Abstract
Upon infection of Escherichia coli by bacteriophage Qβ, the virus-encoded β-subunit recruits host translation elongation factors EF-Tu and EF-Ts and ribosomal protein S1 to form the Qβ replicase holoenzyme complex, which is responsible for amplifying the Qβ (+)-RNA genome. Here, we use X-ray crystallography, NMR spectroscopy, as well as sequence conservation, surface electrostatic potential and mutational analyses to decipher the roles of the β-subunit and the first two oligonucleotide-oligosaccharide-binding domains of S1 (OB1–2) in the recognition of Qβ (+)-RNA by the Qβ replicase complex. We show how three basic residues of the β subunit form a patch located adjacent to the OB2 domain, and use NMR spectroscopy to demonstrate for the first time that OB2 is able to interact with RNA. Neutralization of the basic residues by mutagenesis results in a loss of both the phage infectivity in vivo and the ability of Qβ replicase to amplify the genomic RNA in vitro. In contrast, replication of smaller replicable RNAs is not affected. Taken together, our data suggest that the β-subunit and protein S1 cooperatively bind the (+)-stranded Qβ genome during replication initiation and provide a foundation for understanding template discrimination during replication initiation.
INTRODUCTION
Positive-stranded RNA viruses constitute the largest group of viruses and encompass notable plant, animal and human pathogens such as the tobacco mosaic virus, tomato bushy stunt virus, swine fever virus, hepatitis C virus and polio virus. Upon entry into a host cell, the viral genome functions as an mRNA and hijacks the translation machinery to produce the encoded proteins. One of these is an RNA-dependent RNA polymerase (RdRp), which is dedicated to the production of (+)-stranded genomes via a complementary, (–)-stranded intermediate. The viral RdRps contain the catalytic machinery required for RNA polymerization, while additional tasks during, for example, initiation and termination, may require the involvement of host proteins (1). Apart from interacting with host proteins and the viral RNA, the RdRps of several (+)-RNA viruses have been found to oligomerize with a stimulatory effect on RNA synthesis and viral viability (2–5).
Bacteriophage Qβ is a (+)-RNA virus that infects Escherichia coli. Upon infection, a genome-replicating complex is formed between the virus-encoded RdRp, known as the β-subunit, and translation elongation factors EF-Tu and EF-Ts as well as ribosomal protein S1 of the host cell (6). Structures of the Qβ replicase core complex composed of the β-subunit, EF-Tu and EF-Ts have been determined both in the absence (7,8) and presence (9,10) of a short illegitimate RNA template (i.e. a template that can be copied but not amplified). More recently, the structure of the Qβ replicase core complex bound to the first two domains of ribosomal protein S1 was published (11).
In gram-negative bacteria, S1 is composed of six oligonucleotide-oligosaccharide-binding (OB) domains (OB1–OB6) of approximately 70 amino acids each. The OB domain is a five-stranded, antiparallel β-barrel found in a number of proteins involved in RNA metabolism in both pro- and eukaryotes (12). On the ribosome, S1 is thought to facilitate translation initiation by binding to mRNAs and possibly unwinding their secondary structure (13). The first two OB domains of S1 are essential for binding to the ribosome (14,15), while the four C-terminal domains are thought to be involved in mRNA binding (16).
In the Qβ replicase complex, S1 is needed for the initiation of genomic RNA (+)-strand replication. In vitro, this function can be fulfilled by a fragment of S1 comprising OB1–3 (17). More recently, S1 was found to promote the release of the replication product and the template in their single-stranded forms. This is a thought to facilitate reinitiation and thereby allow for rapid, exponential replication (17) and the first two S1 domains, OB1–2, are sufficient to support the termination function in vitro (17). Within the genomic (+)-RNA, S1 recognizes and binds two internal sites termed the S- and M-site. In vivo, binding of S1 to the S-site causes repression of translation by hindering ribosome binding, while initiation of replication strongly depends on an interaction of S1 with the M-site (18).
The Qβ replicase core complex can form a dimer in solution, which might correspond to the dimer observed in the asymmetric unit of the corresponding crystal structure (7). Here, we show by mutagenesis that the basic residues of the β-subunit that mediate dimerization in the crystal structure via salt bridges are crucial for infectivity, while the corresponding acidic residues play a minor role. We determine the crystal structure of the Qβ replicase holoenzyme complex comprising the viral β-subunit, the first two OB domains of E. coli ribosomal protein S1 (OB1–2) as well as elongation factors EF-Tu and EF-Ts in a new crystal form. The structure reveals a novel dimeric complex (7,11) in which the basic β-subunit residues are located adjacent to the OB2 domain with the potential of participating in RNA binding. Using NMR, we show that OB2 has the ability of transiently interacting with replicative RNA and furthermore, replication assays indicate a possible collaboration between the basic patch of the β-subunit and OB2 during template recognition.
MATERIALS AND METHODS
Site-directed mutagenesis
Point-mutations were introduced into the infectious pQβ7 plasmid (19) and into the expression vector pBAD33-Ts-Tu-TEV-Qβ-3 (20) using the QuikChange Lightning Site-directed Mutagenesis Kit (Agilent Technologies). The presence of the mutations as well as the absence of any unintended mutations were confirmed by DNA sequencing.
Infectivity assay
The ability of Qβ phages, with mutations in the β-subunit, to sustain infection and phage production was analysed in vivo by measuring the number of plaque forming units (PFU).
Forty five microlitres of competent cells (≈108 cfu/ml) of the F− E. coli strain HB101 was transformed with 5 ng of wild-type or mutant pQβ7 plasmids and plated on LB agar supplemented with 0.1 mg/ml ampicillin. Phage titer was determined for serial dilutions of the supernatants of overnight cultures of the transformed cells. From each dilution of phage-containing supernatant, 10 μl was transferred to 100 μl of a suspension of mid-log phase E. coli K603 indicator cells and infection was allowed for 30 min at 37°C. The cell suspension was subsequently mixed with 4 ml molten top agar (10% NaCl, 5% yeast extract, 10% peptone, 0.5% agar, 5% glycerol, 20 μg/ml tetracycline) and plated onto LB tetracycline plates. These plates were incubated overnight at 37°C and on the following day, PFU/mL/OD600 was determined for each mutant and related to the PFU/mL/OD600 determined for the wild-type in the same assay.
Preparation of the Qβ replicase core complex
The Qβ replicase core complex was overproduced as a fusion protein in E. coli BL21(DE3) grown in auto induction medium (10% NaCl, 5% yeast extract, 10% peptone, 34 mg/l chloramphenicol, 25 mM Na2HPO4, 25 mM KH2PO4, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.5% glycerol, 0.05% D(+)-glucose and 0.2% L-arabinose) using the expression vector pBAD33Ts-Tu-TEV-β-3 (7), which allows arabinose-inducible expression. In pBAD33Ts-Tu-TEV-β-3, the genes encoding the subunits of the Qβ replicase core complex have been fused to enable the production of a soluble fusion protein consisting of the following elements in the mentioned order: EF-Ts, EF-Tu, a linker region containing a recognition sequence for the tobacco etch virus (TEV) protease, the β-subunit and finally a purification tag counting six histidines.
The cell pellet containing the overexpressed EF-Ts–EF-Tu–TEV–βS–6×His fusion protein was resuspended in lysis buffer (50 mM Tris pH 8.0, 500 mM NaCl, 5 mM MgCl2, 7 mM 2-mercaptoethanol, 1 mM PMSF) and disrupted by sonication. The cleared lysate was subjected to nickel-chelating affinity chromatography and the fusion protein eluted with a linear gradient of imidazole from 0–500 mM. Relevant fractions were pooled and digested with TEV protease to separate EF-Ts–EF-Tu from the β-subunit. Two volumes of HIC-buffer A (40% (NH4)2SO4, 50 mM Tris; pH 8, 5 mM MgCl2, 0.5 mM DTT, 1 mM EDTA) were added to the protein preparation, which was subsequently loaded on a 9 ml Source-15 ISO column (GE Healthcare Life Sciences). The protein was eluted with a linear gradient from 25–19% (w/v) of (NH4)2SO4. In the final purification step, the protein was separated into a monomeric and a dimeric form by size exclusion chromatography using a Superdex 200 GL 10/300 column (GE Healthcare Life Sciences) equilibrated with gel filtration buffer (20 mM Tris-HCl pH 8, 500 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 7 mM 2-mercaptoethanol).
Preparation of ribosomal protein S1 and its fragment OB1–2
The S1 variant, OB1–2, applied in this study covered amino acid residues 1–171 of E. coli S1 (residues 1–557).
For crystallization and functional assays, ribosomal protein S1 and OB1–2 were overproduced as previously described (17). For 15N-labelling of OB1–2 prior to NMR, 1 l auto-induction minimal medium (0.1 mg/ml ampicillin, 47 mM Na2HPO4, 22 mM KH2PO4, 8.5 mM NaCl, 0.1 mM CaCl2, 2 mM MgSO4, 18 μM FeCl2, 1 ml trace solution (30 mM EDTA, 1 mM ZnSO4, 10 mM MnCl2, 10 mM CaCl2, 1 mM CuSO4, pH 4), 1 g 15NH4Cl (Aldrich or Cambridge Isotope Laboratories), 0.5% glycerol, 0.05% D(+)-glucose, 0.2% α-lactose) was inoculated with 1.25% of an overnight culture and grown overnight.
