Abstract
Cyclic di- and linear oligo-nucleotide signals activate defenses against invasive nucleic acids in animal immunity; however, their evolutionary antecedents are poorly understood. Using comparative genomics, sequence and structure analysis, we uncovered a vast network of systems defined by conserved prokaryotic gene-neighborhoods, which encode enzymes generating such nucleotides or alternatively processing them to yield potential signaling molecules. The nucleotide-generating enzymes include several clades of the DNA-polymerase β-like superfamily (including Vibrio cholerae DncV), a minimal version of the CRISPR polymerase and DisA-like cyclic-di-AMP synthetases. Nucleotide-binding/processing domains include TIR domains and members of a superfamily prototyped by Smf/DprA proteins and base (cytokinin)-releasing LOG enzymes. They are combined in conserved gene-neighborhoods with genes for a plethora of protein superfamilies, which we predict to function as nucleotide-sensors and effectors targeting nucleic acids, proteins or membranes (pore-forming agents). These systems are sometimes combined with other biological conflict-systems such as restriction-modification and CRISPR/Cas. Interestingly, several are coupled in mutually exclusive neighborhoods with either a prokaryotic ubiquitin-system or a HORMA domain-PCH2-like AAA+ ATPase dyad. The latter are potential precursors of equivalent proteins in eukaryotic chromosome dynamics. Further, components from these nucleotide-centric systems have been utilized in several other systems including a novel diversity-generating system with a reverse transcriptase. We also found the Smf/DprA/LOG domain from these systems to be recruited as a predicted nucleotide-binding domain in eukaryotic TRPM channels. These findings point to evolutionary and mechanistic links, which bring together CRISPR/Cas, animal interferon-induced immunity, and several other systems that combine nucleic-acid-sensing and nucleotide-dependent signaling.
INTRODUCTION
In addition to their roles as precursors for nucleic acid biosynthesis, cofactors and energy currency, nucleotides are used as both intra- and extra-cellular signals. Several nucleotides, especially those with cyclic phosphate linkages, are encountered as intracellular signals across the three superkingdoms of life. In addition to being second messengers functioning downstream of extracellular stimuli sensed by surface receptors, they are also produced in direct response to intracellular stimuli. The first identified and best-studied of these, cAMP (1,2), produced in response to different stimuli, binds multiple signaling proteins to regulate several processes, including transcription (3). In animals it is produced upon activation of G-protein-coupled receptors and mediates signaling related to basic metabolic adaptation, as well as specialized processes like learning and memory. In bacteria cAMP regulates developmental and physiological processes (4), including catabolite repression, a phenomenon involving global transcriptional changes to utilize the preferred carbon source (5). A related molecule, cGMP, is also widely utilized as a second messenger. In eukaryotes it plays a role in the global amplification of extracellular signals, contributing to regulation of several processes including ion channel conductance, and in animals has acquired signaling roles in specialized contexts such as ocular phototransduction and smooth muscle relaxation (6). While bacterial cGMP signaling has been implicated in certain developmental signaling processes (e.g. cyst formation in Rhodospirillum (7)), its roles still remain to be investigated in detail (8).
More recently, signaling by other cyclic nucleotides has come to light. The cyclic di-nucleotide c-di-GMP is a major intracellular signaling molecule in bacteria which regulates numerous pathways including the transition between motile single cells and communal biofilms (9), chromosomal replication (10) and polysaccharide synthesis (11–13). The related molecule c-di-AMP has also emerged as a major regulator of global signaling in bacteria, controlling cell-wall synthesis, potassium homeostasis and gene expression (14). It is also generated in response to direct sensing of endogenous branched DNA, making it a checkpoint regulator (15,16). Both eukaryotes and bacteria generate forms of cyclic GMP-AMP (cGAMP). In eukaryotes, 2′-5′ cGAMP is produced upon direct sensing of double-stranded (ds)DNA in the cytoplasm (17). cGAMP then stimulates production of type I interferons, which constitute an important arm of the antiviral response (18). In Vibrio cholerae, formation of 3′-5′ cGAMP regulates virulence (19,20). While the targets and mechanism of activation for cGAMP have been analyzed in some detail in eukaryotes, bacterial cGAMP remains comparatively poorly understood, although in delta-proteobacteria cGAMP has been shown to regulate exoelectrogenesis (21,22).
Like cyclic nucleotides, their linear counterparts play comparable roles as intracellular messengers that convey specific as well as global signals. Guanosine 5′-diphosphate, 3′-diphosphate (ppGpp; known as the alarmone), P1-(adenosine-5′)-P3-(guanosine-3′-diphosphate-5′)-triphosphate (ApppGpp) and P1,P4-diadenosine-5′-tetraphosphate (AppppA) are all produced as intracellular signals in response to stress in bacteria, and some of these have also been reported in eukaryotes. The alarmone regulates the ‘stringent response’ to cellular stress in bacteria (23) and elicits similar responses in plants (24). In vertebrates, linear 2′-5′ oligoadenylates (2′-5′A) ranging from 2 to 30 mers are produced in response to sensing of double-stranded viral RNA and stimulate latent ribonucleases for the degradation of the invading RNA (25). Beyond whole nucleotides, parts thereof might be used as signals. In plants, N6 modified adenines or cytokinins (growth-promoting signaling molecules) are generated via hydrolytic release of the base from corresponding nucleotides, which in turn are derived either from degraded tRNAs or via de novo synthesis (26). A parallel nucleotide-derived signaling molecule was recently demonstrated in Mycobacterium tuberculosis as a potential regulator of its intracellular infectivity (27). Recent work proposes that the TER system, a multi-component anti-bacteriophage and heavy metal resistance network in bacteria, generates a nucleotide-derived modified base that is used as an intracellular signal (28).
Studies on enzymes generating these signaling nucleotides and their derivatives have revealed a web of evolutionary and biochemical connections. All synthetases that use NTPs as substrates to generate the above-mentioned cyclic and linear nucleotides belong to just four distinct superfamilies. The classical adenylyl and guanylyl cyclases (29) and GGDEF domains which generate c-di-GMP (30) belong to a large superfamily of enzymes that also includes most DNA polymerases, reverse transcriptases, viral RNA-dependent RNA polymerases and T7-like DNA-dependent RNA polymerases. Another distinct, large superfamily of nucleotidyltransferases, also including DNA polymerase β (polβ superfamily) (31,32), contains several nucleotide-generating families; namely the CyaA-like bacterial adenylyl cyclases (29,31), the cyclic 2′-5′ GMP-AMP synthase (cGAS), bacterial 3′-5′ cGAMP synthetases typified by the V.cholerae DncV (formerly known as VC0179) (19,20) and 2′-5′A synthetase (oligoadenylate synthetase: OAS). The alarmone-generating RelA/SpoT-like enzymes are highly derived members of this superfamily (31). The characterized c-di-AMP synthetases belong to the DisA superfamily, members of which directly monitor DNA integrity via a fused DNA-binding domain (15,16,33,34). AppppA is generated by amino acyl tRNA synthetases, such as the lysyl tRNA synthetase, in the absence of their usual tRNA substrates (35). Recent work has shown that nucleotide-derived signaling bases, like cytokinins, are generated by NMP ribohydrolases belonging to the vast but poorly-understood SMF/DprA-LOG (SLOG) superfamily (27).
In terms of mechanism of action, some of these signaling nucleotides regulate their targets by direct binding—for instance, the alarmone directly binds the RNA polymerase ω subunit, while the 2′-5′A activates RNaseL via direct binding (8,36). In the case of cAMP and cGMP certain conserved domains, such as the GAF and cNMPBD, serve as sensors within cells (8). While c-di-GMP in bacteria is recognized by protein sensors such as the PilZ domain, it is predominantly sensed using conserved riboswitches (9,11–13). Interestingly, conserved riboswitches are also responsible for the sensing of c-di-AMP generated by DisA in bacteria. Similarly, bacterial cGAMP is sensed by a riboswitch limited in its distribution to deltaproteobacteria, suggesting other potential receptors might be deployed (21,22). In vertebrate defense systems the protein sensor STING detects cGAMP and activates signaling (18). Likewise, the CARF domain was recently proposed as a nucleotide sensor in CRISPR/Cas systems, thereby expanding the role of nucleotide signaling to prokaryotic defense systems (37).
Biochemical and biological studies of classical signaling systems utilizing cNMPs and c-di-GMP along their upstream sensory and downstream signaling cascades has considerably advanced over the past two decades. However, systems centered on other nucleotides, such as the bacterial cGAMP and nucleotide-derived bases, remain less characterized. To better understand these systems we used a comprehensive search strategy based on structural analysis of known nucleotide-generating enzymes combined with comparative genomics. This led to the discovery of a vast network centered on nucleotides and nucleotide-derived bases, encompassing conflict systems acting on non-self nucleic acids, toxin-antitoxin systems and selfish elements. We also identify examples of proteins from these systems being recruited to distinct nucleotide sensor roles in eukaryotes, such as one that might be involved in regulating the transient receptor potential melastatin (TRPM) class of ion channels.
MATERIALS AND METHODS
Iterative sequence profile searches were performed using the PSI-BLAST program (38) against the non-redundant (NR) protein database of National Center for Biotechnology Information (NCBI). Similarity-based clustering for both classification and culling of nearly identical sequences was performed using the BLASTCLUST program (ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html). The HHpred program (39) was used for profile–profile comparisons. Structure similarity searches were performed using the DaliLite program (40). Multiple sequence alignments were built by the MUSCLE (41), KALIGN (42) and PCMA (43) programs, followed by manual adjustments on the basis of profile–profile and structural alignments. Secondary structures were predicted using the JPred program (44). The Pfam database was used as a guide to assign domains to proteins identified through profile searching (45), though the Pfam profiles were often augmented by addition of newly detected divergent members that were not detected by the original models. BLASTCLUST clustering followed by multiple sequence alignment and further sequence profile searches were used to identify domains not present in Pfam. In this way, a comprehensive library of domain architectures covering the entire protein space of the identified systems was constructed. Signal peptides and transmembrane segments were detected using the TMHMM (46) and Phobius (47) programs. Contextual information from prokaryotic gene neighborhoods was retrieved by a custom PERL script that extracts the upstream and downstream genes of the query gene and uses BLASTCLUST to cluster the extracted proteins. This led to identification of all conserved gene neighborhood domain associations depicted in network figures. Domain association networks were rendered using Cytoscape (48). Network edge directionality follows N- to C-terminal positioning for fused domains in a single polypeptide and 3′-5′ order for conserved gene neighborhoods. Domain association counts derived from unique architectures and/or gene neighborhoods at the species level for an organism were used to assign network edge thickness. Networks were constructed with the Kamada–Kawai algorithm (49), with minor manual adjustments made to improve legibility of node connections. Phylogenetic analysis was conducted using an approximately-maximum-likelihood method implemented in the FastTree 2.1 program under default parameters (50). Structural visualization and manipulations were performed using PyMol (http://www.pymol.org). The in-house TASS package, a collection of PERL scripts, was used to automate aspects of large-scale analysis of sequences, structures and genome context.
RESULTS AND DISCUSSION
Identification of a network of novel nucleotide-signaling protein domains
To discover and elucidate components of novel nucleotide-based signaling systems we initially focused on enzymatic domains other than those defining classical second messenger signaling systems, i.e. cNMP and c-di-GMP-centric systems (e.g. cNMP cyclase and GGDEF domains). One such domain is the 3′-5′ cGAMP-generating DncV protein, encoded by the seventh pandemic pathogenesis island-1 (VSP-1) (51) as part of the Vibrio cholerae pathogenesis program (19,52). Little else is known of signaling systems centered on this nucleotide in bacteria. Seeding sensitive iterative sequence profile and hidden Markov model searches with DncV and its orthologs, we detected numerous polβ superfamily proteins. We then clustered the results to isolate DncV-related sequences from other previously-characterized families (31,32) (Materials and Methods). As conserved gene-neighborhood and genomic associations in prokaryotes and domain architectures are a useful tool to elucidate gene functions (53–55), we next constructed a library of such contextual data for DncV-related sequences (Materials and Methods, Supplementary Material).
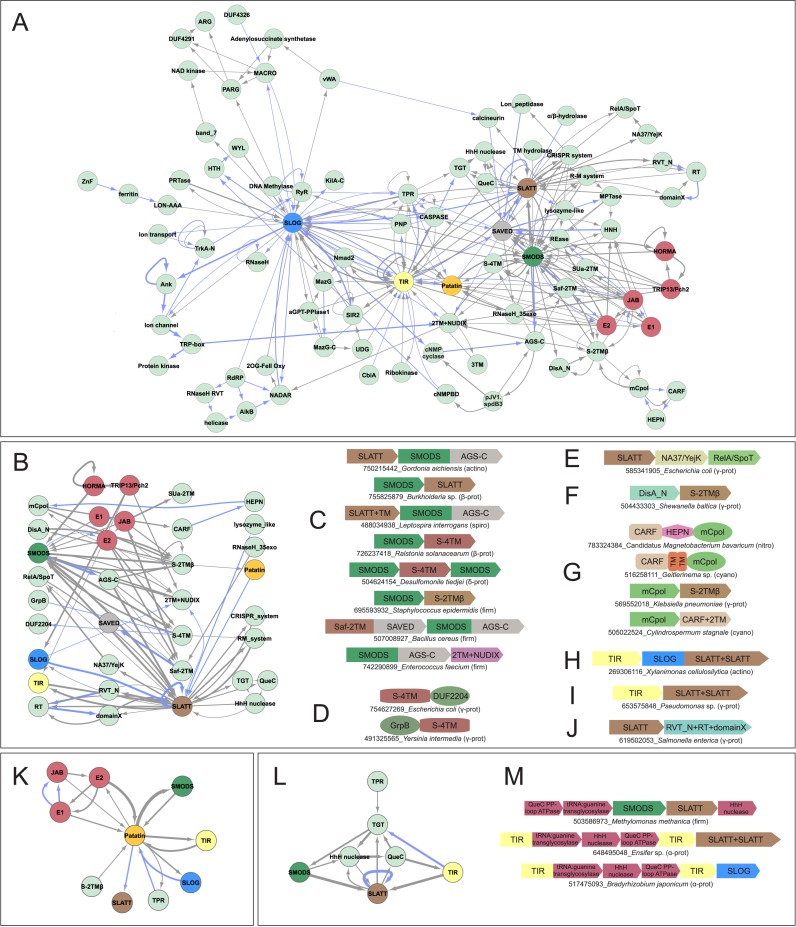
We expanded this library by combining sequence searches with contextual analysis (as above), seeded with domains recovered as having contextual links to DncV-like proteins. Thereby we identified new contextual links for those domains that were independent of DncV-like proteins. Transitively repeating this procedure until we saturated all frequently occurring domains, we obtained a library of potentially functionally-linked proteins. We carried out similar procedures with certain other seeds, such as members of the SLOG superfamily, to extend our analysis to nucleotide-derived signaling bases. The combined findings are displayed as a network in Figure 1A. The nodes of the network are individual domains and usually span phylogenetically distant organisms indicating that they define components of widespread, novel nucleotide-centric signaling systems (for complete list of systems see Supplementary Material). Several nodes, including DncV-like proteins, are hubs, which are shared across multiple systems as defined by conserved gene neighborhoods and architectures (Figure 1A). All domains in the network were subject to in-depth sequence analysis to understand their evolution and structure; hubs in the network are described in detail in the ensuing sections.
