Abstract
CRISPR immunity depends on acquisition of fragments of foreign DNA into CRISPR arrays. For type I-E CRISPR–Cas systems two modes of spacer acquisition, naïve and primed adaptation, were described. Naïve adaptation requires just two most conserved Cas1 and Cas2 proteins; it leads to spacer acquisition from both foreign and bacterial DNA and results in multiple spacers incapable of immune response. Primed adaptation requires all Cas proteins and a CRISPR RNA recognizing a partially matching target. It leads to selective acquisition of spacers from DNA molecules recognized by priming CRISPR RNA, with most spacers capable of protecting the host. Here, we studied spacer acquisition by a type I-F CRISPR–Cas system. We observe both naïve and primed adaptation. Both processes require not just Cas1 and Cas2, but also intact Csy complex and CRISPR RNA. Primed adaptation shows a gradient of acquisition efficiency as a function of distance from the priming site and a strand bias that is consistent with existence of single-stranded adaption intermediates. The results provide new insights into the mechanism of spacer acquisition and illustrate surprising mechanistic diversity of related CRISPR–Cas systems.
INTRODUCTION
CRISPR–Cas systems defend prokaryotic cells from foreign genetic elements such as plasmids and bacteriophages (1,2). A CRISPR–Cas system is composed of a set of Cas proteins and small CRISPR RNAs (crRNAs) encoded by CRISPR loci. These loci consist of arrays of short repeats interspaced by unique ‘spacers’ segments that are often identical to ‘protospacer’ sequences found in phage and plasmid genomes. CRISPR loci are transcribed and processed into CRISPR-derived RNAs (crRNAs) that guide Cas proteins to complementary sequences found in invading genetic parasites (3–6). The lengths of CRISPR arrays can differ significantly in various organisms, from just a few to several hundreds of spacers. A set of spacers reflects cell's potential to mount a defense against genetic parasites with matching protospacers through a process named ‘CRISPR interference’. Complementary base paring between the crRNA-guide and a protospacer, assisted by Cas proteins, triggers degradation of the target (7–11). However, mutations at specific positions of the protospacer result in mismatches that decrease the binding affinity of crRNA-Cas proteins complex and render CRISPR defense inefficient (10–15). These mutations allow viruses to escape detection and productively infect the host (4,11–13).
Three mechanistically different CRISPR–Cas systems have been distinguished based on the presence of specific Cas proteins (16). In addition to base pairing between the crRNA-spacer and the DNA protospacer, target recognition by type I and type II CRISPR–Cas systems requires a protospacer adjacent motif (PAM) (8,9,12,14,17–19). Point mutations in the PAM render CRISPR defense inactive even when there is a perfect match between crRNA spacer and the protospacer (11–13,18).
Acquisition of new spacers into CRISPR loci is called adaptation (3). Spacer acquisition occurs in a polarized manner (at the end of the array closest to promoter) and leads to the synthesis of an additional repeat for every new spacer acquired. While the set of Cas proteins involved in target detection and destruction are diverse, the Cas1 and Cas2 proteins have been shown to be necessary and sufficient for naïve adaption in the type I-E systems (20,21). Cas1 and Cas2 are not required for CRISPR interference (22).
For type I-E CRISPR–Cas system from Escherichia coli, two modes of CRISPR adaptation have been described. Naïve (also called ‘non-primed’) adaptation requires only Cas1 and Cas2; it is generally biased toward mobile DNA, but also leads to acquisition of multiple spacers from host DNA (20). Many spacers acquired during naïve adaptation originate from protospacers without a consensus PAM and therefore do not support interference (20). Primed adaptation requires all Cas proteins and a crRNA recognizing a partially complementary spacer or a spacer with a non-consensus PAM (23). Primed adaptation leads to highly efficient and selective acquisition of spacers with consensus PAM from protospacers located in cis with respect to the priming protospacer (23,24).
In addition to E. coli type I-E CRISPR–Cas system, primed adaptation was described for a type I-B system from an archaeon Haloarcula hispanica (25), and a type I-F system from bacteria Pectobacterium atrosepticum (14). The existence of alternative, non-primed adaptation, was not demonstrated in these cases and in fact it was suggested that the H. hispanica adaptation is strictly dependent on priming (25). On the other hand, recent findings in type II systems suggest that non-primed adaptation in these systems requires the interference protein Cas9 to ensure that spacers are selected from protospacers with correct PAMs (26,27). In this work, we analyze the adaptation process by P. aeruginosa type I-F CRISPR–Cas system transplanted into a heterologous E. coli host. We demonstrate both modes of adaptation and show that in contrast to E. coli, both modes require, in addition to Cas1 and Cas2, intact Csy complex, an ortholog of the E. coli Cascade, and crRNA, which in the case non-primed adaptation does not have to match the target DNA.
EXPERIMENTAL PROCEDURES
Plasmid and strain construction
Escherichia coli strains used are listed in Supplementary Table S1. KD604, KD606, KD628 and KD675 were engineered from the BL21-AI strain using a procedure based on the use of the Red recombinase (28) and contain (KD604, KD606 and KD675) a minimized I-F subtype P. aeruginosa UCBPP-PA14 CRISPR array (two repeats and one spacer) and a 134 bp-long upstream leader region under the control of the T7 RNA polymerase promoter. KD628 contains just a leader (134 bp) and a single repeat. The sequences of KD604 and KD606 arrays spacers are, respectively, ACGCAGTTGCTGAGTGTGATCGATGCCATCAG and ACCGGACCTTCAATCGGCCCTTCGCTGATGGC. KD675 is the same as KD604 but also contains a protospacer with a mismatch at position +1 preceded by a functional GG PAM introduced in it's genome. E. coli ED1a strain with native I-F CRISPR–Cas system is described elsewhere (29) and was a kind gift from Dr. Erick Denamur.
Plasmids pCas (expressing cas1 and cas2–3) and pCsy (expressing csy1, csy2, csy3 and csy4) were described in (10). Mutations in selected cas or csy genes of, were introduced by site-specific mutagenesis with PfuUltra II Fusion HS DNA Polymerase (Agilent Technologies) using oligonucleotides containing desired mutations.
Plasmids pSPA and pSPAmut were generated by cloning double-stranded oligonucleotides containing perfectly matching or A1T mutant protospacer (harbors an A to T substitution at the first position of the seed) sequences and a consensus GG PAM into the EcoN I and Kpn I restriction sites of the pACYCDuet-1 vector. The pSED and pSEDmut plasmids were generated by cloning double-stranded oligonucleotides containing perfectly matching or T1A mutant protospacer sequences and consensus GG PAM in the EcoRV site of the pT7blue vector.
Anti-CRISPR genes were expressed using previously described constructs made in the pHERD30T vector (30), which is compatible with pCas and pCsy. The pHERD30T plasmid encodes gentamicin resistance and anti-CRISPR genes are under control of arabinose inducible promoter.