S1 and OB1–2 were purified as previously described (17). After the first round of nickel-chelating affinity chromatography, relevant fractions were pooled and dialysed overnight at 4°C against a high-salt buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 5 mM 2-mercaptoethanol) or NMR buffer (50 mM phosphate pH 7.0, 200 mM NaCl) in the presence of TEV protease. A second round of nickel-chelating affinity chromatography removed any uncleaved protein as well as unspecifically bound impurities.
Static light scattering
Static light scattering (SLS) was used to determine the oligomeric state of Qβ replicase core complexes in elution fractions from gel filtration. The SLS was measured with a Zetasizer μV (Malvern Instruments Ltd.) connected in-line with the Superdex column on an ÄKTA purifier. Isolated monomeric and dimeric Qβ replicase core complexes were individually run on the gel filtration column and analysed by SLS upon elution to determine their molecular mass. A bovine serum albumine solution of a known concentration was used for calibration. The resulting data were analysed by OmniSEC software version 4.7 (Malvern Instruments Ltd).
Crystallization and structure determination
The complex between the Qβ replicase core complex, EF-Ts–EF-Tu–TEV–βS–6×His, and OB1–2, was obtained by combining the two protein preparations with OB1–2 in a 2-fold molar excess followed by incubation for 30 min on ice. Subsequently, the complex was purified by gel-filtration chromatography as described for the Qβ replicase core complex. Finally, the complex was concentrated to 15 mg/ml using spin filtration. Previously, only the dimeric form was found to crystallize (7) and therefore the dimeric fraction was also used for crystallization of the holoenzyme complex.
Reproducible crystals were obtained using vapour diffusion at 19°C of a mixture of equal volumes of protein complex and crystallization solution composed of 20% PEG 4000, 5% PEG 400, 0.4 M (NH4)2SO4, 0.1 M sodium acetate, pH 7 and 10 mM betaine hydrochloride. Optimization by micro-seeding gave rise to diamond-shaped crystals of approximately 100 μm, which were mounted directly from the drop and flash frozen in liquid nitrogen. Diffraction data were collected at the Swiss Light Source beamline PXIII at a wavelength of 1.00004 Å, and processed with the xia2 -3daii pipeline (21).
The structure was solved by molecular replacement (MR) in phenix.phaser (22) using the coordinates of the monomeric Qβ core complex (PDB ID 3MMP) as a search model. The crystal structure of OB1 fused through its C-terminus to ribosomal protein S2 via a TEV recognition site (23) and the NMR solution structure of OB2 (24) were generously provided prior to publication and used to facilitate chain tracing in Coot (25). Both OB1–2 didomains in the dimer were manually completed in Coot. The refinement was conducted with phenix.refine and non-crystallographic symmetry (NCS) restraints relating the two heterotetramers were used throughout the refinement (26) and the model was validated with the standard Ramachandran criteria of MolProbity (26). Figures were prepared using PyMol (www.pymol.org) and surface potentials were calculated using the APBS PyMol plug-in (27).
Preparation of RNA templates
RQ139 RNA, which is a 139 nt-long variant of RQ135 RNA, and Qβ (+)-RNA templates were prepared by in vitro run-off transcription with the T7 polymerase using the TranscriptAid™ High Yield Transcription Kit (Fermentas). A modified (17) pT7RQ135–1(–) plasmid (28) or plasmid pQβ7 (19), respectively, linearized at the 3′ SmaI site were used as templates for the transcription reactions.
RQ200 RNA, composed of complementary positive and negative strands, was prepared for NMR studies by using the extraordinary ability of the Qβ replicase to replicate this particular template, which was originally isolated as an exponentially amplifying contaminant in replication assays (Figures 1C, 8A and B). Around 20–30 ml reactions were carried out as follows: the replication reaction buffer (125 mM Tris-HCl, pH 8, 5 mM MgCl2, 1 U Ribolock RNase inhibitor (Thermo Scientific), 0.5 mM EDTA, 1 μg/ml Rifampicin) was pre-incubated with 1 nM RQ200, 220 nM Qβ replicase core complex and 220 nM ribosomal protein S1 for 5 min at 30°C. The replication reaction was initiated by the addition of an equimolar rNTP mixture to 0.6 mM each and incubated for 3 h at 30°C. The replication reaction was terminated by addition of NaOAc, pH 5.2 to a concentration of 0.3 M, followed by extraction with an equal volume of a 1:1 phenol (pH 4.7)/chloroform mixture. The aqueous phase was extracted twice with one volume of chloroform and the RNA was precipitated with ethanol. The air-dried RNA precipitate was dissolved in water, and further dialysed against water to remove any remaining salt and ethanol. Lastly, the RNA preparation was distributed in aliquots appropriate for NMR titration experiments and lyophilized.
Figure 1.
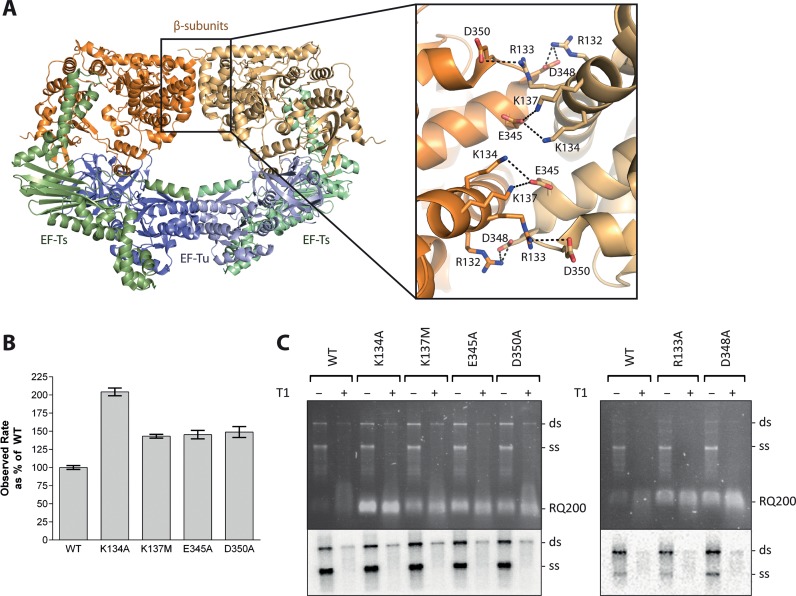
Mutational analysis of residues at the dimerization interface of the Qβ replicase core complex involved in salt-bridge formation. (A) Structure of the dimer of the Qβ replicase core complex consisting of the viral β-subunit (orange) and the host proteins EF-Tu (blue) and EF-Ts (green). The two halves of the dimer are shown in different shades of these colours. The dimerization interface between two β-subunits is highlighted in the inset figure illustrating residues involved in salt bridges. (B) Results of an in vitro replication assay using the RQ139 template. Product formation was followed measuring SYBR green II fluorescence, and the reaction rates were deduced from the exponential phases of the real-time reaction curves and related to the activity of the wild-type Qβ replicase complex (mean set to 100%). Error bars indicate standard deviations for n = 3. Controls included reaction mixtures without Qβ replicase or RNA and reaction mixtures without rNTP to allow quantification of background fluorescence, which was subtracted from the measurements on complete reactions. (C) Results from an in vitro replication assay using the Qβ (+)-RNA template. The reaction was performed in low salt and lasted for 10 min before quenching. Replication products were either digested with T1 ribonuclease prior to phenol/chloroform extraction (+) or not (−). The genomic replication products were separated by agarose gel electrophoresis. The gel was stained with ethidium bromide and photographed under UV light (top). [32P]-labeled product bands were visualized after exposure to a storage phosphor screen (bottom). The positions of single- (ss) and double-stranded (ds) genomic Qβ RNA, as well as a contaminating, replicative RNA (RQ200) are indicated. The lower part of the gel containing RQ200 RNA was removed before exposure to the storage phosphor screen to avoid interference with the signal from the labeled genomic RNA. The assay was repeated three times, and a representative result is shown.
Figure 8.