Figure 1.
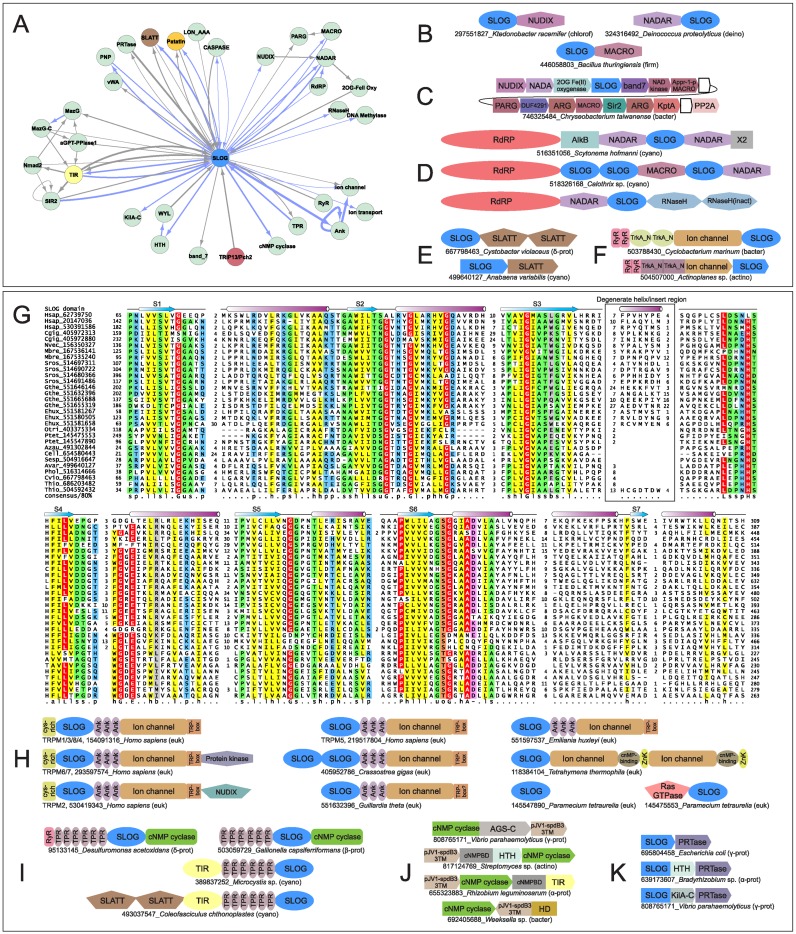
(A) Genome context network overview of identified systems. Networks constructed as described in Materials and Methods, with blue edges representing domains directly fused in the same polypeptide and gray edges representing links in conserved gene neighborhoods. (B) Network depicting the relationship between nucleotide synthesizing and recognition/processing enzymes (left) with sensor/regulatory domains (middle left), TM-containing domains (middle right) and potential secondary effectors (right). (C–J) Representative depictions of conserved domain architectures and gene neighborhoods containing predicted TM pore-forming effectors (for comprehensive list of architectures and neighborhoods, see Supplementary Material). Domain architectures are depicted as adjacent shapes and are not drawn to scale. Gene neighborhoods are depicted as box arrows. Architectures and neighborhoods are labeled with NCBI gene identifier (gi) number and organism name, separated by underscore. Abbreviation of organism lineage is provided to the right of the label in parenthesis. Abbreviations: prot, proteobacteria; bacter, bacteroidetes; actino, actinobacteria; spiro, spirochaetes; firm, firmicutes; nitro, nitrospirae; cyano, cyanobacteria; plancto, planctomycetes; thermo, thermotogae; aquif, aquifex; tener, tenericutes; fuso, fusobacteria; euryarch, euryarchaea; chlorof, chloroflexi; deino, deinococci; euk, eukaryotes. (K) Network centered on the Patatin hub. (L and M) Network and gene neighborhood representation of newly-identified, preQ0-based R-M system.
Overview of hubs in the network
Hub 1: The SMODS domain
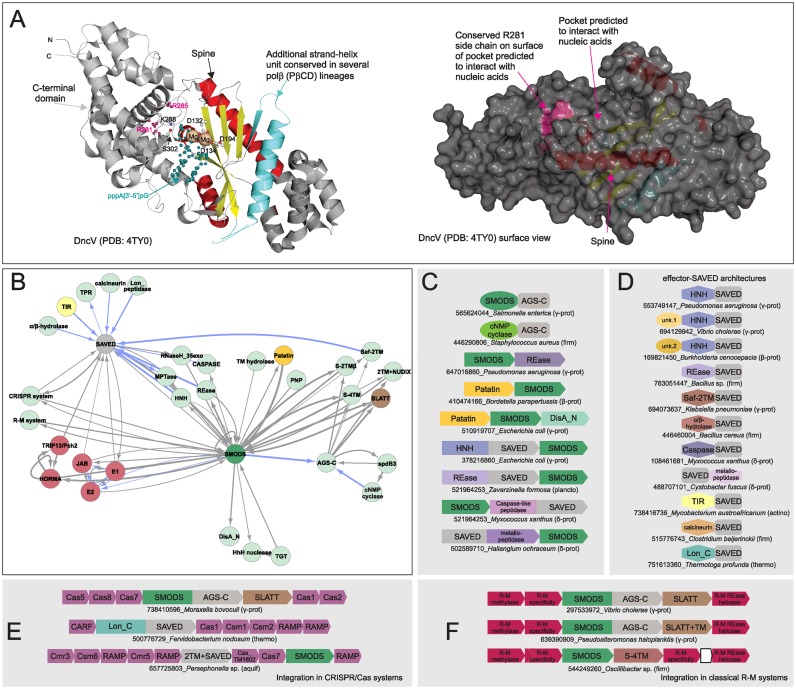
DncV-like proteins are present in most major bacterial lineages and a few archaea (Supplementary Material); however, its sporadic distribution in each lineage is suggestive of dissemination by horizontal gene transfer (HGT). Sequence-similarity searches point to a specific affinity between the DncV-like proteins and the OAS family, with several members of the former group sharing features of the latter. Hence, a subset of the DncV-like proteins are likely to synthesize linear oligonucleotides similar to OAS. Accordingly, we hereafter refer to the DncV-like proteins as the SMODS (Second Messenger Oligonucleotide or Dinucleotide Synthetase) family. The two families are in turn joined in a higher-order clade by the eukaryotic cGAS-like family, which was previously noted to be related to the OAS family (36,56,57). Similarity-based clustering (Materials and Methods) revealed that these three families belong to a large assemblage within the polβ superfamily, which unites several other families, namely the archaeal tRNA CCA-adding enzymes, eukaryotic TRF proteins which add nucleotides to the 3′ end of diverse RNAs, eukaryotic Poly(A)polymerases, NF45/NF90 and NRAP (32,58–60). These are unified by the fusion of the polβ domain to the largely α-helical PβCD domain at the C-terminus (58) and a long N-terminal α-helix in the polβ domain, dubbed the ‘spine’ (36) (Figure 2A).
Figure 2.
(A) Cartoon (left) and surface (right) structure rendering of DncV representative of the SMODS family. Active site residues, predicted oligonucleotide recognition residues, and ligand are rendered as ball-and-stick. Predicted recognition residues (left) and surface patch corresponding to R281 side chain (right) are colored in pink. (B–D) Network and representative domain architectures and gene neighborhoods depicting relationships between SAVED and SMODS domains. See Figure 1 legend for abbreviations and further explanation. (E and F) Representative gene neighborhoods depicting nestling of two-gene systems within classical CRISPR/Cas and R-M systems.
Structural analysis of these families revealed additional features conserved in only a subset of families, including the clade uniting the SMODS, OAS and cGAS-like families (Figure 2A, Supplementary Table S1): (i) a pocket on the ‘backside’ of the nucleotidyltransferase active site, and (ii) two positively-charged residues (lysine or arginine) located in the first helix of the PβCD domain. The same helix also contains a third positively-charged residue (usually lysine) at the opposite end, which points into the active site and interacts with the phosphate group of the nucleotide substrate. In OAS and cGAS these features together with the ‘spine’ are respectively associated with the sensing of dsRNA and dsDNA (17,36). In particular, the second conserved basic residue in the first helix of the PβCD domain might play a role in communicating the sensing of ds-nucleic acids to the active site via the basic residue at the opposite end of this helix. While DncV can form cGAMP in the absence of a double-stranded oligonucleotide in vitro (19), given the conservation of the above features and the depth of the pocket, which houses the nucleic acid in the DncV dimer structure, it is likely that most members of the SMODS family are capable of sensing ds-nucleic acids. It remains unclear if other families of this assemblage, which possess the above features are also similarly regulated by allosteric interactions with nucleic acids. At least in the case of the poorly-characterized eukaryotic NF45/NF90/DZF family (Supplementary Table S1) we propose that such a function is likely: the NF45 protein could function in sensing double-stranded oligonucleotides, consistent with fusions to dsRNA-binding domains and studies linking the family to functional roles in DNA break repair (61), RNA granule assembly (62) and defensive response to the vaccinia virus (63).
Across bacteria several SMODS domains are fused to a previously unknown C-terminal domain, which is also found fused to the C-termini of certain bacterial adenylyl/guanylyl cyclase domains. Hence, we propose the moniker AGS-C (Adenylyl/Guanylyl and SMODS C-terminal) for the domain (Figure 2B, C). AGS-C is predicted to adopt an α+β fold containing a central element with 4–5 contiguous strands (Supplementary Material). It is characterized by multiple well-conserved polar residues, including an absolutely-conserved histidine residue. These features and its independent fusion to structurally unrelated cyclic nucleotide synthetases suggest that AGS-C might act as a sensor for different cyclic nucleotides.
Hub 2: SLOG superfamily proteins
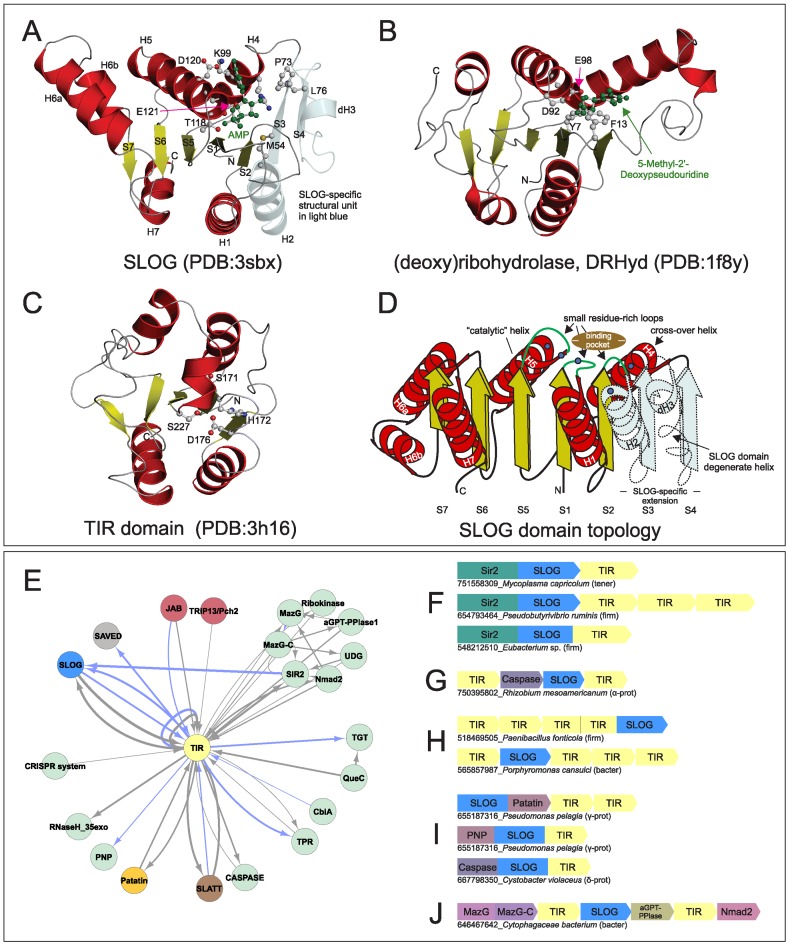
The SLOG superfamily of domains were recently shown in bacteria and plants (26,27) to cleave modified AMPs to release the base as cytokinins, which function as growth-stimulating hormones in plants (26,64–66). Having repeatedly recovered this domain in our contextual linkage searches (Figure 1A), we analyzed it in greater detail. Using similarity-based clustering we identified in the SLOG superfamily a total of 15 families falling into 5 distinct clades (Table 1). Only three of these families have been previously characterized: (i) the classical LOG family which generates cytokinin-like molecules; (ii) the Smf/DprA family which binds single-stranded (ss)DNA and interacts with RecA during transformation and recombination (67); (iii) the molybdenum cofactor-binding (MoCoBD) family (68). We then used available experimental and structural data with alignments constructed for individual families to establish key conserved and lineage-specific features (Table 1). The SLOG domain adopts a three-layered α/β sandwich Rossmannoid fold (69) with a characteristic substrate-binding loop rich in glycine and small residues in the standard location between strand-1 and helix-1 (Figures 2 and 3A–D) (70,71). The SLOG domain is further characterized by (Figure 3A–D, Table 1): (i) two additional loops with small residues C-terminal to strand-2 and strand-5. (ii) A conserved substrate-binding pocket formed predominantly by the region between the crossover helix-4 and helix-5. (iii) Conserved residues derived from helix-2, helix-4 and helix-5 contributing to the active site pocket, which is distinct from other Rossmannoid domains. (iv) Presence of an often degenerate helix (H3) between S3 and S4. (v) Variability of strand-helix units which follow the crossover strand and helix: H6, S7 and H7 are lost in several lineages, while in other families these strands (or S1/H1) are circularly permuted (Table 1).