CRISPR interference and adaptation assays
The BL21-AI-based E. coli cells containing pCas and/or pCsy plasmids were grown at 37°C in LB medium in the presence of 1 mM arabinose and 1 mM IPTG and antibiotics required for maintenance of the plasmids (100 μg/ml of ampicillin for pCsy and 50 μg/ml of spectinomycin for pCas). When cultures reached OD600 0.6–0.8 they were processed to prepare electrocompetent cells using a standard protocol (31). Cells were transformed with 10 ng of target protospacer plasmids or control vector using BioRad MicroPulser using standard E. coli protocol provided by the manufacturer. After 1-h outgrowth in 1 ml of LB medium at 37°C, 10 μl aliquots of serial dilutions of transformation mixtures were deposited onto an LB agar plate containing 34 μg/ml chloramphenicol, 100 μg/ml ampicillin and 50 μg/ml of spectinomycin. The plate was incubated at 37°C overnight and growth results recorded. Each experiment was conducted at least in triplicate.
Electrocompetent E. coli ED1a cells prepared after growth in LB (in the absence of inducers) and after transformation were processed as above and plated on LB plates with 100 μg/ml ampicillin.
To induce adaptation in BL21-AI-based strains, clones of cells transformed with selected plasmids were grown in LB supplemented with 1 mM arabinose and 1 mM IPTG for a total of 72 hours at 37°C on an orbital shaker (180 rpm). Every 24 h aliquots of cultures were diluted (1:500) into fresh medium. The adaptation was detected by PCR with a pair of primers, one (forward) annealing in the leader sequence and another (reverse) at the CRISPR array spacer. For KD628, the reverse primer annealed downstream of the single CRISPR repeat present in this strain. After amplification, reaction products were analyzed by agarose gel electrophoresis.
KD604 cells carrying pCas and pCsy were transformed individually with anti-CRISPR containing plasmids and transformants were selected on plates containing 30 μg/ml gentamicin. 100 μg/ml ampicillin, and 50 μg/ml of spectinomycin were added for pCas and pCsy maintenance. After the transformation, passaging was performed as described above, with 30 μg/ml gentamicin included throughout the duration of the 72-h experiment to maintain anti-CRISPR plasmids.
For ED1a cells the procedures were the same except that no inducers were added to the media and a different primer set was used for amplification.
KD475 cells were transformed with pCas and pCsy plasmids. To monitor adaptation, the overnight culture of transformed cells were diluted 200-fold with LB supplemented with 1 mM arabinose, 1 mM IPTG, 100 μg/ml ampicillin and 50 μg/ml of spectinomycin. After overnight incubation adaptation was detected by PCR as described above.
Northern blot analysis
RNA purification and Northern blot analysis of crRNAs from ED1a cells was performed using appropriate oligonucleotide probes exactly as described in (32).
High throughput sequence data analysis
High-throughput sequence analysis was made with MiSeq Illumina system. The data were preprocessed and analyzed using ShortRead (33) and BioStrings (34) Bioconductor packages. Sequences located between two CRISPR repeats were considered as spacers. They were mapped on genome and all plasmids presented in the certain sample with no mismatches allowed. To assign non-unique spacers from common regions of pCas and pCsy plasmids the following procedure was used. First, spacers originating from unique regions of each plasmid were counted. The ratio of the number of such unique spacers, normalized for the length of each plasmid, was taken as a measure of mean spacer acquisition efficiency from each plasmid. The non-unique spacers that could have originated from either plasmid were next assigned to either pCas or pCsy based on this measure. R scripts were used for statistical analysis and Circos (35) was used for graphical representation of the data.
RESULTS
Transplantation of the P. aeruginosa I-F CRISPR–Cas system into a heterologous E. coli host
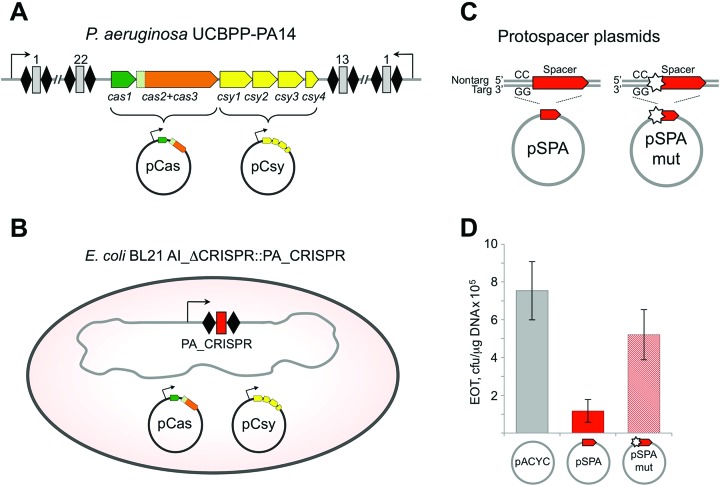
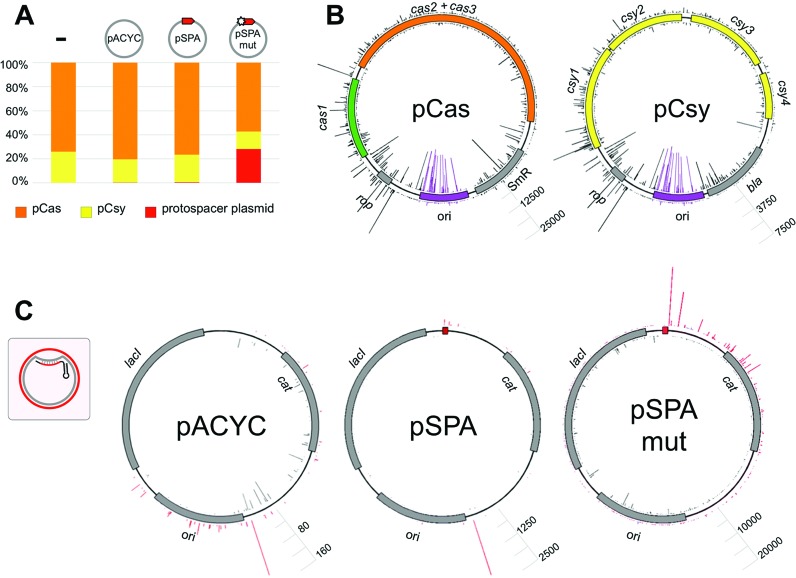
The genetic structure of the P. aeruginosa UCBPP-PA14 type I-F CRISPR–Cas system locus is shown in Figure 1A. Previously, very inefficient spacer acquisition by this system was detected during infection with a lytic phage (13). We wondered if more efficient spacer acquisition could be obtained by transplanting the P. aeruginosa UCBPP-PA14 type I-F CRISPR–Cas system in a heterologous host. A minimized P. aeruginosa CRISPR array consisting of two repeats, one spacer, and a leader sequence was inserted into the chromosome of an E. coli strain (KD604) lacking an endogenous CRISPR–Cas system (Figure 1B). This strain was transformed with two compatible plasmids, one expressing P. aeruginosa genes cas1–3 (pCas) and another expressing csy1–4 (pCsy) (Figure 1A and B). To determine if the transplanted type I-F immune system from P. aeruginosa is capable of interference in E. coli we transformed induced KD604 cells harboring pCas and pCsy with a plasmid containing a protospacer flanked by a GG consensus PAM (pSPA); a plasmid with a mutant version of this protospacer with a single mismatch in the first position of the seed region (pSPAmut), or a control plasmid with no protospacer (pACYC) (Figure 1C). The efficiency of transformation (EOT) of pSPA amounted to ∼15% of EOT of the pACYC vector (Figure 1D). For pSPAmut, EOT was decreased less, to ∼70% of pACYC vector EOT (Figure 1D). Thus, albeit the effects are modest, the type I-F immune system from P. aeruginosa is capable of interfering with plasmid transformation in E. coli (KD604) and this immune response relies on complementary base pairing between the crRNA-spacer sequence and the protospacer. The weak level of interference may be caused by a heterologous background (and therefore indicate that an additional factor is required, or be a simple cause of a particular spacer-protospacer pair chosen).
