HUVECs (Figures 5B and 5E). Loss of the EC cobblestone pattern was not, however, rescued in either PIK3CA-variant or TIE2-p.Leu914Phe HUVECs (Figure 5C). This could be because PI3K activation, in addition to leading to changes in the AKT/mTOR pathway, leads to wider changes in kinase activity in pathways including MAPK and ErbB;26 only a subset of these changes are probably reversed by rapamycin.
We previously showed that TIE2-mediated VMs from mouse transplant models and affected individuals have features (revealed by histology and electron microscopy) that correspond well to HUVEC-intrinsic phenotypes observed in vitro.2, 8 These include an uneven endothelial layer with prominent cellular elongations and abnormal perivascular ECM characterized by a wide basement membrane with randomly oriented, isolated collagen fibrils and irregularly distributed vSMCs.2 Downregulation of PDGF-B, which we demonstrated ex vivo in VM tissue sections from affected individuals,7 most likely contributes to defective recruitment of vSMCs. Downregulation of ANGPT2 might contribute to decreased sprouting (with increased lumen expansion)2 by abnormal ECs, whereas ECM alterations are associated with upregulation of serine proteases, including tissue plasminogen activator (tPA/PLAT) and urokinase-type plasminogen activator (uPA).2 Induction of the plasmin/plasminogen cascade (tPA and uPA upregulation with Plasminogen Activator Inhibitor 1 (PAI1) downregulation2) might also contribute to increased fibrinolysis and elevated levels of D-dimers in individuals with VM.1, 22, 23 PIK3CA inhibition reverses all abnormal features of PIK3CA- and TIE2-variant HUVECs. It therefore represents an attractive potential therapy for TIE2- and PIK3CA-mediated disease, which together account for the majority (82%) of unifocal VMs.
Acknowledgments
We are grateful to all the participants and their family members. These studies were partially supported by funding from the Belgian Science Policy Office Interuniversity Attraction Poles (BELSPO-IAP) program through the project IAP P7/43-BeMGI; the National Institutes of Health, Program Project P01 AR048564 (both to M.V.); the Fonds de la Recherche Scientifique (FRS-FNRS) under Grant T.0026.14 (to M.V.); and the Academy of Finland (136880 and Centre of Excellence Program 2012–2017 to L.E.). We also thank la Communauté Française de Wallonie-Bruxelles and la Lotterie Nationale, Belgium for their support. N.L. is a “Chercheur Qualifié du FRS-FNRS.” We thank the genetics platform of Université Catholique de Louvain for access to the IonTorrent Next Generation Sequencer. The authors thank Ms. Liliana Niculescu for secretarial help.
Published: December 3, 2015
Footnotes
Supplemental Data include one figure and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.11.011.
Accession Numbers
The accession number for the 108 sequences (TEK and PIK3CA coding exons from 87 tissue samples and 21 blood samples) reported in this paper is PRJEB11395, available at European Nucleotide Archive.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://www.1000genomes.org/
European Nucleotide Archive accession number, http://www.ebi.ac.uk/ena/data/view/PRJEB11395
Exome Aggregation Consortium (ExAC), http://exac.broadinstitute.org/
Functional Analysis though Hidden Markov Models (FATHMM), http://fathmm.biocompute.org.uk/
Genome of the Netherlands (GoNL), http://www.nlgenome.nl/
Highlander, http://sites.uclouvain.be/highlander/
Mutation Assessor, http://mutationassessor.org/
Mutation Taster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP), http://evs.gs.washington.edu/EVS/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
Polyphen2, http://genetics.bwh.harvard.edu/pph2/
SIFT, http://sift.jcvi.org/
Supplemental Data
References