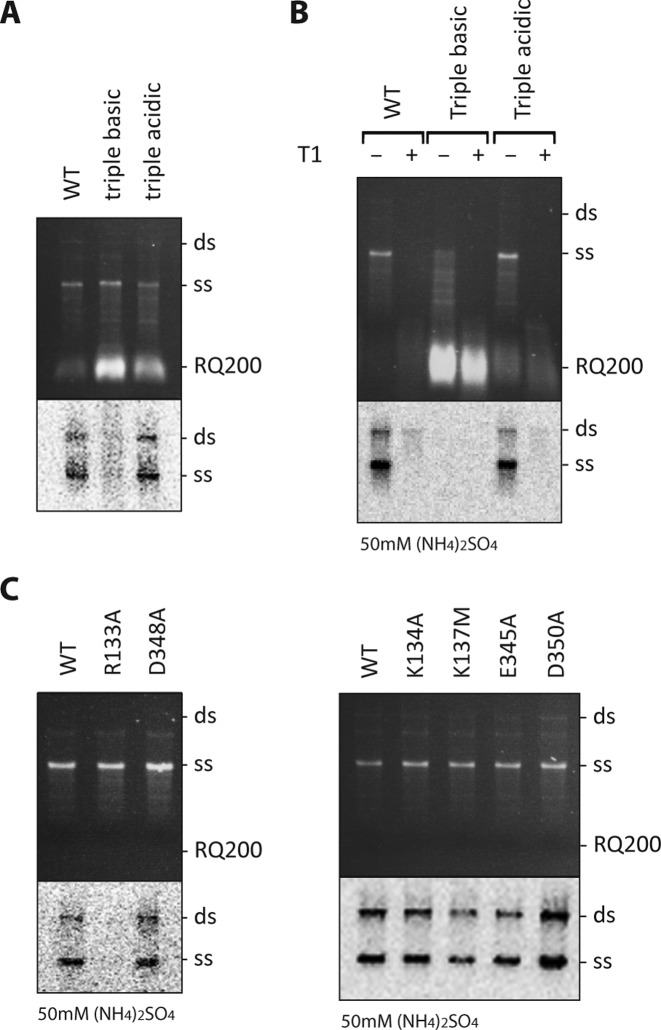
In vitro replication of the Qβ (+)-RNA genome by Qβ replicase complex triple-point mutants R133A/K134A/K137M and E345A/D348A/D350A located in the β-subunit. (A and B) The assays were carried out for 10 min in a buffer containing 0 or 50 mM (NH4)2SO4 as indicated. In B, replication products were either digested with T1 ribonuclease prior to phenol/chloroform extraction (+) or not (−). The mutants R133A/K134A/K137M and E345A/D348A/D350A are denoted "triple basic" and "triple acidic", respectively. (C) Contribution of individual amino acids forming basic and acidic patches on the β-subunit to the replication characteristics observed for the triple-point mutants at 50 mM (NH4)2SO4. The replication products were separated by agarose gel electrophoresis. The gels were stained with ethidium bromide and photographed under UV light (upper panels). [32P]-labelled product bands were visualized after drying and exposure to a storage phosphor screen (lower panels). The positions of single- (ss) and double-stranded (ds) genomic RNA, as well as a contaminating, replicative RNA (RQ200) are indicated. The lower part of the gels containing RQ200 RNA were removed before exposure to the storage phosphor screen to avoid interference with the signal from the labeled genomic RNA. A wild-type Qβ replicase core complex was prepared and tested along with each set of mutants. All assays were repeated at least three times, and representative gels are shown.
NMR analysis of RNA binding by OB1–2
RNA-binding studies were performed using 0.2 mM uniformly 15N-labelled samples of the OB1–2 fragment in which aliquots of the lyophilized RNA were dissolved. At each titration point, the sample was transferred from the NMR tube to a vial with the lyophilized RQ200 RNA and mixed before being returned to the same NMR tube. 1H-15N heteronuclear single-quantum correlation (HSQC) spectra were recorded for [RQ200] / [OB1–2] molar ratios of 0, 0.1, 0.25, 0.5 and 1 at 1H frequency of 700 MHz at 25°C, with a standard triple resonance probe with pulse field gradients. The recording parameters were kept identical throughout the titration experiment.
Replication assay employing SYBR green II RNA staining
The replication activities of Qβ replicase core complexes carrying mutations in the β-subunit were measured by monitoring the binding of SYBR green II to the product. Replication reactions, each with a total volume of 100 μl reactions consisting of a reaction base (125 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 1 U Ribolock RNase inhibitor, 0.5 mM EDTA, 1 μg/ml Rifampicin, 1xSYBR Green II (Invitrogen), 660 nM protein S1) were prepared and for each reaction, the Qβ replicase core complex and the RNA template were added to final concentrations of 220 nM and 1 nM, respectively. Individual reaction mixtures performed in triplicate were then transferred into 96-well Microtest™ flat bottom polystyrene plates (Becton Dickinson) and incubated at 30°C for 10 min. The reactions were synchronized by cooling the plate to 5°C on a metal block prior to the addition of rNTP to a final concentration of 400 μM. Then, the plate was transferred back to 30°C and SYBR Green II fluorescence was measured every 33 s on a BMG PolarStar Plate Reader with excitation filter 485 nm and emission filter 535 nm.
Replication assay employing radiolabelling of the product
Replication reactions were carried out in 10 μl reaction buffer (100 mM Tris-HCl, рН 7.6, 10 mM MgCl2, 1 mM EDTA, 0.1 v/v% Triton X-100) at 30°C in the presence of 1 mM of each rNTP, 1 MBq/ml [α-32P] UTP, 0.01 μM Qβ (+)-RNA, 0.01 μM Hfq and 0.3 μM ribosomal protein S1. Where indicated, 50 mM (NH4)2SO4 was included in the reaction. After 1 min incubation at 30°C, 0.1 μM wild-type or mutant Qβ replicase core complex was added and replication continued for 10 min. Where indicated, single-stranded RNA product was digested by addition of 100 U ribonuclease T1 (Thermo Scientific) for 15 s at 30°C. Reactions were stopped by addition of 4 μl 60 mM EDTA on ice and the RNA was isolated by phenol/chloroform extraction (1:1, pH 4.5). The RNA was recovered by centrifugation and analysed by electrophoresis using a 1% agarose gel stained with ethidium bromide. Next, the gel was dried onto a Biodyne B transfer membrane (Pall Life Sciences) and visualized by autoradiography on a Personal Molecular Imager™ (Bio-Rad). Escherichia coli Hfq was overproduced using the intein system (Impact-CN™, New England Biolabs) as described by Link et al. (29).
RESULTS
Basic residues at the Qβ replicase core complex dimerization interface are critical for infection
In the crystal structure of the Qβ replicase core complex (7), the adjecent β-subunits interact via a symmetric network of salt bridges (Figure 1A). We disrupted the interface by neutralizing negative or positive charges to understand the importance of dimerization for virus replication. The mutations E345A, D348A, D350A, R132M/R133A (double mutant), K134A and K137M were introduced into the gene encoding the β-subunit in the infectious plasmid, pQβ7, which contains the sequence of the complete Qβ genome (19). Transformation of E. coli with this plasmid results in an artificial infection that allows the efficiency of infection of the mutant Qβ variants to be determined by counting the number of plaque forming units. Using this approach, we find that the effect of a mutation appears to depend on the charge of the mutated residue: removal of basic residues completely abolished infection, while neutralization of acidic residues reduced infectivity to 1–20% of the wild-type phage (Table 1).
Table 1. Effect of β-subunit mutations on infectivity. The table shows relative infectivity efficiencies in percentage of wild type (100%), which on average gave rise to 5.3 × 108 PFU/ml/OD600. All assays were performed in duplicate at least three times. Average values with standard deviations are provided.
| β-subunit species | Infectivity (% of wild type) |
|---|---|
| Wild type | 100 ± 20 |
| R132M/R133A | 0a |
| K134A | 0a |
| K137M | 0a |
| E345A | 1.05 ± 1.2 |
| D348A | 17.01 ± 14 |
| D350A | 3.97 ± 2.1 |
aNo plaques were observed even for the undiluted sample, i.e. the mutant was not infectious.
The mutations studied in the infectivity assay were introduced into the pBAD33Ts-Tu-TEV-β-3 expression vector to assess whether the defects observed during infection were a result of impaired RNA-replication ability. Purified mutant Qβ replicase core complexes were assessed for their ability to amplify in vitro the short replicative RNA template, RQ139 (17) by measuring the fluorescence increases caused by RNA-binding of SYBR green II (Figure 1B). Surprisingly, the mutant Qβ replicase complexes were 50–100% more active than the wild-type protein indicating that a reduction in the catalytic replication potential of the mutants could not explain the observed infectivity defects.
The replication activities of the mutants were also investigated using the native Qβ (+)-RNA template, while following product formation via [32P]-UTP incorporation (Figure 1C). None of the mutants displayed any major replication defects using the genomic (+)-RNA template. However, we observed a strong competition by a contaminating replicative RNA, referred to as RQ200, in particular for the mutant K134A. This is a well-known phenomenon for replication reactions carried out in low salt (30).
Exponential replication of the Qβ genome during infection is based on the release of product and template in their single-stranded form. This process is supported by ribosomal protein S1 (17), and a defect preventing strand separation could potentially lead to a decreased infectivity. Thus, one half of each replication reaction was treated with single-strand specific ribonuclease T1 prior to phenol/chloroform extraction to reveal the true amount of double-stranded product, which can otherwise form post termination via annealing of single-stranded template and product. None of the mutants displayed any abnormal release of template and product (Figure 1C).
Thus, the inhibition of infectivity caused by mutating basic residues of the β subunit (Table 1) could neither be attributed to defects in the catalytic capacity (Figure 1B) nor to problems with product release (Figure 1C).
