Abstract
CRISPR–Cas (clustered regularly interspaced short palindromic repeats-CRISPR associated) systems allow bacteria to adapt to infection by acquiring ‘spacer’ sequences from invader DNA into genomic CRISPR loci. Cas proteins use RNAs derived from these loci to target cognate sequences for destruction through CRISPR interference. Mutations in the protospacer adjacent motif (PAM) and seed regions block interference but promote rapid ‘primed’ adaptation. Here, we use multiple spacer sequences to reexamine the PAM and seed sequence requirements for interference and priming in the Escherichia coli Type I-E CRISPR–Cas system. Surprisingly, CRISPR interference is far more tolerant of mutations in the seed and the PAM than previously reported, and this mutational tolerance, as well as priming activity, is highly dependent on spacer sequence. We identify a large number of functional PAMs that can promote interference, priming or both activities, depending on the associated spacer sequence. Functional PAMs are preferentially acquired during unprimed ‘naïve’ adaptation, leading to a rapid priming response following infection. Our results provide numerous insights into the importance of both spacer and target sequences for interference and priming, and reveal that priming is a major pathway for adaptation during initial infection.
INTRODUCTION
In bacteria and archaea, CRISPR arrays and Cas proteins comprise an RNA-guided immune system that silences mobile genetic elements including viruses and horizontally transferred DNA (1,2). CRISPR–Cas immune systems proceed through three major steps. First, a short segment of invader DNA is inserted as a new spacer following the first repeat of the CRISPR array, a process called adaptation (3,4). Second, the CRISPR array is transcribed and processed into short CRISPR RNAs (crRNAs), each containing a different spacer sequence (5,6). Finally, crRNAs guide Cas effector proteins to DNA or RNA targets containing protospacers that match the crRNA spacer sequence, and a Cas endonucleolytic activity cleaves the target leading to its destruction, a process called CRISPR interference (7,8).
Based on phylogenetic analysis, CRISPR–Cas systems are clustered into two classes and five basic types (Types I–V), which are further divided into 16 subtypes (Types I-A to I-F and I-U, Types II-A to II-C, and Types III-A to III-D, Types IV and V) (9). Cas1 and Cas2, the only two proteins that are conserved in all CRISPR–Cas systems, form a stable heterocomplex that is required for spacer acquisition (9–13). The mechanisms for CRISPR interference vary between each type, and subtype-specific interference Cas proteins are common (9). Class 1 (Types I, III and IV) systems utilize large, multi-protein surveillance complexes as crRNA effectors and in some cases require a separate Cas endonuclease (14–16), while Class 2 (Types II, V) systems require only a single protein that acts as both crRNA effector and Cas endonuclease (17–20) (for a recent review see (7)).
Escherichia coli K12 contains a Type I-E CRISPR–Cas system, which utilizes Cas1 and Cas2 for spacer acquisition and the crRNA-effector complex Cascade and the signature endonuclease Cas3 for interference (Figure 1A) (10,13,14,21–27). In this system, adaptation can occur through two different spacer acquisition processes, termed ‘naïve’ and ‘primed’ adaptation (11,28). Naïve adaptation requires only Cas1 and Cas2 (29), and occurs when a bacterium acquires new spacers from mobile genetic elements that it has not previously encountered. In contrast, primed adaptation also requires the interference machinery and acts as a positive feedback loop, in which spacers already present against a target promote acquisition of new spacers from the same target (11,28).
Figure 1.
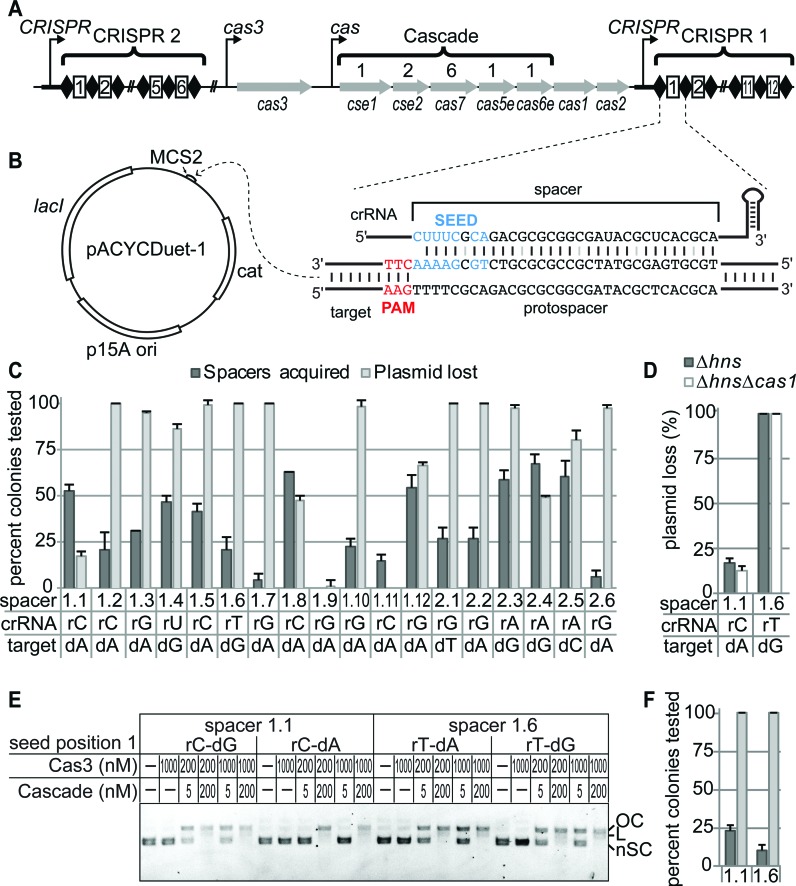
Seed mutations do not completely block CRISPR interference. (A) CRISPRs and cas operon from E. coli K12. Genes and stoichiometry of Cascade proteins are indicated. (B) Construct design for priming targets for 18 endogenous E. coli K12 spacers. PAM/protospacer sequences were inserted into pACYCDuet-1. CRISPR 1 spacer 1 and target sequence containing a mismatch at the first position of the seed are shown. (C) Spacers acquisition and plasmid loss rates for each endogenous spacer against a target with a mismatch at the first seed position after 24 h growth. Position 1 of the crRNA spacer sequence and the corresponding mismatched nucleotide in the target are labeled for each spacer. Spacers are named based on their CRISPR and position within the CRISPR (e.g. CRISPR 1 spacer 1 is 1.1, CRISPR 2 spacer 1 is 2.1 etc.). (D) Plasmid loss rates of spacer 1.1 and 1.6 targets with seed position 1 mismatches in E. coli BW25113 Δhns and BW25113 ΔhnsΔcas1 after 24 h growth. (E) In vitro Cascade-mediated Cas3 degradation of pACYCDuet-1 target plasmids. Cascade (Cse1 at 1 μM and Cse2–Cas6e-crRNA complex at indicated concentration) bearing spacer 1.1 or 1.6 crRNA and Cas3 concentrations are indicated. The first base pair of the crRNA spacer and target is labeled for each plasmid tested. Plasmid DNA is labeled as follows: OC – open circle; L – linear; nSC – negatively supercoiled. (F) Spacer acquisition and plasmid loss rates in E. coli BW25113 ∆hnsfor bona fide spacer 1.1 and 1.6 targets after 24 h growth, colored as in (C).
Primed adaptation was identified as a mechanism that allows the host to overcome invader escape from the CRISPR interference pathway (11). During CRISPR interference, the Cascade crRNA spacer base pairs to the target-strand of the protospacer, and the non-target strand is displaced forming an R-loop (Figure 1B) (21). Cas3 is then recruited by the Cse1 subunit of Cascade and processively degrades the DNA (25–27). Cascade is thought to initially recognize targets based on the presence of a correct protospacer adjacent motif (PAM) sequence prior to interrogating the adjacent sequence for complementarity to the crRNA, in a mechanism that is analogous to other Types I and II crRNA effectors (Figure 1B) (30–33). R-loop formation proceeds directionally away from the PAM, and the first base pairs that form during target binding comprise a seed sequence that nucleates target binding (Figure 1B) (34,35). Due to their importance in Cascade–dsDNA binding, spontaneous mutations in the PAM and seed regions disrupt high-affinity target binding and can lead to invader escape from CRISPR interference (31,36). However, in some cases these mutations promote primed adaptation, which allows the host to combat this escape by rapidly acquiring new functional spacers against other regions of the invader DNA (11). In addition, primed adaptation enables the CRISPR–Cas system to rapidly respond to any mobile genetic element with imperfect sequence homology to an existing spacer, providing an important strategy for combating closely related families of viruses and plasmids.
A recent high-throughput plasmid loss assay of a randomized PAM and protospacer library indicated that up to five mutations in the PAM and seed can still promote primed spacer acquisition (36). If single mutations within the PAM or seed dramatically decrease the binding affinity of Cascade, it is unclear how Cascade may recognize these types of targets in order to promote primed spacer acquisition. Recent single-molecule FRET studies of Cascade–DNA binding revealed that Cascade adopts a non-canonical binding mode when bound to a target with an incorrect PAM or containing partial complementarity in the seed, suggesting that priming targets are bound by Cascade, but in an alternative binding mode as compared to bona fide targets (37).
Genome editing studies using the Type II crRNA effector endonuclease Cas9 have revealed that targets with PAM and seed mutations can still be cleaved by Cas9, but that this mutational tolerance is dependent on the spacer sequence of the crRNA (38–42). In contrast, the effects of spacer sequence on Type I-E interference and priming activity have not been studied systematically. Here, we have investigated the priming efficiency of a large variety of spacer sequences in E. coli K12 against targets with a single mutation in the seed. Surprisingly, the spacers display extremely varied activities against seed mutant protospacers for both priming and direct interference. For many of the spacers tested, single seed mutations did not block direct interference or promote priming, suggesting that the level of priming and seed mismatch tolerance may be dependent on spacer sequence. To test the importance of spacer sequence on PAM or seed mutation tolerance, we have developed a high-throughput assay that distinguishes between direct interference and priming activity. Our results reveal spacer-specific differences in CRISPR activity against both PAM and seed mutant targets, including several mutations that completely block CRISPR activity from one spacer but allow robust CRISPR activity from another. We find that Type I-E CRISPR–Cas systems can avoid priming against self sequences based on the presence of a self-signal in the CRISPR repeat. In addition, PAM authentication for targets with PAM sequences that promote priming occurs following Cascade binding but prior to Cas3 degradation. Intriguingly, many of the functional PAMs identified in our study are preferentially acquired during unprimed ‘naïve’ adaptation, leading to a rapid priming response following infection. These findings reveal the importance of both the spacer and target sequence in CRISPR–Cas activity, and highlight the sophistication of the adaptive immune response in the Type I-E system.