Figure 1.
The type I-F CRISPR–Cas system from P. aeruginosa interferes with plasmid transformation in a heterologous background. (A) Organization of the type I-F CRISPR–Cas P. aeruginosa system is schematically presented. The system consists of two CRISPR arrays (black rhombi indicate repeats, numbered gray rectangles are spacers) and a set of cas and csy genes. The cas genes (cas1 and cas2+cas3) and csy genes (csy1–4) were cloned into expression vectors pCas and pCsy, respectively. (B) An E. coli KD604 cell transformed with pCas and pCsy plasmids is schematically shown. KD604 does not have a CRISPR–Cas system of its own but carries a CRISPR array containing two P. aeruginosa type I-F repeats, a single spacer, and an upstream leader sequence, and an upstream promoter. (C) pACYC-based protospacer plasmids pSPA and pSPAmut used in this work are schematically illustrated. The plasmids carry a protospacer matching the spacer present in the KD604 genomic CRISPR array bordered by a consensus GG PAM. In pSPAmut the first position of the protospacer carries a mutation introducing a mismatch with the KD604 spacer. (D) Efficiency of transformation (EOT) of pSPA, pSPAmut and a control vector with no protospacer (pACYC) was determined in induced KD604 cells co-expressing P. aeruginosa cas and csy genes from pCas and pCsy, respectively. Bars represent mean EOT values obtained in three independent experiments with standard deviations are shown.
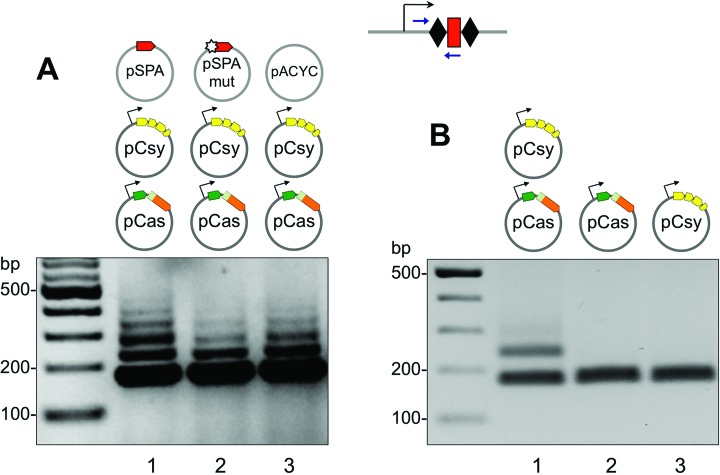
We next set out to determine if the transplanted immune system from P. aeruginosa was capable of incorporating new spacer sequences into the minimal CRISPR locus. E. coli (KD604) cells were transformed with pSPA, pSPAmut or pACYC and cultured in LB-media supplemented with arabinose and IPTG to induce expression of the cas and csy genes. Aliquots of cell cultures were subjected to PCR with a pair of primers amplifying the CRISPR cassette and the proximal leader sequence. Acquisition of new spacers is reflected by the appearance of PCR products longer than the 176 bp fragment amplified from the starting cells genomic DNA. Robust spacer acquisition was detected in cells transformed with pSPA, pSPAmut or pACYC (Figure 2A). In fact, these plasmids were not necessary for adaptation, since spacer acquisition was detectable in cells containing just pCas and pCsy plasmids (Figure 2B, lane 1). However, no new spacer acquisition occurred in cells that contained either pCas or pCsy plasmids alone (Figure 2B, lanes 2 and 3).
Figure 2.
Spacer acquisition by the P. aeruginosa CRISPR–Cas system in a heterologous background. (A) Results of a CRISPR adaptation experiment in KD604 cells transformed with pCas, pCsy and indicated protospacer plasmids or control vector are shown. The leader-proximal end of CRISPR cassette was amplified using a primer pair schematically shown in Figure 1B; amplification products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. (B) The experiment was performed as in (A) in the absence of protospacer plasmid and in the presence of only pCas, only pCsy or both plasmids, as indicated.
Genetic requirements for spacer acquisition by the I-F CRISPR–Cas system from P. aeruginosa
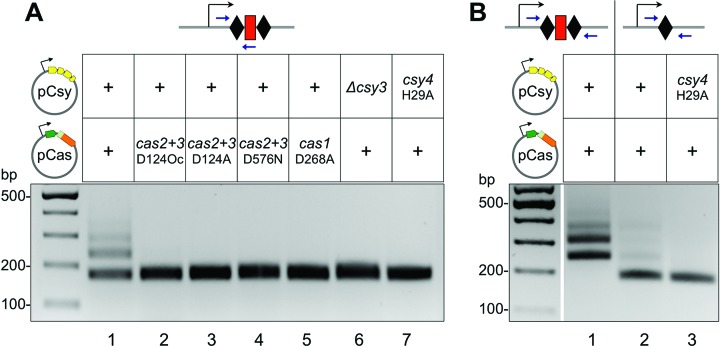
The effects of mutations in P. aeruginosa cas and csy genes on spacer acquisition were determined. Introducing an alanine at position 268 of Cas1 (Cas1D268A) abolished adaptation (Figure 3A, lane 5). This is an expected result since D268 is a conserved metal coordinating residue and substitution of the corresponding residue in E. coli Cas1 also abolishes adaptation (23). A specific feature of type I-F systems is a fusion of cas2 and cas3 homologs, which are encoded on separate genes in other CRISPR–Cas systems (16). A D124Oc mutation in the cas2+cas3 gene that introduced an ochre stop codon instead of aspartate codon at position 124, after the cas2 portion of the fused gene, abolished spacer acquisition (Figure 3A, lane 2). Point mutations introducing single amino acid substitutions in the endonuclease (D124A) and helicase (D576N) domains of the Cas3 protein also prevented spacer acquisition (Figure 3A, lanes 3 and 4). Deleting the csy3 gene (lane 6), or mutating the catalytic residue His29 of Csy4 nuclease (lane 7) that is needed for generation of mature crRNA also abolished spacer acquisition (37). We conclude that spacer acquisition by the transplanted P. aeruginosa CRISPR–Cas system requires both Cas1 and Cas2 and the Csy proteins.
Figure 3.
Active site mutations or gene deletions perturb new spacer acquisition by the type I-F CRISPR–Cas system. (A) Results of a CRISPR adaptation experiment with KD604 cells transformed with pCas and pCsy expressing wild-type cas/csy genes (lane 1) or various mutant versions of these genes (see text for details). (B) Results of a CRISPR adaptation experiment with KD604 cells transformed with pCas and pCsy expressing wild-type cas/csy genes (lane 1) or with KD628 cells harboring a single CRISPR repeat (schematically shown at the top of the figure) transformed with wild-type pCas and pCsy or indicated mutant plasmid combination are shown.