Mutations at the dimer interface do not disrupt dimer formation between Qβ replicase core complexes in vitro
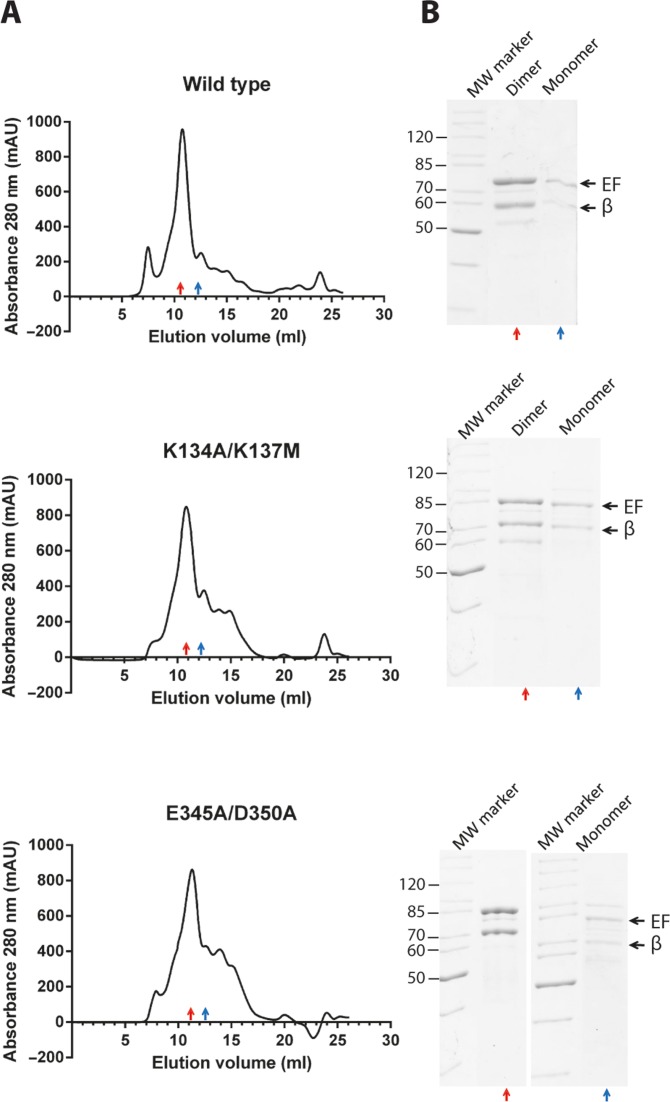
Next, we used size exclusion chromatography to assess whether dimer formation in vitro was affected by mutations introduced at the dimer interface of the Qβ replicase core complex. On the column used, the Qβ replicase core complex monomer elutes at 12.94 ml and the dimer elutes at 10.96 ml (Supplementary Figure S1). The molecular masses of the protein molecules in these fractions were measured by static light scattering (SLS) to 123 kDa and 248 kDa, respectively, in good agreement with the theoretical mass of the recombinant Qβ replicase core complex (140 kDa and 280 kDa, respectively).
Next, the ratios between the dimeric and monomeric forms of both recombinant wild-type and mutant Qβ replicase core complexes were examined by gel filtration. The double-mutant Qβ replicase core complexes, K134A/K137M and E345A/D350A, were used for these experiments, since they were expected to generate a weaker dimer interface than the corresponding single-point mutants that we previously analysed for impact on infectivity (Table 1). However, no significant variation in the elution profile between wild-type and the two double-mutant Qβ replicase core complexes could be detected (Figure 2) indicating that the interface mutations introduced did not cause dissociation of the dimer. Thus, the infectivity loss of the Qβ mutants (Table 1) is unlikely to be from disruption of the β:β interface and dissociation of Qβ replicase core complex dimer in agreement with the differential effects of mutating basic (complete loss of infectivity) and acidic residues (partial loss of infectivity).
Figure 2.
Gel filtration analysis of monomeric and dimeric forms of Qβ replicase core complexes. (A) TEV-digested Qβ replicase core complexes were subject to analytical gel filtration on a Superdex 200 10/30 GL column. The following Qβ replicase core complex variants were analysed: wild-type (upper panels), double-mutant K135A/K137M (middel panels) and double-mutant E345A/D350A (lower panels). Red and blue arrows indicate the dimer and the monomer peaks, respectively, as verified by static light scattering (see Supplementary Figure S1). (B) A 12% SDS-PAGE analysis of the indicated dimer and monomer peak fractions from the gel filtration confirms the presence of the Qβ replicase core complex is these fractions. The upper and lower black arrows indicate the positions of the EF-Ts–EF-Tu fusion protein and the β-subunit, respectively.
Crystal structure of the Qβ replicase holoenzyme complex bound to OB1–2
To gain further insight into the conformational states of the Qβ RdRp complex we determined a new structure of the dimeric form of the Qβ replicase holoenzyme complex bound to the first two OB domains of ribosomal protein S1, OB1–2. The structure is in space group P1211 and was determined by molecular replacement using the core complex structure as search model (PDB ID 3MMP) and subsequently refined to R/Rfree of 21.4/26.8% (Table 2). A representative example of the electron density map is provided in Supplementary Figure S2.
Table 2. Data processing and refinement statistics for the Qβ replicase core complex bound to OB1–2.
| Native | |
|---|---|
| Data collection | |
| Space group | P1211 |
| Cell dimensions | |
| a, b, c (Å) | 99.73, 115.45, 178.52 |
| α, β, γ (°) | 90, 96.14, 90 |
| Resolution (Å) | 67.3–3.2 (3.3–3.2) |
| Rmerge (%) | 9.6 (59.4) |
| Rmeas (%) | 12.8 (79.7) |
| Mean I / σI | 12.6 (2.2) |
| Completeness (%) | 98.7 (99.5) |
| Redundancy | 3.9 (3.8) |
| Unique reflections | 65 128 |
| CC1/2 | 99.6 (66.6) |
| Refinement | |
| Resolution (Å) | 59.8–3.2 Å |
| No. reflections | 65 128 |
| Rwork / Rfree (%) | 21.4/26.8 |
| No. of atoms | |
| Protein | 21 130 |
| Water | 0 |
| R.m.s. deviations from idealist | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.546 |
| Ramachandran statistics (%) | |
| Favoured | 93.9 |
| Allowed | 5.3 |
| Outliers | 0.8 |
Values for the highest resolution shell are given in brackets.
The asymmetric unit of the new structure contains eight polypeptide chains that can be described as a homo-dimer of Qβ holoenzyme heterotetramers. An overview of a single Qβ replicase holoenzyme complex is presented in Figure 3A, while folds of the two OB domains are shown in Figure 3B and C. The structure is complete except for residues constituting the linker 76–80 and loop 139–145 of S1, the switch I region 42–63 of EF-Tu and the disordered loop 520–532 of the β-subunit. Finally, a few residues are missing from the termini of the Qβ replicase core complex.
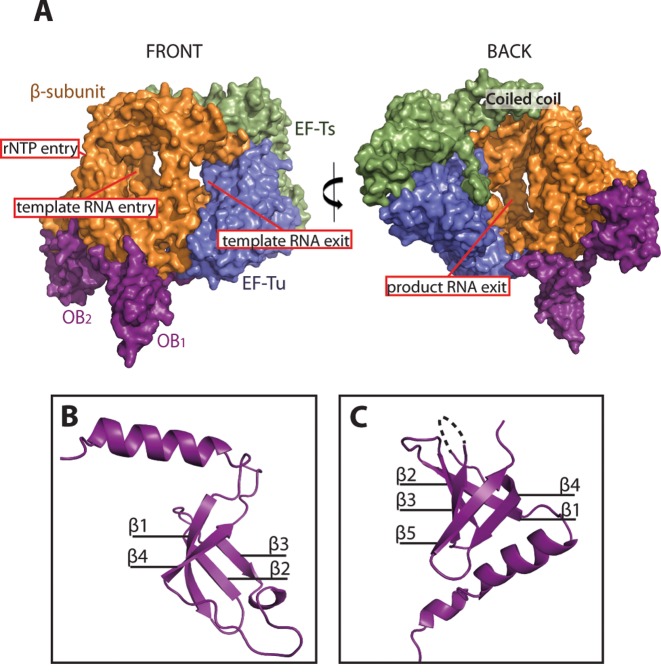
Figure 3.
Overall structure of the monomeric form of the Qβ replicase core complex bound to the first two domains, OB1–2, of ribosomal protein S1. The OB1–2 domains bind at distant sites on the β-subunit and are connected by an extended 15 amino-acid linker region, which is only partly visible in the electron density. (A) Surface representation of the monomeric Qβ replicase complex. The following colour code is applied: β-subunit (orange), EF-Tu (blue), EF-Ts (green) and OB1–2 (magenta). The front and back of the complex are related by an approximately 180° vertical rotation. (B) Structure of the OB1 domain illustrating the presence of a flexible N-terminal helix packing against the β-subunit. The orientation of the domain matches closely the back view of the complex. The four β-strands are denoted β1–4. (C) Structure of the OB2 domain having the classical OB fold and a helix, which is part of the connecting linker between OB1 and OB2. The five β-strands are denoted β1–5. The missing density for loop L34 between β3 and β4 is indicated by a dotted line.
The overall conformation of the Qβ replicase core complex (Figure 3A) is similar to the previously determined structures (7–10). The first two OB domains of S1 (Figure 3B and C) interact extensively with the fingers domain of the β-subunit (Supplementary Figure S3A). Details of this interaction are presented in Supplementary Figure S3B.