MATERIALS AND METHODS
Bacterial strains
Strains, plasmids and oligonucleotides used in this study are listed in Supplementary Tables S1–S4. E. coli K12 BW25113 was used as background for all constructed strains (43,44). Individual gene deletion strains for hns, cas3, cse1, cas1 were obtained from the Keio collection (44). The cas3, cse1 or cas1 deletions were moved from BW25113 Δcas3::kan, BW25113 Δcse1::kan or BW25113 Δcas1::kan into BW25113 Δhns or E. coli X019 using P1 phage transduction. Kanamycin resistance cassettes were removed for all strains using pCP20 (45), with the exception of the cas3::kan cassette in strain X030 (Supplementary Table S1). The X019 strain, in which spacers 1.2–1.12 and 2.2–2.6 were deleted was created using lambda Red recombinase, as previously described (43). DNA fragments containing kanamycin resistance cassette flanked by FRT sites with 50-bp homology to CRISPR 1 or CRISPR 2-adjacent sequences were created using primers XCY302–303 and XCY304–305, respectively (Supplementary Table S3).
Plasmid construction
All protospacers used in this study were ligated to pACYCDuet-1 or pCDF-1b between BgIII and XhoI or NcoI and NotI sites as indicated (Supplementary Tables S2 and S3). For recombinant Cascade expression with different spacer sequences, CRISPRs bearing spacers 1.1, 1.6, 1.9 or 2.1 were generated using pWUR547 (21) as template with primers indicated in Supplementary Tables S2 and S3 through ‘round-the-horn (RTH) cloning (http://openwetware.org/wiki/'Round-the-horn_site-directed_mutagenesis). For recombinant Cascade expression and purification, the N-terminal Streptactin-tag in Cse2 of pWUR480 (14) plasmid was changed to a His6-tag using HL005 and HL006 through RTH cloning to produce pDGS010. For recombinant Cas3 expression and purification, the cas3 gene was amplified from the E. coli K12 genome using XCY001 and XCY002 and cloned into pSV272, encoding an N-terminal His6-MBP (maltose-binding protein) tag. For the Cas1 and Cas2 expression plasmid pX288, PCR product of native tac promoter controlled Cas1-Cas2 was cloned using XCY255 and XCY256 in pACYCDuet-1.
For the pACYC-GFP-tac plasmid, a tac promoter (46) controlled GFP gene was PCR amplified from psfGFP using XCY413 and XCY417 and cloned into pACYCDuet-1 (Supplementary Tables S2 and S3). Our preliminary results indicated that the use of a native tac promoter for GFP expression slowed E. coli growth, changing the GFP+ and GFP- cells ratio through a non-CRISPR related mechanism. To solve this problem, the native tac promoter was changed to a weaker tac-derived promoter (47). The resulting plasmid (pACYC-GFP-pro3) did not increase the doubling time of E. coli compared to E. coli bearing empty pACYCDuet-1. Competition assays between E. coli X019 bearing either empty pACYCDuet-1 or pACYC-GFP-pro3 further indicated that GFP expression does not affect the ratio of GFP- and GFP+ cells, as ratios between the two strains remained constant after 24 h growth without selection. To decrease the half-life of GFP, the protease-sensitive SsrA peptide tag AANDENYALAA (48) was added to the C-terminus of GFP using XCY449 and XCY452 with pACYC-GFP-pro3 as template through RTH cloning. This final plasmid is referred to as pACYC-GFP throughout the text.
CRISPR-like plasmid pRep-Sp8-Rep was created by cloning a PCR amplicon containing the first two repeats and one spacer (spacer #8) from E. coli PIM5 (28) into the PciI site of plasmid pGFP-Kan (36) (Supplementary Tables S2 and S3 for plasmids and primers). This plasmid then served as a template to create derivative plasmids pSp8-Rep (containing the distal repeat) and pRep-Sp8 (containing the proximal repeat). Plasmids were PCR amplified using Phusion DNA polymerase, 5′ phosphorylated using T4 polynucleotide kinase, end-to-end ligated, and transformed to E. coli XL1 Blue. pRep-Sp8 then served as a template for a series of 16 derivative plasmids (Supplementary Table S2), using primers listed in Supplementary Table S3. Plasmids sequences were confirmed by Sanger sequencing at GATC Biotech (Konstanz, Germany).
pACYC-Cas3-C85Venus-Cse1-N155Venus (referred as pACYC-BiFC in the paper) was created by Gibson Assembly using primers XCY479–XCY486 (Supplementary Table S3) (49). Cas3 contains a C-terminal fusion of the C-terminal Venus fragment and Cse1 contains a C-terminal fusion of the N-terminal fragment of Venus. The expression vector was assembled from 4 separate PCR products amplified using either pACYC-Cas3-Cse1 or mVenus-pBAD vector (a gift from Michael Davidson (Addgene plasmid # 54845)) as template.
Plasmid-loss and spacer acquisition experiments
Plasmids were introduced into E. coli BW25113 derived strains via heat shock and single colonies were used to inoculate initial cultures. All strains were grown for 24 h (sub-cultured at 12 h) in 2 ml LB in 15 ml tubes at 37ºC with shaking at 200 rpm. For passaging, 20 μl of culture was sub-cultured into 2 ml LB. When indicated, further periods of incubation were performed at the same conditions. E. coli cultures were diluted 250 000-fold and 10 μl of the final dilution was plated on LB plates (1.5% agar) without antibiotic. After 6 h, 35–50 colonies on these plates were replicated onto LB plates supplemented with chloramphenicol to check for plasmid loss. For each sample, 16 colonies on the no antibiotic plates were picked randomly to analyze spacer acquisition by colony PCR using Taq DNA polymerase. Newly acquired spacers in CRISPR 1 or CRISPR 2 were detected by PCR using primers XCY076–077 or XCY152–153, respectively (Supplementary Table S3). PCR products were visualized on 2% agarose gels stained with SYBR Safe (Thermo Fisher Scientific). All experiments were performed for three individual cultures. Plasmid loss and spacer acquisition rates reflect the average of these three biological replicates, and errors are the standard deviation between replicates.
Plasmid loss experiments to assess autopriming were performed in E. coli strain PIM5 (28), which is a derivative of BW25113 Δhns. Plasmids were transformed into PIM5 by electroporation, and strains were grown for 48 h in non-selective liquid media. Plasmid loss was assessed on non-selective plates by scoring fluorescence of the colony resulting from the presence of the GFP plasmid. Non-fluorescent colonies were analyzed by colony PCR for the integration of new spacers in CRISPR 1 and 2, and sequenced to confirm the strand bias that is typical for priming in E. coli strains with Type I-E CRISPR–Cas systems (11).
Protein expression and purification
Cascade lacking Cse1 (Cse2–Cas6e) was expressed in BL21(DE3) cells using pDGS010, pWUR404 and the appropriate CRISPR expression plasmid (spacer 1.1: pX238, spacer 1.6: pX230, spacer 1.9: pX503, spacer 2.1: pX569, Supplementary Table S2) in 1 l LB media supplemented with ampicillin, chloramphenicol and streptomycin. Cultures were grown to 0.5 OD600 at 37°C, and induced overnight at 16°C with 0.5 mM IPTG. His6-tagged Cse2–Cas6e was purified using HisPur Ni-NTA affinity resin (Thermo Fisher Scientific). The eluent was concentrated to ∼1 ml, then purified by size exclusion chromatograph using a Superdex 200 column (GE Life Sciences) in a buffer containing 20 mM Tris (pH 7.5), 100 mM NaCl, 5% glycerol and 1 mM TCEP. Cse1 was expressed in BL21(DE3) using the EcCse1-pSV272 expression vector, and purified as previously described (30). Cas3 was expressed in BL21(DE3) using the Cas3-pSV272 expression vector (Supplementary Table S2) and purified as described previously with the following modifications (25). During the whole Cas3 purification process, 1mM TCEP was added in all buffers. To maintain the activity of Cas3, the purification process was completed in one day. Briefly, after lysis and affinity purification using HisPur Ni-NTA resin, His6-MBP-Cas3 was purified on a Superdex 200 column. The purified His6-MBP-Cas3 protein was cleaved by tobacco etch virus protease for 3 h at 4°C. The cleaved sample was flowed through a Ni-NTA column, concentrated to 1 ml, and finally purified on a Superdex 200 column.
DNA binding and cleavage assays
All binding assays were performed in binding buffer: 20 mM Tris (pH 7.5), 100 mM NaCl, and 5% glycerol. All cleavage assays were performed in reaction buffer: 10 mM HEPES (pH 7.5), 100 mM KCl, 5% (v/v) glycerol, 2 mM ATP, 100 μM CoCl2, and 10 mM MgCl2. Concentrations indicated for Cascade in Figure 1E, 6A–C, Supplementary Figures S2A and S7B–D are for the Cse2–Cas6e complex, as Cse1 was held at a constant concentration to ensure complete formation of the Cascade complex (30). Cse2–Cas6e at indicated concentrations and 1000 nM Cse1 were pre-incubated for 20 min at 37°C to form the Cascade complex. Samples were cooled on ice for 1 min prior to initiating binding or cleavage reactions. For Cascade–DNA binding, Cascade was incubated with 2 nM target plasmid, and samples were incubated at 37°C for 30 min prior to electrophoresis on a 0.8% agarose gel stained with SYBR Safe run at 15 V at 4°C for 18 h. For Cas3 cleavage, Cascade was incubated with 2 nM target plasmid at 37°C for 15 min. Cas3 was added at the indicated concentration to initiate plasmid digestion. Reactions were incubated at 37°C for 30 min and terminated by the addition of 20 mM EDTA. Proteins were removed by phenol extraction. Reactions were analyzed by electrophoresis on a 1% agarose gel stained with SYBR Safe.
Figure 6.
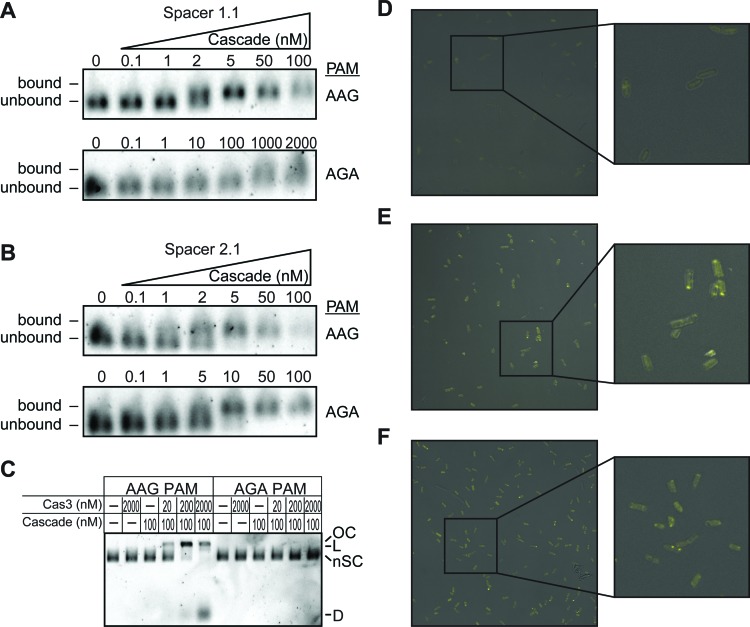
Priming PAM blocks Cascade-mediated Cas3 cleavage but not Cas3-Cascade association. (A and B) Electrophoretic mobility shift assay for Cascade binding to (A) spacer 1.1 and (B) 2.1 targets with AAG or AGA PAMs. Cse2–Cas6e concentration is varied, and concentrations are labeled for each sample. Cse1 concentration was held constant at 1 μM to ensure complete formation of the Cascade complex. (C) Cascade-mediated Cas3 cleavage of spacer 2.1 targets with AAG or AGA PAMs. Plasmid DNA is labeled as follows: OC – open circle; L – linear; nSC – negatively supercoiled; D – degraded. (D–F) Confocal micrographs for BiFC experiments detecting interactions between Cse1 and Cas3. (D) E. coli BW25113 Δcse1Δcas3 grown with pACYC-BiFC and empty pCDF-1b plasmid. (E) E. coli BW25113 Δcse1Δcas3 grown with pACYC-BiFC and pCDF containing spacer 2.1 target with an AAG PAM. (F) E. coli BW25113 Δcse1Δcas3 grown with pACYC-BiFC and pCDF containing spacer 2.1 target with an AGA PAM.