Table 1. Shared features of the SLOG superfamily.
| Secondary structure elements and conserved residue positions common to SLOG superfamily | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Higher order clade | Family name (pdbid) | S1 small residue loop | S1 loop binding pocket resi. | S2 small residue loop | H2 binding pocket residue | S3 loop residues | H3/dH3 | H4 binding pocket residue | S5 small residue loop | H5 binding pocket residues | H6/H6a-H6b; presence/absence of S7 and H7 |
| LOG proper | classic LOG (2Q4D) | GS | - | GGG | M | PxxL | dH3 | RK | PGG | TxE/DE | H6a-H6b; S7 and H7 present |
| Moco-binding (2IZ5) | GsG | - | sGG | M | P- | dH3 | R | GxG | TxxE | H6; S7 and H7 present | |
| Genome tandem LOG (1WEK) | GSs | - | GGGsG | M | PF | dH3 | RK | PGG | TxDE | H6a-H6b; S7 and H7 present | |
| DUF4478/3412 fusion (4NPA) | GG | H | GCG | M | P- | dH3 | R | PGG | TxEE | H6a-H6b; S7 and H7 present | |
| Sir2/TIR-associating | STALD (Sir2/TIR-Associating LOG-Smf/DprA) | SGs | p | sGxG | h | PF | dH3 | R | GSR/K | xxxE | H6-H6b?; cpS7 and cpH7 present |
| LDcluster2 | SxS | D | GG- | h | P- | H3 | oxMR | GG- | xxEE | H6; H7 present, S7 possibly absent | |
| LDcluster3 | SxS | R | GGH | h | Qo | dH3 | SxxxMR | GG- | xxxE | H6; H7 present, S7 possibly absent | |
| LDcluster4 | GSG | - | TGss | h | P- | dH3 | R | sG- | TxxE | H6; S7 and H7 present | |
| TPALS (TIR/PNP-Associating LOG-DprA/Smf) | GS | - | -ss | DW | polar patch | H3 | Q | GGG | —p | H6; S7 and H7 present | |
| YpsA proper | YpsA (2NX2) | oG | R | ss- | GxD/E | PF | H3 | a | D/Ns | Txxx | H6, S7, H7 absent |
| cpYpsA (3IMK) | NxAGs (cpS1,H1) | R | SGGQ | GxD | P- (dS3) | dH3 | RTxxN | — | GoxxT | H6; S7 and H7 absent | |
| YAcAr (YspA, cpYpsA related) | sGs | - | Gs | GxD/E | – | dH3 | RN | SG- | oxxx | H6, S7, H7 absent | |
| LSDAT proper | LSDAT (LOG-Smf/DprA in TRPM) prokaryote | GGA | - | ssGT | h | – | dH3 | WxxE | sGG | TxxE/D | H6a-H6b; S7 and H7 present |
| LSDAT (LOG-Smf/DprA in TRPM) eukaryote | GG | - | ssG | - | – | dH3 | ExxxR | -GG | —- | H6a-H6b; S7 and H7 present | |
| SMF/DprA | Smf/DprA (4LJR) | Gs | R | SGxA | GxD | Px[6]Y | H3 | RN/D | — | —- | ee-H6; S7 and H7 present |
Abbreviations: o, serine/threonine; s, small residue; h, long hydrophobic residue; x, any residue; d, degenerate secondary structure element; a, aromatic; ?, presence of element not clear from alignment; cp, circularly-permuted element; -, element or residue not conserved; ee, extended loop; resi, residue.
Figure 3.
(A–C) Cartoon renderings of SLOG (A), DRHyd (B) and TIR (C) domains. Secondary structure elements specific to SLOG (A) are colored in light blue and rendered as transparent. Strands in the SLOG domain, N- and C-termini, and conserved residues are labeled. (D) Topology diagram of idealized SLOG domain, with strands and helices depicted as boxed arrows and coils, respectively. (E–J) Network and representative gene neighborhoods of systems containing the TIR domain. See Figure 1 for abbreviations and further explanation.
These features have multiple implications for the SLOG superfamily (Figure 3A–D, Table 1): (i) nucleotide binding, whether of DNA/RNA oligonucleotides as observed in the DprA/Smf family or NMPs as in the LOG family, is a likely ancestral and pervasive feature of the superfamily. (ii) In some cases when alternative ligands were acquired, as in the MocoBD family, they often share structural features with nucleotides, like the pterin phosphate moiety, which is accommodated similar to the sugar phosphate in other families (68). (iii) Several uncharacterized families in the SLOG superfamily display conserved residues, which could catalyze a reaction on a nucleotide or nucleotide-derived substrate similar to the classical LOG proteins. These observations suggest that the SLOG superfamily is likely to encompass dual functions, with certain versions serving as (oligo)nucleotide sensors and others as enzymes that operate on nucleotides (Figure 1A).
Hub 3: TIR domain
Prokaryotic TIR domains (72) repeatedly emerged in our contextual analysis with links to other hubs in the network. TIR domains are best-known for their overlapping roles in innate immunity and apoptosis in various eukaryotes (73,74), where they are believed to act as adaptors mediating protein-protein interactions (75). While bacterial TIR proteins have long been recognized (73,76,77), their biochemistry remains poorly understood. Initially, bacterial TIR domains were thought to be virulence factors disrupting innate immunity in eukaryotic cells; however, this notion has since been persuasively repudiated (72). Additionally, computational analyses have suggested that at least a subset of bacterial TIR domains are likely to function as enzymes that operate on nucleic acids (60,78,79). Moreover, it displays a Rossmannoid fold with conserved residues in the typical active site positions of enzymatic versions of this fold (Figure 3D) (80). Profile–profile comparisons (see Materials and Methods) indicated a specific relationship between the TIR domain and another Rossmannoid domain, the catalytic domain of the (deoxy)ribohydrolase (DRHyd) superfamily (some members included in Pfam Clan CL0498), which unites enzymes hydrolyzing the bond between bases and the pentose sugar in nucleotides/ nucleosides (Figure 3A–C, Supplementary Material). The active site residues of these enzymes are positioned similar to the conserved residues in the TIR domain and the two domains adopt a comparable trimeric configuration. Moreover, these two Rossmannoid domains show further specific structural relationships to the SLOG superfamily, which also recovers the DRHyd superfamily and vice versa in profile–profile searches (Figure 3A–C). These observations suggest that TIR domains are likely to perform functions similar to representatives of the DRHyd and SLOG superfamilies, either as ligand-binding sensors which recognize (oligo)nucleotides or as enzymes processing them.
Hub 4: SLATT domain
A previously-uncharacterized superfamily of domains with two transmembrane (TM) helices was found to frequently link the SMODS and SLOG domains in the contextual network (Figure 1A). This domain has representatives in most major bacterial lineages, some eukaryotes and Nucleo-Cytoplasmic Large DNA viruses (NCLDVs). While a subset of the superfamily is detected by the DUF4231 model (Domain of Unknown Function (81)) from the Pfam database (45), a major fraction of this family as defined by us was not captured by this model. Multiple alignments revealed a conserved core for the domain consisting of a pair of N-terminal TM helices and a largely helical C-terminal cytoplasmic region (Supplementary Material). We accordingly term the expanded superfamily the SLATT (for SMODS and LOG-Smf/DprA-Associating Two TM) domain. Clustering analysis identified seven monophyletic families of SLATT domains, of which five are critical components of systems we uncovered (Supplementary Material). The TM helices often contain family-specific polar residues that are likely to form an intramembrane aqueous channel that might facilitate transport of molecules across the membrane. Of the two families of SLATT domains that do not seem to occur in nucleotide-centric systems, one tends to be encoded by solo genes bereft of genomic context in bacteria. The other occurs in several fungi and is typically lineage-specifically expanded in them (Supplementary Material).
Hub 5: SAVED, a potential nucleotide sensor domain fused to diverse effectors
We uncovered a previously uncharacterized domain with strong operonic linkage to genes encoding SMODS enzymes. Strikingly, it seldom occurred by itself, instead showing fusions to various domains that we interpret as being effector domains (see below and Figure 2B, D). Hence, we named this domain the SAVED domain, for SMODS-associated and fused to various effector domains. The SAVED domain is predicted to adopt an α+β fold featuring a central 4–5 strand β-sheet and multiple well-conserved residues including a characteristic histidine. The position of the conserved histidine within the central β-sheet and the secondary structure parallels the above-mentioned AGS-C domain, suggestive of a potential distant relationship between them (Supplementary Material). Moreover, given its strong operonic and/or phyletic pattern correlation with the SMODS domain, it appears that the SAVED domain, like the AGS-C domain, might bind cGAMP or a linear oligonucleotide (similar to 2′-5′A) generated by the former enzymes.
The SAVED domain is fused to domains such as (Figures 2B, D and 4A): (i) an HNH endonuclease domain sometimes accompanied by a further N-terminal fusion to an uncharacterized domain; (ii) a restriction endonuclease (REase) domain; (iii) a TIR domain; (iv) a calcineurin-like phosphoesterase domain (82); (v) a Lon peptidase domain with a serine-lysine active site (83); (vi) a metallopeptidase (MPTase) domain; (vii) a caspase-like peptidase (84); (viii) a JAB deubiquitinating peptidase domain; (ix) an α/β-hydrolase domain; (x) a 2TM module named SAF-2TM (for SAVED-fused 2TM), distinct from the above-mentioned SLATT domains. Except for the MPTase domain, which is fused to the C-terminus, the rest of the above domains are fused to the N-terminus of the SAVED domain. This architectural theme where the constant and well-conserved SAVED domain is combined with unrelated variable domains (Figure 2D), which are often found in other inter-genomic or inter-organismic conflict systems, parallels polymorphic toxin proteins with their constant parts and variable toxin domains (85,86). This suggests that the fused domains are likely to act as effectors enzymatically targeting nucleic acids, proteins or membranes (the 2-TM domain being a potential pore-forming toxin (87)).
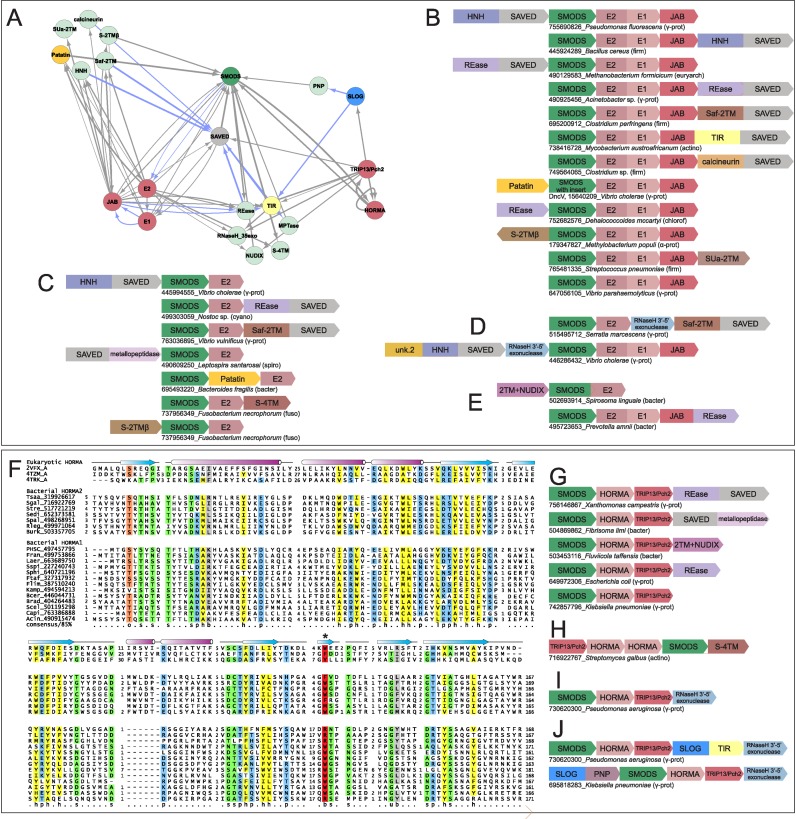
Figure 4.
(A–E) Network and conserved gene neighborhoods representing systems containing the Ubl conjugation systems combined with two-gene systems. See Figure 1 for abbreviations and further explanation. (F) Multiple sequence alignment of bacterial HORMA domains. Sequences in alignment are labeled to the left by organism abbreviation and gi number, separated by underscore. The first three sequences are known eukaryotic HORMA domains with experimentally determined structures labeled by Protein Databank (pdb) id and chain letter, separated by underscore. Bottom line provides amino acid residue consensus conservation for individual columns in the alignment, color-coordinated as follows: o, hydroxyl-bearing colored in orange; s, small colored in green; p, polar colored in blue; h, hydrophobic colored in yellow; b, big colored in gray; u, tiny colored in green. Conserved tryptophan crucial to the conserved HORMA binding pocket is marked with ‘*’ and colored in red. (G–J) Network and representative gene neighborhoods for systems containing the HORMA-TRIP13/Pch2 dyad combined with two-gene systems.
Hub 6: Mutually exclusive, functionally equivalent Ubiquitin-conjugation and HORMA-TRIP13/Pch2 systems
Two self-contained systems of domains constitute hubs in the network, which display comparable connectivity to partner nodes while never being connected to themselves (Figures 1A and 4A). This suggests that, while these two systems are biochemically distinct, they are likely to perform comparable functions with respect to their partners in the network. The more common of the two is a prokaryotic ubiquitin (Ub)-conjugation system (Figure 4B–E). A spectrum of prokaryotic Ub systems, containing either a complete or a partial complement of the domains present in eukaryotic Ub-conjugation systems, have been described in prokaryotes (88–91). Those found in the current network contain the Ub-ligase E2 and usually also the E1-ligase, but lack RING-like E3 adaptor ligases (89,92,93). They further usually contain a JAB deubiquitinase (DUB) which removes Ub from the substrate (Figure 4B) (88,89). Notably, while several prokaryotic Ub systems contain operonically-linked Ub-like (Ubl) proteins (88,89), they are absent in the systems considered here. Absence of the Ubl and occasionally the E1 ligase in the systems (Figure 4C) is likely compensated for by utilization of another Ubl/E1 encoded in the genome. Consistent with this, there is evidence that ThiS and/or MoaD-like Ubls and ThiF/MoeB-like E1s might be utilized in conjugation even in the absence of operonic couplings of Ubls and E1 in prokaryotic genomes (94–98).