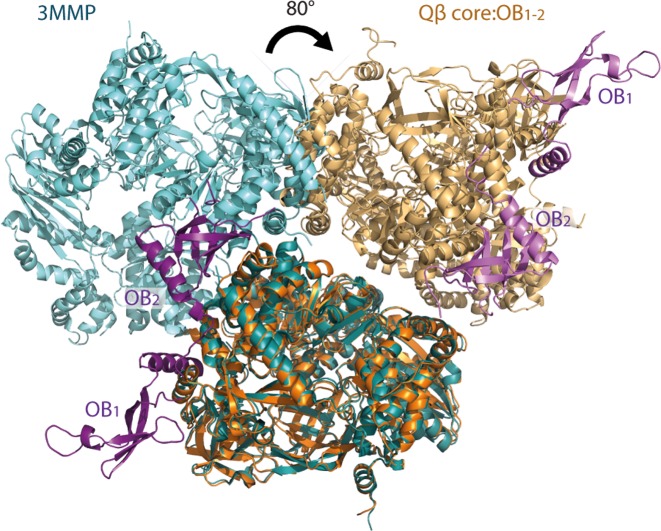
Interestingly, the dimer interface in the asymmetric unit of the intact holoenzyme is different from that observed for the core complex alone (7) as well as the dimeric form of the holoenzyme complex published earlier (11) (Supplementary Figure S4). The interaction between the β-subunits observed in the isolated Qβ replicase core complex is impossible in the OB1–2-bound structure, since the OB2 domain binds very close to the patch of the β-subunit that mediates dimerisation of the Qβ replicase core complex (Figure 4).
Figure 4.
Binding of OB1–2 to the Qβ replicase core complex dimer alters the dimerization interface. Cartoon representation of the dimeric Qβ replicase core complex (PDB ID 3MMP; dark and light turquoise for each monomer, respectively) superimposed on the dimeric OB1–2-bound Qβ replicase complex (this study; purple: OB1–2; orange: Qβ replicase core complex). Binding of OB1–2 causes a tilt of approximately 80° of one half of the Qβ replicase core complex dimer as measured from the centre of each Qβ replicase core complex. Thereby, the interface between the Qβ replicase core complexes in the dimer is dramatically altered.
We have noticed that the binding of the OB1–2 domain to the β-subunit leads to a displacement of the EF-Tu–EF-Ts subcomplex towards the β-subunit, whereby the cavity forming the RNA exit channel becomes narrower (Supplementary Figure S5). The largest shift in the structure relative to the complex without OB1–2 is found in α-helices 1, 2 and 3 of EF-Ts, which are displaced by up to 6.5 Å in our structure.
The basic residues of the β-subunit interface and OB2 form a coherent putative RNA-binding surface
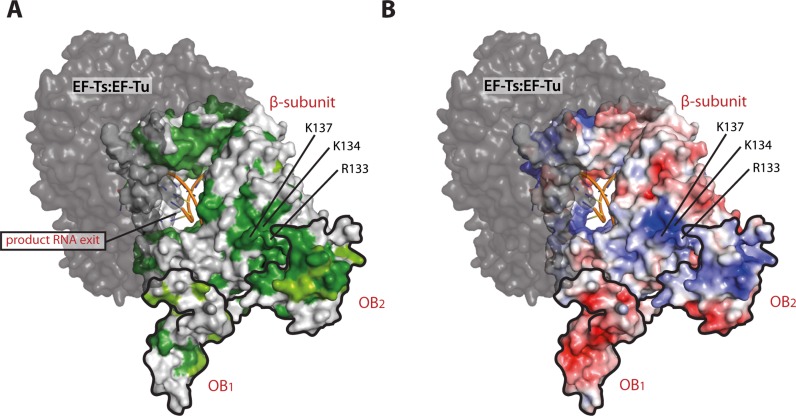
We used ConSurf (34) to display conservation of the β-subunit among Alloleviviruses on the surface of the structure (Figure 5A). Interestingly, a small but strictly conserved patch was identified adjacent to the OB2 binding site and overlapping with the dimer interface observed in the Qβ replicase core complex structure.
Figure 5.
Conservation analysis and assessment of electrostatic surface potential of the β-subunit and OB1–2. (A) Surface representation of the β-subunit and OB1–2 coloured according to conservation among Allolevivirus RNA-dependent RNA polymerases and bacterial ribosomal S1 proteins. Dark green: strictly conserved, lighter green: highly conserved and grey: not conserved. RNA product and template strands (from PDB ID 3AVY) are illustrated for orientation and OB1 and OB2 are contoured in black. (B) The electrostatic surface potentials are calculated using the APBS PyMol plugin and mapped onto the separate OB domains and the β-subunit. These are shown from negative kbT/ec = –5 (red) to positive kbT/ec = 5 (blue), where kb, T and ec are the Boltzmann's constant, absolute temperature and the charge of an electron, respectively. The view is identical to the view presented in A. In both panels, the EF-Ts–EF-Tu entity is shown in dark grey.
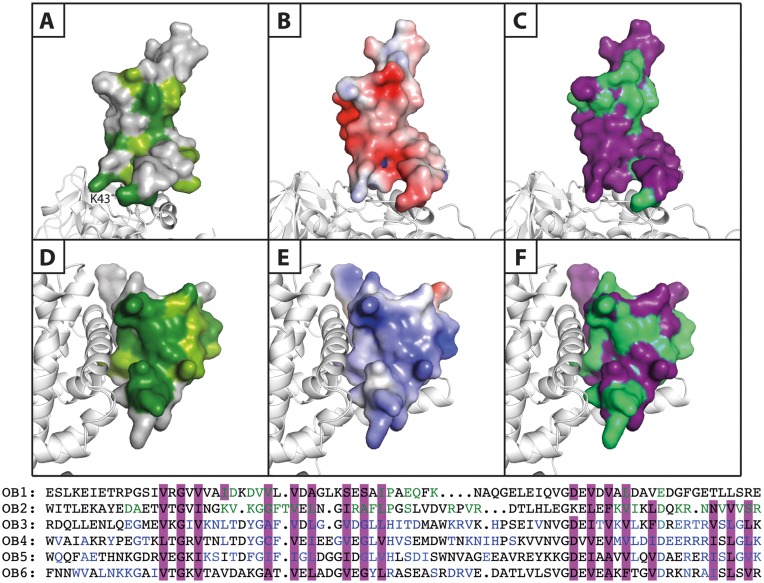
Analysis of the conservation of OB1 (Figures 5A and 6A) and OB2 (Figures 5A and 6D) furthermore reveals remarkable differences between the β-barrels of the two domains. OB1 presents only strictly conserved residues at the β-subunit interface, in the interior of the β-barrel as well as in the sharp turns of the β-strands, where conserved glycine residues maintain the tertiary structure. On the contrary, OB2 possesses a rather well-defined, highly conserved surface flanking the β-subunit interface, which was mutated in this study. This surface is neither involved in binding the Qβ replicase core complex, nor required for preserving the β-barrel fold. Furthermore, mapping of the electrostatic potential on the surface of OB2 reveals a significant build-up of positive charge forming a clearly defined basic area on the β-barrel (Figures 5B and 6E), while the OB1 domain is predominantly acidic (Figure 6B).
Figure 6.
Comparison of the properties of the OB1 and OB2 β-barrels. Surface representation of the two N-terminal OB domains of S1 bound to the β-subunit presented in white cartoon. Panels A–C illustrate OB1, while panels D–F show OB2. (A and D) Conservation scores of OB1 and OB2 generated using the ConSurf server. Dark green: strictly conserved, lighter green: highly conserved and grey: not conserved. (B and E) Electrostatic surface potentials calculated using the APBS Pymol plugin and mapped onto the separate OB domains. These are shown from negative kbT/ec = –4 (red) to positive kbT/ec = 4 (blue), where kb, T and ec are the Boltzmann's constant, absolute temperature and the charge of an electron, respectively. (C and F) A sequence alignment of the six OB domains of ribosomal protein S1 (OB1–6) was prepared (lower part of the figure) and the residues involved in formation of the β-barrel are highlighted in purple, while the residues with an involvement in general RNA-binding in OB3–6 are coloured blue. Based on this information, the putative RNA-binding residues in OB1 and OB2 are coloured green and mapped onto the OB domains (purple).
These two basic areas on OB1–2 and the β-subunit form a continuous positively charged surface that may likely constitute an RNA-binding site (Figure 5B). This idea is supported by sequence alignment of the six OB domains, OB1–6, of E. coli S1 (Figure 6, bottom). Here, putative RNA-binding residues in OB2 (Figure 6F) were predicted based on their alignment with residues in OB3–6 shown previously to be involved in RNA binding (35–37). On the contrary, OB1 did not display similar features and therefore is unlikely to possess a conserved RNA-binding surface (Figure 6C).
Domain OB2 is capable of interacting with replicative RNA
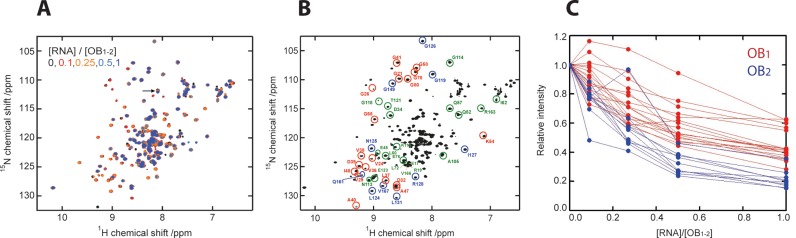
NMR titration experiments were conducted with a replicative RNA of approximately 200 nucleotides to experimentally access the predicted RNA-binding properties of OB1–2. This RNA, termed RQ200, is a legitimate template for the Qβ replicase as observed in replication assays, where it can outcompete genome replication (Figures 1C, 8A and B).