Generation of PAM and seed libraries
To avoid sequence bias, initial libraries were constructed in DH5α by RTH cloning using pACYC-GFP as template (see Supplementary Table S3 for primers). Primer locations were designed to avoid complementarity between the overhanging degenerate sequence and the template. However, this library design did not result in an unbiased library for the original spacer 1.1 seed library, so an alternative method was used for spacer 1.1 and 2.1 6MM seed library creation. A 24-bp protospacer (position 9 to position 32) of spacer 1.1 (XCY573 and XCY574) or spacer 2.1 (XCY577 and XCY578) (Supplementary Table S3) was ligated into pACYC-GFP to create pX735 and pX737, and these plasmids were used as templates for RTH cloning of the libraries. All primers were phosphorylated using polynucleotide kinase prior to PCR. Primers were used to PCR amplify the pACYC-GFP, pX735 or pX737 backbone and PCR products were purified. PCR products were ligated and transformed into E. coli DH5α. For each library, over 30,000 transformants were isolated. All colonies were resuspended in LB, the bacteria were pelleted, and plasmids were extracted using a Promega Wizard Plus SV Miniprep DNA Purification kit. This procedure yielded the five original libraries, PAM of spacer 1.1, seed of spacer 1.1, PAM of spacer 2.1, and two seed libraries of spacer 2.1. All libraries were prepared in triplicate from three separate ligations and DH5α transformations. High-throughput plasmid loss, priming assays and sequencing were performed for all three biological replicates.
High-throughput plasmid loss and priming assays
All original libraries were transformed into X019, X019 Δcse1 and X019 Δcas1 and plated onto LB plates with chloramphenicol yielding around 30 000 colonies. All colonies were resuspended using 1 ml LB. After adjusting the concentration of the resuspended bacteria to OD600 of ∼5.0, 20 μl of the culture was used to inoculate 2 ml LB without antibiotic for five growth cycles, with sub-culturing every 6 or 12 h. Next, 40 μl of the each culture was used to inoculate 4 ml LB supplemented with chloramphenicol. These cultures were grown at 37°C with shaking at 200 rpm for 12 h and plasmids were extracted, yielding an additional 12 plasmid libraries. These 12 plasmid libraries and the four original plasmid libraries were transformed into X019 and cultured for two cycles, sub-cultured at 6 or 12 h. The cultures were diluted 100-fold and analyzed by BD FACSAria III flow cytometer. For each culture, 100 000 GFP+ and GFP- cells were sorted. The average percentage of GFP-cells for three biological replicates are reported in Figure 2D, and errors reflect the standard deviation between the three replicates. Spacer acquisition was analyzed for the genomic DNA of the sorted GFP- cells by PCR amplification using XCY076 and XCY077 for CRISPR 1 and XCY152 and XCY153 for CRISPR 2. PCR products were analyzed on a 2% agarose gel stained with SYBR Safe, and intensity of PCR bands was measured using ImageQuant TL (GE Life Sciences). Spacer acquisition rates were measured as the intensity of extended CRISPR PCR products relative to the intensity of total PCR product. The relative intensity of CRISPR 1 and CRISPR 2 were averaged to determine the relative spacer acquisition for each sample. Spacer acquisition rates reported in Figure 2E are the average from three separate biological replicates and error bars reflect standard deviation between the three replicates.
Figure 2.
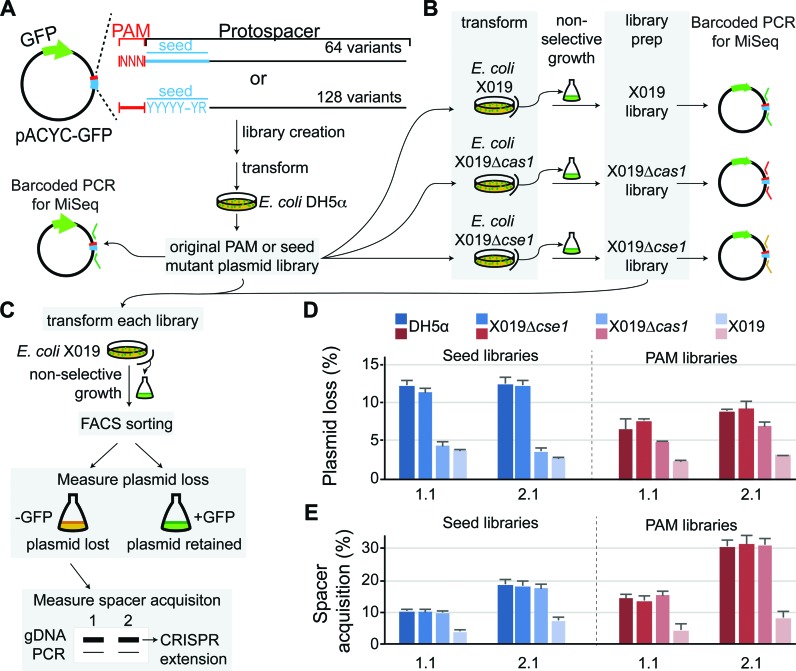
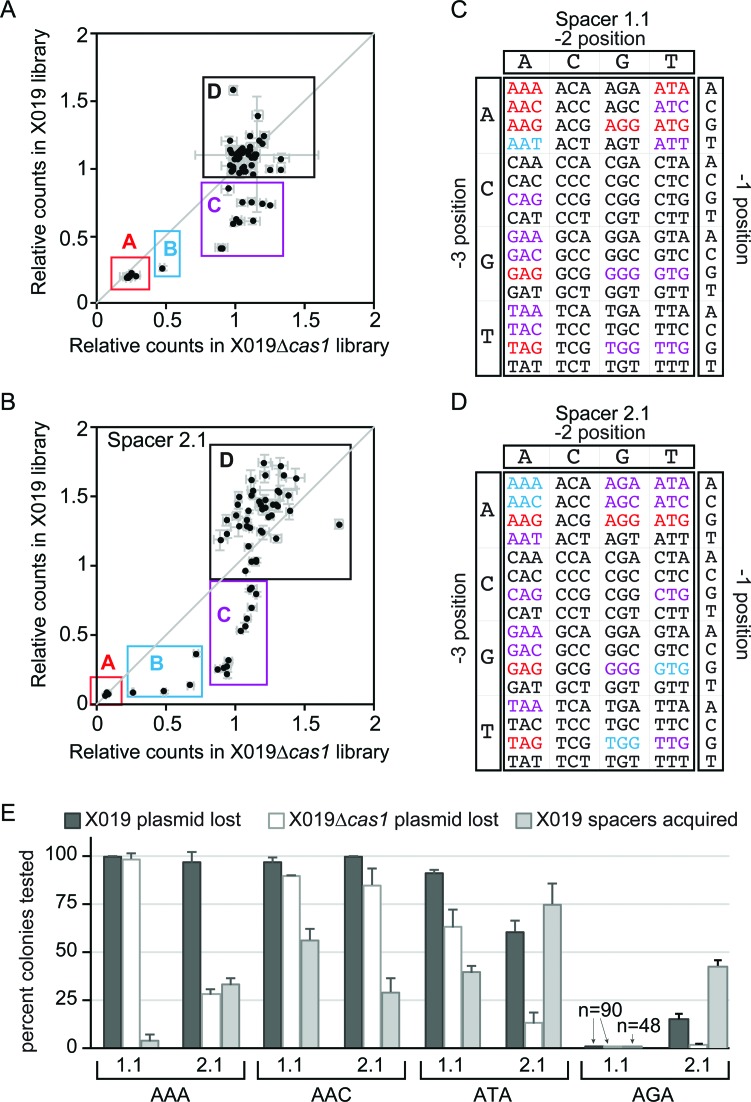
High-throughput screen for CRISPR activity of PAM and seed mutants. (A–C) Experimental design for high-throughput screen. (A) PAM and seed library construction. PAM libraries contained completely degenerate sequences at the -3, -2 and -1 positions of the target, resulting in 64 possible sequences. Seed libraries contained two potential sequences at each positions 1–5 and 7–8 of the target, resulting in 128 possible sequences. (B) The original libraries were transformed to E. coli X019, X019 Δcas1 and X019 Δcse1 and libraries were prepared for each strain after an extended growth period in non-selective media. These libraries were used for barcoded PCR as experimental samples for high-throughput sequencing analysis. (C) All libraries were transformed to E. coli X019 and grown for 2 cycles of 6–12 h. Cells were sorted by FACS to measure rates of plasmid loss and the genomic DNA of GFP- cells were used for PCR of CRISPRs to determine rates of spacer acquisition. (D) Plasmid loss rates for libraries created using this high-throughput experimental design, as measured by percent of GFP- cells for ∼100 000 cells tested. (E) Spacer acquisition rates for the libraries.
MiSeq Illumina sequencing
The PAM and seed libraries extracted from DH5α, X019, X019 Δcse1, and X019 Δcas1 were amplified by PCR using Phusion DNA polymerase using a pair of primers containing unique 6-nt barcodes to differentiate between libraries and replicates (Supplementary Table S4). The 100–120 bp PCR fragments were analyzed by 2% agarose gel electrophoresis and absorbance at 260 nm. Based on the gel analysis and absorbance reading, equal quantities were mixed and pooled. The mixed samples were run on a 2% agarose gel, the band was excised and purified using a Promega Wizard Gel and PCR Clean-up kit. Samples were analyzed on an Agilent 2100 Bioanalyzer and a Qubit Fluorometer (Thermo Fisher Scientific) to determine DNA size and concentration. Samples were prepared for Illumina Sequencing using the TruSeq Nano DNA Sample Preparation kit (v3) for 1× 150 bp (single-end). To increase the diversity of sequences, samples were spiked with ∼30% of a PhiX Control v3 adapter-ligated library. Samples were sequenced on an Illumina MiSeq at the Iowa State University DNA Facility.
Plasmid libraries were sequenced in three separate MiSeq runs (Supplementary Table S5). MiSeq run 1 contained the three replicates each for the PAM and seed libraries for spacer 2.1. MiSeq run 2 contained three replicates for the PAM library for spacer 1.1 and three replicates for an incomplete seed library for spacer 1.1, which had 42 sequences with fewer than 100 reads in all libraries. Analysis of this library is included in Supplemental Data File 1, although this analysis was omitted in the main text. MiSeq run 3 contained three replicates each for the redesigned spacer 1.1 seed library and the spacer 2.1 6MM library.