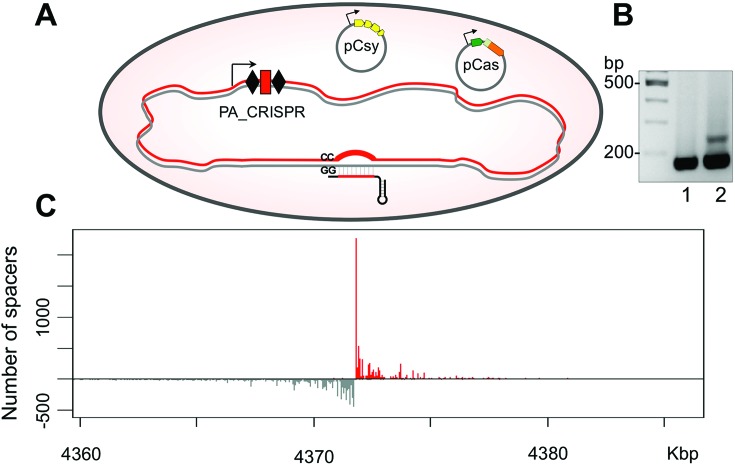
To determine if adaptation depends on specific crRNA, an additional E. coli strain (KD606) containing a different spacer in the engineered P. aeruginosa CRISPR locus was constructed. Neither KD604 nor KD606 spacers have detectable similarity to pCas or pCsy sequences or to the E. coli genome (at least 13 mismatches, longest stretch of complementarity 8 nucleotides). Just like in the case of the KD604 strain, robust spacer acquisition was observed when both pCas and pCsy were introduced into E. coli (KD606), but no adaptation was detected when only one plasmid was present (data not shown). When E. coli (KD628) cells harboring just a leader and a single CRISPR repeat were transformed with pCas and pCsy plasmids, very weak adaptation (compared to adaptation observed in cells containing two repeats and one spacer) was observed (Figure 3B), indicating that removal of a spacer or one of the two repeats, both of which should affect crRNA production, inhibits adaptation. This residual adaptation was abolished when combined with the H29A mutation in csy4. Overall, we conclude that the P. aeruginosa CRISPR–Cas system is capable of robust adaptation in the apparent absence of pre-existing matches between crRNA spacer and the target. In E. coli, such a ‘naïve’ adaptation requires just the Cas1 and Cas2 proteins (20). However, in the case of P. aeruginosa CRISPR–Cas system, proteins that constitute the Csy effector complex are also required. The presence of crRNA strongly stimulates spacer acquisition.
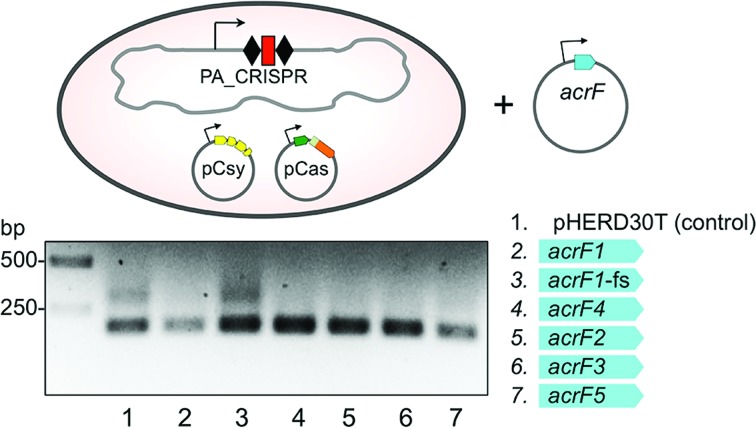
Anti-CRISPR proteins prevent spacer acquisition by P. aeruginosa I-F CRISPR–Cas system
Several P. aeruginosa phages encode anti-CRISPR proteins that prevent CRISPR-mediated immunity (30,38). These inhibitors allow a phage to infect cells despite the presence of previously acquired phage-derived spacers (38–40). Anti-CRISPRs are diverse proteins that target different components of the Csy complex or the Cas3 protein (40). To determine if anti-CRISPR proteins also affect spacer acquisition we transformed E. coli (KD604) cells harboring pCas and pCsy plasmids with compatible plasmids expressing distinct anti-CRISPR proteins and monitored spacer acquisition (Figure 4). Spacer acquisition was inhibited in the presence of each of the five different anti-CRISPR proteins tested (acrF1–5, Figure 4). A plasmid bearing a frame-shift mutation in the beginning of the acrF1 anti-CRISPR gene that inactivates its anti-interference function (40) also inactivated the anti-acquisition function. We conclude that anti-CRISPR proteins that inhibit CRISPR interference in P. aeruginosa by targeting either the Csy complex or Cas3, also inhibit spacer acquisition. This supports the results of our genetic analysis that demonstrate the requirement of these proteins for adaptation in the Type I-F CRISPR–Cas system.
Figure 4.
Diverse anti-CRISPR proteins encoded by P. aeruginosa bacteriophages inhibit spacer acquisition. KD604 cells carrying pCas and pCsy expressing wild-type cas/csy genes were transformed with compatible pHERD30T-based plasmids expressing various anti-CRISPR proteins (AcrFs) listed on the right (acrF1-fs carries a frame-shift mutation). Expression of plasmid-borne gene was induced and spacer acquisition was monitored by PCR amplification.
Two modes of spacer acquisition by P. aeruginosa subtype I-F CRISPR–Cas system
To determine the origin of spacers acquired by E. coli (KD604) cells containing pCas and pCsy–with or without pSPA, pSPAmut or pACYC–PCR fragments corresponding to expanded CRISPR arrays were subjected to high throughput sequencing. Since most spacers mapped back to plasmids rather than the KD604 genome (<1% of all spacers), spacers from the genome were excluded from further analysis.
In cells harboring pCas and pCsy only, the ratio of unique spacers originating from pCas and pCsy was ∼4 to 1 (Figure 5A). The two plasmids are of similar size and have the same origin of replication. The nature of this bias, which is highly reproducible, is thus unknown and requires further investigation. In the presence of either pSPA or pACYC, <1% of all spacers were acquired from these additional plasmids (Figure 5A). In contrast, in cells harboring pSPAmut ∼30% of acquired spacers were derived from this plasmid (Figure 5A). Thus, an imperfect spacer match with the target strongly stimulates spacer selection from DNA sequences in cis, a hallmark of primed adaption. Increased efficiency of spacer selection from pSPAmut did not affect the ratio of spacers acquired from pCas and pCsy (Figure 5A).
Figure 5.