1H-15N HSQC spectra were recorded from a sample containing uniformly 15N labelled OB1–2 (19 kDa) and unlabelled RQ200 RNA (64 kDa) at various concentrations (Figure 7A). Interestingly, addition of RNA decreased the signal intensity of many protein resonances without changing their chemical shifts. The disappearance of OB1–2 resonances is rationalized by the relatively large size of the RNA ligand, which adds significantly to the mass of OB1–2 upon binding and leads to a slow tumbling and faster relaxation of transverse magnetization (38). Thus, the RNA-bound state ultimately becomes NMR-invisible. Figure 7B presents the resonance assignments of domain OB1–2 obtained by superimposing our spectrum on the OB1–2 1H-15N HSQC spectrum from Giraud et al. (24). A comparison of these with the titration spectra and plotting of the signal intensity at each step of the titration revealed a clear pattern: the signal intensities arising from residues of the OB2 domain decrease fast to near zero, while the signal intensities belonging to residues of the OB1 domain only decrease slowly (Figure 7C). The latter is unlikely to be a result of specific OB1 RNA-binding, but is most likely caused by the slow tumbling of the entire OB1–2 molecule upon interaction with RQ200. Thus, the signals cluster in two groups corresponding to each OB domain suggesting that only OB2 binds RNA.
Figure 7.
OB2 binds a legitimate RNA template. The capacities of OB1 and OB2 for RNA binding were studied by NMR in a titration experiment. (A) Superposition of the OB1–21H-15N HSQC spectra at each step of the RNA titration steps. The signals are coloured according to [RNA] / [OB1–2] ratio as indicated. Addition of RQ200 RNA decreased the signal intensity of many OB1–2 resonances without changing their chemical shifts. Only the resonance marked by an arrow displayed a 15N chemical shift. (B) Partial resonance assignments based on Giraud et al. (24). The resonances marked in red (belonging to OB1) and the resonances marked in blue (belonging to OB2) are included in the analysis presented in C. (C) Representation of the signal intensities at the individual titration steps relative to the initial signal intensity at 0:1 [RNA] / [OB1–2].
In summary, the OB2 domain appears to be capable of binding the RNA template, whereas the OB1 domain is not, in agreement with the surface and conservation analyses (Figures 5 and 6). The specific residues involved in the interaction cannot be determined from these experiments, as the high molecular mass of the protein:RNA complex also causes line broadening of signals from residues that are not directly at the binding interface.
Basic residues of the β-subunit dictate the specificity for genomic Qβ RNA
β-subunit residues at the patches of positive and negative charge located next to the putative RNA-binding site of OB2 (Figure 5B) were finally neutralized as separate entities by site-directed mutagenesis, and the replication activities of the resulting triple-point mutants, R133A/K134A/K137M and E345A/D348A/D350A, were examined (Figure 8A and B). Buffers with both low and high concentrations of salt (0 and 50 mM (NH4)2SO4, respectively) were used since high salt is known to favour the replication of genomic (+)-RNA in a strictly S1-dependent manner, while at low salt concentrations, contaminating RNAs can compete with the genomic RNA for binding to the Qβ replicase complex in a reaction, which is less dependent on S1 (17). In both low (Figure 8A) and high salt (Figure 8B), genomic RNA replication by the R133A/K134A/K137M (triple basic) mutant was abolished, while the E345A/D348A/D350A (triple acidic) mutant showed no defects. Instead, the R133A/K134A/K137M mutant amplified the contaminating RQ200 RNA template with very high efficiency showing that the catalytic capacity of the enzyme is maintained. Examination of the replication activities of the corresponding single-point mutants in a high-salt buffer (Figure 8C) revealed that the shortcomings of the R133A/K134A/K137M mutant in genomic (+)-RNA replication may primarily be caused by the R133A mutation. This was not evident during replication in low salt (Figure 1C), where all single-point mutants demonstrated replication capacities similar to the wild type. Expectedly, replication of the small contaminating RQ200 was strongly decreased in high salt. However, the R133A/K134A/K137M mutant was found to support surprisingly efficient replication of RQ200 in 50 mM (NH4)2SO4 (Figure 8B).
In brief, the mutation of acidic residues on the β-subunit does not affect RNA replication of the genomic RNA, while the mutation of basic residues has a profound effect on this activity, especially at high salt concentrations, where the dependency on S1 is strong. Specifically, mutation of basic residues causes a strong preference for replicating smaller RNAs in 50 mM (NH4)2SO4.
DISCUSSION
Infectivity loss is not caused by dimer disruption
The Qβ replicase core complex separates into monomeric and dimeric fractions during purification (7). In this study, the identities of these fractions were confirmed by SLS (Supplementary Figure S1). The dimeric Qβ replicase core complex crystallizes into a dimer, which is primarily supported by symmetric salt bridges between two β-subunits giving rise to a buried surface area of 1770 Å2 (7). Biologically significant oligomerization of RdRps has been observed in other positive-stranded RNA viruses, where it leads to cooperative RNA binding and polymerization (39–41). Thus, we decided to investigate the functional relevance of the Qβ replicase core complex dimer by neutralizing acidic and basic residues involved in salt bridge formation. These studies indicated that disruption of the dimer could not explain the infectivity loss (Table 1), since the effects of removing negative and positive charges were distinctly different. This was substantiated by gel filtration analysis of two double-mutants (Figure 2), which showed no appreciable change in the ratios between the monomeric and dimeric forms. Curiously, all single-point mutants had increased replication activities as compared to the wild type when using the short RQ139 RNA template (Figure 1B), while no or only minor defects were observed during replication of the genomic Qβ (+)-RNA (Figure 1C). None of these effects on RNA replication activity were of a magnitude required to explain the infectivity loss.
Structural studies of a dimeric Qβ replicase holoenzyme complex was undertaken to shed new light on the role of replicase complex dimerization and the interaction with S1. The resulting structure highlighted other interesting features as well of which global conformational changes occurring upon binding of OB1–2 will be described in the next section, while the subsequent section presents a comparison between S1 binding to the ribosome versus the β-subunit. In the remaining part of the discussion, we will link the structure to the central theme of the study as it leads us to comprehend how the crucial basic residues of the β-subunit play a role during template recognition, which may be unrelated to the dimerization observed in the Qβ replicase core complex.
Template channel properties may be adjusted upon binding of OB1–2
Structures of a number of RNA-bound Qβ replicase core complexes have been determined, representing an initiation complex and various stages during elongation (9). No major structural rearrangements were identified in comparison to the unbound core complex, although the Qβ replicase complex is expected to undergo a structural change during initiation as a means of securing processivity (42). Likewise, structures of the holoenzyme complex as presented here and in (11) revealed no large conformational changes upon binding of the OB1–2 domain to the Qβ replicase core complex. However, we have noticed a displacement of the EF-Tu:EF-Ts subunits towards the interior of the RNA exit channel, which suggests a role for flexibility of this part of the complex (Supplementary Figure S5).
The observed plasticity may be related to the expected transition during initiation, which is a specific characteristic of legitimate templates and is supposed to ensure their exponential amplification (42). Flexibility may serve to adjust the tunnel diameter and/or help the template RNA to exit, which is in line with the general observation that viral RdRps appear to have an inherently flexible template channel, which allows for conformational adjustments during template binding and translocation (43).
S1 interacts with the β-subunit and the ribosome in similar ways
The S1 fragment, OB1–2, was found to bind primarily to the β-subunit of the Qβ replicase core complex, which complements early observations that the holoenzyme complex can dissociate into two entities: the EF-Tu–EF-Ts complex and a β-subunit·S1 complex (44,45). It is well established that the OB1–2 domains of S1 are required for binding of the protein to both the ribosome and the β-subunit (14,23,31–33). Recent studies have narrowed this further down and identified the N-terminal 18 residues of OB1 as essential for the interaction with the ribosome (23). This explains why our crystallization trials of the Qβ holoenzyme complex were unsuccessful when employing a truncated form of OB1–2 lacking the first 20 amino acids.
The N-terminal helix of S1 makes a substantial contribution towards the binding to the core complex with a total buried surface area of 718 Å2. A prominent contribution of the N-terminal α-helix of OB1 is also observed during binding of S1 to the small ribosomal subunit (23). Pentaerythritol propoxylate (PEP), which was used as a precipitant during crystallization of the Qβ replicase core complex (7), binds to the same hydrophobic groove as the N-terminal α-helix of S1 (Supplementary Figure S6A).
Similar mechanisms of interactions are employed upon binding of S1 to the β-subunit and the ribosome (compare Supplementary Figure S6B and SC). The strictly conserved Phe5 of S1 mediates binding via a network of T-shaped and parallel π-stacking interactions. The finding that S1 binds to the ribosome and the Qβ replicase core complex with similar affinities (46) also reflects similarities in the binding mechanism. Interestingly, residues 16–19 could not be modelled in the S2-bound form of S1 and the N-terminal α-helix only extends to residue 14, while the N-terminal helix covers residues 5–18 in the replicase-bound form leading to an enlargement of the interaction surface (Supplementary Figure S6B and SC). Furthermore, OB2 is also involved in the binding of S1 to the β-subunit, while only OB1 is needed for the interaction with ribosomal protein S2 (23). Our results imply that a direct competition between the S2 protein and the Qβ replicase core complex for S1 binding may contribute to the tight regulation of the conflicting processes of translation and replication in the Qβ phage infected cell.