Analysis of MiSeq data
Sequences from MiSeq output files were demultiplexed and sorted into separate files for each library and replicate based on the presence of specific pairs of barcodes at both ends of the read using a bash script (Supplementary Table S4). Reads corresponding to the target (forward reads) and non-target (reverse reads) strand of the protospacer were sorted separately. To determine read counts for all possible sequences in each library, the resulting files were searched for the 64 or 128 possible PAM/protospacer sequences for each PAM or seed library, respectively. An output file was generated for each replicate of each library containing the counts for each PAM/protospacer search sequence in the forward and reverse direction (compiled in Supplemental Data File 1). Forward read counts for highly depleted sequences in the X019 Δcas1 and X019 library were systematically higher than reverse read counts for the same sequences. This phenomenon does not appear to be a result of the demultiplexing strategy, as demultiplexing using alternative methods (fastx-multx command from ea-utils package (50) or sabre (https://github.com/najoshi/sabre)) produced very similar results. Overall trends in sequence depletion are the same between forward and reverse reads, although the absolute value of counts differs. Therefore, forward and reverse read counts were summed and treated as total read counts for each sequence. Read counts between samples were normalized by calculating a scaling factor based on the sample with the highest number of sequences. For seed libraries, sequences with anomalously high read counts (>2-fold greater than the DH5α reference library following normalization) in the X019 or X019 Δcas1 were omitted when calculating the scaling factor. Normalized read counts from three biological replicates for each library were averaged and standard deviations were determined. To determine the relative number of counts for each sequence in the experimental libraries, average read counts for X019 Δcse1, X019 Δcas1, and X019 libraries were divided by the average read count for the DH5α reference library. Standard deviations were propagated and are reported as errors for relative counts in Figures 3B–C, 4A–B, Supplementary Figures S4 and S6.
Figure 3.
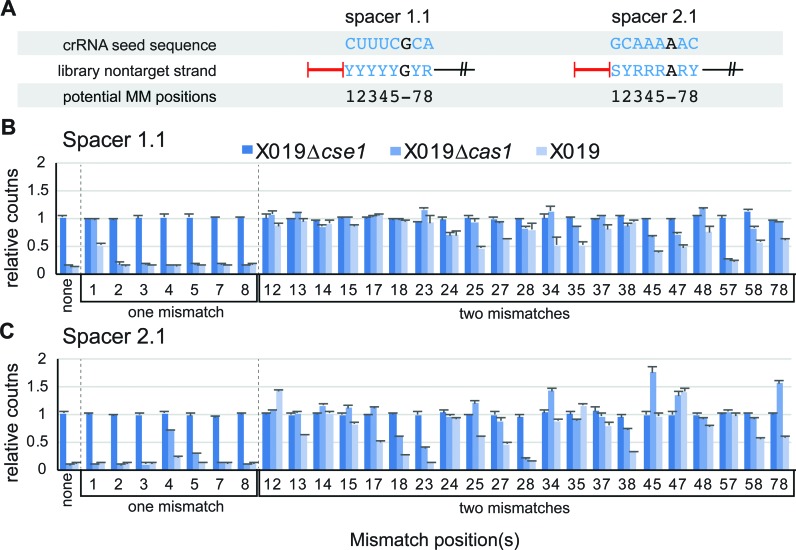
One or two mismatches in the seed can be tolerated for CRISPR activity. (A) Seed libraries tested in this study. The crRNA seed sequence and corresponding region of the non-target strand of the protospacer are shown. Degenerate DNA labels: Y – cytosine or thymine; R – adenine or guanine; S – guanine or cytosine. (B and C) Counts of sequences with one or two mismatches in the seed sequence for X019 Δcse1, X019 Δcas1, and X019 relative to the reference DH5α library. (B) Spacer 1.1 seed libraries. (C) Spacer 2.1 seed libraries. Mismatch position(s) are labeled for each set of data.
Figure 4.
Direct interference and priming can be promoted by a large set of PAM sequences. (A and B) Scatter plots for relative counts of 64 PAM/Protospacer sequence for libraries extracted from E. coli X019 versus E. coli X019 Δcas1 for (A) spacer 1.1 targets and (B) spacer 2.1 targets. Counts are relative to the E. coli DH5α reference library. (C and D) PAM sequences colored by groups as defined in (A and B) for (C) spacer 1.1 targets and (D) spacer 2.1 targets. Red: Group A, blue: Group B, purple: Group C, black: Group D. (E) Plasmid loss and spacer acquisition rates for spacer 1.1 and 2.1 targets with AAA, AAC, ATA or AGA PAMs after 24 h growth.
Fluorescence microscopy
BiFC experiments were performed in E. coli X030 (Supplementary Table S1) carrying pACYC-BiFC and empty pCDF-1, pCDF-1b bearing spacer 2.1 AAG target, or pCDF-1b bearing spacer 2.1 AGA target. Single colonies were grown at 37°C in LB containing chloramphenicol (34 ug/ml) until OD600 reached 0.05. Cultures were shifted to 18°C for 6 h to ensure that plasmid loss of the pCDF-1b bearing spacer 2.1 AAG target would occur slowly, allowing for fluorescence to be observed. Cells were adjusted to OD600 0.5 and re-suspended in phosphate buffer (pH 7.2) and 5 μl of the cells were applied to poly-l-lysine covered microscope slides, and analyzed using a Leica SP5 X MP confocal/multiphoton microscope system with an inverted microscope front end, with a 40× oil immersion objective and an argon laser as the excitation source (514 nm) and detection at 530–600 nm.
Analysis of naïve PAMs and simulation of adaptation
A data set generated by Yosef et al. was used to analyze the frequency of PAMs of targets for spacers acquired through naïve adaptation (29). Spacer sequences and genomic or plasmid locations were reported in the original paper. PAM sequences were extracted from the genomic (NCBI Reference sequence NC_012947.1) or plasmid sequences (reported in (10)) using the BEDtools getfasta tool (51).
In scenario 1, pX288 (pACYC-Cas1–2) and empty pCDF-1b were co-transformed into the X019 Δcse1 E. coli strain. In scenario 2, pX288 and priming plasmid (pCDF-1b bearing a spacer 2.1 target with an AGA PAM) were co-transformed into the X019 strain. In scenario 3, pX288 and empty pCDF-1b were co-transformed into the X019 strain. Single colonies were grown to saturation (OD600 of 3.5) for two cycles in LB supplemented with chloramphenicol to maintain pX288. The 5′-end of CRISPR 1 was PCR amplified from genomic DNA isolated from each culture using XCY076 and XCY077 (Supplementary Table S3) and visualized on a 2% agarose gel stained with SYBR Safe to test for spacer acquisition. The relative amount of each band corresponding to a different number of acquired spacers was determined by densitometry using ImageQuant TL software. Cultures were performed in triplicate and the average amount of product is plotted in Figure 7B, with error bars reflecting the standard deviation between replicates.
Figure 7.
Naïve adaptation triggers a rapid priming response. (A) Analysis of PAM sequences for spacers acquired in Yosef et al. naïve adaptation study (29). In the study, spacers were acquired from the E. coli genomic DNA or a plasmid borne by host. Percentage of reads for spacers derived from sequences with AAG PAMs or other functional or nonfunctional PAMs identified in our study are plotted. Total distribution of each type of PAM in each source DNA are also plotted. (B) Quantified PCR product resulting from newly acquired spacer from three adaptation scenarios following two cycles of growth. Scenario 3 products with significant differences (P < 0.005 based on unpaired two-tailed t-test, n= 3 cultures) compared to scenario 1 are marked with an asterisk. (C and D) Model for adaptation during initial encounter of invader DNA. Naïve adaptation, requiring only the adaptation machinery (orange), allows for integration of spacers against the previously unencountered virus. Spacers may be against targets with PAMs that promote (C) interference or (D) priming by the interference machinery (blue). Cascade bearing newly acquired spacers can bind targets with (C) interference or (D) priming PAMs and recruit Cas3. PAM licensing at this step elicits a (C) target degradation or (D) priming response, although rare occurrences of the alternative mechanism are also possible for each type of target.
RESULTS
Mutations in the seed region do not abolish CRISPR interference
To date, only a limited number of spacers have been used to study priming in E. coli, and it is unknown how spacer sequence may affect direct interference or priming efficiency in the presence of PAM or seed mutations. To test a larger pool of spacers, we investigated the priming efficiency of the 18 endogenous spacers present in the two CRISPR loci of E. coli K12 against targets with mutations at the first seed position (Figure 1A and B). For each spacer, we created a target plasmid containing a 35-bp sequence comprising an AAG PAM and a 32-bp protospacer with a mismatch at the first seed position, which has previously been shown to block interference and promote priming (11,31,36) (Figure 1B). We then tested the rates of plasmid loss and spacer acquisition for each target plasmid in the CRISPR active strain E. coli BW25113 Δhns (28,52–55) after 24 h growth without antibiotic selection. All 18 target plasmids are stable in a cse1 deletion strain (BW25113 ΔhnsΔcse1), which knocks out Cascade DNA-binding activity and CRISPR interference, indicating that any observed plasmid loss in BW25113 Δhns is the result of CRISPR interference.
Surprisingly, the 18 spacers display extremely varied activities for both plasmid loss and spacer acquisition (Figure 1C). Three spacers (spacers 1.1, 1.8 and 2.4) displayed higher rates of spacer acquisition than plasmid loss, while most of the remaining spacers exhibited higher rates of plasmid loss than spacers acquired. Several plasmids (targets for spacers 1.2, 1.6, 1.7, 2.1 and 2.2) were lost in all colonies tested. For half of the spacers (1.2, 1.5, 1.6, 1.7, 1.10, 2.1, 2.2, 2.3 and 2.6), target plasmid loss appears to be largely independent of spacer acquisition, as plasmids were lost in >98% of colonies before even 30% of bacteria had acquired new spacers. Two spacers (1.9 and 1.11) displayed low CRISPR activity against both mutant and bona fide targets (AAG PAM and no mutations in protospacer) (Figure 1C, Supplementary Figure S1A), but this low activity may be due to defects in Cascade assembly due to high G-C content (spacer 1.9) or mutations in the repeat (spacer 1.11) (Supplementary Figure S1B and C). Overall, these results suggest that the majority of endogenous E. coli K12 spacers can direct interference against targets containing a mutation at the first seed position, in contrast to numerous reports that base pairing at seed position 1 is strictly required for Cascade-directed target binding and interference (11,31,36).
To verify that the observed plasmid loss is based on direct CRISPR interference from the original spacer and not from newly acquired spacers, we created a cas1 deletion strain (BW25113 ΔhnsΔcas1), which maintains the interference pathway but knocks out spacer acquisition activity. We chose to further analyze mutant targets for two spacers (1.1 and 1.6) that displayed substantially different rates of plasmid loss in our initial assay (Figure 1C). Plasmid loss for these targets is similar in the Δhns and ΔhnsΔcas1 strains, confirming that the observed CRISPR interference is independent of new spacer acquisition and is directed by the original spacer (Figure 1D).