The second system of domains, constituting the alternative to the above hub, combines the first-identified prokaryotic homologs of the eukaryotic HORMA domain (Figure 4F) (99) with a prokaryotic homolog of the TRIP13/Pch2 family of the AAA+ superfamily of P-loop NTPases (100,101) (Figures 1A and 4G–J). In eukaryotes, homologs of the two domains play complementary roles during meiosis: HORMA proteins form a multimeric scaffolding complex via peptide-capture interactions mediated by a conserved binding cleft in the HORMA to ‘coat’ the chromosomal axis, while the TRIP13/Pch2 ATPase is involved in depletion of these complexes in regions where synaptonemal complexes form (102,103). Conservation of the peptide-binding pocket with the characteristic tryptophan in prokaryotic HORMA domains (Figure 4F, Supplementary Material), along with the strict operonic association with the TRIP13/Pch2 ATPase, suggest that the functional association between the two, which is observed in eukaryotes, is also preserved in bacteria.
While biochemical properties of these hubs are not immediately suggestive of cyclic-/oligo- nucleotide generation or recognition, their strong connectivity to the SMODS and SAVED domains suggests that they are likely to dynamically interact with components directly involved in a nucleotide-related function (see below).
Hub 7: Patatin domain
The patatin domain shows a strong connection in the network to the SMODS domain and also less-prominent but consistent connections to the TIR and SLATT domains (Figure 1A, K). Patatin belongs to the α/β hydrolase fold (104) and functions as a phospholipase, involved in cleavage of fatty acyl moieties from lipids at the cell membrane (105). When linked to the SMODS domain, it typically occurs mutually exclusively with SAVED-containing effectors, suggesting that it might operate as a membrane-targeting effector that is directly regulated by the SMODS partner.
Functional reconstruction of prokaryotic nucleotide-dependent systems
Analysis of the over 3000 prokaryotic conserved gene neighborhoods (Supplementary Material) represented by the network (Figure 1A), together with sequence analysis of individual components, allowed us to discern several ‘syntactical’ features in their organization. Based on these we reconstructed several potential functional themes for these nucleotide-dependent systems and accordingly organize them for further discussion (Supplementary Table S2, Material).
Systems with two primary components
The simplest systems we identified consist of two genes tightly-coupled both in terms of physical distance in the genome and the breadth of the prokaryotic tree traversed by the gene pair. Based on the proteins encoded by the gene pairs we discerned two broad functional themes: (i) a cyclic- or oligo-nucleotide-generating enzyme coupled with another protein, which might belong to one of several unrelated superfamilies and (ii) a nucleotide-binding or processing domain similarly coupled to a protein which might be drawn from different, unrelated superfamilies. This pattern of gene-coupling is one of the defining characteristics of type-II toxin-antitoxin systems (T-A), wherein one of the components has a deleterious effect on the host (the toxin) while the other (the antitoxin) counteracts or controls the effects of the toxin (106). Indeed, this model helps interpret certain key features that we observe in these gene pairs. Sequence analysis revealed that the second gene in the pair, which encodes a protein drawn from one of several unrelated families, shows all hallmarks of being a toxin or effector (see below). However, the key variation on the basic T-A-like theme is the signaling aspect of the first component of the system, which either generates, binds or processes a nucleotide. In light of this, we interpret these as being nucleotide-regulated signaling systems, which generate or process a nucleotide in response to a stimulus, and this nucleotide in turn activates the coupled effector protein. We discuss below examples of each of the above themes to illustrate the striking diversity of components within them.
Systems with a nucleotide-synthesizing enzyme
All of these systems are united by one of the two components being a nucleotidyltransferase, which is predicted to generate a signaling nucleotide using NTP substrates (Figure 1B). The most common of these enzymes are members of the SMODS clade, contributing to its emergence as a hub in the network (Figures 1A–C and 2B–C). In addition to SMODS, we also infrequently found two other functionally uncharacterized families of the polβ fold, GrpB and Pfam DUF2204 (Figure 1B, D) (32), in similar systems with two components, suggesting that they too might generate an uncharacterized nucleotide product. Further, we also identified systems with a divergent variant of the polβ fold, which is specifically related to the nucleotide-generating domain of the RelA/SpoT proteins, suggesting that it might synthesize a molecule similar to the alarmone ppGpp (Figure 1B, E) (107). Other systems of this type display unrelated nucleotide-generating enzymes: some display the DisA-N domain suggesting that they likely synthesize c-di-AMP, similar to previously characterized DisA-N domains (Figure 1B, F) (16). Finally, we found one further conserved domain in such systems, which profile–profile comparisons showed to contain a minimal version of the polymerase palm domain (with the RRM fold) with a specific relationship to the catalytic domain of the CRISPR polymerases (frequently referred to as Cmr2 or Cas10) (Figure 1B, G)(108,109). As they conserve structure and sequence features required for nucleotidyltransferase activity we named this the mCpol (minimal CRISPR polymerase) domain. Given that these nucleotidyltransferase domains share a specific relationship to the GGDEF domains that generate c-di-GMP, it is conceivable that both mCpol and regular CRISPR polymerases generate cyclic nucleotides like c-di-AMP (especially given that the active site of crystallized CRISPR polymerase domains contain an ATP (110)).
Despite this diversity of nucleotidyltransferases, these systems can be placed in two thematic categories:
Those coupled primarily to enzymatic effector domains with intracellular targets. This thematic category is dominated by systems in which a SMODS protein is linked in an operon to a protein with a SAVED domain fused to one of at least 10 distinct effector domains (described above, Figure 2B–D). The other frequently occurring variant features effectors with Patatin or REase domains that however lack the SAVED domain (Figure 2C). These are most frequently operonically combined with a SMODS enzyme but occasionally also to a DisA-N c-di-AMP-generating enzyme (Figure 2C). Certain CRISPR polymerase-related mCpol proteins might also constitute such systems when they are fused to divergent CARF (another previously predicted nucleotide-sensor domain with a Rossmann fold (111)), wHTH, and predicted RNase HEPN domains (Figure 1G) (37,79). The nucleotide generated by the nucleotidyltransferase could be sensed by the SAVED (Figure 2C, D), CARF (Figure 1G) or directly (in case of Patatin and certain REase domains) (Figure 2C) to unleash the activity of the effector domain to then target nucleic acids, proteins and lipids.
Those coupled to potential pore-forming effectors. Here, in place of an intracellular effector protein, the second gene encodes a SLATT domain (also a hub in the network) or another TM protein (Figure 1B–H). Like the SLATT domain, the other linked TM proteins also typically contain polar residues within their predicted TM-segments (Supplementary Material). Together with the functional analogy to the above theme, this suggests that the TM proteins function as pore-forming effectors, which are gated on the intracellular face of the membrane by nucleotides generated by the associated nucleotide-generating enzyme. The most common arrangement combines a SMODS-clade enzyme with a SLATT superfamily protein (Figure 1C). A variant of this system combines the SLATT domain with the RelA/SpoT-like enzyme in place of the SMODS (Figure 1E). This type typically contains a third component, a protein of the NA37/YejK superfamily, which binds DNA (112,113). In several cases the RelA/SpoT-like enzyme might also occur with just the NA37/YejK independently of the SLATT protein. The presence of NA37/YejK proteins in these systems suggests that, like the previously reported DisA systems, these might be activated by sensing DNA, perhaps with unusual structural or compositional features, which in turn induces the RelA/SpoT-like enzyme to generate a nucleotide that triggers a response either via the linked SLATT protein and/or a version of the stringent response.
The S-4TM (for SMODS-associating 4TM) is another novel TM domain, which in addition to the SMODS enzymes might be coupled to other polβ enzymes of the GrpB and DUF2204 families (Figure 1C, D). Another family of 2TM proteins, unrelated to the SLATT proteins, is linked to SMODS, DisA-N and mCpol nucleotide-generating enzymes in different systems. These proteins are characterized by a novel C-terminal intracellular domain S-2TMβ (for SMODS-associating 2TM, β-strand rich) with seven predicted β-strands, suggestive of a lipocalin-like β-barrel-like structure (Figure 1C, F, G, Supplementary Material). We predict that this domain probably serves as a sensor that binds the nucleotide generated by the linked generating enzyme. Finally, in a further system SMODS enzymes are coupled to a 2TM protein with an intracellular region containing a divergent version of the NUDIX domain with certain distinctive features in the substrate-interacting region (Figure 1C, Supplementary Material) (97). An additional characteristic of these systems is the fusion of the predicted nucleotide-binding AGS-C domain to the SMODS domain. NUDIX enzymes cleave nucleotide diphosphate bonds (114); hence, as has been observed for certain eukaryotic ion-channels, it is conceivable that they cleave such bonds in the nucleotides synthesized by the operonically-linked nucleotide-generating enzymes to regulate flux through the pore formed by the associated 2TM protein.
Both of these thematic categories of systems are conceived as being comparable to the well-studied eukaryotic systems with OAS and cGAS (36,57). Given that SMODS proteins conserve the cognate nucleic-acid-recognition elements we predict that they too recognize invasive nucleic acids from bacteriophages or plasmids and synthesize nucleotide signals in response. The synthesized nucleotide in turn binds the effector and activates it to either target cellular components, thereby depriving the invader of an opportunity to replicate, or prevents the same by programmed cell death (115). Alternatively, they could directly target the nucleic acids or proteins of the invader. Moreover, a small subset of systems contains both SMODS and DisA-N proteins suggesting that in these cases more than one nucleotide signal might be deployed (Figure 2C, Supplementary Material). Presence of the mCpol domain in these systems (Figure 1G) also suggests that the related CRISPR polymerase in the CRISPR/Cas Cmr complex operates by generating comparable cyclic/oligo-nucleotide signals (111,116). Further, the observed linkage of mCpol to the CARF domain implies that the latter domain might similarly sense nucleotide signals in classical CRISPR/Cas systems, which are rife with CARF domain proteins (37). In light of this proposal, the HD-phosphoesterase domain, which is fused to the CRISPR polymerase domain in CRISPR/Cas systems, might provide a means of terminating the signal by hydrolyzing the nucleotide. This would be comparable to the activity of the cNMP phosphodiesterases with HD domains in classical cNMP signaling (117,118).
Systems with a nucleotide-binding or processing protein
While these usually share the two-gene operon architecture with the above-described systems, they differ from them in lacking a nucleotide-synthesizing enzyme. Nevertheless, presence of potential nucleotide-binding domains, as well as effectors shared with the above systems suggest that these systems also operate in a nucleotide-dependent fashion. Here too the same dichotomy is observed as above, with some containing predicted intracellular effectors and others TM effectors. Most of these systems are centered on TIR domains, which are predicted to bind/process nucleotides or their derivatives in these prokaryotic contexts (see above, Figures 1A,B, and 3E–J). A prominent type of system with a potential intracellular effector combines a TIR domain protein with another protein that contains a SLOG domain fused to a distinctive version of the sirtuin (Sir2) domain that lacks the Zn-ribbon insert of the classical versions (Figure 3F, Table 1: Sir2/TIR-associating clade). While classical Sir2 domains are thought to function as protein deacylases (119), other studies have repeatedly implicated Sir2 as a potential effector in conflict systems, which targets DNA, perhaps via ADP-ribosylation or even nuclease activity (60,120). Hence, we posit that the Sir2 domain functions as the effector, while the SLOG domain is the potential sensor of the nucleotide or its derivative (Figure 3F).
We also observed few variations on this theme: the first of these replaces the Sir2 protein with a peptidase of the caspase superfamily, which presumably functions as the effector (Figure 3G). It is linked to a SLOG domain from a clade different from those linked to Sir2, and a TIR protein, both encoded by separate genes. The second of these displays a gene coding for a protein with a SLOG domain fused to either of two distinct families of the SLATT domain occasionally combined with a gene for a TIR domain (Figure 1H, Supplementary Table S2). Yet another unites two genes respectively coding for SLATT proteins belonging to different families (Figure 1I, Supplementary Table S2), which are predicted to function as pore-forming effectors, and a TIR protein. Other related systems depart from this basic architecture by often combining one to four evolutionarily distant TIR domains with two distinct families of SLOG domains (Figure 3H, Table 1: Sir2/TIR-associating clade), either as multiple tandem genes in an operon or via fusion into a gene coding for a single polypeptide. Some of these systems additionally include a gene coding for a patatin domain (Figure 3I). In some the TIR and the SLOG domains are fused to caspase or purine nucleoside phosphorylase domains (Figure 3I). As proposed for the above systems, the linked enzymatic domains could function as effectors.
The exact mechanism of action of these systems is less clear than those with nucleotide-generating synthetases. Given our proposal that at least a subset of the TIR domains might bind and/or process (oligo)nucleotides or their derivatives, they could sense such molecules generated by other cellular processes or modified nucleotides generated by bacteriophages and restriction systems (see below). They could bind these to relay a signal to their effector components. Additionally, the SLOG domains are also known or predicted to bind and/or process nucleotides to release a free base and could act as further nucleotide-sensors or regulators of signaling by processing the nucleotide. In those systems where there are multiple TIR domains, in addition to nucleotide-recognition, they could also form multimeric complexes as part of the response.
Multi-component systems combinatorically derived from core systems with two components
These are derived systems where a basic system with two components, no different from those described above, is combined with other systems, each with several additional components. The latter systems too might occur independently. However, the multi-component combination travels as a distinct evolutionarily mobile unit between different prokaryotes indicating that the combination of these otherwise independent systems can have special functional consequences. Hence, we describe them below in greater detail with an emphasis on functional predictions for the combined units.
Systems with Ubl-conjugation or HORMA-TRIP13/Pch2 components
In their most common manifestation, these systems combine a basic two-gene system with a SMODS enzyme and an effector with one of the two mutually exclusive multi-protein network hubs described above (Ubl-conjugation or HORMA-TRIP13/Pch2) (Figures 1A and 4A). The HORMA-TRIP13/Pch2-containing systems come in two types with either a single or two divergent HORMA proteins (Figure 4G, H). The SMODS component of these systems also display distinct sequence affinities depending on whether the basic system is combined with a Ubl-conjugation or the HORMA-TRIP13/Pch2 system: those associated with the former share specific sequence features with the V. cholerae DncV cyclase suggesting that they are likely to synthesize a cGAMP signal, while the HORMA-TRIP13/Pch2-associated versions share specific sequence features with OAS, suggesting they might generate 2′-5′A-like oligonucleotides. Both types of systems sometimes feature a 3′-5′ exonuclease of the RNaseH fold, which is not found in the standalone two-gene systems. This 3′-5′ exonuclease might either play the role of the sole effector or act in combination with the other effector encoded by the operon (Figure 4D, I, J). While the SAVED domain is most commonly found as the predicted effector-linked nucleotide sensor in these systems (Figure 4B, C, G), variant versions (always linked to HORMA-TRIP13/Pch2) instead feature the SLOG domain in its place (Figure 4J). This clade of SLOG domains was previously unknown, and lack the residues typical of catalytic versions of the superfamily (Table 1: TPALS family, Supplementary Material), thereby supporting the idea that they are nucleotide sensors rather than active enzymes. These SLOG domains are fused to TIR or a nucleoside phosphorylase domain (Figure 4J) (121), which could act as effectors in these systems (Supplementary Table S2).