The origin and distribution of spacers acquired by the P. aeruginosa CRISPR–Cas system. DNA fragments corresponding to expanded CRISPR cassettes shown in Figure 2A (lanes 1–3) and Figure 2B lane 1 were subjected to Illumina sequencing. Spacer sequences were extracted from filtered reads and mapped back to their origin. (A) Bar graph showing the origin of plasmid-derived spacers. Spacers originating from pCas are shown in orange, pCsy – in yellow, and pSPA, pSPAmut or pACYC – in red. (B) Mapping of spacers acquired by cells containing pCas and pCsy on donor plasmids. The cas genes are shown in green and orange, csy genes in yellow, antibiotic resistance and repressor of primer (rop) genes in gray, replication origins – in purple. The heights of gray and purple bars indicate the efficiency (number of times) of spacer from this position was observed. Bars protruding inside and outside of plasmid circles represent spacers derived from different strands of DNA. The height of bars corresponding to most frequently acquired protospacers in both plasmids is made the same for easier comparisons. Scale bars allow to access spacer acquisition efficiencies for each plasmid (number of reads). Purple bars indicate spacers originating from ori regions. Grey bars indicate spacers originating from the rest of each plasmid. (C) Mapping of spacers acquired from pSPA, pSPAmut, or pACYC vector control in cells expressing the cas and csy genes. Where present, a protospacer matching crRNA is shown as a small red box. The inset schematically shows the structure of the R-loop formed by crRNA. Targeted strand is shown in gray, non-targeted – in red. Bars showing spacers originating from each of these strands are colored accordingly. Scale bars indicate spacer acquisition efficiency for each plasmid (number of reads).
We next examined the distribution of donor protospacers and efficiency of their use (determined by the number of corresponding spacer reads) for each plasmid (Figure 5B and C). Both the distribution and the efficiency of use of donor protospacers in pCas and pCsy were found to be highly reproducible and independent on the presence of pSPA, pSPAmut or pACYC (Figure 5B, correlation coefficients for spacer distributions between different samples or between biological replicates of the same sample being 0.82 or higher). Spacers derived from pCas originated from both strands and from every part of the plasmid. The overall efficiency of spacer selection was considerably higher for the ori region and the adjacent rop gene (Figure 5B). 27% of all spacers originated from the ori. A strong strand bias for selection of these spacers was observed, with 95.5% of ori spacers selected from the ‘inner’ strand of the plasmid as shown in Figure 5B. The remaining 73% of spacers were selected with equal efficiency from both strands of the plasmid (53% from the ‘inner’ strand). A similar pattern was observed for pCsy: equal efficiency of spacer acquisition from both strands throughout the plasmid backbone with strong strand bias at the ori. It should be noted that the ori sequences of pCas and pCsy are highly similar and so most spacers derived from these regions are not unique. The same distribution of acquired spacers from pCas and pCsy was observed in KD606 cells (a correlation coefficient with KD604 of 0.85 for pCas and 0.89 for pCsy) indicating that spacer acquisition preferences do not depend on spacer of pre-existing crRNA. This, and the absence of strand bias for spacers acquired from most of pCas and pCsy sequence is consistent with naïve, non-primed adaptation. The only exception is the ori region, where a strong strand bias as well as increased overall level of spacer selection efficiency is observed.
The pattern of spacer selection from pACYC, pSPA, and pSPAmut is shown in Figure 5C. Spacers from pACYC were acquired from both strands of the plasmid with most actively used protospacers located at or close to the ori region. In the case of pSPA plasmid, the distribution was similar, with a modest bias for selection of protospacers in the vicinity of the protospacer matching KD604 crRNA. In the case of pSPAmut most spacers originated from one strand of the plasmid and there was a strong preference for selection of protospacers ‘to the right’, i.e. downstream of the protospacer matching the KD604 spacer (Figure 5C). A strong (96%) bias for spacer acquisition from the non-targeted strand in this region was observed (Figure 5C). Protospacers located closer to the priming spacer appeared to be used more efficiently than the ones located further away, revealing a gradient in protospacer selection efficiency as a function of distance from the priming site. Protospacers in the area located upstream of the priming spacer were used much less efficiently. Spacers from this area were more efficiently selected from the strand targeted by the KD604 crRNA spacer (97% bias).
Of more than 4000 unique plasmid-derived spacers, 97.76% matched protospacers containing consensus GG PAM. This value was the same for pCas and pCsy-derived spacers, which were acquired in the absence of priming, and for pSPAmut spacers acquired in the course of primed adaptation (Supplementary Table S2). Most spacers that appeared to originate from protospacers with non-consensus PAMs were ‘derived’ from regions containing a consensus PAM by 1–2 nucleotide upstream or downstream shifting or by insertion into CRISPR array in an opposite orientation. Similar aberrant spacers were previously observed in the E. coli subtype I-E system (41,42). When such ‘derived’ spacers were considered as originating from parental protospacers with consensus PAM, the preference for GG PAM increased to more than 99% for both pCas and pCsy and for pSPAmut. In E. coli subtype I-E system, a strong bias toward protospacers with consensus, interference-proficient PAMs is indicative of primed adaptation (23).
Spacer acquisition during targeting of the E. coli genome
To obtain a more detailed view of donor protospacer selection upstream and downstream of the priming site an E. coli KD675 strain, a derivative of KD604 was created. KD675 contains a partially matching protospacer (the same as in the pSPAmut plasmid, above) and PAM in its genome (Figure 6A). We therefore expected primed adaption to be initiated from the genomic protospacer under conditions of cas and csy genes expression. KD675 cells were transformed with pCas and pCsy plasmids, induced, and grown in presence of antibiotics to reduce a fraction of the cells with plasmid-derived spacers in the culture. Despite self-targeting, induced KD675 cultures continued to grow normally, consistent with low levels of interference observed during plasmid transformation (Figure 1D). As a result, while only modest level of spacer acquisition was observed (Figure 6B), only 30% of spacers originated from pCas and pCsy, the remainder were from the KD675 genome. Spacers acquired from pCas and pCsy had a distribution similar to that shown in Figure 5A (correlation coefficients of 0.85 or more). Spacers from the KD675 genome originated from either side of the priming site. The efficiency of spacer selection decreased as the distance from the priming spacer increased but remained above background levels at distances as large as 5,000 bp (Figure 6C). In the downstream direction, in agreement with data obtained with the pSPAmut plasmid, spacers were selected predominantly from the non-targeted strand where the protospacer matching the crRNA spacer is located. In the upstream direction, spacers were selected from the opposite, targeted strand. Though on each side of the priming site the strand bias was very strong (∼98% spacers acquired from one of the two strands), when all spacers were considered together there was no overall strand bias (Supplementary Table S3) since spacers upstream and downstream of the priming site are selected from different strands.
Figure 6.
New spacers are acquired from regions of the genome that are distributed around the priming site. (A) An E. coli KD675 cell transformed with pCas and pCsy plasmids is schematically shown. KD675 does not have a CRISPR–Cas system of its own but carries a CRISPR array containing two P. aeruginosa type I-F repeats, a single spacer, and an upstream leader sequence, and an upstream promoter. The spacer targets a protospacer in KD675 own genome. The structure of the R-loop formed upon the recognition of protospacer by self-targeting crRNA is shown, with targeted strand shown in grey, non-targeted – in red. (B) Results of a CRISPR adaptation experiment with KD675 cells transformed with pCas and pCsy plasmids. No spacer acquisition was observed in the absence of inducers (lane 1), the cells acquired new spacers after induction of cas and csy genes expression (lane 2). (C) Mapping of acquired spacers on a region of KD675 genome within ∼25 kb of the priming site. Spacers acquired from the non-targeted strand are shown in red. Spacers acquired from the targeted strand are shown in gray.