It is possible that differences in the amounts of individual crRNAs in the cell may affect the observed variations in plasmid loss in our in vivo assay. To address this possibility, we next examined whether the differences in CRISPR interference observed in vivo for spacer 1.1 and 1.6 mutant targets could be recapitulated in vitro. To test this, we purified Cascade bearing a crRNA with either spacer 1.1 or 1.6 and performed Cascade-mediated Cas3 cleavage assays for bona fide and seed position 1 mutant targets (Figure 1E and Supplementary Figure S2A). For each target, cleavage was measured at varied Cascade (5, 50 and 200 nM) or Cas3 (200 and 1000 nM) concentrations (Figure 1E and Supplementary Figure S2A). We observe similar cleavage activity for both bona fide targets and the spacer 1.6 target with an rT-dG mismatch, even at low Cascade concentration (5 nM), suggesting that Cascade affinity is not perturbed substantially for this mutant target (Figure 1E). In contrast, the spacer 1.1 rC-dA mismatch target plasmid appears to be completely intact at both 5 and 50 nM Cascade and is only cleaved at the higher concentration (200 nM) regardless of Cas3 concentration, suggesting that Cascade has a greatly reduced affinity for this target (Figure 1E and Supplementary Figure S2A). Accordingly, in the E. coli X019 strain, in which the cellular concentrations of Cascade bearing spacer 1.1 is increased by deleting all spacers except for 1.1 and 2.1 (Supplementary Figure S2B), CRISPR interference against the mutant spacer 1.1 target is increased substantially in comparison to BW25113 Δhns (Supplementary Figure S2C). Priming does not increase for the X019 strain (Supplementary Figure S2D), suggesting that the increased rate of interference may preclude a concurrent increase in spacer acquisition. Together, these data indicate that defects in Cascade binding can inhibit CRISPR interference, as has been previously proposed (31), but that seed position 1 mutants do not significantly disrupt Cascade binding for all spacer sequences.
Previous reports have indicated that single point mutations throughout the seed (positions 1–5, 7 and 8 of the protospacer) inhibit Cascade-target binding and direct interference (31,36). To determine whether the mismatch tolerance we observed at seed position 1 is position-specific, we tested the effect of single mutations at each position of the seed region for spacers 1.1 and 1.6 on plasmid loss and spacer acquisition. For all targets, we observe >90% plasmid loss, and several mutant targets were lost in all colonies tested (Supplementary Figure S2E). Spacer acquisition for all mutant targets was substantially lower than plasmid loss (Supplementary Figure S2E), although we observed overall higher rates of priming for spacer 1.1 targets than for spacer 1.6. Together, these results indicate that single seed mutations are not sufficient to completely block CRISPR interference for these spacers, and that the efficiency of priming may be dependent on spacer sequence.
Bona fide targets can promote spacer acquisition through priming
Based on our initial results, we found that spacer acquisition could occur for all seed mutations, even those that do not significantly inhibit direct interference. These results indicate that bona fide protospacers may also promote primed spacer acquisition. To test this, we measured the rate of spacer acquisition of bona fide targets for spacers 1.1 and 1.6 in E. coli BW25113 Δhns, which can acquire spacers through naïve or primed acquisition, and E. coli BW25113 ΔhnsΔcse1, which can only acquire spacers through naïve acquisition, although this acquisition is very rare and unlikely to be observed over the timespan of our experiment (28). Interestingly, we observe spacer acquisition for both bona fide target plasmids in the Δhns strain (10–25% of colonies tested) (Figure 1F) but no spacer acquisition in the ΔhnsΔcse1 strain (not shown). These data indicate that spacers were acquired through priming against the bona fide targets in the Δhns strain, and not through naïve acquisition. It is possible that the plasmid may develop escape mutations over the course of the experiment, and that these mutant targets could lead to the observed priming. However, we observe reproducible variability in the amount of priming for the two targets (Figure 1F), indicating that priming is not caused by random mutations but is instead directed from the bona fide targets. Together with our seed mutation experiments, these results reveal that targets that can undergo direct interference can also promote priming.
High-throughput screen for PAM and seed mutation tolerance
We next wanted to establish a high-throughput system to study spacer sequence-specific tolerance of seed or PAM mutations by the CRISPR interference machinery. In order to study a large number of mutant sequences, we developed a high-throughput method to detect CRISPR-dependent loss of a plasmid expressing green fluorescence protein (GFP) using flow cytometry (Supplementary Figure S3). Based on this system, we created a novel high-throughput screening method to determine the effects of both seed and PAM mutations on priming and direct interference for multiple spacer sequences (Figure 2A–C). Our screen utilizes two E. coli strains, X019 Δcas1 and X019, to distinguish between targets that can be lost through direct interference and targets that can be lost through both direct interference and priming, respectively. In comparison to the control strain X019 Δcse1, libraries of PAM and seed mutants grown in X019 Δcas1 and X019 will be depleted of sequences that are functional, while non-functional sequences may be unchanged or enriched.
Target libraries for spacer 1.1 and 2.1 were created by randomizing the PAM or the seed (Figure 2A). A completely degenerate library was created for the PAM (64 possible sequences). To ensure complete coverage of seed mutations (16,384 possible sequences), we created a limited degenerate library in which positions 1–5, 7–8 of the protospacer had two possible sequences, either the correct sequence or a mismatch, resulting in 128 possible sequences (Figure 2A). For all libraries, we performed a first round selection process by transforming and growing the libraries in the control X019 Δcse1 strain, and the two experimental strains, X019 Δcas1 and X019 (Figure 2B). Following this selection process, libraries were extracted from each strain and were then subjected to a second round of CRISPR interference in E. coli X019, to verify the success of the first round selection (Figure 2C).
For both plasmid loss and spacer acquisition (Figure 2D-E), there is no difference between the original libraries and libraries extracted from X019 Δcse1, which indicates that plasmid distribution did not change through non-CRISPR-related mechanisms and any changes in the X019 and X019 Δcas1 libraries resulted from the CRISPR–Cas immune system. Interestingly, differences in the rate of plasmid loss between the X019 Δcas1 and X019 libraries are larger for the PAM mutant libraries than the seed mutant libraries (Figure 2D), suggesting that there are several PAM mutations that can block interference but still enable priming, while seed mutations that promote priming are more likely to also be tolerated for direct interference.
As expected, GFP- cells grown with the X019 Δcas1 libraries exhibited high rates of spacer acquisition, similar to those observed for the original DH5α and X019 Δcse1 libraries that contain all functional sequences (Figure 2E). This indicates that the X019 Δcas1 libraries still contain sequences that can promote priming. In contrast, GFP- cells from the X019 library had very low rates of spacer acquisition (Figure 2E), indicating that the majority of sequences that promote priming were lost during the first round of growth. Therefore, comparison of sequences depleted from the X019 Δcse1 and X019 libraries should distinguish which sequences can be lost through direct interference and which can only be lost through priming.
High-throughput sequencing of libraries
We next sequenced PCR amiplicons of the PAM–protospacer region for each library using MiSeq Illumina sequencing (Figure 2A-B). To determine the relative depletion or enrichment of sequences in each library, counts from the X019 Δcse1, X019 Δcas1 and X019 libraries were normalized to the original DH5α library (Figures 3 and 4, Supplementary Figures S4 and S6). As expected based on our second round analysis of the libraries, the original DH5α library and X019 Δcse1 libraries are highly similar, while both the X019 Δcas1 and X019 libraries have many sequences that are highly depleted. In addition, the X019 Δcas1 and X019 libraries have many sequences that are enriched relative to the original libraries, as is expected given the depletion of several sequences. Several highly enriched sequences were also observed, especially for the seed mutant libraries. This enrichment appears to be sequence specific, as it occurred consistently across three biological replicates (Supplementary Figures S4 and S6). These sequences may have had a higher transformation efficiency in the X019 or X019 Δcas1 strains, or they may have been relatively stabilized in one of the strains during growth.
For spacer 2.1 we created an additional seed library containing a mismatch at position 6 of the protospacer (hereafter called spacer 2.1 6MM) (Supplementary Figure S4A, D, G, J). This position has been shown previously to have no effect on interference, as the crRNA of Cascade does not base pair with every sixth nucleotide of the protospacer target strand (23,36). Consistently, this library has similar depletion profiles to the seed library with the correct sequence at position 6 (Figure 3C, Supplementary Figure S4A). Differences between the two libraries may be due to variations in the timing of growth cycles for the two libraries. The overall similarity between the datasets indicates that the high-throughput method presented here is largely reproducible.
One or two seed mutations allow CRISPR activity
Analysis of seed sequence depletion for the spacer 1.1 and 2.1 libraries reveals that almost all sequences with one or two mismatches are functional (Figure 3, Supplementary Figure S4A), while three or more mismatches in the seed render the CRISPR immune system nearly completely inactive (Supplementary Figure S4B–J). As predicted based on the second round growth experiments with the seed mutant libraries (Figure 2D and E), several functional seed mutant sequences are highly depleted in both the X019 and X019 Δcas1 strain, indicating that many seed mismatches that promote priming do not completely block direct interference. For both spacers, a single seed mutation does not block interference, with two exceptions: position 1 for spacer 1.1, as observed in our initial experiments (Figures 1A and 3B); and position 4 for spacer 2.1, which we confirmed in an individual assay (Figure 3C, Supplementary Figures S4A, S5). These defects are compounded by the addition of another seed mismatch, as very little depletion is observed in either X019 or X019 Δcas1 libraries for sequences with two mismatches where one of the mismatches is at position 1 for spacer 1.1 or at position 4 for spacer 2.1 (Figure 3B and C, Supplementary Figure S4A). Combined with our in vitro studies of spacer 1.1 (Figure 1E, Supplementary Figure S2A), this result implies that single seed mismatches that inhibit direct interference reduce the affinity of Cascade for the target, and that a second mismatch in the seed blocks Cascade binding completely, such that even priming is not observed.
In contrast, combinations of seed mutations that do not block interference individually generally allow some degree of direct interference and often promote efficient priming, based on abundance of these sequences in X019 Δcas1 and X019, respectively (Figure 3B and C). Notably, the spacer 1.1 target with mutations at positions 5 and 7 is highly depleted in both strains, but the spacer 2.1 target with mismatches at the same positions is stable in both strains (Figure 3B-C). Similarly, the targets with mutations at positions 2 and 8 are depleted from the spacer 2.1 libraries but not the spacer 1.1 libraries (Figure 3B and C). These data suggest that up to two seed mismatches can be tolerated for direct interference, and provide further evidence that the determinants for position-specific mismatch tolerance is dependent on spacer sequence.