There are other subtle architectural features in the organization of these systems: first, there are instances of fusions between otherwise discrete components; e.g. occasionally, Ubl-system components are observed fused to the effector proteins and SMODS is observed to be fused to the E2 domain (Figure 4E). Second, these gene neighborhoods display certain syntactical conservation in terms of ordering of the genes (Figure 4B, C, G, J, Supplementary Material). Together with the tendency of these systems to travel as a unit across the bacterial tree, these observations suggest that the components are assembled into a single complex, with their synthesis in a certain order facilitating proper assembly. In functional terms, these multicomponent systems could be interpreted thusly:
The Ubl-conjugation or HORMA-TRIP13/Pch2 components are ‘co-effectors’. In this scenario they are activated either directly by the nucleotide generated by the SMODS enzyme or through physical interaction between the SMODS and components in the formed complex and act in conjunction with the effector encoded by the basic two-gene core to reinforce its action. Thus, the Ubl-conjugation components can be conceived as modifying invader- or host-proteins via ligation of an Ubl tag thereby targeting them for degradation; the JAB protein would help in cleaving off this Ubl tag at the cellular protein-degradasome, after which the Ubl could be ‘recycled’ for further rounds of conjugation. Similarly, in this proposal the HORMA domain undergoes a switch, mediated by the ATPase action of the TRIP13/Pch2 component, allowing it to capture peptides from the host or invader proteins.
In an alternative although not mutually exclusive scenario, the Ubl-conjugation or HORMA-TRIP13/Pch2 components could function as modulators that help contain the effectors unleashed by nucleotide-production after they have completed their role in the ongoing conflict. Consistent with this, fusions between the JAB domain and the effector component or the SMODS and the E2 ligase (Figure 4E) suggest that components of the basic two-gene system might be targeted for modification and subsequent degradation. Similarly, in this scenario the HORMA domains might capture peptides from components encoded by the basic two-gene system and perhaps render them suitable for degradation in conjunction with the ATPase action of TRIP13/Pch2.
Combinations with CRISPR/Cas systems
Proteins with the effector and SAVED domains are found independently of the SMODS domain lodged within CRISPR/Cas systems in several distant bacterial lineages (Figure 2E, Supplementary Material). Likewise certain SLATT-SMODS systems are also lodged within CRISPR/Cas systems (Figure 2E). This embedding again strengthens the above proposal that the CRISPR/Cas systems are activated by nucleotide second messengers. Consistent with this, in most genomes with such combinations at least one CRISPR-polymerase is predicted to be catalytically active, suggesting that they might generate the nucleotide to activate the embedded effector proteins in addition to the standard CRISPR/Cas effectors in the form of the CARF domain proteins.
Combinations with diverse R-M and DNA-modification systems
The basic two-gene systems (e.g. certain SLATT-SMODS systems) are often nested within classical restriction-modification (R-M) systems (Figure 2F). Even more striking is the sporadic but phyletically widespread system coupling a basic two-gene system, coding for a SLATT protein and either a SMODS or TIR domain protein, with a distinctive three-gene system (Figure 1L, M). This latter system codes for a QueC-like PP-loop ATPase, a divergent member of the nucleic acid guanine transglycosylase family (TGT) and an endoDNase domain related to Helix-hairpin-Helix (HhH) DNA glycosylases. Recent research has identified this divergent member of the TGT family as an enzyme that catalyzes the transfer of a deazaguanine base, PreQ0, into DNA in place of guanine (122). Similarly, QueC in this system was identified as the enzyme which likely synthesizes PreQ from free 7-carboxy-7-deazaguanine available in the cell as a precursor for tRNA metabolism. The HhH-endoDNase/DNA-glycosylase component of the system is predicted to act as the enzyme targeting foreign DNA not containing the PreQ0 modification, analogous to the methylation-based restriction in prototypical R-M systems. Another distinctive combination is seen in the form of a subset of the TIR-SLOG systems from diverse bacterial lineages being associated with a Nmad2 domain protein, the MazG nucleotide pyrophosphohydrolase fused to the MazG-C domain (122) and a α-glutamyl/putrescinyl thymine phosphorylase (aG/PT-pyrophoshorylase) domain (Figure 3J, Supplementary Table S2). This combination of components was recently reported as constituting a nucleotide modification pathway for biosynthesis of hypermodified thymines in DNA (122), which might again be used in distinguishing self from non-self DNA in bacteriophage-host conflicts.
In all the above cases, the basic two-gene system is interpreted as functioning in conjunction with the associated R-M system, either as a force-multiplier or as a backup, which can facilitate the induction of dormancy or cell suicide in the event of the failure of the associated R-M system (115). Indeed, such couplings of R-M systems with HEPN domain RNases and Abi-like counter-phage systems have been previously reported and observed to function as back-ups for the former systems (79,123).
Disparate systems sharing certain components with the above systems
A Novel retroelement with diversity-generating potential
This system displays the same two-gene architecture as the earlier-described systems coding for SLATT proteins; however, here the SLATT genes are remarkably combined with a distinctive reverse transcriptase (RT) gene (Figure 1B, J; Supplementary Table S2). These SLATT proteins are distinguished by the presence of a third TM segment after the C-terminal helical cytoplasmic tail. Moreover, these SLATT domains display rapid sequence divergence relative to all other prokaryotic SLATT domains (Supplemental Material). The RT domain, also rapidly evolving, is most closely related in terms of domain architectural features and sequence affinities to RTs observed in bacterial retroelements, including group-II introns (124) and diversity-generating retroelements (DGRs) (125). These two retroelements, along with this novel system, share a C-terminal fusion to the so-called ‘domain X’, which is thought to bind RNA during reverse transcription (126,127). However, like the DGRs, this novel system does not contain the HNH endonuclease found fused in many group-II intron RTs. Phylogenetic analysis reveals that this system is highly mobile and is seen in diverse proteobacteria, firmicutes, bacteroidetes and fusobacteria (Supplementary Material). Elements from different strains of the same species or even same genome often fail to group together in phylogenetic analyses, suggesting independent acquisition due to hypermobility (Supplementary Figure S1). These observations indicate that the system defines a highly-mobile selfish retroelement.
Drawing analogy to the aforementioned retroelements and retrons, which use the RT domain in the production of multicopy single-stranded DNA in certain bacterial genomes (128), we propose a mechanism of dispersal for this novel retroelement: RNA copies of the element are likely transcribed and translated by host machinery. The resulting RNA transcript is then reverse-transcribed into DNA by the RT domain and transported out of host bacteria via pores formed by the SLATT protein. Specificity in DNA transport could be mediated by the distinctive C-terminal region of these SLATT domains. Interestingly, given the rapid sequence divergence of both components of this system, it is likely that the RT is error-prone and has potential to generate diversity. It is possible that diversification of the SLATT protein by this mechanism might confer some advantage to the host cells.
Systems linked to NAD utilization and ADP-ribosylation
Several biological conflicts involve NAD-dependent pathways: toxins from T-A systems, polymorphic toxin systems and related conflict systems ADP-ribosylate proteins and nucleic acids using NAD as the substrate (86,129). RNAs cleaved by toxins often have 2′ phosphates, which are repaired by the KptA enzyme using NAD as a substrate generating an ADP-ribose derivative as a byproduct (130). Members of a distinct clade of the SLOG superfamily (Table 1; YspA clade) show fusions to NUDIX, NADAR and MACRO domains (Figure 5A, B), which are involved in cleaving ADP-ribose adducts or processing ADP-ribose derivatives for clearance (129,131). Members of one these clades also occur as part of previously-described giant operons, which combine several genes that utilize NAD and process ADP-ribose derivatives (Figure 5C) (129,131). Similarly, they are also found as domains in previously-described large proteins fused to C-termini of bacterial RNA-dependent RNA Polymerase (RdRP) modules along with Macro and NADAR domains (Figure 5D) (132). Hence, these SLOG domains might act both as sensors and processing enzymes that act on ADP-ribose or NAD. These systems could be potentially deployed in response to ADP-ribosylation of cellular components by toxins or, in the case of those containing KptA, for RNA repair. The enigmatic proteins with RdRP modules might likewise have a role in a hitherto unknown RNA-repair mechanism.
Figure 5.
(A–F) Network and conserved gene neighborhoods representing systems centered on the SLOG domain. See Figure 1 for abbreviations and further explanation. (G) Multiple sequence alignment of SLOG domain family found in TRPM ion and predicted prokaryotic membrane channels (LSDAT family in Table 1). See Figure 4 for explanation of multiple sequence alignment and abbreviations. Conserved residues are colored in red with letters shaded in white. Additional abbreviations: -, negatively-charged residue; l, aliphatic residue. (H) Representative domain architectures of the SLOG domain in TRPM ion channels and other eukaryote contexts. (I) Representative domain architectures of SLOG domains predicted to be part of ‘conventional’ second messenger signaling systems. (J) Representative gene neighborhoods of pJV1-spdB3-based signaling systems. (K) Gene neighborhoods representing predicted signaling switch mediated by SLOG and PRTase domains.
Nucleotide-centric systems in non-conflict contexts
Despite their intrinsic diversity, all above-described systems can be broadly interpreted as participating in biological conflicts. This general functional theme distinguishes such systems from other previously well-studied nucleotide-based signaling systems with primarily homeostatic and environment-sensing functions for the cell. Nevertheless, our analysis uncovered several instances where components related to those from above systems have been deployed in apparently non-conflict contexts, in both prokaryotes and eukaryotes. We describe the most notable of these below.
Prokaryotic and eukaryotic systems regulating ion flux and membrane transport
Several bacterial proteins with fusions of SLOG and SLATT domains are encoded by solo genes independently of their TIR partners (Figure 5E). Hence, it is possible that these have roles other than in conflicts. Again in bacteria, the SLOG domain is combined either in operons or via direct fusion to a novel 4TM domain (e.g. gi: 503733372 from Nitrosomonas). This TM domain is also fused to two tandem Rossmann fold TrkA-N domains, which bind NAD+ (133), and/or the poorly-understood RyR domains, which are often observed fused to calcium channels in eukaryotes (Figure 5F) (134). Iterative sequence searches revealed that it is distantly related to eukaryotic ion channel domains, suggesting that this TM domain might function as a novel type of bacterial ion channel.
PSI-BLAST searches initiated with one of the families of SLOG domains fused to the SLATT domain in bacteria (Table 1: LSDAT clade) also recovered homologs from diverse eukaryotes (see alignment, Figure 5G). Strikingly, the domain mapped to the N-terminal region of the TRPM family of ion channels (Figure 5H) (e.g. query gi: 499640127, Anabaena variabilis recovers gi: 109730277, Homo sapiens; PSI-BLAST e-value: 7e-11; iteration: 1). These channels have been extensively studied in animals including humans and are monovalent cation channels, also accommodating divalent cations with varying specificity (135–137). Previous studies have revealed complex domain architectures for the TRPM family but no known domain had been found in the large conserved N-terminal region where we identified the SLOG domain (138–141). Thus, we can now present the core domain architecture of the TRPM family as: an N-terminal cytoplasmic SLOG domain followed by three divergent ankyrin repeats (consistent with ankyrin repeats previously reported in other classes of TRP channels (142)), the 6TM ion channel domain and the so-called C-terminal cytoplasmic ‘TRP-box’ motif (Figure 5H). In several members of the TRPM family there is an additional cysteine-rich cytoplasmic domain fused at the extreme N-terminus. Further, we found that the core architecture (SLOG+3Ankyrin+ion-channel) is found not just in animals and choanoflagellates (143) but is also present in the algae such as the cryptomonad Guillardia and the haptophyte Emiliania (Figure 5H). This suggests a potentially deeper evolutionary origin for the TRPM proteins than previously thought. While ciliate versions of this clade of SLOG domains are often standalone domains, they are also found fused to a distinct ion channel or Ras-like GTPase domains (Figure 5H).
Multiple independent fusions of SLOG domains to different ion-channel and SLATT domains (Figure 5A, E, F, H) in both bacteria and eukaryotes indicate that this domain is widely recruited to regulate flux across membranes. Above-discussed contextual connections across different systems and different families of SLOG domains strongly suggest a role for it in binding nucleotides or their derivatives. Possibilities suggested by the contextual links and previous experimental results are AMP, NAD or ADP-ribose or its derivatives. This is especially notable in the context of TRPM channel regulation. A multiple sequence alignment of eukaryotic SLOG domains reveals conservation of the nucleotide-binding pocket; however, unlike most prokaryotic representatives of this clade of SLOG domains, these lack conservation of the predicted catalytic residues (Figure 5G). This suggests that they are more likely to function as sensors rather than nucleotide-processing enzymes. While an universal TRPM ligand has not been identified, a range of soluble ligands have been linked to gating and regulation of TRPM channels, including ADP-ribose, cyclic ADP-ribose (cADPR), NAADP, cAMP, H2O2 and phosphatidylinositol (4,5)-bisphosphate (PIP2) (135–137). ADP-ribose, cADPR and NAADP are generally believed to act via the C-terminal cytoplasmic Nudix domain found in some TRPM channels (e.g. TRPM2). However, in light of our discovery of a SLOG domain in all TRPM proteins it is possible that nucleotide-derived ligands have a more general role in regulating these channels.
Miscellaneous signaling systems
The systems described so far are largely self-contained, hardly showing links to previously well-characterized signaling networks dependent on cAMP/cGMP. Nevertheless, we did recover some less-frequent but phyletically-widespread links to these conventional cNMP signaling systems. One of these features a SLOG domain (belonging to the same clade as those found linked to ADP-ribose processing enzymes) fused to TPR repeats and either a cNMP-generating cyclase or a TIR domain (Figure 5I). Occasionally, these might also contain a further fusion to RyR or SLATT domains (Figure 5I). It is possible that the SLOG and RyR domains (if present) are sensors for specific nucleotide-derived ligands, and are in regulatory interplay with the activities of the fused cyclase and TIR domains.