Spacer acquisition by subtype I-F CRISPR–Cas system of E. coli
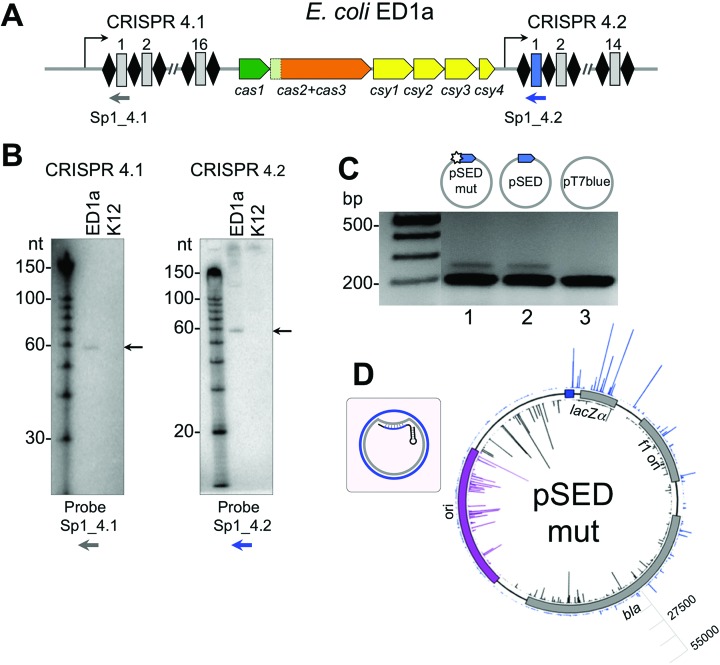
While most strains of E. coli contain a type I-E CRISPR–Cas system, the ED1a stain of E. coli contains a type I-F system (Figure 7A) (18,29). Northern blot analysis performed using probes specific for the first spacer from each of the two ED1a CRISPR arrays revealed a distinct transcript with an apparent size of ∼60 nt for both arrays, suggesting that crRNA is expressed and processed under laboratory growth conditions (Figure 7B). To monitor the function of ED1a CRISPR–Cas system, we transformed the cells with plasmids containing protospacers complementary to the first spacer in the endogenous CRISPR 4.2 array. No difference in transformation efficiency was detected for a pSED plasmid containing a protospacer with a consensus GG PAM as compared pSEDmut plasmid containing protospacer with a mismatch in the seed region or pT7blue vector control. However, upon prolonged cultivation in the absence of antibiotics the pSED plasmid was lost from the culture, while pSEDmut and the pT7blue vector were maintained (data not shown). When transformed cultures were cultivated and analyzed for CRISPR array expansion, PCR fragments corresponding to expanded CRISPR arrays were observed in cultures of cells containing both pSED and pSEDmut, but not in cultures harboring pT7blue (Figure 7C). Thus, the ED1a cells apparently undergo primed adaption, however, it proceeds with the same efficiency from targets with completely complementary or partially complementary protospacers.
Figure 7.
Primed adaptation by the type I-F CRISPR–Cas system from E. coli ED1a. (A) Organization of the type I-F CRISPR–Cas system from the E. coli ED1a is schematically presented. See Figure 1A legend for details. (B) Northern blot analysis of RNA purified from E. coli ED1a or K12 (does not contain a type I-F CRISPR–Cas system) with probes complementary to first spacers in both arrays (schematically shown as a gray or a blue arrow in panel A). (C) E. coli ED1a cells transformed with plasmid pSED (carrying a fully matching protospacer and a GG PAM), plasmid pSEDmut (containing single spacer-protospacer mismatch at position +1), or the pT7blue vector control (no protospacer). PCR products from the leader end of CRISPR locus 4.2 were separated by electrophoresis and visualized by ethidium bromide staining. (D) Mapping of spacers acquired by E. coli ED1a cells transformed with pSPAmut. The priming protospacer is shown as a blue box. The inset schematically shows the structure of the R-loop formed by crRNA. Targeted strand is shown in gray, non-targeted – in blue. Bars showing spacers originating from each of these strands are colored accordingly. Bars showing spacers originating from the ori are colored purple. Scale bar indicates spacer acquisition efficiency for each plasmid (number of reads).
High-throughput sequencing of acquired spacers was performed and spacer sequences were mapped. An identical result was obtained when spacers acquired in cultures harboring either pSED or pSEDmut were analyzed. In both cases, 98% spacers were plasmid-derived. The remaining spacers originated from the bacterial genome. The distribution of donor protospacers and the efficiency of spacer selection from the pSEDmut plasmid is shown in Figure 7D and overall statistics is given in Supplementary Table S4. As can be seen from Figure 7D, a gradient of spacer acquisition was also revealed in this case with strand biases upstream and downstream of the priming site matching those observed for the type I-F P. aeruginosa system.
DISCUSSION
The principal finding of this work is the demonstration that both naïve adaptation and primed adaptation are operational in P. aeruginosa type I-F system. However, in marked contrast to the situation in type I-E system in E. coli, the cas1 and cas2 genes are not sufficient for naive adaptation. In fact, in the type I-F system from P. aeruginosa, both modes of adaptation require intact Csy effector complex and a crRNA.
By definition, primed adaptation in E. coli also requires a crRNA that contains mismatches with a protospacer in the target DNA (23). These mismatches have been shown to reduce but not abolish binding by the Cascade (CRISPR-associated complex for anti-viral defense). Residual interaction with the mutated protospacer recruits Cas1 and Cas2 to target DNA, resulting in primed adaptation. A very similar process must be occurring during primed adaptation by the P. aeruginosa I-F system, when crRNA with a spacer partially matching a protospacer guides the Csy complex (a functional ortholog of Cascade), and Cas2–3 and Cas1 to the target, leading to efficient spacer acquisition (14). Consistent with previous results, our analysis reveals that spacers are acquired from both sides of the priming site. Apparently, the adaptation machinery proceeds bi-directionally from the priming site with efficiency of spacer selection decreasing as the distance from the priming site increases. Experiments with an E. coli strain containing a spacer complementary to the E. coli chromosome reveal that the P. aeruginosa I-F system adaptation machinery is apparently highly processive and able to acquire spacers from protospacers located several thousands base pairs away from the priming site.
Strand bias is a hallmark of primed adaptation in E. coli: spacers from the strand opposite to the one targeted by the Cascade are selected ∼10 times more frequently than from the targeted strand (23). Overall, primed adaptation by the P. aeruginosa I-F system does not reveal such a bias. However, when spacers selected from both sides of the priming site are considered separately a very strong strand bias is observed. Upstream of the priming site, spacers are selected from the strand targeted by the priming crRNA. In the downstream direction the strand bias is reversed and spacers are predominantly selected from non-targeted strand protospacers. A similar strand bias was earlier detected for spacers acquired during primed adaptation by the type I-F system from Pectobacterium atrosepticum (14) and is also revealed during primed adaptation by the E. coli type I-F system studied in this work. A strong strand bias that changes its direction at the priming site is most consistent with existence of single-stranded intermediates formed at both sides (and on different strands) of the priming site. New spacers could be selected from these intermediates upon the recognition of PAMs by the adaptation machinery. If such intermediates were double stranded, PAM sequences in both strands would have been recognized, abolishing the strand bias.