The β-subunit and S1 collaborate during RNA recognition
The functions of S1 during genome replication by the Qβ RdRp complex implicate the requirement for RNA binding. The roles of S1 during initiation and termination can be fulfilled by OB1–3 and OB1–2 in a low salt buffer, respectively, which contradicts the traditional view that only domains OB3–6 are relevant for RNA binding (16). More recent data, however, indicate that the RNA-binding properties of the individual domains of S1 may be diverse and optimized for the translation of mRNAs with different structural characteristics. In particular, domains OB1–3 were found to be essential for the recognition of a pseudoknot structure located in the ribosomal binding site of the E. coli rpsO mRNA (15). Additionally, an OB1–2-less S1 fragment was incapable of binding of a structured RNA and lacked the unwinding activity (33). Furthermore, a contribution of OB1–2 towards the cooperative binding of mRNA and tmRNA by OB3 has been observed (47).
Collectively, these findings by others together with our new structure and mutagenesis results, prompted us to revisit the RNA-binding potential of OB1–2. A surface analysis of conservation and charge identified a patch of particular interest at the boundary between the β-subunit and OB2, which corresponded to the basic residues that were found to be crucial for infection (Figure 5). A structure-based alignment considering general RNA-binding residues of OB3–6 supported a possible role of OB2 in RNA binding (Figure 6). NMR studies confirmed that OB2 does indeed interact with a replicative RNA template (Figure 7).
S1 ensures the release of a single-stranded product during termination to allow exponential amplification (17). The two N-terminal OB domains were found to be sufficient to support this function of S1. It has been difficult to reconcile how the product strand release mediated by S1 can occur without—at least transient—RNA interaction. Here, we provide the first piece of evidence that OB2 is in fact able to interact with RNA. Yet, the more specific details of how OB2 contributes to the product strand release during termination are not revealed by our or previous structures. Our mutagenesis studies indicate that the basic patch on the β-subunit is unlikely to have any influence on this step, since no effect on the ratio between released single- and double-stranded RNA could be observed when assaying single-point mutants (Figure 1C).
Replication of the genomic Qβ (+)-RNA template was completely abolished for the R133A/K134A/K137M replicase complex mutant, but the contaminating RQ200 RNA template was amplified with very high efficiency implying preservation of the catalytic properties of this mutant (Figure 8). A similar tendency was seen for the K134A mutant (Figure 1C) explaining the increased replication rate observed when amplifying the short RQ139 template (Figure 1B). The preference for smaller, contaminating templates displayed by the R133A/K134A/K137M mutant is rather unusual at 50 mM (NH4)2SO4 (30) and indicates a defect in S1-dependent template recognition. None of the individual mutations had a similar effect on the efficiency of subgenomic replication (Figure 8C). This supports the predicted collaboration between the residues of the basic patch and ribosomal protein S1 during RNA binding. Salt inhibits the initiation of replication (17) by suppressing binding of the replicase at an internal site of the Qβ (+)-RNA known as the ‘M site’ (49) via electrostatic interactions (50). Protein S1 stabilizes the replicase binding to the M site (49,51) thereby making the initiation step fairly insensitive to salt. Our mutagenesis studies revealed that the basic patch created by Arg133, Lys134 and Lys137 in the β-subunit is important for mediating Qβ (+)-RNA recognition and replication in 50 mM (NH4)2SO4. Accordingly, involvement of this area in the interaction with the Qβ (+)-RNA may confer the salt-sensitivity to the Qβ replicase core complex, as well as the decreased replication of competing RNA species and increased preference for genomic RNA by the wild-type replicase holoenzyme.
Previously, gel retardation assays have failed to detect binding of OB1–2 to M-site RNA (11) or RQ135 (17). This kind of assay is inherently dissociative and therefore only suitable for complexes with Kd values below the μM range (48). Thus, this method may be unsuitable for detection of the OB2-RNA interaction, which seems to be transient. It is plausible that the contiguous basic surfaces of the β-subunit (harbouring Arg133, Lys134 and Lys137) and OB2 mediate RNA binding, in cooperation with the OB3 domain. In a recent study, the OB3 domain of S1 was shown to be the primary binder of the M-site of the (+)-RNA genome during initiation of replication by the Qβ replicase complex. Importantly, very little binding was observed with OB3–6 alone and domains OB1–2 were found to be required for a strong interaction between S1 and the M-site (11).
The inability of the R133A/K134A/K137M mutant to recognise Qβ RNA may well explain the infectivity loss demonstrated for the corresponding single-point mutants (Table 1). Similarly, C-terminal deletion mutants of the β-subunit were previously found to abolish infection due to their inability to distinguish Qβ RNA from cellular RNA (52).
CONCLUSION
Contrary to our working hypothesis stating a specific, functional role for dimerization of the Qβ replicase core complex, we found that the key importance of the basic residues Arg133, Lys134 and Lys137 during viral infection arose from their involvement in the recognition of the Qβ (+)-RNA during replication initiation. We found that this basic β-subunit patch is positioned next to a potential RNA-binding site on OB2, and our NMR studies support a possible role for OB2 in RNA binding. Thus, we hypothesize that OB2 and the basic β-subunit patch form a cavity involved in template recognition, most likely in collaboration with OB3. The S1 protein was previously believed to be the main element responsible for recognition of the genomic RNA by the Qβ replicase holoenzyme complex via binding to the M-site (18,49). We show that the β-subunit participates in the Qβ (+)-RNA recognition as well, probably also via M-site binding.
ACCESSION NUMBER
Coordinates and structure factors have been deposited in the Protein Data Bank, under the accession code 4R71.
Supplementary Material
Acknowledgments
Karen Margrethe Nielsen is acknowledged for technical assistance. The scientific staff at the synchrotron sites where crystals were tested (Diamond, ESRF) and the data were collected (Swiss Light Source) are acknowledged for their support. We are grateful to Dr Isabella Moll, Dr Max F. Perutz Laboratories, University of Vienna, Austria and Christina Sizun, CNRS, Gif-sur-Yvette, France for sharing the structures of OB1 fused to ribosomal protein S2 (23) and the NMR solution structure of OB1–2 (24), respectively, prior to publication. The plasmid, pTYB11-Hfq, for overproduction of E. coli Hfq was kindly donated by Dr Poul Valentin-Hansen, University of Southern Denmark, Odense, Denmark.
Footnotes
Present addresses:
Heidi Gytz, Department of Biochemistry, McGill University, Montreal, Quebec H3G 1Y6, Canada.
Durita Mohr, Eurofins Steins Laboratorium, 6600 Vejen, Denmark.
Paulina Seweryn, Department of Molecular Biology, MPI, 37077 Göttingen, Germany.
Bosene Chelchessa, Biogen, 3400 Hillerød, Denmark.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Augustinus Foundation [10-3197], Lundbeck Foundation [R77-A6781], Novo Nordisk Foundation (to C.R.K.); Danish National Research Foundation ‘Centre for mRNP biogenesis and metabolism’ (to D.E.B.); Russian Science Foundation [14-14-00350 to A.B.C]; EMBO Long-Term Fellowship [ALTF 687-2013 to Y.Y.]. Funding for open access charge: Funding from private Danish foundations.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ahlquist P., Noueiry A.O., Lee W.M., Kushner D.B., Dye B.T. Host factors in positive-strand RNA virus genome replication. J. Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chinnaswamy S., Murali A., Li P., Fujisaki K., Kao C.C. Regulation of de novo-initiated RNA synthesis in hepatitis C virus RNA-dependent RNA polymerase by intermolecular interactions. J. Virol. 2010;84:5923–5935. doi: 10.1128/JVI.02446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang S., Sun D., Liang H., Wang J., Li J., Guo L., Wang X., Guan C., Boruah B.M., Yuan L., et al. Cryo-EM Structure of Influenza Virus RNA Polymerase Complex at 4.3 A Resolution. Mol. Cell. 2015;57:925–935. doi: 10.1016/j.molcel.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 4.Cevik B. The RNA-dependent RNA polymerase of Citrus tristeza virus forms oligomers. Virology. 2013;447:121–130. doi: 10.1016/j.virol.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 5.Tellez A.B., Wang J., Tanner E.J., Spagnolo J.F., Kirkegaard K., Bullitt E. Interstitial contacts in an RNA-dependent RNA polymerase lattice. J. Mol. Biol. 2011;412:737–750. doi: 10.1016/j.jmb.2011.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumenthal T., Carmichael G.G. RNA replication: function and structure of Qbeta-replicase. Annu. Rev. Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 7.Kidmose R.T., Vasiliev N.N., Chetverin A.B., Andersen G.R., Knudsen C.R. Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins. Proc. Natl. Acad. Sci. U.S.A. 2010;107:10884–10889. doi: 10.1073/pnas.1003015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeshita D., Tomita K. Assembly of Q{beta} viral RNA polymerase with host translational elongation factors EF-Tu and -Ts. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15733–15738. doi: 10.1073/pnas.1006559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeshita D., Tomita K. Molecular basis for RNA polymerization by Qbeta replicase. Nat. Struct. Mol. Biol. 2012;19:229–237. doi: 10.1038/nsmb.2204. [DOI] [PubMed] [Google Scholar]