In another distinct system, the AGS-C domain, which is normally found fused to SMODS enzymes, is instead fused to a cNMP-generating cyclase domain (Figure 2C). Alternatively, genes encoding related cNMP-generating cyclases might be combined with a gene encoding a cNMP-binding domain (cNMPBD) fused to either an HTH or TIR domain (Figure 5J). Across phylogenetically diverse bacteria these cyclase genes linked to a gene coding for a novel 3TM protein belonging to the pJV1-spdB3 family (Figure 5J), implicated in intramycelial plasmid DNA transfer in Streptomyces (144–146). In some gene-neighborhoods, the pJV1-spdB3 domain is fused to HD domain phosphoesterases, which could hydrolyze cNMPs, while also associating with cNMP cyclases and other genes with potential roles in extracellular DNA (eDNA) recognition or processing (L Aravind, AM Burroughs, personal observations). Consistent with the conservation of multiple cytoplasmic positively-charged residues (Supplementary Material), its contextual associations, and role in DNA-transfer pJV1-spdB3 might serve as a membrane-anchored DNA receptor. The associated cyclic nucleotide-related domains could then signal the presence of DNA via a nucleotide signal. Despite recovery of these links to cNMP-signaling systems, it should be stressed that these systems seldom show links to dominant domains in conventional cNMP signaling pathways like PAS or GAF domains.
Finally, an enigmatic two-gene system encodes proteins respectively containing the SLOG domain and a phosphoribosyltransferase (PRTase) domain. The SLOG domain is often additionally fused to either of two distinct DNA-binding domains, a helix-turn-helix or a KilA-C domain (147) (Figure 5K). PRTase domains catalyze synthesis of nucleotides from 5-phospho-ribose 1-diphosphate and a free base, a reaction which is exactly the opposite of that known to be catalyzed by certain SLOG domains. Hence, it is conceivable that this pair constitutes a signaling switch with the PRTase domain generating a nucleotide, which is then recognized by the SLOG domain (and perhaps eventually hydrolyzed by it to terminate the signal). The DNA-binding domains fused to the SLOG domain could regulate transcription in a ligand-regulated fashion.
Evolutionary and general functional implications
Our analysis has uncovered an extensive network of systems dominated by themes related to biological conflicts (Figure 1A). More directly, these include inter-genomic conflicts between invasive entities (phages, plasmids and conjugative transposons) and host genomes, or inter-organismal conflicts. In a more general sense, biological conflicts also include situations such as selfish elements fostering their own mobility, or conditions precipitating decisions between continued growth and dormancy/sacrifice of self via cell-death for the good of kin (e.g. during attacks by rivals using antibiotics, host immune attack or stress). Thus, we can now interpret the role of the archetypal SMODS protein DncV in V.cholerae virulence (19) in a broader context. More generally, these prokaryotic systems appear to have been enormously elaborated to include a striking diversity of nucleotide-dependent systems that also includes the CRISPR/Cas systems. Specifically, the CRISPR polymerase, which was poorly understood, with proposed functions in amplification of CRISPR transcripts (148), untemplated RNA modification (108) and cyclase activity (111,116,149), can now be seen as generating a nucleotide signal to activate the associated CARF effectors(37). Identification of mCpol, the ancestral version of the CRISPR polymerase, and its association with CARF domains suggest that minimal mCpol-CARF units combined with other mobile elements including the RAMPs and the Cas1-Cas2 dyad to give rise to the classical Type-I and Type-III CRISPR systems(115).
These systems also reinforce the deeper evolutionary connection between nucleic acid polymerases and synthetases generating nucleotide signals. On multiple occasions the latter enzymes appear to have emerged in the same superfamilies as the former, even across unrelated folds—e.g. the polβ-like fold (SMODS, cGAS, OAS, RelA/SpoT) (31,32) and RRM-like polymerase palm fold (mCpol and CRISPR polymerase) (108). It is interesting to note that some of the synthetases for signaling nucleotides are specifically related to enzymes involved in RNA repair: Thg1 involved in tRNA 5′ end repair is related to mCpol and CRISPR polymerase, and the RNA 3′ nucleotidyltranferases (e.g. tRNA CCA-adding enzyme) are related to SMODS, cGAS and OAS. Though the DisA-N domain, another cyclic dinucleotide generating enzyme, is unrelated to any known cellular polymerase domain, it still acts in response to sensing branched DNA (16). Another notable aspect of our study is the identification of potential nucleotide-binding and processing domains (TIR and SLOG superfamily), some members of which might play biochemically distinct but functionally equivalent roles as the above synthetases and nucleotide sensors in comparable systems. This suggests that not just nucleotides but also fragments derived from them might be used as equivalent signals. Even here we see potential evolutionary links to direct interactions with nucleic acids: (i) Members of one of the most conserved clades of SLOG domains (Smf/DprA) directly binds ssDNA as part of the transformation process (67,150). (ii) The classical SLOG family releases modified adenines as part of tRNA degradation (26,27,151). (iii) TIR domains have also been previously implicated as potential effectors that might operate on DNA (60,78).
This raises the possibility that cyclic/oligo-nucleotide-generating synthetases, SLOG and TIR domains might have all originally emerged in the context of nucleic-acid-related interactions. In the case of the polymerases it is conceivable that their ancestral role was nucleic acid repair when under attack by effectors of selfish elements. This might have allowed their cyclic/oligo-nucleotide byproduct to be channelized as an activating signal for immune response, thereby leading to the emergence of dedicated signaling synthetases that retained their ancestral nucleic-acid sensing capability. The ancestral role in sensing incoming DNA or degraded tRNA might have allowed SLOG domains and probably also TIR domains (due their predicted nucleic-acid-sensing capacity) to be similarly recruited, albeit in a new role dependent on their nucleotide-binding/processing capacity. In mechanistic terms incorporation of such a nucleotide (or derivative) signal provides a means for: (i) an additional level of control which would not be possible in the case of a direct response; (ii) potential for signal amplification; (iii) setting a response threshold that allows robust discrimination of signal from noise. These would be especially useful when effector systems, which are expensive in terms of production and deleterious/fatal for the host cell, are deployed.
While bacterial homologs of animal polβ cyclase enzymes (OAS and cGAS) involved in interferon-associated antiviral immunity have been previously recognized (57), their significance was poorly understood. Our work shows that not only are their bacterial cognates more extensive than previously reported but that OAS and cGAS can be seen as being a subset of the larger diversity of such systems observed in prokaryotes. In light of this it is likely they were acquired early in animal evolution via lateral transfer from bacteria and utilized ‘as is’ in a similar capacity as nucleotide-signal generating enzymes in the defensive response against invasive nucleic acids. In contrast, SLOG domains in TRPM ion-channels and the HORMA-Pch2 dyad are examples of a more derived use of components from prokaryotic systems. Bacterial HORMA and Pch2 proteins are nearly always linked together in a single operon. Further, the bacterial HORMA domains represent the minimal version of this domain without any of the additional C-terminal extensions of their eukaryotic counterparts. These features suggest that they are the precursors of the eukaryotic counterparts. Eukaryotic HORMA domains are central to both meiosis and mitosis, with a HORMA-Pch2 dyad being critical for the former process (102,103). Given the role of the Pch2 AAA+ ATPase in clearing HORMA protein assemblies at sites of synaptonemal complexes (103), it is likely that the acquisition of this pair from one of the above-described bacterial systems was a key factor in the origin of eukaryotic meiosis. This again adds to the growing evidence that conflict systems derived from bacterial endosymbionts in the stem eukaryote contributed major components for the emergence of quintessentially eukaryotic processes (152–154).
CONCLUSIONS
Identification of this network and reconstruction of diverse systems encompassed by it not only adds a new layer to nucleotide-centric signaling but also helps clarify certain obscure evolutionary and functional aspects. Importantly, it establishes nucleotide/nucleotide-derived signals as an overarching principle unifying diverse biological conflict systems from the three superkingdoms of life. Thus, previously known systems as disparate as animal 2′-5′ OA and 2′-5′ cGAMP signaling and prokaryotic CRISPR/Cas systems are brought together with the new ones reported here under a single mechanistic umbrella. This opens new avenues for both investigation of basic biology and development of new biotechnology. Given the function of DncV, an archetype for these novel systems, further investigation might offer alternatives in managing bacterial virulence. The extensive but underappreciated spread of these signaling systems and their effectors might also provide unexplored regulatory handles that can be harnessed for biotechnological purposes. In a similar vein, the novel retroelement recovered in this study with its potential for diversity generation might also provide a means of mutagenizing and exporting DNA generated by reverse-transcription. Finally, discovery of a potential universal ligand-binding domain for the TRPM channels opens up several opportunities to better understand this important class of signaling proteins, which have been implicated in several human diseases (155–161). Given their role in numerous sensory pathways, manipulation of this ligand-binding domain might also offer a means to control these channels in hitherto unexpected ways.
AVAILABILITY
The supplementary information is also available from the following FTP site: ftp://ftp.ncbi.nih.gov/pub/aravind/temp/SMODS_SLOG/Supplementary_Material.html.
Supplementary Material
Acknowledgments
This work is supported by the funds of the Intramural Research Program of National Library of Medicine at the National Institutes of Health, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Library of Medicine, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sutherland E.W., Rall T.W. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J. Biol. Chem. 1958;232:1077–1091. [PubMed] [Google Scholar]
- 2.Rall T.W., Sutherland E.W. Formation of a cyclic adenine ribonucleotide by tissue particles. J. Biol. Chem. 1958;232:1065–1076. [PubMed] [Google Scholar]
- 3.Gancedo J.M. Biological roles of cAMP: variations on a theme in the different kingdoms of life. Biol. Rev. Camb. Philos. Soc. 2013;88:645–668. doi: 10.1111/brv.12020. [DOI] [PubMed] [Google Scholar]
- 4.Diaz M.R., King J.M., Yahr T.L. Intrinsic and Extrinsic Regulation of Type III Secretion Gene Expression in Pseudomonas Aeruginosa. Front. Microbiol. 2011;2:89. doi: 10.3389/fmicb.2011.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorke B., Stulke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 6.Lucas K.A., Pitari G.M., Kazerounian S., Ruiz-Stewart I., Park J., Schulz S., Chepenik K.P., Waldman S.A. Guanylyl cyclases and signaling by cyclic GMP. Pharmacol. Rev. 2000;52:375–414. [PubMed] [Google Scholar]
- 7.Marden J.N., Dong Q., Roychowdhury S., Berleman J.E., Bauer C.E. Cyclic GMP controls Rhodospirillum centenum cyst development. Mol. Microbiol. 2011;79:600–615. doi: 10.1111/j.1365-2958.2010.07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalia D., Merey G., Nakayama S., Zheng Y., Zhou J., Luo Y., Guo M., Roembke B.T., Sintim H.O. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 2013;42:305–341. doi: 10.1039/c2cs35206k. [DOI] [PubMed] [Google Scholar]
- 9.Ryjenkov D.A., Simm R., Romling U., Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 10.Lori C., Ozaki S., Steiner S., Bohm R., Abel S., Dubey B.N., Schirmer T., Hiller S., Jenal U. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature. 2015;523:236–239. doi: 10.1038/nature14473. [DOI] [PubMed] [Google Scholar]
- 11.De N., Navarro M.V., Raghavan R.V., Sondermann H. Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J. Mol. Biol. 2009;393:619–633. doi: 10.1016/j.jmb.2009.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitney J.C., Colvin K.M., Marmont L.S., Robinson H., Parsek M.R., Howell P.L. Structure of the cytoplasmic region of PelD, a degenerate diguanylate cyclase receptor that regulates exopolysaccharide production in Pseudomonas aeruginosa. J. Biol. Chem. 2012;287:23582–23593. doi: 10.1074/jbc.M112.375378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross P., Weinhouse H., Aloni Y., Michaeli D., Weinberger-Ohana P., Mayer R., Braun S., de Vroom E., van der Marel G.A., van Boom J.H., et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 14.Commichau F.M., Dickmanns A., Gundlach J., Ficner R., Stulke J. A jack of all trades: the multiple roles of the unique essential second messenger cyclic di-AMP. Mol. Microbiol. 2015;97:189–204. doi: 10.1111/mmi.13026. [DOI] [PubMed] [Google Scholar]
- 15.Bejerano-Sagie M., Oppenheimer-Shaanan Y., Berlatzky I., Rouvinski A., Meyerovich M., Ben-Yehuda S. A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell. 2006;125:679–690. doi: 10.1016/j.cell.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 16.Witte G., Hartung S., Büttner K., Hopfner K.P. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol. Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X., Wu J., Du F., Xu H., Sun L., Chen Z., Brautigam C.A., Chen Z.J. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 2014;6:421–430. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao T.S., Fitzgerald K.A. The cGAS-STING pathway for DNA sensing. Mol. Cell. 2013;51:135–139. doi: 10.1016/j.molcel.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies B.W., Bogard R.W., Young T.S., Mekalanos J.J. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato K., Ishii R., Hirano S., Ishitani R., Nureki O. Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase. Structure. 2015;23:843–850. doi: 10.1016/j.str.2015.01.023. [DOI] [PubMed] [Google Scholar]