PAM tolerance is highly dependent on spacer sequence
For the PAM libraries, the counts for X019 Δcas1 and X019 relative to DH5α were plotted as a scatter plot, allowing for direct comparison of sequences that promote both direct interference and priming (X019) versus sequences that only promote direct interference (X109 Δcas1) (Figure 4A and B). Based on their relative amount of CRISPR activity, PAMs were divided into four groups (Figure 4A-D). Group A contains bona fide PAM sequences that are highly depleted in both strains. Group B contains PAMs that do not fully block direct interference, resulting in some depletion in X019 Δcas1 (<0.9 relative to DH5α reference library), but not as significantly as for X019. Group C contains sequences that block direct interference but promote priming, resulting in depletion in X019 but no change in X019 Δcas1 (>0.9 relative to DH5α reference library). Group D contains sequences that are stable in both strains (>0.9 relative to DH5α reference library). Notably, sequences within groups A-C prefer adenine residues at the -3 and -2 positions and a guanine residue at the -1 position (Figure 4C and D). In contrast, PAMs with cytosine residues at positions -3 or -2 are highly represented in group D, indicating that a C at either position disrupts CRISPR–Cas activity (Figure 4C and D).
In both libraries, group A contains five previously identified PAMs that can promote interference (AAG, AGG, ATG, GAG and TAG) located near the origin of the plot, indicating that targets with these PAM sequences are lost in both strains through direct interference as expected (Figure 4A-D and Supplementary Figure S6) (26,36). Surprisingly, for spacer 1.1, there are three additional PAM sequences (AAA, AAC and ATA) located near the origin of the plot, suggesting that spacer 1.1 can tolerate eight different PAM sequences for interference (Figure 4A, C and Supplementary Figure S6A). In individual assays, all three spacer 1.1 targets can be lost through direct interference in X019 Δcas1 (Figure 4E), and the AAA PAM target displays the accompanying low rate of spacer acquisition observed for bona fide targets (Figures 1F and 4E). Strikingly, for spacer 2.1, the AAA and ATA targets are relatively stable in X019 Δcas1 but promote high efficiency priming (Figure 4E), suggesting that, in contrast to spacer 1.1, direct interference is strongly inhibited for these targets. Thus, similar to seed mutations, spacer sequence appears to dictate whether mutant PAM sequences can be tolerated for direct interference or mainly promote priming.
We additionally observed several PAM sequences that are functional for one spacer but have no activity for the other spacer. For example, several Group C PAMs (ATT and TAC for spacer 1.1, AGA, AGC and CTG for spacer 2.1) are nonfunctional for the other spacer (Figure 4A–D, Supplementary Figure S6). To validate this observation, we created target plasmids for both spacers containing an AGA PAM and tested their CRISPR activity in individual assays (Figure 4E). Consistent with the high-throughput experiment, we observe no CRISPR activity against the spacer 1.1 target containing an AGA PAM (Figure 4E). In contrast, spacer 2.1 can promote efficient priming against targets containing an AGA PAM, although direct interference in X019 Δcas1 is completely blocked (Figure 4E). These data demonstrate that some potentially functional PAM sequences can be rendered completely nonfunctional based on the spacer sequence, and may explain differences in functional PAMs identified by high-throughput screens in ours and previous studies (36).
A self-signal protects the host genome from autopriming
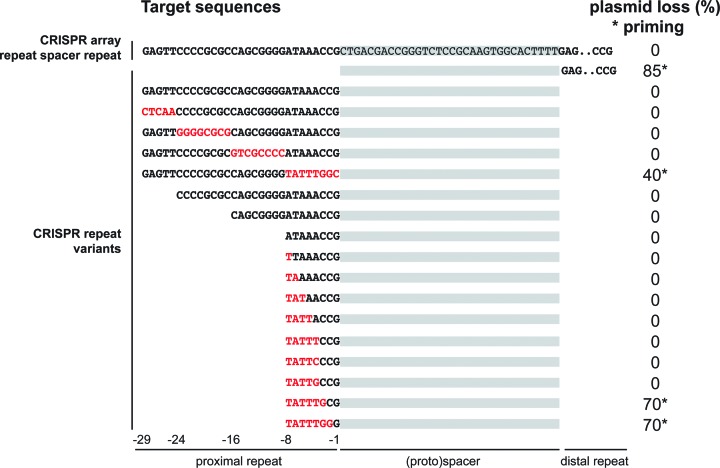
Our high-throughput results indicate that cytosine residues at the -3 and -2 positions of the PAM inhibit both interference and priming (Figure 4C and D). In the E. coli genome, the final three nucleotides of the repeat are CCG, suggesting that the repeat sequence may encode an inactive ‘PAM’ sequence to prevent Cascade binding to the spacer template strand. To investigate if repeat-derived nucleotides at PAM positions indeed protect the CRISPR array from suicidal autopriming, we constructed a CRISPR-like target plasmid containing a non-transcribed repeat – spacer – repeat sequence (Figure 5). This plasmid was introduced in E. coli cells that contained the same spacer in the genome, and checked for stable maintenance and priming in non-selective media. As expected, the CRISPR-like target plasmid showed no priming. However, when we deleted the proximal repeat of the target plasmid, we regained priming behavior of the plasmid (Figure 5). We then constructed a series of proximal CRISPR repeat variants, including blocks of mutations and truncations from the 5′ end of the repeat. Assessing their priming behavior revealed that only when repeat nucleotides at positions -3 to -1 (i.e. CCG) were altered, the plasmid was subject to priming. This means that the presence of repeat nucleotides directly adjacent to the spacer protects CRISPR arrays from self-priming and therefore from autoimmunity.
Figure 5.
Repeat nucleotides at the PAM (i.e. CCG at position -3, -2, -1) protect the CRISPR array from priming. The sequence of the distal repeat is shown in truncated form in the upper two sequences. The sequence of the spacer is shaded in gray in the top sequence. Red sequences indicate mutated nucleotides.
Differential interference against AGA PAM targets based on spacer sequence
Our results indicate that spacer sequence can greatly influence the activity of the interference machinery against targets with altered PAM or seed sequences. For example, our observation that an AGA PAM completely blocks CRISPR activity against spacer 1.1 targets suggests that Cascade binding is blocked, while spacer 2.1 AGA PAM targets may still be bound by Cascade to promote priming, but not degraded by Cas3 to block interference (Figure 4E). To test this, we purified Cascade with crRNAs bearing spacer 1.1 or spacer 2.1, and tested binding of their respective targets containing AGA PAMs using electrophoretic mobility shift assays (EMSA) (Figure 6A–B). Spacer 2.1 Cascade can still bind the AGA PAM target and there is little difference between the affinities for the AGA PAM target and the AAG PAM target (Figure 6B). In contrast, for spacer 1.1 Cascade, AGA PAM target binding is only observed at very high concentrations of Cascade (1–2 μM), and is likely due to non-specific DNA binding that is often observed at high concentrations of Cascade (Figure 6A) (21,26,30). These data reveal that spacer sequence can have a major impact on the ability of Cascade to bind targets with incorrect PAM sequences.
After confirming that Cascade can bind a spacer 2.1 target with an AGA PAM, we next investigated whether Cas3 can degrade this Cascade-bound target in vitro. Strikingly, the spacer 2.1 AGA PAM target is not cleaved at any concentration of Cas3, even up to 2 μM (Figure 6C). These results are consistent with the complete lack of direct interference observed in vivo against the AGA PAM target (Figure 4E). Overall, these in vitro results for a priming PAM are in sharp contrast to the seed mutant targets we have tested, in particular the spacer 1.1 position 1 mutant target, which, similar to the spacer 2.1 AGA PAM target, promotes priming but strongly inhibits direct interference (Figures 1C and 4E). For the PAM mutant target, Cascade binding affinity is not greatly affected, but Cascade-mediated Cas3 cleavage is completely blocked (Figure 6B and C). For the seed mutant target, inhibition of interference appears to be caused by a defect in Cascade binding, but Cas3 cleavage appears to be unaffected for Cascade-bound target (Figure 1E, Supplementary Figure S2A).
Cas3 can be recruited to, but does not cleave, a priming PAM target in vivo
We next wanted to determine whether the lack of Cascade-mediated degradation for the spacer 2.1 AGA target occurs because Cas3 cannot be recruited to Cascade bound to this target, or instead because Cas3 can be recruited but cannot initiate degradation. To investigate this, we used bimolecular fluorescence complementation (BiFC) experiments to monitor the interaction between Cascade and Cas3 in vivo (Figure 6D–F). Previously, BiFC has been used to show that Cascade cannot interact with Cas3 in the absence of an invading DNA sequence containing a correct PAM and protospacer (26). Cas3 was only recruited to Cascade by the Cse1 subunit in the presence of a target DNA. We used a similar experimental design, in which we fused Cse1 with N-terminal Venus and Cas3 with C-terminal Venus in a single expression vector (pACYC-BiFC). Each fragment of Venus is non-fluorescent, but fluorescence is reconstituted upon interaction between Cas3 and Cse1, which brings the two Venus fragments into proximity.
For BiFC experiments, we co-transformed pACYC-BiFC with various target plasmids in E. coli X030 (hns, cse1 and cas3 genes deleted). E. coli containing a non-target plasmid (empty pCDF-1b) were non-fluorescent, indicating that a target plasmid is necessary for Cas3 and Cse1 interaction (Figure 6D). As expected, fluorescent signal is observable in E. coli carrying the bona fide target plasmid, indicating that Cascade has bound to the target and recruited Cas3 (Figure 6E). Interestingly, the AGA target plasmid also induces fluorescence at similar levels to those observed with the AAG target (Figure 6F). These results indicate that Cas3 can still be recruited to the Cascade-AGA target complex, but in a manner that does not lead to target degradation.
Naïve spacer acquisition leads to rapid primed adaptation
During naïve adaptation, spacers are acquired at a relatively low rate, and many spacers are acquired from protospacer locations containing PAMs that differ from the canonical AAG sequence (11,12,29,56,57). We wondered whether functional PAMs are enriched for sequences acquired through naïve acquisition. We compared PAM sequences from a previously published set of spacers acquired through naïve adaptation with functional PAMs identified in this study (Figure 7A and Supplementary Figure S7A) (29). In the previous study, spacers could be acquired against both the genomic DNA and a Cas1–Cas2 expression plasmid. Although functional PAMs are ∼1.8-fold less abundant than nonfunctional PAMs in both source DNAs, spacers derived from sequences with functional PAMs are acquired ∼2.5-fold more frequently (Figure 7A). Interestingly, functional PAMs are nearly equally divided between AAG and non-AAG sequences among the acquired spacers. We also note that some PAM sequences, especially the highly enriched ACG, have been identified in another study as priming PAMs, but were not identified in our high-throughput study potentially due to differences in spacer sequences used in the two studies (36).
Given the high proportion of functional PAMs observed in the previous data set, we hypothesized that naïve acquisition should rapidly trigger a primed adaptation response to previously unencountered invader DNA. To test this hypothesis, we simulated three adaptation scenarios: (i) invasion by a previously unencountered DNA (empty plasmid) with only naïve adaptation (cse1 deletion strain); (ii) invasion by a previously encountered DNA (spacer 2.1 AGA target plasmid) with a primed adaptation response; (iii) invasion by a previously unencountered DNA with both naïve and primed adaptation responses (Figure 7B, Supplementary Figure S7B). In all three scenarios, we increased the rate of naïve acquisition through constitutive overexpression of Cas1 and Cas2 using an expression plasmid (29).
