Abstract
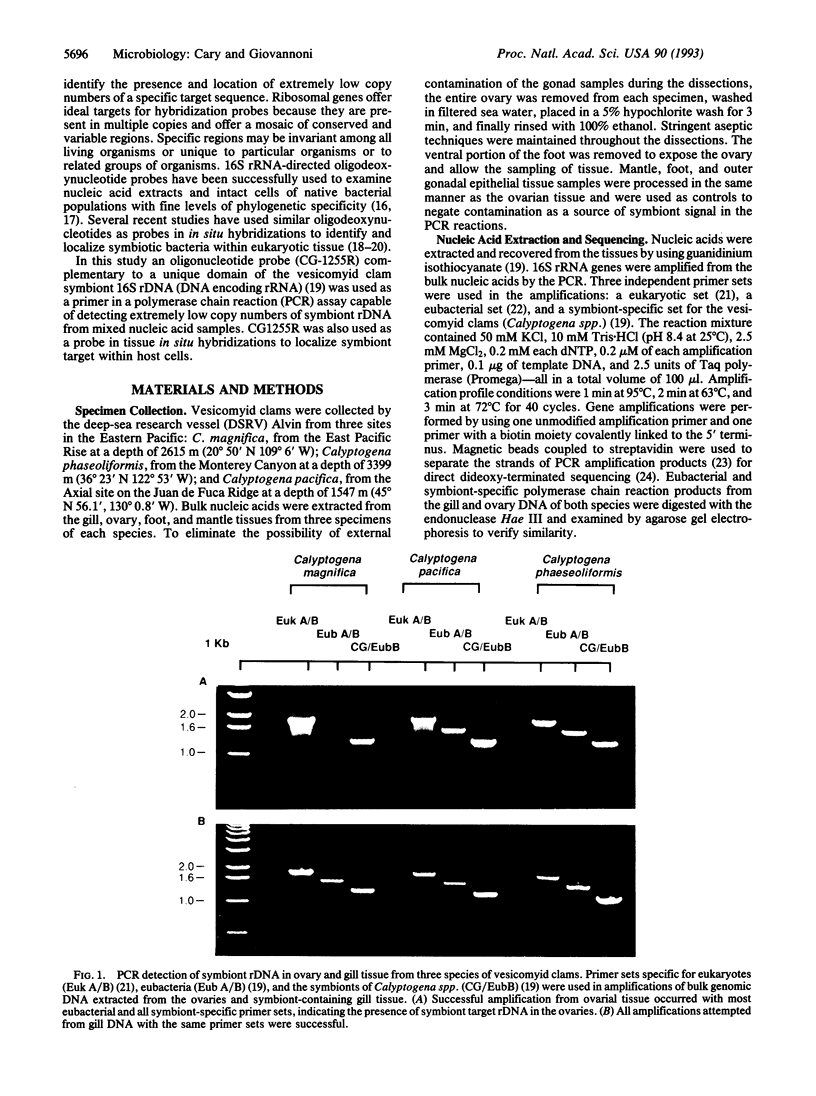
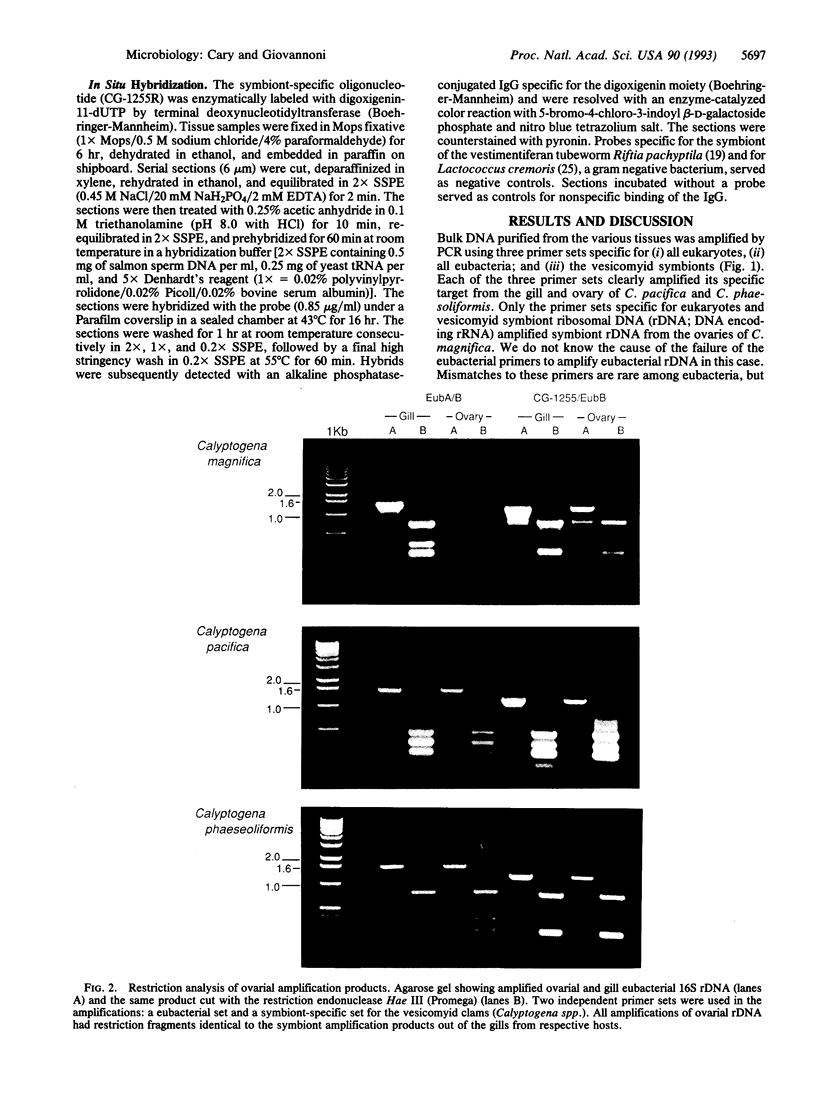
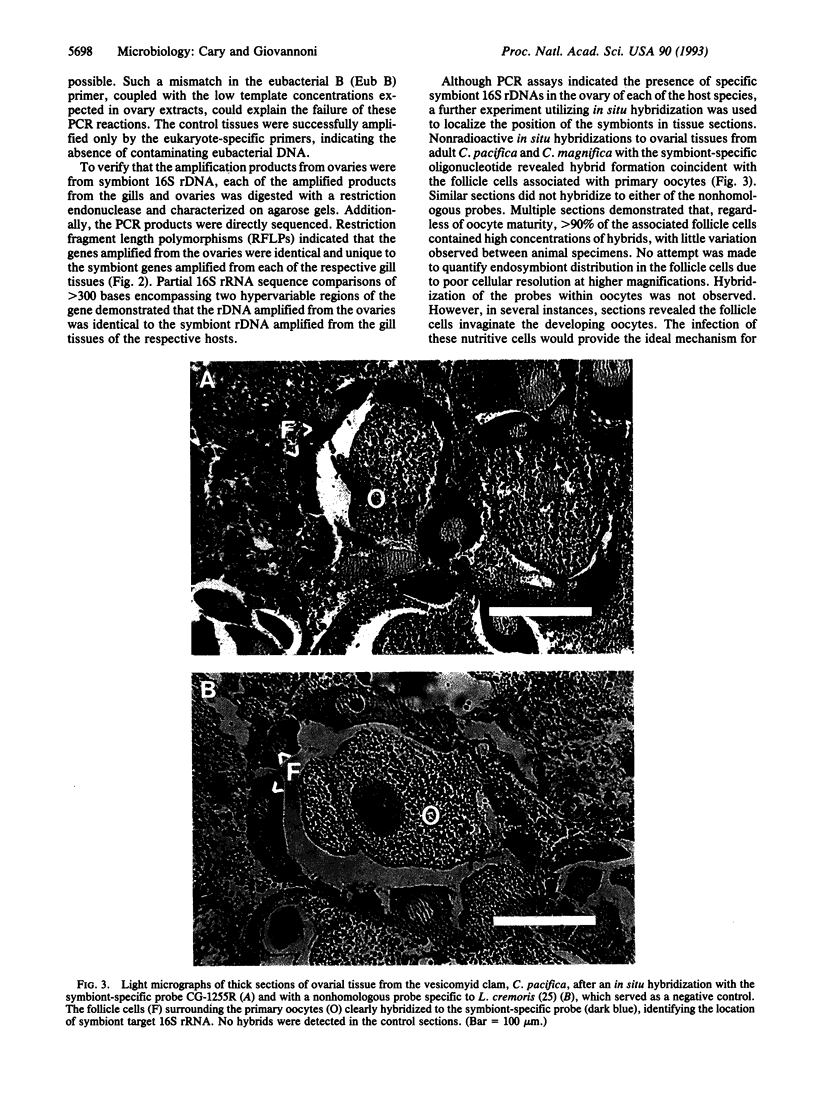
Vesicomyid clams are conspicuous fauna at many deep-sea hydrothermal-vent and cold-seep habitats. All species examined have specialized gill tissue harboring endosymbiotic bacteria, which are thought to provide the hosts' sole nutritional support. In these species mechanisms of symbiont inheritance are likely to be key elements of dispersal strategies. These mechanisms have remained unresolved because the early life stages are not available for developmental studies. A specific 16S rRNA-directed oligodeoxynucleotide probe (CG1255R) for the vesocomyid endosymbionts was used in a combination of sensitive hybridization techniques to detect and localize the endosymbionts in host germ tissues. Symbiont-specific polymerase chain reaction amplifications, comparative gene sequencing, and restriction fragment length polymorphisms were used to detect and confirm the presence of symbiont target in tissue nucleic acid extracts. Nonradioactive in situ hybridizations were used to resolve the position of the bacterial endosymbionts in host cells. Symbiont 16S rRNA genes were consistently amplified from the ovarial tissue of three species of vesicomyid clams: Calyptogena magnifica, C. phaseoliformis, and C. pacifica. The nucleotide sequences of the genes amplified from ovaries were identical to those from the respective host symbionts. In situ hybridizations to CG1255R labeled with digoxigenin-11-dUTP