- 21.Nelson J.W., Sudarsan N., Phillips G.E., Stav S., Lunse C.E., McCown P.J., Breaker R.R. Control of bacterial exoelectrogenesis by c-AMP-GMP. Proc. Natl. Acad. Sci. U.S.A. 2015;112:5389–5394. doi: 10.1073/pnas.1419264112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kellenberger C.A., Wilson S.C., Hickey S.F., Gonzalez T.L., Su Y., Hallberg Z.F., Brewer T.F., Iavarone A.T., Carlson H.K., Hsieh Y.F., et al. GEMM-I riboswitches from Geobacter sense the bacterial second messenger cyclic AMP-GMP. Proc. Natl. Acad. Sci. U.S.A. 2015;112:5383–5388. doi: 10.1073/pnas.1419328112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magnusson L.U., Farewell A., Nystrom T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Tozawa Y., Nomura Y. Signalling by the global regulatory molecule ppGpp in bacteria and chloroplasts of land plants. Plant Biol. (Stuttg.) 2011;13:699–709. doi: 10.1111/j.1438-8677.2011.00484.x. [DOI] [PubMed] [Google Scholar]
- 25.Mashimo T., Simon-Chazottes D., Guenet J.L. Innate resistance to flavivirus infections and the functions of 2′-5′ oligoadenylate synthetases. Curr. Topics Microbiol. Immunol. 2008;321:85–100. doi: 10.1007/978-3-540-75203-5_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., Sakakibara H., Kyozuka J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 27.Samanovic M.I., Tu S., Novak O., Iyer L.M., McAllister F.E., Aravind L., Gygi S.P., Hubbard S.R., Strnad M., Darwin K.H. Proteasomal control of cytokinin synthesis protects Mycobacterium tuberculosis against nitric oxide. Mol. Cell. 2015;57:984–994. doi: 10.1016/j.molcel.2015.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anantharaman V., Iyer L.M., Aravind L. Ter-dependent stress response systems: novel pathways related to metal sensing, production of a nucleoside-like metabolite, and DNA-processing. Mol. Biosystems. 2012;8:3142–3165. doi: 10.1039/c2mb25239b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mock M., Crasnier M., Duflot E., Dumay V., Danchin A. Structural and functional relationships between Pasteurella multocida and enterobacterial adenylate cyclases. J. Bacteriol. 1991;173:6265–6269. doi: 10.1128/jb.173.19.6265-6269.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pei J., Grishin N.V. GGDEF domain is homologous to adenylyl cyclase. Proteins. 2001;42:210–216. doi: 10.1002/1097-0134(20010201)42:2<210::aid-prot80>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 31.Aravind L., Koonin E.V. DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuchta K., Knizewski L., Wyrwicz L.S., Rychlewski L., Ginalski K. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 2009;37:7701–7714. doi: 10.1093/nar/gkp854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oppenheimer-Shaanan Y., Wexselblatt E., Katzhendler J., Yavin E., Ben-Yehuda S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011;12:594–601. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campos S.S., Ibarra-Rodriguez J.R., Barajas-Ornelas R.C., Ramirez-Guadiana F.H., Obregon-Herrera A., Setlow P., Pedraza-Reyes M. Interaction of apurinic/apyrimidinic endonucleases Nfo and ExoA with the DNA integrity scanning protein DisA in the processing of oxidative DNA damage during Bacillus subtilis spore outgrowth. J. Bacteriol. 2014;196:568–578. doi: 10.1128/JB.01259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tshori S., Razin E., Nechushtan H. Amino-acyl tRNA synthetases generate dinucleotide polyphosphates as second messengers: functional implications. Topics Curr. Chem. 2014;344:189–206. doi: 10.1007/128_2013_426. [DOI] [PubMed] [Google Scholar]
- 36.Hornung V., Hartmann R., Ablasser A., Hopfner K.P. OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat. Rev. Immunol. 2014;14:521–528. doi: 10.1038/nri3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makarova K.S., Anantharaman V., Grishin N.V., Koonin E.V., Aravind L. CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front. Genet. 2014;5:102. doi: 10.3389/fgene.2014.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holm L., Kaariainen S., Rosenstrom P., Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lassmann T., Frings O., Sonnhammer E.L. Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 2009;37:858–865. doi: 10.1093/nar/gkn1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pei J., Sadreyev R., Grishin N.V. PCMA: fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics. 2003;19:427–428. doi: 10.1093/bioinformatics/btg008. [DOI] [PubMed] [Google Scholar]
- 44.Cole C., Barber J.D., Barton G.J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 47.Kall L., Krogh A., Sonnhammer E.L. Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamada T., Kawai S. An algorithm for drawing general undirected graphs. Inf. Process. Lett. 1989;31:7–15. [Google Scholar]
- 50.Price M.N., Dehal P.S., Arkin A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PloS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rahman M.H., Biswas K., Hossain M.A., Sack R.B., Mekalanos J.J., Faruque S.M. Distribution of genes for virulence and ecological fitness among diverse Vibrio cholerae population in a cholera endemic area: tracking the evolution of pathogenic strains. DNA Cell Biol. 2008;27:347–355. doi: 10.1089/dna.2008.0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lowden M.J., Skorupski K., Pellegrini M., Chiorazzo M.G., Taylor R.K., Kull F.J. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:2860–2865. doi: 10.1073/pnas.0915021107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aravind L. Guilt by association: contextual information in genome analysis. Genome Res. 2000;10:1074–1077. doi: 10.1101/gr.10.8.1074. [DOI] [PubMed] [Google Scholar]
- 54.Dandekar T., Snel B., Huynen M., Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 55.Overbeek R., Fonstein M., D'Souza M., Pusch G.D., Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lohofener J., Steinke N., Kay-Fedorov P., Baruch P., Nikulin A., Tishchenko S., Manstein D.J., Fedorov R. The Activation Mechanism of 2′-5′-Oligoadenylate Synthetase Gives New Insights Into OAS/cGAS Triggers of Innate Immunity. Structure. 2015;23:851–862. doi: 10.1016/j.str.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 57.Kranzusch P.J., Lee A.S., Berger J.M., Doudna J.A. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 2013;3:1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rogozin I.B., Aravind L., Koonin E.V. Differential action of natural selection on the N and C-terminal domains of 2′-5′ oligoadenylate synthetases and the potential nuclease function of the C-terminal domain. J. Mol. Biol. 2003;326:1449–1461. doi: 10.1016/s0022-2836(03)00055-x. [DOI] [PubMed] [Google Scholar]
- 59.Martin G., Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burroughs A.M., Ando Y., Aravind L. New perspectives on the diversification of the RNA interference system: insights from comparative genomics and small RNA sequencing. Wiley Interdiscip. Rev. RNA. 2014;5:141–181. doi: 10.1002/wrna.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shamanna R.A., Hoque M., Lewis-Antes A., Azzam E.I., Lagunoff D., Pe'ery T., Mathews M.B. The NF90/NF45 complex participates in DNA break repair via nonhomologous end joining. Mol. Cell. Biol. 2011;31:4832–4843. doi: 10.1128/MCB.05849-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shiina N., Nakayama K. RNA granule assembly and disassembly modulated by nuclear factor associated with double-stranded RNA 2 and nuclear factor 45. J. Biol. Chem. 2014;289:21163–21180. doi: 10.1074/jbc.M114.556365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langland J.O., Kao P., Jacobs B.L. Regulation of IL-2 gene expression and nuclear factor-90 translocation in vaccinia virus-infected cells. J. Interferon. Cytokine Res. 2003;23:489–500. doi: 10.1089/10799900360708614. [DOI] [PubMed] [Google Scholar]
- 64.Kuroha T., Tokunaga H., Kojima M., Ueda N., Ishida T., Nagawa S., Fukuda H., Sugimoto K., Sakakibara H. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell. 2009;21:3152–3169. doi: 10.1105/tpc.109.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chickarmane V.S., Gordon S.P., Tarr P.T., Heisler M.G., Meyerowitz E.M. Cytokinin signaling as a positional cue for patterning the apical-basal axis of the growing Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U.S.A. 2012;109:4002–4007. doi: 10.1073/pnas.1200636109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tokunaga H., Kojima M., Kuroha T., Ishida T., Sugimoto K., Kiba T., Sakakibara H. Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 2012;69:355–365. doi: 10.1111/j.1365-313X.2011.04795.x. [DOI] [PubMed] [Google Scholar]
- 67.Mortier-Barriere I., Velten M., Dupaigne P., Mirouze N., Pietrement O., McGovern S., Fichant G., Martin B., Noirot P., Le Cam E., et al. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell. 2007;130:824–836. doi: 10.1016/j.cell.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 68.Fischer K., Llamas A., Tejada-Jimenez M., Schrader N., Kuper J., Ataya F.S., Galvan A., Mendel R.R., Fernandez E., Schwarz G. Function and structure of the molybdenum cofactor carrier protein from Chlamydomonas reinhardtii. J. Biol. Chem. 2006;281:30186–30194. doi: 10.1074/jbc.M603919200. [DOI] [PubMed] [Google Scholar]
- 69.Rao S.T., Rossmann M.G. Comparison of super-secondary structures in proteins. J. Mol. Biol. 1973;76:241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- 70.Burroughs A.M., Allen K.N., Dunaway-Mariano D., Aravind L. Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J. Mol. Biol. 2006;361:1003–1034. doi: 10.1016/j.jmb.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 71.Aravind L., Anantharaman V., Koonin E.V. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: implications for protein evolution in the RNA. Proteins. 2002;48:1–14. doi: 10.1002/prot.10064. [DOI] [PubMed] [Google Scholar]
- 72.Spear A.M., Loman N.J., Atkins H.S., Pallen M.J. Microbial TIR domains: not necessarily agents of subversion. Trends Microbiol. 2009;17:393–398. doi: 10.1016/j.tim.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 73.Rock F.L., Hardiman G., Timans J.C., Kastelein R.A., Bazan J.F. A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. U.S.A. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kopp E.B., Medzhitov R. The Toll-receptor family and control of innate immunity. Curr. Opin. Immunol. 1999;11:13–18. doi: 10.1016/s0952-7915(99)80003-x. [DOI] [PubMed] [Google Scholar]
- 75.Narayanan K.B., Park H.H. Toll/interleukin-1 receptor (TIR) domain-mediated cellular signaling pathways. Apoptosis. 2015;20:196–209. doi: 10.1007/s10495-014-1073-1. [DOI] [PubMed] [Google Scholar]
- 76.Aravind L., Dixit V.M., Koonin E.V. The domains of death: evolution of the apoptosis machinery. Trends Biochem. Sci. 1999;24:47–53. doi: 10.1016/s0968-0004(98)01341-3. [DOI] [PubMed] [Google Scholar]
- 77.Koonin E.V., Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 2002;9:394–404. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- 78.Iyer L.M., Abhiman S., Aravind L. MutL homologs in restriction-modification systems and the origin of eukaryotic MORC ATPases. Biol. Direct. 2008;3:8. doi: 10.1186/1745-6150-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anantharaman V., Makarova K.S., Burroughs A.M., Koonin E.V., Aravind L. Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct. 2013;8:15. doi: 10.1186/1745-6150-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu Y., Tao X., Shen B., Horng T., Medzhitov R., Manley J.L., Tong L. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 81.Bateman A., Coggill P., Finn R.D. DUFs: families in search of function. Acta Crystallogr. F Struct. Biol. Crystallization Commun. 2010;66:1148–1152. doi: 10.1107/S1744309110001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aravind L., Koonin E.V. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 1998;26:3746–3752. doi: 10.1093/nar/26.16.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang N., Gottesman S., Willingham M.C., Gottesman M.M., Maurizi M.R. A human mitochondrial ATP-dependent protease that is highly homologous to bacterial Lon protease. Proc. Natl. Acad. Sci. U.S.A. 1993;90:11247–11251. doi: 10.1073/pnas.90.23.11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aravind L., Koonin E.V. Classification of the caspase-hemoglobinase fold: detection of new families and implications for the origin of the eukaryotic separins. Proteins. 2002;46:355–367. doi: 10.1002/prot.10060. [DOI] [PubMed] [Google Scholar]
- 85.Zhang D., Iyer L.M., Aravind L. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 2011;39:4532–4552. doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang D., de Souza R.F., Anantharaman V., Iyer L.M., Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Doehlemann G., Reissmann S., Assmann D., Fleckenstein M., Kahmann R. Two linked genes encoding a secreted effector and a membrane protein are essential for Ustilago maydis-induced tumour formation. Mol. Microbiol. 2011;81:751–766. doi: 10.1111/j.1365-2958.2011.07728.x. [DOI] [PubMed] [Google Scholar]
- 88.Iyer L.M., Burroughs A.M., Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 2006;7:R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burroughs A.M., Iyer L.M., Aravind L. Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system. Mol. Biosystems. 2011;7:2261–2277. doi: 10.1039/c1mb05061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Burroughs A.M., Iyer L.M., Aravind L. The natural history of ubiquitin and ubiquitin-related domains. Front Biosci. (Landmark Ed.) 2012;17:1433–1460. doi: 10.2741/3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burroughs A.M., Iyer L.M., Aravind L. Structure and evolution of ubiquitin and ubiquitin-related domains. Methods Mol. Biol. 2012;832:15–63. doi: 10.1007/978-1-61779-474-2_2. [DOI] [PubMed] [Google Scholar]
- 92.Nunoura T., Takaki Y., Kakuta J., Nishi S., Sugahara J., Kazama H., Chee G.J., Hattori M., Kanai A., Atomi H., et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011;39:3204–3223. doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spang A., Saw J.H., Jorgensen S.L., Zaremba-Niedzwiedzka K., Martijn J., Lind A.E., van Eijk R., Schleper C., Guy L., Ettema T.J. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521:173–179. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Humbard M.A., Miranda H.V., Lim J.M., Krause D.J., Pritz J.R., Zhou G., Chen S., Wells L., Maupin-Furlow J.A. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature. 2010;463:54–60. doi: 10.1038/nature08659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shigi N. Posttranslational modification of cellular proteins by a ubiquitin-like protein in bacteria. J. Biol. Chem. 2012;287:17568–17577. doi: 10.1074/jbc.M112.359844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Begley T.P., Xi J., Kinsland C., Taylor S., McLafferty F. The enzymology of sulfur activation during thiamin and biotin biosynthesis. Curr. Opin. Chem. Biol. 1999;3:623–629. doi: 10.1016/s1367-5931(99)00018-6. [DOI] [PubMed] [Google Scholar]