As expected, naïve adaptation is significantly slower than priming, based on the number and amount of spacers acquired in the scenario 1 versus scenario 2 cultures, respectively (Figure 7B and Supplementary Figure S7B). Scenario 3 mimics an actual naïve infection event, in which both adaptation and interference activities are functional. Initially, the only CRISPR–Cas response against the previously unencountered DNA is naïve adaptation, and accordingly spacer acquisition after one cycle of growth is very low (Supplementary Figure S7B). The slow naïve response observed in the scenario 1 culture is exacerbated in the scenario 3 culture, as spacers acquired against the genomic DNA or Cas1–Cas2 expression plasmid are not permitted when the interference machinery is intact. However, over time, adaptation in the scenario 3 culture overtakes the rate of the scenario 1 culture, based on the amount of spacer acquisition observed after a second cycle of growth (Figure 7B and Supplementary Figure S7B). When priming is active, bacteria can acquire multiple spacers more rapidly, as evidenced by the statistically significant increase in product for 2 and 3 acquired spacers in the scenario 3 analysis versus scenario 1 (Figure 7B). Together, these results suggest that preferential uptake of spacers against targets with any type of functional PAM during naïve adaptation may lead to a rapid priming response to infection.
DISCUSSION
In this study, we have reexamined the sequence requirements for CRISPR interference and primed spacer acquisition in the Type I-E CRISPR–Cas immune system. Our results reveal that single point mutations within the seed sequence of target protospacers do not completely block CRISPR interference, and in some cases are highly tolerated. Similarly, some non-canonical PAM sequences can be tolerated for direct interference, especially those with adenine residues at the -3 or -2 position or a guanine residue at the -1 position. Suprisingly, the crRNA spacer sequence has a significant effect on the ability of the interference machinery to recognize protospacers with seed mutations or non-canonical PAM sequences. Intriguingly, this result suggests that spacers that are highly tolerant of mutations throughout the seed may be more effective for CRISPR–Cas immunity, as they may provide a broader range of immunity against potential escape mutants or related invaders. In the future, it will be interesting to evaluate whether bacteria preferentially incorporate spacers with higher tolerance of mutated seed or PAM sequences. At present, the determinants for these spacer sequence-specific effects are unclear and a more systematic study of spacer/target sequence variations will be necessary to decipher the code for mutational tolerance by CRISPR–Cas systems.
Our results differ from previous studies, which have shown that single point mutations in the protospacer seed or PAM sequences block the CRISPR interference pathway (31,36,58). In many cases, these mutations lead to primed spacer acquisition (11,28), making it difficult to deconvolute indirect interference driven by newly acquired spacers from direct interference driven by the original spacer. By using a cas1 deletion strain, we decoupled direct interference from priming, definitively demonstrating the extent to which seed and PAM mutations can be tolerated for interference. In addition, differences between ours and previous studies may be due to variations in experimental design between studies. For example, phage-infectivity and plasmid-based assays have previously been observed to yield differing results when assessing CRISPR interference (58,59). Seed or PAM mutations that are tolerated in plasmid-based assays may sufficiently block CRISPR interference allowing for phage escape at high multiplicities of infection, including conditions found in the environment. Going forward, it will be important to assess how mutational tolerance may vary based on the outcome of invasion by different types of mobile genetic elements.
Self-sequence avoidance in the Type I-E system
To prevent autoimmunity, CRISPR–Cas immune systems must distinguish non-self protospacer sequences from identical self-spacer sequences present in the CRISPR array. Type III-A systems accomplish this self vs. non-self recognition based on differential complementarity between the crRNA and sequences flanking the target (60). Types I and II CRISPR–Cas immune systems utilize PAM sequences to differentiate targets from non-targets (17,30,58), but a mechanism for avoidance of self sequences has not been previously identified. Our high-throughput PAM mutant screen revealed that cytosine residues at the -3 and -2 position of the PAM prevents both interference and priming. Consistent with this observation, we found that the final 3 nucleotides of the preceding repeat (CCG) prevent priming against a spacer in the context of a CRISPR array. The position of this 3-nucleotide protective element is identical to where PAMs reside in target DNA and indicates that PAM recognition is actually more sophisticated than previously thought (30,58). Apart from authenticating bona fide targets for direct interference, many PAM sequences are eligible for priming (34,56, this work). The repeat-PAM, however, abolishes both pathways by providing a genuine self-signal, and this leaves the CRISPR array exempt from detrimental autopriming.
Mechanistic insights into CRISPR–Cas immunity
Overall, the large number of functional PAMs identified in our study, including several noncanonical PAMs that allow some degree of direct interference, suggests that Cascade-target binding requirements are less stringent than previously thought. We found that Cascade binding affinity for targets with noncanonical PAMs is significantly affected by the spacer sequence, and that some targets with priming PAMs can be bound with high affinity. When Cascade binds priming PAM targets, Cas3 is recruited but does not cleave the DNA. Similarly, a recent single-molecule study of the Streptococcus thermophilus Type I-E Cascade found that low-affinity, non-permissive PAM targets with induced R-loops were cleaved at a very low rate by Cas3 (34), and bulk biochemical studies have indicated that targets with mutated PAM sequences cannot be cleaved by Cas3 (25,27,37). Notably, for high-affinity priming PAM targets, PAM discrimination apparently occurs following target binding, but prior to target cleavage. Thus, targets with noncanonical PAMs may be bound with high affinity by Cascade, but a second PAM authentication step prevents interference from occurring.
The molecular basis for PAM authentication during target cleavage is currently unknown. It is possible that interactions between Cascade or Cas3 and the PAM sequence may differ for interference and priming PAMs, resulting in alternative conformations that lead to the two different immune reactions. This idea is consistent with recent single-molecule FRET studies of Cascade-target binding, which revealed that Cascade engages priming targets in a noncanonical binding mode (37). However, the mode of binding observed in that study was relatively short-lived, and it is unclear whether the high-affinity Cascade-target interaction we observe for the spacer 2.1 AGA target is analogous to this binding event. Our identification of a high-affinity priming PAM target should enable further study into the conformations that promote priming and the mechanism of PAM authentication during target cleavage.
The mechanisms of interference and priming appear to exist in a dynamic equilibrium in which targets that block interference promote priming, and targets that allow interference inhibit priming. In general, target sequences that are tolerated for direct interference lead to inefficient priming, including bona fide targets and several of the seed and PAM mutants tested in our study. It is possible that this inefficiency is caused by loss of the target plasmid through rapid target degradation through CRISPR interference, thus limiting the amount of invader DNA from which to acquire new spacers. In this case, the equilibrium between interference and priming favors interference and disfavors priming, whereas mutations that block interference push the equilibrium toward priming. Consistently, the short-lived noncanonical binding mode observed by single-molecule FRET for priming targets was also observed for bona fide targets (37), suggesting that Cascade can sample an alternative binding mode that promotes priming for any type of target. The noncanonical binding mode may occur relatively rarely for high-affinity targets that can adopt the interference binding mode, including bona fide targets and targets with seed or PAM mutations that are tolerated for direct interference.
Priming is a major mechanism for adaptation
Primed spacer acquisition allows bacteria to mount a rapid defense against infection, even in the presence of PAM or protospacer mutations that could allow an invader DNA to escape CRISPR interference. In addition, the promiscuity of Cascade in recognizing priming PAMs may have implications during the early stages of CRISPR–Cas adaptation. We showed that naïve acquisition can lead to a rapid priming response, suggesting that previously identified ‘incorrect’ PAMs of sequences acquired during naïve adaptation are likely actually functional PAMs that promote priming (11,12,29,56,57). These data suggest that the Cas1–Cas2 acquisition complex, like Cascade, may recognize a broad range of PAM sequences that can promote either interference or priming in the absence of Cascade and Cas3. Notably, different adaptation specificities have been observed for the E. coli K12 and O157:H7 Type I-E variants, which preferentially acquire protospacers with AAG and ATG PAMs, respectively (57). These observations suggest that the closely related Cas1-Cas2 homologs (85% identity) in these Type I-E variants bind alternative functional PAMs with variable affinities, resulting in different PAM preferences. A recent crystal structure of the E. coli K12 Cas1-Cas2 complex bound to a PAM-containing protospacer reveals the structural basis for preferential AAG PAM recognition, and will be important for guiding future studies of functional PAM selectivity (62).
Together with previous results, our findings suggest that naïve adaptation is a mechanism for acquiring spacers for both interference and priming, while primed adaptation is a mechanism for acquiring only interference spacers (11,56). This two-tiered strategy may be of particular importance for adaptation against invaders with depleted canonical PAM sequences, as has been observed for several bacteriophages with hosts containing CRISPR–Cas systems (63). During initial infection, a naïve adaptation strategy in which only protospacers with canonical PAM sequences were acquired would severely limit the ability of the host to mount an effective defense. Instead, the CRISPR–Cas system overcomes this limitation by initially acquiring any sequence adjacent to a functional PAM during naïve adaptation, then honing the system for interference through priming. Priming may, in fact, be the major mechanism for adaptation in Type I systems. In the Haloarcula hispanica Type I-B system, priming is strictly required for adaptation and naïve adaptation has not been observed (64). Similar to our observations, this system has relaxed stringency for priming PAM sequences, maximizing the adaptation capacity during an invasion event (61). The Pectobacterium atrosepticum Type I-F system contains a Cas2-Cas3 fusion, physically linking the naïve and primed acquisition machinery and resulting in extremely robust priming in this organism (65). It is possible that the Type I-E system also uses priming as the main mechanism for adaptation, and that naïve acquisition is simply a means to enable priming.
Model for priming and interference dynamics in E. coli
Overall, our results suggest a model for adaptation during the initial encounter of an invader DNA (Figure 7C and D). The Cas1-Cas2 complex initially acquires new spacers from any region of a replicating invader (66), preferentially incorporating sequences that are adjacent to functional PAMs. Cascade bearing the resulting crRNAs can bind to these targets and recruit Cas3. Prior to target degradation, the PAM is authenticated through an unknown mechanism, and this leads to either interference (Figure 7C) or priming (Figure 7D). Depending on the PAM sequence, the frequency of these two responses will vary, with bona fide PAM targets leading to a majority of interference and few priming events (Figure 7C) and priming PAM targets leading to rapid spacer acquisition and infrequent interference events (Figure 7D). Thus, the initial spacers acquired during naïve acquisition allow the CRISPR–Cas system to simultaneously mount two defensive responses to infection.
Supplementary Material
Acknowledgments
We thank Shawn Rigby and Christine Deal of the Iowa State Flow Cytometry Facility, Michael Baker of the Iowa State DNA Facility, and Margie Carter of the Iowa State Confocal and Multiphoton Facility for assistance with data collection. We thank Hayun Lee and other members of the Sashital lab for helpful discussions and experimental suggestions. The sfGFP template was generously donated by Brenden Hawk and Yeon-Kyun Shin, pKD46 and pCP20 plasmids were obtained from The Coli Genetic Stock Center (CGSC), and the mVenus-pBAD vector was a gift from Michael Davidson (Addgene plasmid # 54845). Keio collection strains were generously donated by Thomas Bobik and CGSC.