- 1.Dompmartin A., Vikkula M., Boon L.M. Venous malformation: update on aetiopathogenesis, diagnosis and management. Phlebology. 2010;25:224–235. doi: 10.1258/phleb.2009.009041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nätynki M., Kangas J., Miinalainen I., Sormunen R., Pietilä R., Soblet J., Boon L.M., Vikkula M., Limaye N., Eklund L. Common and specific effects of TIE2 mutations causing venous malformations. Hum. Mol. Genet. 2015;24:6374–6389. doi: 10.1093/hmg/ddv349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vikkula M., Boon L.M., Carraway K.L., 3rd, Calvert J.T., Diamonti A.J., Goumnerov B., Pasyk K.A., Marchuk D.A., Warman M.L., Cantley L.C. Vascular dysmorphogenesis caused by an activating mutation in the receptor tyrosine kinase TIE2. Cell. 1996;87:1181–1190. doi: 10.1016/s0092-8674(00)81814-0. [DOI] [PubMed] [Google Scholar]
- 4.Wouters V., Limaye N., Uebelhoer M., Irrthum A., Boon L.M., Mulliken J.B., Enjolras O., Baselga E., Berg J., Dompmartin A. Hereditary cutaneomucosal venous malformations are caused by TIE2 mutations with widely variable hyper-phosphorylating effects. Eur. J. Hum. Genet. 2010;18:414–420. doi: 10.1038/ejhg.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Limaye N., Wouters V., Uebelhoer M., Tuominen M., Wirkkala R., Mulliken J.B., Eklund L., Boon L.M., Vikkula M. Somatic mutations in angiopoietin receptor gene TEK cause solitary and multiple sporadic venous malformations. Nat. Genet. 2009;41:118–124. doi: 10.1038/ng.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soblet J., Limaye N., Uebelhoer M., Boon L.M., Vikkula M. Variable somatic TIE2 mutations in half of sporadic venous malformations. Mol. Syndromol. 2013;4:179–183. doi: 10.1159/000348327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uebelhoer M., Nätynki M., Kangas J., Mendola A., Nguyen H.L., Soblet J., Godfraind C., Boon L.M., Eklund L., Limaye N., Vikkula M. Venous malformation-causative TIE2 mutations mediate an AKT-dependent decrease in PDGFB. Hum. Mol. Genet. 2013;22:3438–3448. doi: 10.1093/hmg/ddt198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boscolo E., Limaye N., Huang L., Kang K.T., Soblet J., Uebelhoer M., Mendola A., Natynki M., Seront E., Dupont S. Rapamycin improves TIE2-mutated venous malformation in murine model and human subjects. J. Clin. Invest. 2015;125:3491–3504. doi: 10.1172/JCI76004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorpe L.M., Yuzugullu H., Zhao J.J. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samuels Y., Wang Z., Bardelli A., Silliman N., Ptak J., Szabo S., Yan H., Gazdar A., Powell S.M., Riggins G.J. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 11.Samuels Y., Waldman T. Oncogenic mutations of PIK3CA in human cancers. Curr. Top. Microbiol. Immunol. 2010;347:21–41. doi: 10.1007/82_2010_68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J.H., Huynh M., Silhavy J.L., Kim S., Dixon-Salazar T., Heiberg A., Scott E., Bafna V., Hill K.J., Collazo A. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat. Genet. 2012;44:941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindhurst M.J., Parker V.E., Payne F., Sapp J.C., Rudge S., Harris J., Witkowski A.M., Zhang Q., Groeneveld M.P., Scott C.E. Mosaic overgrowth with fibroadipose hyperplasia is caused by somatic activating mutations in PIK3CA. Nat. Genet. 2012;44:928–933. doi: 10.1038/ng.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivière J.B., Mirzaa G.M., O’Roak B.J., Beddaoui M., Alcantara D., Conway R.L., St-Onge J., Schwartzentruber J.A., Gripp K.W., Nikkel S.M., Finding of Rare Disease Genes (FORGE) Canada Consortium De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat. Genet. 2012;44:934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rios J.J., Paria N., Burns D.K., Israel B.A., Cornelia R., Wise C.A., Ezaki M. Somatic gain-of-function mutations in PIK3CA in patients with macrodactyly. Hum. Mol. Genet. 2013;22:444–451. doi: 10.1093/hmg/dds440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maclellan R.A., Luks V.L., Vivero M.P., Mulliken J.B., Zurakowski D., Padwa B.L., Warman M.L., Greene A.K., Kurek K.C. PIK3CA activating mutations in facial infiltrating lipomatosis. Plast. Reconstr. Surg. 2014;133:12e–19e. doi: 10.1097/01.prs.0000436822.26709.7c. [DOI] [PubMed] [Google Scholar]