- 97.Burroughs A.M., Balaji S., Iyer L.M., Aravind L. Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold. Biol. Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Burroughs A.M., Iyer L.M., Aravind L. Natural history of the E1-like superfamily: implication for adenylation, sulfur transfer, and ubiquitin conjugation. Proteins. 2009;75:895–910. doi: 10.1002/prot.22298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aravind L., Koonin E.V. The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem. Sci. 1998;23:284–286. doi: 10.1016/s0968-0004(98)01257-2. [DOI] [PubMed] [Google Scholar]
- 100.Iyer L.M., Leipe D.D., Koonin E.V., Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 101.Neuwald A.F., Aravind L., Spouge J.L., Koonin E.V. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 102.Kim Y., Rosenberg S.C., Kugel C.L., Kostow N., Rog O., Davydov V., Su T.Y., Dernburg A.F., Corbett K.D. The chromosome axis controls meiotic events through a hierarchical assembly of HORMA domain proteins. Develop. Cell. 2014;31:487–502. doi: 10.1016/j.devcel.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wojtasz L., Daniel K., Roig I., Bolcun-Filas E., Xu H., Boonsanay V., Eckmann C.R., Cooke H.J., Jasin M., Keeney S., et al. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 2009;5:e1000702. doi: 10.1371/journal.pgen.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rydel T.J., Williams J.M., Krieger E., Moshiri F., Stallings W.C., Brown S.M., Pershing J.C., Purcell J.P., Alibhai M.F. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry. 2003;42:6696–6708. doi: 10.1021/bi027156r. [DOI] [PubMed] [Google Scholar]
- 105.Mignery G.A., Pikaard C.S., Park W.D. Molecular characterization of the patatin multigene family of potato. Gene. 1988;62:27–44. doi: 10.1016/0378-1119(88)90577-x. [DOI] [PubMed] [Google Scholar]
- 106.Hayes F., Melderen L. Toxins-antitoxins: diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 2011;46:386–408. doi: 10.3109/10409238.2011.600437. [DOI] [PubMed] [Google Scholar]
- 107.Atkinson G.C., Tenson T., Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PloS One. 2011;6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Anantharaman V., Iyer L.M., Aravind L. Presence of a classical RRM-fold palm domain in Thg1-type 3′- 5′ nucleic acid polymerases and the origin of the GGDEF and CRISPR polymerase domains. Biology Direct. 2010;5:43. doi: 10.1186/1745-6150-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Makarova K.S., Aravind L., Grishin N.V., Rogozin I.B., Koonin E.V. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Osawa T., Inanaga H., Numata T. Crystal structure of the Cmr2-Cmr3 subcomplex in the CRISPR-Cas RNA silencing effector complex. J. Mol. Biol. 2013;425:3811–3823. doi: 10.1016/j.jmb.2013.03.042. [DOI] [PubMed] [Google Scholar]
- 111.Cocozaki A.I., Ramia N.F., Shao Y., Hale C.R., Terns R.M., Terns M.P., Li H. Structure of the Cmr2 subunit of the CRISPR-Cas RNA silencing complex. Structure. 2012;20:545–553. doi: 10.1016/j.str.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee C., Marians K.J. Characterization of the nucleoid-associated protein YejK. J. Biol. Chem. 2013;288:31503–31516. doi: 10.1074/jbc.M113.494237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Murphy L.D., Rosner J.L., Zimmerman S.B., Esposito D. Identification of two new proteins in spermidine nucleoids isolated from Escherichia coli. J. Bacteriol. 1999;181:3842–3844. doi: 10.1128/jb.181.12.3842-3844.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McLennan A.G. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Makarova K.S., Anantharaman V., Aravind L., Koonin E.V. Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol. Direct. 2012;7:40. doi: 10.1186/1745-6150-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhu X., Ye K. Crystal structure of Cmr2 suggests a nucleotide cyclase-related enzyme in type III CRISPR-Cas systems. FEBS Lett. 2012;586:939–945. doi: 10.1016/j.febslet.2012.02.036. [DOI] [PubMed] [Google Scholar]
- 117.Ryan R.P., McCarthy Y., Andrade M., Farah C.S., Armitage J.P., Dow J.M. Cell-cell signal-dependent dynamic interactions between HD-GYP and GGDEF domain proteins mediate virulence in Xanthomonas campestris. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5989–5994. doi: 10.1073/pnas.0912839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ryan R.P., Fouhy Y., Lucey J.F., Crossman L.C., Spiro S., He Y.W., Zhang L.H., Heeb S., Camara M., Williams P., et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6712–6717. doi: 10.1073/pnas.0600345103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 119.Shore D. The Sir2 protein family: A novel deacetylase for gene silencing and more. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14030–14032. doi: 10.1073/pnas.011506198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Iyer L.M., Makarova K.S., Koonin E.V., Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mao C., Cook W.J., Zhou M., Koszalka G.W., Krenitsky T.A., Ealick S.E. The crystal structure of Escherichia coli purine nucleoside phosphorylase: a comparison with the human enzyme reveals a conserved topology. Structure. 1997;5:1373–1383. doi: 10.1016/s0969-2126(97)00287-6. [DOI] [PubMed] [Google Scholar]
- 122.Iyer L.M., Zhang D., Burroughs A.M., Aravind L. Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic Acids Res. 2013;41:7635–7655. doi: 10.1093/nar/gkt573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Klaiman D., Steinfels-Kohn E., Krutkina E., Davidov E., Kaufmann G. The wobble nucleotide-excising anticodon nuclease RloC is governed by the zinc-hook and DNA-dependent ATPase of its Rad50-like region. Nucleic Acids Res. 2012;40:8568–8578. doi: 10.1093/nar/gks593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bonen L., Vogel J. The ins and outs of group II introns. Trends Genet. 2001;17:322–331. doi: 10.1016/s0168-9525(01)02324-1. [DOI] [PubMed] [Google Scholar]
- 125.Medhekar B., Miller J.F. Diversity-generating retroelements. Curr. Opin. Microbiol. 2007;10:388–395. doi: 10.1016/j.mib.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Matsuura M., Saldanha R., Ma H., Wank H., Yang J., Mohr G., Cavanagh S., Dunny G.M., Belfort M., Lambowitz A.M. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 1997;11:2910–2924. doi: 10.1101/gad.11.21.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Centron D., Roy P.H. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob. Agents Chemother. 2002;46:1402–1409. doi: 10.1128/AAC.46.5.1402-1409.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lampson B.C., Inouye M., Inouye S. Retrons, msDNA, and the bacterial genome. Cytogenet Genome Res. 2005;110:491–499. doi: 10.1159/000084982. [DOI] [PubMed] [Google Scholar]
- 129.de Souza R.F., Aravind L. Identification of novel components of NAD-utilizing metabolic pathways and prediction of their biochemical functions. Mol. Biosystems. 2012;8:1661–1677. doi: 10.1039/c2mb05487f. [DOI] [PubMed] [Google Scholar]
- 130.Spinelli S.L., Kierzek R., Turner D.H., Phizicky E.M. Transient ADP-ribosylation of a 2′-phosphate implicated in its removal from ligated tRNA during splicing in yeast. J. Biol. Chem. 1999;274:2637–2644. doi: 10.1074/jbc.274.5.2637. [DOI] [PubMed] [Google Scholar]
- 131.Aravind L., Zhang D., de Souza R.F., Anand S., Iyer L.M. The natural history of ADP-ribosyltransferases and the ADP-ribosylation system. Curr. Topics Microbiol. Immunol. 2015;384:3–32. doi: 10.1007/82_2014_414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Iyer L.M., Aravind L. Insights from the architecture of the bacterial transcription apparatus. J. Struct. Biol. 2012;179:299–319. doi: 10.1016/j.jsb.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schlosser A., Hamann A., Bossemeyer D., Schneider E., Bakker E.P. NAD+ binding to the Escherichia coli K(+)-uptake protein TrkA and sequence similarity between TrkA and domains of a family of dehydrogenases suggest a role for NAD+ in bacterial transport. Mol. Microbiol. 1993;9:533–543. doi: 10.1111/j.1365-2958.1993.tb01714.x. [DOI] [PubMed] [Google Scholar]
- 134.Ponting C.P. Novel repeats in ryanodine and IP3 receptors and protein O-mannosyltransferases. Trends Biochem. Sci. 2000;25:48–50. doi: 10.1016/s0968-0004(99)01513-3. [DOI] [PubMed] [Google Scholar]
- 135.Venkatachalam K., Montell C. TRP channels. Annu. Rev. Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kraft R., Harteneck C. The mammalian melastatin-related transient receptor potential cation channels: an overview. Pflugers Arch. 2005;451:204–211. doi: 10.1007/s00424-005-1428-0. [DOI] [PubMed] [Google Scholar]
- 137.Fleig A., Penner R. The TRPM ion channel subfamily: molecular, biophysical and functional features. Trends Pharmacol. Sci. 2004;25:633–639. doi: 10.1016/j.tips.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 138.Phelps C.B., Gaudet R. The role of the N terminus and transmembrane domain of TRPM8 in channel localization and tetramerization. J. Biol. Chem. 2007;282:36474–36480. doi: 10.1074/jbc.M707205200. [DOI] [PubMed] [Google Scholar]
- 139.Mei Z.Z., Jiang L.H. Requirement for the N-terminal coiled-coil domain for expression and function, but not subunit interaction of, the ADPR-activated TRPM2 channel. J. Membr. Biol. 2009;230:93–99. doi: 10.1007/s00232-009-9190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pertusa M., Gonzalez A., Hardy P., Madrid R., Viana F. Bidirectional modulation of thermal and chemical sensitivity of TRPM8 channels by the initial region of the N-terminal domain. J. Biol. Chem. 2014;289:21828–21843. doi: 10.1074/jbc.M114.565994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kuhn F.J., Kuhn C., Naziroglu M., Luckhoff A. Role of an N-terminal splice segment in the activation of the cation channel TRPM2 by ADP-ribose and hydrogen peroxide. Neurochem. Res. 2009;34:227–233. doi: 10.1007/s11064-008-9755-0. [DOI] [PubMed] [Google Scholar]
- 142.Montell C. Physiology, phylogeny, and functions of the TRP superfamily of cation channels. Sci. STKE. 2001;90:re1. doi: 10.1126/stke.2001.90.re1. [DOI] [PubMed] [Google Scholar]
- 143.Mederos y Schnitzler M., Waring J., Gudermann T., Chubanov V. Evolutionary determinants of divergent calcium selectivity of TRPM channels. FASEB J. 2008;22:1540–1551. doi: 10.1096/fj.07-9694com. [DOI] [PubMed] [Google Scholar]
- 144.Servin-Gonzalez L., Sampieri A.I., Cabello J., Galvan L., Juarez V., Castro C. Sequence and functional analysis of the Streptomyces phaeochromogenes plasmid pJV1 reveals a modular organization of Streptomyces plasmids that replicate by rolling circle. Microbiology. 1995;141:2499–2510. doi: 10.1099/13500872-141-10-2499. [DOI] [PubMed] [Google Scholar]
- 145.Huang C.H., Chen C.Y., Tsai H.H., Chen C., Lin Y.S., Chen C.W. Linear plasmid SLP2 of Streptomyces lividans is a composite replicon. Mol. Microbiol. 2003;47:1563–1576. doi: 10.1046/j.1365-2958.2003.03403.x. [DOI] [PubMed] [Google Scholar]
- 146.Yang Y., Kurokawa T., Takahama Y., Nindita Y., Mochizuki S., Arakawa K., Endo S., Kinashi H. pSLA2-M of Streptomyces rochei is a composite linear plasmid characterized by self-defense genes and homology with pSLA2-L. Biosci. Biotechnol. Biochem. 2011;75:1147–1153. doi: 10.1271/bbb.110054. [DOI] [PubMed] [Google Scholar]
- 147.Iyer L.M., Koonin E.V., Aravind L. Extensive domain shuffling in transcription regulators of DNA viruses and implications for the origin of fungal APSES transcription factors. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-research0012. RESEARCH0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Makarova K.S., Grishin N.V., Shabalina S.A., Wolf Y.I., Koonin E.V. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sorek R., Lawrence C.M., Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 150.Smeets L.C., Bijlsma J.J., Kuipers E.J., Vandenbroucke-Grauls C.M., Kusters J.G. The dprA gene is required for natural transformation of Helicobacter pylori. FEMS Immunol. Med. Microbiol. 2000;27:99–102. doi: 10.1111/j.1574-695X.2000.tb01418.x. [DOI] [PubMed] [Google Scholar]
- 151.Kamada-Nobusada T., Sakakibara H. Molecular basis for cytokinin biosynthesis. Phytochemistry. 2009;70:444–449. doi: 10.1016/j.phytochem.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 152.Aravind L., Anantharaman V., Zhang D., de Souza R.F., Iyer L.M. Gene flow and biological conflict systems in the origin and evolution of eukaryotes. Front. Cell. Infect. Microbiol. 2012;2:89. doi: 10.3389/fcimb.2012.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Aravind L., Burroughs A.M., Zhang D., Iyer L.M. Protein and DNA modifications: evolutionary imprints of bacterial biochemical diversification and geochemistry on the provenance of eukaryotic epigenetics. Cold Spring Harb. Perspect. Biol. 2014;6:a016063. doi: 10.1101/cshperspect.a016063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang D., Iyer L.M., Burroughs A.M., Aravind L. Resilience of biochemical activity in protein domains in the face of structural divergence. Curr. Opin. Struct. Biol. 2014;26:92–103. doi: 10.1016/j.sbi.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nilius B., Owsianik G., Voets T., Peters J.A. Transient receptor potential cation channels in disease. Physiol. Rev. 2007;87:165–217. doi: 10.1152/physrev.00021.2006. [DOI] [PubMed] [Google Scholar]
- 156.Nilius B., Voets T., Peters J. TRP channels in disease. Sci. STKE. 2005;295:re8. doi: 10.1126/stke.2952005re8. [DOI] [PubMed] [Google Scholar]
- 157.Plato C.C., Galasko D., Garruto R.M., Plato M., Gamst A., Craig U.K., Torres J.M., Wiederholt W. ALS and PDC of Guam: forty-year follow-up. Neurology. 2002;58:765–773. doi: 10.1212/wnl.58.5.765. [DOI] [PubMed] [Google Scholar]
- 158.Hermosura M.C., Nayakanti H., Dorovkov M.V., Calderon F.R., Ryazanov A.G., Haymer D.S., Garruto R.M. A TRPM7 variant shows altered sensitivity to magnesium that may contribute to the pathogenesis of two Guamanian neurodegenerative disorders. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11510–11515. doi: 10.1073/pnas.0505149102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schlingmann K.P., Sassen M.C., Weber S., Pechmann U., Kusch K., Pelken L., Lotan D., Syrrou M., Prebble J.J., Cole D.E., et al. Novel TRPM6 mutations in 21 families with primary hypomagnesemia and secondary hypocalcemia. J. Am. Soc. Nephrol. 2005;16:3061–3069. doi: 10.1681/ASN.2004110989. [DOI] [PubMed] [Google Scholar]
- 160.Schlingmann K.P., Weber S., Peters M., Niemann Nejsum L., Vitzthum H., Klingel K., Kratz M., Haddad E., Ristoff E., Dinour D., et al. Hypomagnesemia with secondary hypocalcemia is caused by mutations in TRPM6, a new member of the TRPM gene family. Nat. Genet. 2002;31:166–170. doi: 10.1038/ng889. [DOI] [PubMed] [Google Scholar]
- 161.Walder R.Y., Landau D., Meyer P., Shalev H., Tsolia M., Borochowitz Z., Boettger M.B., Beck G.E., Englehardt R.K., Carmi R., et al. Mutation of TRPM6 causes familial hypomagnesemia with secondary hypocalcemia. Nat. Genet. 2002;31:171–174. doi: 10.1038/ng901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.