Author contributions: X.C.Y. and D.G.S. conceived the project, and X.C.Y. performed all experiments with the exception of the autopriming experiment. X.C.Y., D.G.S., A.S.S. and A.J.S. performed high-throughput sequencing data analysis and computational analysis of the Yosef et al. naïve acquisition dataset. K.S. and S.J.J.B. conceived the autopriming experiments and O.M. performed autopriming experiments. X.C.Y. and D.G.S. wrote the manuscript with input from A.S.S., A.J.S., K.S. and S.J.J.B.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Startup funds (to D.G.S.) from Iowa State University College of Liberal Arts and Sciences and the Roy J. Carver Charitable Trust; National Institutes of Health [GM10407]; Russian Science Foundation [14-14-00988]; Ministry of Education and Science of Russian Federation [14.B25.31.0004 to K.S.]; Nertherlands Organisation for Scientific Research NWO VIDI [864.11.005]; European Research Council Starting [639707 to S.J.J.B.]. Funding for open access charge: Start-up funds to Dipali Sashital from the Iowa State University College of Liberal Arts and Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1.Barrangou R., Marraffini L.A. CRISPR–Cas systems: Prokaryotes upgrade to adaptive immunity. Mol. Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Oost J., Westra E.R., Jackson R.N., Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat. Rev. Microbiol. 2014;12:479–92. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heler R., Marraffini L.A., Bikard D. Adapting to new threats: The generation of memory by CRISPR–Cas immune systems. Mol. Microbiol. 2014;93:1–9. doi: 10.1111/mmi.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiro R., Goren M.G., Yosef I., Qimron U. CRISPR adaptation in Escherichia coli subtypeI-E system. Biochem. Soc. Trans. 2013;41:1412–1415. doi: 10.1042/BST20130109. [DOI] [PubMed] [Google Scholar]
- 5.Hochstrasser M.L., Doudna J.A. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem. Sci. 2015;40:58–66. doi: 10.1016/j.tibs.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Charpentier E., Richter H., van der Oost J., White M.F. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR–Cas adaptive immunity. FEMS Microbiol. Rev. 2015;39:428–441. doi: 10.1093/femsre/fuv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson R.N., Wiedenheft B. A conserved structural chassis for mounting versatile CRISPR RNA-guided immune responses. Mol. Cell. 2015;58:722–728. doi: 10.1016/j.molcel.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plagens A., Richter H., Charpentier E., Randau L. DNA and RNA interference mechanisms by CRISPR–Cas surveillance complexes. FEMS Microbiol. Rev. 2015;39:442–463. doi: 10.1093/femsre/fuv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J.J., Charpentier E., Haft D.H., et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yosef I., Goren M.G., Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datsenko K.A., Pougach K., Tikhonov A., Wanner B.L., Severinov K., Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 12.Nuñez J.K., Kranzusch P.J., Noeske J., Wright A.V., Davies C.W., Doudna J.a. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat. Struct. Mol. Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nunez J.K., Lee A.S.Y., Engelman A., Doudna J.A. Integrase-mediated spacer acquisition during CRISPR–Cas adaptive immunity. Nature. 2015;519:193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brouns S.J.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J.H., Snijders A.P.L., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P. RNA-Guided RNA Cleavage by a CRISPR RNA-Cas Protein Complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatoum-Aslan A., Samai P., Maniv I., Jiang W., Marraffini L.A. A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J. Biol. Chem. 2013;288:27888–27897. doi: 10.1074/jbc.M113.499244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., et al. Cpf1 Is a single RNA-guided endonuclease of a class 2 CRISPR–Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K., et al. Discovery and functional characterization of diverse class 2 CRISPR–Cas systems. Mol. Cell. 2015 doi: 10.1016/j.molcel.2015.10.008. doi:10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jore M.M., Lundgren M., van Duijn E., Bultema J.B., Westra E.R., Waghmare S.P., Wiedenheft B., Pul U., Wurm R., Wagner R., et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 22.Jackson R.N., Golden S.M., van Erp P.B.G., Carter J., Westra E.R., Brouns S.J.J., van der Oost J., Terwilliger T.C., Read R.J., Wiedenheft B. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014;345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulepati S., Héroux A., Bailey S. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–1484. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao H., Sheng G., Wang J., Wang M., Bunkoczi G., Gong W., Wei Z., Wang Y. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014;515:147–150. doi: 10.1038/nature13733. [DOI] [PubMed] [Google Scholar]
- 25.Mulepati S., Bailey S. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA Target. J. Biol. Chem. 2013;288:22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westra E.R., van Erp P.B.G., Künne T., Wong S.P., Staals R.H.J., Seegers C.L.C., Bollen S., Jore M.M., Semenova E., Severinov K., et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hochstrasser M.L., Taylor D.W., Bhat P., Guegler C.K., Sternberg S.H., Nogales E., Doudna J.a. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc. Natl. Acad. Sci. U.S.A. 2014;111:6618–6623. doi: 10.1073/pnas.1405079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swarts D.C., Mosterd C., van Passel M.W.J., Brouns S.J.J. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yosef I., Shitrit D., Goren M.G., Burstein D., Pupko T., Qimron U. DNA motifs determining the efficiency of adaptation into the Escherichia coli CRISPR array. Proc. Natl. Acad. Sci. U.S.A. 2013;110:14396–14401. doi: 10.1073/pnas.1300108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sashital D.G., Wiedenheft B., Doudna J.A. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol. Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Semenova E., Jore M.M., Datsenko K.A., Semenova A., Westra E.R., Wanner B., van der Oost J., Brouns S.J.J., Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rollins M.F., Schuman J.T., Paulus K., Bukhari H.S.T., Wiedenheft B. Mechanism of foreign DNA recognition by a CRISPR RNA-guided surveillance complex from Pseudomonas aeruginosa. Nucleic Acids Res. 2015;43:2216–2222. doi: 10.1093/nar/gkv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutkauskas M., Sinkunas T., Songailiene I., Tikhomirova M.S., Siksnys V., Seidel R. Directional R-loop formation by the CRISPR–Cas surveillance complex cascade provides efficient off-target site rejection. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.067. doi:10.1016/j.celrep.2015.01.067. [DOI] [PubMed] [Google Scholar]
- 35.Szczelkun M.D., Tikhomirova M.S., Sinkunas T., Gasiunas G., Karvelis T., Pschera P., Siksnys V., Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl. Acad. Sci. U.S.A. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fineran P.C., Gerritzen M.J.H., Suárez-Diez M., Künne T., Boekhorst J., van Hijum S.a F.T., Staals R.H.J., Brouns S.J.J. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E1629–E1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blosser T.R., Loeff L., Westra E.R., Vlot M., Kunne T., Sobota M., Dekker C., Brouns S.J.J., Joo C. Two distinct DNA binding modes guide dual roles of a CRISPR–Cas protein complex. Mol. Cell. 2015;58:60–70. doi: 10.1016/j.molcel.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotech. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A., Liu D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotech. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu X., Scott D.A., Kriz A.J., Chiu A.C., Hsu P.D., Dadon D.B., Cheng A.W., Trevino A.E., Konermann S., Chen S., et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu Y., Foden J.a, Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR–Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cherepanov P.P., Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 46.de Boer H.A., Comstock L.J., Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc. Natl. Acad. Sci. U. S. A. 1983;80:21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Mey M., Maertens J., Lequeux G.J., Soetaert W.K., Vandamme E.J. Construction and model-based analysis of a promoter library for E. coli: an indispensable tool for metabolic engineering. BMC Biotechnol. 2007;7:34. doi: 10.1186/1472-6750-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keiler K.C., Waller P.R., Sauer R.T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 49.Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A. 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 50.Aronesty E. Comparison of sequencing utility programs. Open Bioinforma. J. 2013;7:1–8. [Google Scholar]
- 51.Quinlan A.R. BEDTools: the Swiss-Army tool for genome feature analysis. Curr. Protoc. Bioinformatics. 2014;47:11.12.1–11.12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westra E.R., Pul Ü., Heidrich N., Jore M.M., Lundgren M., Stratmann T., Wurm R., Raine A., Mescher M., Van Heereveld L., et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 2010;77:1380–1393. doi: 10.1111/j.1365-2958.2010.07315.x. [DOI] [PubMed] [Google Scholar]
- 53.Edgar R., Qimron U. The Escherichia coli CRISPR system protects from lambda lysogenization, lysogens, and prophage induction. J. Bacteriol. 2010;192:6291–6294. doi: 10.1128/JB.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pougach K., Semenova E., Bogdanova E., Datsenko K.A., Djordjevic M., Wanner B.L., Severinov K. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol. Microbiol. 2010;77:1367–1379. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pul Ü., Wurm R., Arslan Z., Geißen R., Hofmann N., Wagner R. Identification and characterization of E. coli CRISPR–Cas promoters and their silencing by H-NS. Mol. Microbiol. 2010;75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 56.Savitskaya E., Semenova E., Dedkov V., Metlitskaya A., Severinov K. High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA Biol. 2013;10:716–725. doi: 10.4161/rna.24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diez-Villasenor C., Guzman N.M., Almendros C., Garcia-Martinez J., Mojica F.J.M. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR–Cas I-E variants of Escherichia coli. RNA Biol. 2013;10:792–802. doi: 10.4161/rna.24023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Westra E.R., Semenova E., Datsenko K.A., Jackson R.N., Wiedenheft B., Severinov K., Brouns S.J.J. Type I-E CRISPR–Cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet. 2013;9:e1003742. doi: 10.1371/journal.pgen.1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Semenova E., Kuznedelov K., Datsenko K.A., Boudry P.M., Savitskaya E.E., Medvedeva S., Beloglazova N., Logacheva M., Yakunin A.F., Severinov K. The Cas6e ribonuclease is not required for interference and adaptation by the E. coli type I-E CRISPR–Cas system. Nucleic Acids Res. 2015;43:6049–6061. doi: 10.1093/nar/gkv546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marraffini L.A., Sontheimer E.J. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li M., Wang R., Xiang H. Haloarcula hispanica CRISPR authenticates PAM of a target sequence to prime discriminative adaptation. Nucleic Acids Res. 2014;42:7226–7235. doi: 10.1093/nar/gku389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J., Li J., Zhao H., Sheng G., Wang M., Yin M., Wang Y. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR–Cas systems. Cell. 2015 doi: 10.1016/j.cell.2015.10.008. doi:10.1016/j.cell.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 63.Kupczok A., Bollback J.P. Motif depletion in bacteriophages infecting hosts with CRISPR systems. BMC Genomics. 2014;15:663. doi: 10.1186/1471-2164-15-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li M., Wang R., Zhao D., Xiang H. Adaptation of the Haloarcula hispanica CRISPR–Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 2014;42:2483–2492. doi: 10.1093/nar/gkt1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Richter C., Dy R.L., McKenzie R.E., Watson B.N.J., Taylor C., Chang J.T., McNeil M.B., Staals R.H.J., Fineran P.C. Priming in the Type I-F CRISPR–Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014;42:8516–8526. doi: 10.1093/nar/gku527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Levy A., Goren M.G., Yosef I., Auster O., Manor M., Amitai G., Edgar R., Qimron U., Sorek R. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.