It was suggested that in the case of the type I-B system from Haloarcula hispanica, where adaptation strictly requires priming, the Cas3 protein activity is responsible for generation of single-stranded intermediates for spacer selection (25). Our data are consistent with this model, however, the direction of strand bias in H. hispanica is opposite to the one observed in I-F systems. One can speculate that this difference might be caused by opposing directionality of Cas3 helicase/nuclease action in these systems.
The transplanted P. aeruginosa I-F system was capable of primed adaptation only when CRISPR interference was inactivated by a mutation that introduced a mismatch between crRNA spacer and the priming protospacer. Interestingly, primed adaptation by the E. coli I-F system occurred with equal efficiency whether there was a mismatch or full match between crRNA spacer and the priming site. However, it should be noted that CRISPR interference by the E. coli I-F system even at conditions of full match between a crRNA spacer and target protspacer studied here was very weak. It is possible that the outcome of Cascade-crRNA protein complex interaction with a protospacer, i.e. interference or primed adaptation, may be determined not so much by the presence of mismatches between the spacer and protospacer but by the overall stability and/or life time of the complex, which could be affected by the sequence of the spacer-protospacer heteroduplex.
The P. aeruginosa type I-F CRISPR–Cas system is capable of robust adaptation in the absence of pre-existing matches between a crRNA and the target. In E. coli type I-E system, such ‘naïve’ adaptation requires just the Cas1 and Cas2 proteins (20). However, in the case of P. aeruginosa CRISPR–Cas system, all Cas and Csy proteins are required for this process. Naïve adaptation is also strongly stimulated by the crRNA. However, the sequence of crRNA spacer appears to be unimportant, since the KD604 and KD606 strains contain different crRNAs and yet exhibit the same spacer acquisition preferences. This suggests that the role of crRNA may be limited to stabilization of the Csy complex and that the Csy complex is involved in protospacer selection in a process that is independent of crRNA-guided base paring to the target.
The observation that phage-encoded anti-CRISPR proteins inhibit both interference and acquisition supports the genetic data presented here and provides further mechanistic links between these two processes. It has recently been shown that distinct anti-CRISPR proteins operate through diverse mechanisms, by preventing DNA-binding by the Csy complex or blocking the recruitment of the Cas2–3 protein (40). The ability of each anti-CRISPR tested to inhibit both acquisition and interference reveals a redundancy that is likely beneficial in the phage response to CRISPR–Cas systems.
In E. coli (type I-E), overexpression of Cas1 and Cas2 results in more than 60% of spacers derived from protospacers with non-consensus PAM (41,42). Spacers derived from protospacers not flanked by a PAM are unable to elicit an interference response, but could induce primed adaptation (23,43). In contrast, both primed and naïve adaptation by the P. aeruginosa system result in almost absolute selectivity for protospacers with consensus PAM, therefore leading to crRNAs capable of interference. Presumably, the higher level of selectivity towards consensus PAM is due to the involvement of the Csy complex in PAM recognition (14). Interestingly, recent analysis of spacer acquisition by Type II CRISPR–Cas systems revealed that in addition to ‘professional’ adaptation proteins Cas1 and Cas2, the Cas9 protein, which is an crRNA-guided endonuclease involved in target DNA cleavage, is also required for adaptation and determines selection of protospacers with correct PAMs (26,27). Involvement of PAM recognizing proteins from the interference pathway in selection of donor protospacers during CRISPR adaptation appears to be a common strategy, which ensures that newly acquired spacers will result in crRNAs capable of eliciting direct degradation of the foreign target.
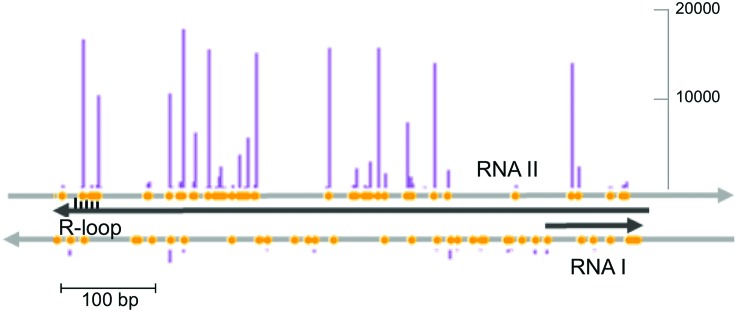
The P. aeruginosa type I-F CRISPR–Cas system clearly prefers to acquire spacers from the ColEI origin of replication during non-primed spacer adaptation. Spacer acquisition from ori is highly biased to one strand, suggesting that it is driven by a specific structural feature of the origin itself. An extended RNA-DNA duplex and an R-loop are formed on ColEI when RNA polymerase transcribes RNA II, a transcript used to prime plasmid replication. A second RNA, RNA I, is transcribed in the opposite direction and is used to control the number of RNA II molecules (44). The structure of the ColEI origin is schematically shown in Figure 8 along with distribution of GG PAMs and spacers acquired from this region of pCas plasmid. Very similar distributions were observed in experiments involving KD604 and KD606 cells that contain unrelated crRNA spacers. In fact, the distribution of spacers acquired from the ColE1 origin of pSED plasmids by the E. coli ED1a type I-F CRISPR–Cas system is also very similar (correlation coefficient of 0.8). Thus, spacers are acquired from the strand complementary to RNA II. The mechanistic reasons for this bias remain to be elucidated. The E. coli type I-E Cas3 protein is involved in copy number control of ColE1 plasmids (45), while naïve spacer acquisition by this system is targeted to genomic replication termination region (46), which in case of unidirectional ColE1 replication coincides with ori. These observations suggest that there may be a deep link between naïve CRISPR adaptation and replication that remains to be elucidated. Such a link could initially target CRISPR adaptation machinery to actively replicating foreign DNA, while priming would allow additional protective spacers to be specifically acquired at a later point.
Figure 8.
The distribution of spacers acquired by the P. aeruginosa CRISPR–Cas system at the ColE1 origin of replication. The two strands to the ColE1 origin are shown as grey arrows. Black arrows show origin transcripts. The position of the R-loop at the 3′-terminal part of RNA II is indicated. Yellow circles indicate GG sequences (PAMs) in both strands. The purple vertical bars show the number of spacers acquired. The distribution of acquired spacers for the pCas plasmid from KD604 (see Figure 5B) is shown. Very similar distributions were obtained with KD606 cells and with ED1a E. coli that acquired spacers from ColE1 plasmid pSED (Figure 7D). Scale bar indicates spacer acquisition efficiency (number of reads).
Supplementary Material
Acknowledgments
We thank Michael Terns for advice.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [GM10407 to K.S.]; Russian Science Foundation [14-14-00988]; Ministry of Education and Science of Russian Federation project [14.B25.31.0004 to K.S.]; Russian Foundation for Basic Research Foundation [14-04-00916 to E.E.S.]; Canadian Institutes of Health Research [MOP-130482 to A.D]; MSU Agricultural Experimental Station (to B.W.); J.B.-D. was supported by a CIHR Canada Graduate Scholarship Doctoral Award and an Ontario Graduate Scholarship award. Funding for open access charge: Russian Science Foundation grant.
Conflict of interest statement. None declared.