- 17.Kurek K.C., Luks V.L., Ayturk U.M., Alomari A.I., Fishman S.J., Spencer S.A., Mulliken J.B., Bowen M.E., Yamamoto G.L., Kozakewich H.P., Warman M.L. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am. J. Hum. Genet. 2012;90:1108–1115. doi: 10.1016/j.ajhg.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luks V.L., Kamitaki N., Vivero M.P., Uller W., Rab R., Bovée J.V., Rialon K.L., Guevara C.J., Alomari A.I., Greene A.K. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J. Pediatr. 2015;166 doi: 10.1016/j.jpeds.2014.12.069. 1048–54.e1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keppler-Noreuil K.M., Sapp J.C., Lindhurst M.J., Parker V.E., Blumhorst C., Darling T., Tosi L.L., Huson S.M., Whitehouse R.W., Jakkula E. Clinical delineation and natural history of the PIK3CA-related overgrowth spectrum. Am. J. Med. Genet. A. 2014;164A:1713–1733. doi: 10.1002/ajmg.a.36552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boscolo E., Coma S., Luks V.L., Greene A.K., Klagsbrun M., Warman M.L., Bischoff J. AKT hyper-phosphorylation associated with PI3K mutations in lymphatic endothelial cells from a patient with lymphatic malformation. Angiogenesis. 2015;18:151–162. doi: 10.1007/s10456-014-9453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osborn A.J., Dickie P., Neilson D.E., Glaser K., Lynch K.A., Gupta A., Dickie B.H. Activating PIK3CA alleles and lymphangiogenic phenotype of lymphatic endothelial cells isolated from lymphatic malformations. Hum. Mol. Genet. 2015;24:926–938. doi: 10.1093/hmg/ddu505. [DOI] [PubMed] [Google Scholar]
- 22.Dompmartin A., Acher A., Thibon P., Tourbach S., Hermans C., Deneys V., Pocock B., Lequerrec A., Labbé D., Barrellier M.T. Association of localized intravascular coagulopathy with venous malformations. Arch. Dermatol. 2008;144:873–877. doi: 10.1001/archderm.144.7.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dompmartin A., Ballieux F., Thibon P., Lequerrec A., Hermans C., Clapuyt P., Barrellier M.T., Hammer F., Labbé D., Vikkula M., Boon L.M. Elevated D-dimer level in the differential diagnosis of venous malformations. Arch. Dermatol. 2009;145:1239–1244. doi: 10.1001/archdermatol.2009.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graupera M., Guillermet-Guibert J., Foukas L.C., Phng L.K., Cain R.J., Salpekar A., Pearce W., Meek S., Millan J., Cutillas P.R. Angiogenesis selectively requires the p110alpha isoform of PI3K to control endothelial cell migration. Nature. 2008;453:662–666. doi: 10.1038/nature06892. [DOI] [PubMed] [Google Scholar]
- 25.Furet P., Guagnano V., Fairhurst R.A., Imbach-Weese P., Bruce I., Knapp M., Fritsch C., Blasco F., Blanz J., Aichholz R. Discovery of NVP-BYL719 a potent and selective phosphatidylinositol-3 kinase alpha inhibitor selected for clinical evaluation. Bioorg. Med. Chem. Lett. 2013;23:3741–3748. doi: 10.1016/j.bmcl.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Wu X., Renuse S., Sahasrabuddhe N.A., Zahari M.S., Chaerkady R., Kim M.S., Nirujogi R.S., Mohseni M., Kumar P., Raju R. Activation of diverse signalling pathways by oncogenic PIK3CA mutations. Nat. Commun. 2014;5:4961. doi: 10.1038/ncomms5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
- 28.Ory D.S., Neugeboren B.A., Mulligan R.C. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.