were performed on ovarial tissue from each of the vesicomyid clams. Detection of hybrids localized the symbionts to follicle cells surrounding the primary oocytes. These results suggest that vesicomyid clams assure successful, host-specific inoculation of all progeny by using a transovarial mechanism of symbiont transmission.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann R., Springer N., Ludwig W., Görtz H. D., Schleifer K. H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991 May 9;351(6322):161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- Cary S. C., Warren W., Anderson E., Giovannoni S. J. Identification and localization of bacterial endosymbionts in hydrothermal vent taxa with symbiont-specific polymerase chain reaction amplification and in situ hybridization techniques. Mol Mar Biol Biotechnol. 1993 Feb;2(1):51–62. [PubMed] [Google Scholar]
- Cavanaugh C. M., Gardiner S. L., Jones M. L., Jannasch H. W., Waterbury J. B. Prokaryotic Cells in the Hydrothermal Vent Tube Worm Riftia pachyptila Jones: Possible Chemoautotrophic Symbionts. Science. 1981 Jul 17;213(4505):340–342. doi: 10.1126/science.213.4505.340. [DOI] [PubMed] [Google Scholar]
- Distel D. L., DeLong E. F., Waterbury J. B. Phylogenetic characterization and in situ localization of the bacterial symbiont of shipworms (Teredinidae: Bivalvia) by using 16S rRNA sequence analysis and oligodeoxynucleotide probe hybridization. Appl Environ Microbiol. 1991 Aug;57(8):2376–2382. doi: 10.1128/aem.57.8.2376-2382.