REFERENCES
- 1.Makarova K.S., Grishin N.V., Shabalina S.A., Wolf Y.I., Koonin E.V. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaya D., Davison M., Barrangou R. CRISPR–Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 3.van der Oost J., Jore M.M., Westra E.R., Lundgren M., Brouns S.J. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 2009;34:401–407. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 5.Pougach K., Semenova E., Bogdanova E., Datsenko K.A., Djordjevic M., Wanner B.L., Severinov K. Transcription, transcript processing and function of E. coli CRISPR locus. Mol. Microbiol. 2010;77:1367–1379. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jore M.M., Lundgren M., van Duijn E., Bultema J.B., Westra E.R., Waghmare S.P., Wiedenheft B., Pul U., Wurm R., Wagner R., et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 8.Mulepati S., Bailey S. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J. Biol. Chem. 2013;288:22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westra E.R., van Erp P.B., Kunne T., Wong S.P., Staals R.H., Seegers C.L., Bollen S., Jore M.M., Semenova E., Severinov K., et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiedenheft B., van Duijn E., Bultema J.B., Waghmare S.P., Zhou K., Barendregt A., Westphalb W., Heckc A.J.R., Boekemad E.J., Dickmane M.J., et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Semenova E., Jore M.M., Datsenko K.A., Semenova A., Westra E.R., Wanner B., van der Oost J., Brouns S.J., Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deveau H., Barrangou R., Garneau J.E., Labonté J., Fremaux C., Boyaval P., Romero D.A., Horvath P., Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cady K.C., Bondy-Denomy J., Heussler G.E., Davidson A.R., O'Toole G.A. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J. Bacteriol. 2012;194:5728–5738. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter C., Dy R.L., McKenzie R.E., Watson B.N., Taylor C., Chang J.T., McNeil M.B., Staals R.H., Fineran P.C. Priming in the Type I-F CRISPR–Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014;42:8516–8526. doi: 10.1093/nar/gku527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiedenheft B., Sternberg S.H., Doudna J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 16.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J., Charpentier E., Horvath P., Moineau S., Mojica F.J., Wolf Y.I., Yakunin A.F., et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mojica F.J., Díez-Villaseñor C., García-Martínez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 18.Almendros C., Guzmán N.M., Díez-Villaseñor C., García-Martínez J., Mojica F.J. Target motifs affecting natural immunity by a constitutive CRISPR–Cas system in Escherichia coli. PLoS One. 2012;7:e50797. doi: 10.1371/journal.pone.0050797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rollins M.F., Schuman J.T., Paulus K., Bukhari H.S., Wiedenheft B. Mechanism of foreign DNA recognition by a CRISPR RNA-guided surveillance complex from Pseudomonas aeruginosa. Nucleic Acids Res. 2015;43:216–222. doi: 10.1093/nar/gkv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yosef I., Goren M.G., Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arslan Z., Hermanns V., Wurm R., Wagner R., Pul Ü. Detection and characterization of spacer integration intermediates in type I-E CRISPR–Cas system. Nucleic Acids Res. 2014;42:7884–7893. doi: 10.1093/nar/gku510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brouns S.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J., Snijders A.P., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datsenko K.A., Pougach K., Tikhonov A., Wanner B.L., Severinov K., Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 24.Swarts D.C., Mosterd C., van Passel M.W., Brouns S.J. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M., Wang R., Zhao D., Xiang H. Adaptation of the Haloarcula hispanica CRISPR–Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 2014;42:2483–2492. doi: 10.1093/nar/gkt1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei Y., Terns R.M., Terns M.P. Cas9 function and host genome sampling in Type II-A CRISPR–Cas adaptation. Genes Dev. 2015;29:356–361. doi: 10.1101/gad.257550.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heler R., Samai P., Modell J.W., Weiner C., Goldberg G.W., Bikard D., Marraffini L.A. Cas9 specifies functional viral targets during CRISPR–Cas adaptation. Nature. 2015;519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Touchon M., Rocha E.P.C. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One. 2010;5:e11126. doi: 10.1371/journal.pone.0011126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bondy-Denomy J., Pawluk A., Maxwell K.L., Davidson A.R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–432. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller E.M., Nickoloff J.A. Escherichia coli electrotransformation. Methods Mol. Biol. 1995;47:105. doi: 10.1385/0-89603-310-4:105. [DOI] [PubMed] [Google Scholar]
- 32.Pougach K., Severinov K. Use of semi-quantitative Northern blot analysis to determine relative quantities of bacterial CRISPR transcripts. Methods Mol. Biol. 2012;905:73–86. doi: 10.1007/978-1-61779-949-5_6. [DOI] [PubMed] [Google Scholar]
- 33.Morgan M., Anders S., Lawrence M., Aboyoun P., Pagès H., Gentleman R. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25:2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pages H., Aboyoun P., Gentleman R., DebRoy S. Biostrings: String objects representing biological sequences, and matching algorithms. 2012 R package version 2.24.1. [Google Scholar]
- 35.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiedenheft B., Zhou K., Jinek M., Coyle S.M., Ma W., Doudna J.A. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 37.Haurwitz R.E., Jinek M., Wiedenheft B., Zhou K., Doudna J.A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pawluk A., Bondy-Denomy J., Cheung V.H.W., Maxwell K.L., Davidson A.R. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR–Cas system of Pseudomonas aeruginosa. mBio. 2014;5:896–914. doi: 10.1128/mBio.00896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiedenheft B. In defense of phage: viral suppressors of CRISPR-mediated adaptive immunity in bacteria. RNA Biol. 2013;10:886–890. doi: 10.4161/rna.23591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bondy-Denomy J., Garcia B., Strum S., Du M., Rollins M.F., Hidalgo-Reyes Y., Wiedenheft B., Maxwell K.L., Davidson A.R. Multiple mechanisms for CRISPR–Cas inhibition by anti-CRISPR proteins. Nature. 2015 doi: 10.1038/nature15254. doi:10.1038/nature15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shmakov S., Savitskaya E., Semenova E., Datsenko K.A., Severinov K. Pervasive generation of oppositely-oriented spacers during CRISPR adaptation. Nucleic Acids Res. 2014;42:5907–5916. doi: 10.1093/nar/gku226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yosef I., Shitrit D., Goren M.G., Burstein D., Pupko T., Qimron U. DNA motifs determining the efficiency of adaptation into the Escherichia coli CRISPR array. Proc. Natl. Acad. Sci. U.S.A. 2013;110:14396–14401. doi: 10.1073/pnas.1300108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fineran P.C., Gerritzen M.J., Suárez-Diez M., Künne T., Boekhorst J., van Hijum S.A., Staals R.H., Brouns S.J. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1629–1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kornberg A., Baker T.A. DNA Replication. 2nd edn. University Science Books; 2005. [Google Scholar]
- 45.Ivančić-Baće I., Radovčić M., Bočkor L., Howard J.L., Bolt E.L. Cas3 stimulates runaway replication of a ColE1 plasmid in Escherichia coli and antagonises RNaseHI. RNA Biol. 2013;10:770–778. doi: 10.4161/rna.23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levy A., Goren M.G., Yosef I., Auster O., Manor M., Amitai G., Edgar R., Qimron U., Sorek R. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.