- 10.Takeshita D., Yamashita S., Tomita K. Mechanism for template-independent terminal adenylation activity of Qbeta replicase. Structure. 2012;20:1661–1669. doi: 10.1016/j.str.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Takeshita D., Yamashita S., Tomita K. Molecular insights into replication initiation by Qbeta replicase using ribosomal protein S1. Nucleic Acids Res. 2014;42:10809–10822. doi: 10.1093/nar/gku745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theobald D.L., Mitton-Fry R.M., Wuttke D.S. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajnsdorf E., Boni I.V. Multiple activities of RNA-binding proteins S1 and Hfq. Biochimie. 2012;94:1544–1553. doi: 10.1016/j.biochi.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Byrgazov K., Manoharadas S., Kaberdina A.C., Vesper O., Moll I. Direct interaction of the N-terminal domain of ribosomal protein S1 with protein S2 in Escherichia coli. PLoS One. 2012;7:e32702. doi: 10.1371/journal.pone.0032702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duval M., Korepanov A., Fuchsbauer O., Fechter P., Haller A., Fabbretti A., Choulier L., Micura R., Klaholz B.P., Romby P., et al. Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol. 2013;11:e1001731. doi: 10.1371/journal.pbio.1001731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subramanian A.R. Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 1983;28:101–142. doi: 10.1016/s0079-6603(08)60085-9. [DOI] [PubMed] [Google Scholar]
- 17.Vasilyev N.N., Kutlubaeva Z.S., Ugarov V.I., Chetverina H.V., Chetverin A.B. Ribosomal protein S1 functions as a termination factor in RNA synthesis by Qbeta phage replicase. Nat. Commun. 2013;4:1781–1787. doi: 10.1038/ncomms2807. [DOI] [PubMed] [Google Scholar]
- 18.Miranda G., Schuppli D., Barrera C., Hausherr C., Sogo J., Weber H. Recognition of bacteriophage Qbeta plus strand RNA as a template by Qbeta replicase: role of RNA interactions mediated by ribosomal proteins S1 and host factor. J. Mol. Biol. 1997;267:1089–1103. doi: 10.1006/jmbi.1997.0939. [DOI] [PubMed] [Google Scholar]
- 19.Shaklee P.N., Miglietta J.J., Palmenberg A.C., Kaesberg P. Infectious positive- and negative-strand transcript RNAs from bacteriophage Q beta cDNA clones. Virology. 1988;163:209–213. doi: 10.1016/0042-6822(88)90250-4. [DOI] [PubMed] [Google Scholar]
- 20.Kita H., Cho J., Matsuura T., Nakaishi T., Taniguchi I., Ichikawa T., Shima Y., Urabe I., Yomo T. Functional Qbeta replicase genetically fusing essential subunits EF-Ts and EF-Tu with beta-subunit. J. Biosci. Bioeng. 2006;101:421–426. doi: 10.1263/jbb.101.421. [DOI] [PubMed] [Google Scholar]
- 21.Winter G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 2010;43:186–190. [Google Scholar]
- 22.McCoy A. Solving structures of protein complexes by molecular replacement with Phaser. Acta Cryst. D Biol. Cryst. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byrgazov K., Grishkovskaya I., Arenz S., Coudevylle N., Temmel H., Wilson D., Djinovic-Carugo K., Moll I. Structural basis for the interaction of protein S1 with the Escherichia coli ribosome. Nucleic Acids Res. 2015;43:661–673. doi: 10.1093/nar/gku1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giraud P., Crechet J., Uzan M., Bontems F., Sizun C. Resonance assignments of the ribosome binding domain of E. coli ribosomal protein S1. Biomol. NMR Assign. 2015;9:107–111. doi: 10.1007/s12104-014-9554-2. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P., Lohkamp B., Scott W., Cowtan K. Features and development of Coot. Acta Cryst. D Biol. Cryst. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams P., Afonine P., Bunkoczi G., Chen V., Davis I., Echols N., Headd J., Hung L., Kapral G., Grosse-Kunstleve R., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D Biol. Cryst. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker N., Sept D., Joseph S., Holst M., McCammon J. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morozov I., Ugarov A., Chetverin A., Spirin A. Synergism in replication and translation of messenger RNA in a cell-free system. Proc. Natl. Acad. Sci. U.S.A. 1993;90:9325–9329. doi: 10.1073/pnas.90.20.9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Link T., Valentin-Hansen P., Brennan R. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biebricher C.K., Eigen M., McCaskill J.S. Template-directed and template-free RNA synthesis by Q beta replicase. J. Mol. Biol. 1993;20:175–179. doi: 10.1006/jmbi.1993.1271. [DOI] [PubMed] [Google Scholar]
- 31.Guerrier-Takada C., Subramanian A.R., Cole P.E. The activity of discrete fragments of ribosomal protein S1 in Qbeta replicase function. J. Biol. Chem. 1983;258:13649–13652. [PubMed] [Google Scholar]
- 32.Laughrea M., Moore P. Physical properties of ribosomal protein S1 and its interactions with the 30S ribosomal subunit of Escherichia coli. J. Mol. Biol. 1977;112:399–421. doi: 10.1016/s0022-2836(77)80189-7. [DOI] [PubMed] [Google Scholar]
- 33.Thomas J., Boublik M., Szer W., Subramanian A.R. Nucleic acid binding and unfolding properties of ribosomal protein S1 and the derivatives S1-F1 and m1-S1. Eur. J. Biochem. 1979;102:309–314. doi: 10.1111/j.1432-1033.1979.tb06293.x. [DOI] [PubMed] [Google Scholar]
- 34.Celniker G., Ashkenazy N., Glaser F., Martz E., Mayrose I., Pupko T., Ben-Tal N. ConSurf: Using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem. 2013;53:199–206. [Google Scholar]
- 35.Aliprandi P., Sizun C., Perez J., Mareuil F., Caputo S., Leroy J.L., Odaert B., Laalami S., Uzan M., Bontems F. S1 ribosomal protein functions in translation initiation and ribonuclease RegB activation are mediated by similar RNA-protein interactions: an NMR and SAXS analysis. J. Biol. Chem. 2008;283:13289–13301. doi: 10.1074/jbc.M707111200. [DOI] [PubMed] [Google Scholar]
- 36.Bycroft M., Hubbard T., Proctor M., Freund S., Murzin A. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binidng fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 37.Salah P., Bisaglia M., Aliprandi P., Uzan M., Sizun C., Bontems F. Probing the relationship between Gram-negative and Gram-positive S1 proteins by sequence analysis. Nucleic Acids Res. 2009;37:5578–5588. doi: 10.1093/nar/gkp547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foster M., McElroy C., Amero C. Solution NMR of large molecules and assemblies. Biochemistry. 2007;46:331–340. doi: 10.1021/bi0621314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen J., Long A., Schultz S. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 40.Pata J., Schultz S., Kirkegaard K. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA. 1995;1:466–477. [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Q., Hockman M., Staschke K., Johnson R., Case K., Lu J., Parsons S., Zhang F., Rathnachalam R., Kirkegaard K., et al. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 2002;76:3865–3872. doi: 10.1128/JVI.76.8.3865-3872.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ugarov V.I., Demidenko A.A., Chetverin A.B. Qbeta replicase discriminates between legitimate and illegitimate templates by having different mechanisms of initiation. J. Biol. Chem. 2003;278:44139–44146. doi: 10.1074/jbc.M305992200. [DOI] [PubMed] [Google Scholar]
- 43.te Velthuis A.J.W. Common and unique features of viral RNA-dependent polymerases. Cell. Mol. Life Sci. 2014;71:4403–4420. doi: 10.1007/s00018-014-1695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kondo M., Gallerani R., Weissmann C. Subunit structure of Q-beta replicase. Nature. 1970;228:525–527. doi: 10.1038/228525a0. [DOI] [PubMed] [Google Scholar]
- 45.Landers T.A., Blumenthal T., Weber K. Function and structure in ribonucleic acid phage Q beta ribonucleic acid replicase. The roles of the different subunits in transcription of synthetic templates. J. Biol. Chem. 1974;249:5801–5808. [PubMed] [Google Scholar]
- 46.Cole P.E., Sawchyn I., Guerrier-Takada C. Q beta replicase containing altered forms of ribosomal protein S1. J. Biol. Chem. 1982;257:12929–12934. [PubMed] [Google Scholar]
- 47.McGinness K., Sauer R. Ribosomal protein S1 bindsmRNA and tmRNA similarly but plays distinct roles in translation of these molecules. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13454–13459. doi: 10.1073/pnas.0405521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryder S., Recht M., Williamson J. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol. Biol. 2008;488:99–115. doi: 10.1007/978-1-60327-475-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer F., Weber H., Weissmann C. Interactions of Q beta replicase with Q beta RNA. J. Mol. Biol. 1981;153:631–660. doi: 10.1016/0022-2836(81)90411-3. [DOI] [PubMed] [Google Scholar]
- 50.Privalov P.L., Dragan A.I., Crane-Robinson C. Interpreting protein/DNA interactions: distinguishing specific from non-specific and electrostatic from non-electrostatic components. Nucleic Acids Res. 2011;39:2483–2491. doi: 10.1093/nar/gkq984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuppli D., Miranda G., Qiu S., Weber H. A branched stem-loop structure in the M-site of bacteriophage Qbeta RNA is important for template recognition by Qbeta replicase holoenzyme. J. Mol. Biol. 1998;283:585–593. doi: 10.1006/jmbi.1998.2123. [DOI] [PubMed] [Google Scholar]
- 52.Inokuchi Y., Kajitani M. Deletion analysis of Qbeta replicase. J. Biol. Chem. 1997;272:15339–15345. doi: 10.1074/jbc.272.24.15339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.