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel D. L., Lane D. J., Olsen G. J., Giovannoni S. J., Pace B., Pace N. R., Stahl D. A., Felbeck H. Sulfur-oxidizing bacterial endosymbionts: analysis of phylogeny and specificity by 16S rRNA sequences. J Bacteriol. 1988 Jun;170(6):2506–2510. doi: 10.1128/jb.170.6.2506-2510.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S. J., Britschgi T. B., Moyer C. L., Field K. G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990 May 3;345(6270):60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- Hultman T., Bergh S., Moks T., Uhlén M. Bidirectional solid-phase sequencing of in vitro-amplified plasmid DNA. Biotechniques. 1991 Jan;10(1):84–93. [PubMed] [Google Scholar]
- Medlin L., Elwood H. J., Stickel S., Sogin M. L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988 Nov 30;71(2):491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- Rau G. H. Hydrothermal Vent Clam and Tube Worm 13C/12C: Further Evidence of Nonphotosynthetic Food Sources. Science. 1981 Jul 17;213(4505):338–340. doi: 10.1126/science.213.4505.338. [DOI] [PubMed] [Google Scholar]
- Salama M., Sandine W., Giovannoni S. Development and application of oligonucleotide probes for identification of Lactococcus lactis subsp. cremoris. Appl Environ Microbiol. 1991 May;57(5):1313–1318. doi: 10.1128/aem.57.5.1313-1318.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Flesher B., Mansfield H. R., Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988 May;54(5):1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]